Abstract
The nuclear retinoic acid receptor RARγ2 undergoes proteasome-dependent degradation upon ligand binding. Here we provide evidence that the domains that signal proteasome-mediated degradation overlap with those that activate transcription, i.e. the activation domains AF-1 and AF-2. The AF-1 domain signals RARγ2 degradation through its phosphorylation by p38MAPK in response to RA. The AF-2 domain acts via the recruitment of SUG-1, which belongs to the 19S regulatory subunit of the 26S proteasome. Blocking RARγ2 degradation through inhibition of either the p38MAPK pathway or the 26S proteasome function impairs its RA-induced transactivation activity. Thus, the turnover of RARγ2 is linked to transactivation.
Keywords: degradation/p38MAPK/phosphorylation/proteasome/RARγ/transcription
Introduction
The highly pleiotropic effects of retinoic acid (RA), the physiologically active derivative of vitamin A, are mediated by two families of ligand-dependent transcriptional regulators, the RA nuclear receptors (RARs) and the retinoid X nuclear receptors (RXRs) that function as RAR/RXR heterodimers both in vitro and in vivo (Kastner et al., 1995; Mangelsdorf and Evans, 1995; Chambon, 1996; Chiba et al., 1997; Mark et al., 1999). There are three RARs (α, β and γ) and three RXRs (α, β and γ), and for each isotype there are at least two main isoforms differing only in their N-terminal region. RARs bind both all-trans and 9-cis RA, while RXRs bind 9-cis RA only (Chambon, 1996).
As with most nuclear receptors, RARs and RXRs contain two transactivation functions (AFs) (Chambon, 1996). The AF-1 domain, located at the N-terminal end (A/B region) of the receptor, is ligand-independent, while the ligand-activated AF-2 domain that overlaps the ligand binding domain (LBD) is located in the C-terminal region E. AF-2 requires the integrity of a highly conserved amphipatic α-helix, the AF-2 AD core that corresponds to the LBD helix 12. Binding of an agonistic ligand results in a transconformation of the LBD that involves helix 12 and creates a new surface for binding of coactivators (Moras and Gronemeyer, 1998; Chen, 2000; Glass and Rosenfeld, 2000). The AF-1 domain contains conserved serine residues which belong to consensus phosphorylation sites for proline-dependent protein kinases such as cyclin-dependent kinases (CDKs) and the mitogen-activated protein kinases (MAPKs) (Morgan 1997; Pearson et al. 2001; and references therein). Unliganded RARα1 and RARγ2 are constitutively phosphorylated at these sites (Rochette-Egly et al., 1997; Bastien et al., 2000) by cdk7/cyclin H associated to TFIIH, a general transcription factor also involved in DNA repair (Egly, 2001). Phosphorylation of these sites appears to be required for RAR transactivation (Keriel et al., 2002).
Nuclear hormone receptors have been recently reported to be degraded by the ubiquitin–proteasome pathway upon ligand binding (Hauser et al., 2000; Lange et al., 2000; Lonard et al., 2000). In the case of ER and PR, this ligand-dependent degradation has been correlated to transcriptional activity (Lonard et al., 2000; Shen et al., 2001). In addition, phosphorylation of the N-terminal A/B region by MAPKs has been shown to play an essential role in the degradation of PR, whereas the integrity of the AF-2 domain is required for ER degradation. It has also been suggested that the proteasomal SUG-1 subunit could be recruited by liganded nuclear receptors to selectively specify their own degradation (vom Baur et al., 1996).
In the presence of retinoic acid, RARα1 and RARγ2 are also degraded by the ubiquitin–proteasome pathway (Kopf et al., 2000). How these receptors are targeted for degradation, and whether their degradation is linked to their ability to activate transcription is however unknown. In the present study, we have investigated whether the targeted degradation of RARγ2 might be linked to its transactivation capacity by examining whether its activation functions are also involved in its downregulation. We show that the activation domains AF-1 and AF-2 cooperate in the RA-induced RARγ2 degradation. The RA-liganded AF-2 domain acts through the recruitment of the proteasomal SUG-1 subunit while the AF-1 domain requires to be phosphorylated. In that respect, we show that the phosphorylation of the serine residues located in the AF-1 domain is markedly increased in response to RA through a RA-induced activation of p38MAPK. Finally, we show that blocking RARγ2 degradation in the presence of ubiquitin proteasome inhibitors, p38MAPK inhibitors, or through competition of endogenous proteasomal SUG-1, impairs RA-induced transcriptional activation by RARγ2.
Results
Proteasome-dependent degradation is required for activation of transcription by RA-liganded RARγ2
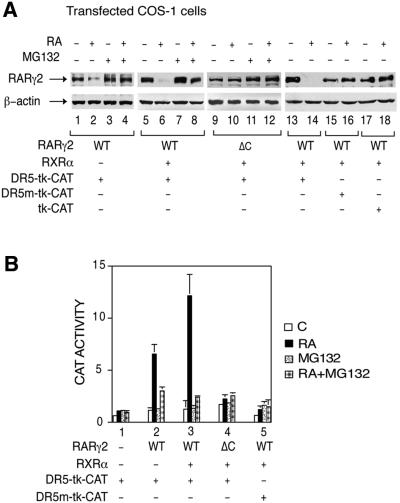
To address the role of proteasome-mediated degradation in RARγ2-activated transcription, COS-1 cells cotransfected with a RARγ2 expression vector and a CAT reporter gene controlled by a DR5 RA-responsive element (DR5-tk-CAT) were treated with RA and the proteasome inhibitor MG132, or their respective vehicles. RA treatment resulted in a significant decrease in the amount of RARγ2 that was maximal at 48 h (Figure 1A, lane 2; data not shown). Cotransfection of RXRα increased the degradation of liganded RARγ2 (Figure 1A, lane 6), as well as its ability to induce CAT activity (Figure 1B, compare lanes 2 and 3), in agreement with the known heterodimer requirement for both processes (Kopf et al., 2000).
Fig. 1. RA-induced RARγ2 degradation and transactivation are reversed by the proteasome inhibitor MG132 and require engagement of the receptor at a RARE. (A) COS-1 cells cotransfected with the DR5-tk-CAT reporter construct and the expression vectors for mRARγ2 (WT or ΔC) and RXRα, were treated for 48 h with vehicle or 1 × 10–6 M RA. When mentioned, MG132 was added 15 h before harvesting. In lanes 15 and 16 the DR5 element of the CAT reporter gene was mutated and in lanes 17 and 18 the DR5 element was deleted. WCEs were immunoblotted with RPγ(F) (upper panels) or actin antibodies (lower panels). (B) COS-1 cells were cotransfected with the DR5-tk-CAT reporter construct and the expression vector for mRARγ2, in the absence (–) or presence (+) of RXRα as indicated. In lane 5 the DR5 responsive element was mutated. The cells were treated as in (A) and analyzed for CAT activity. The results are the mean ± SD of three independent experiments.
MG132 prevented RARγ2 degradation (Figure 1A, lanes 4 and 8), confirming that proteasome mediates the degradation of the receptor (Kopf et al., 2000). MG132 did not alter RARγ2 levels in the absence of RA (Figure 1A, compare lanes 1 and 3, and 5 and 7), nor β-actin levels (Figure 1A, lower panel). Interestingly, MG132 also abrogated the induction of CAT activity (Figure 1B). The basal level of CAT activity observed in the absence of RARγ2 and/or in the absence of RA was unaltered by the proteasome inhibitor (Figure 1B, lane 1). Similarly, the activity of the β-galactosidase gene, which was added to allow normalization of the results according to transfection efficiency, was not affected (data not shown). Note that similar results were obtained with lactacystin, a specific proteasome inhibitor (data not shown).
Importantly, RARγ2 was not significantly degraded when cotransfected with a reporter gene in which the DR5 RA responsive element had been deleted (the tk-CAT reporter gene; Figure 1A, lanes 17 and 18) or mutated (the DR5 mut-tk-CAT reporter gene; Figure 1A, lanes 15 and 16), even in the presence of cotransfected RXRα. Moreover, a transcriptionally inactive deletion mutant of RARγ2 lacking the DNA binding domain (RARγΔC) was also resistant to the RA-induced degradation (Figure 1A, lanes 9–12). Altogether, these data demonstrate that RARγ2 degradation and transactivation processes are coupled, and indicate that degradation of the receptor requires its engagement in transcription.
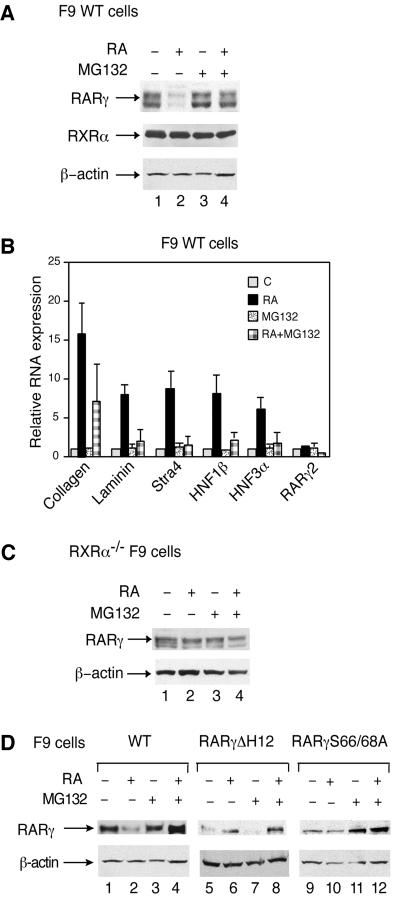
The effects of proteasome inhibitors were also tested in F9 embryocarcinoma cells, on both degradation of endogenous RARγ2 and RA-induced RARγ2-mediated transcription of endogenous RA-target genes. (for a review, see Rochette-Egly and Chambon, 2001). In these cells, maximal degradation of RARγ2 occurs at 48 h of RA treatment (Kopf et al., 2000; see also Figure 2A, lane 2), when the transactivation of RA-target genes (collagen IV, laminin B1, Stra4, HNF1β and HNF3α) is maximal and reaches a plateau (Figure 2B). Note that the heterodimerization partner RXRα was not degraded (Figure 2A, lane 2). However, RA-dependent degradation of RARγ2 requires the presence of RXRα (Kopf et al., 2000), and accordingly RXRα–/– F9 cells (Clifford et al., 1996) did not show any evidence of RARγ2 degradation in response to RA (Figure 2C). In F9 cells, MG132 and lactacystin abrogated the RA-induced decrease in RARγ2 levels (Figure 2A, compare lanes 2 and 4; data not shown), but did not alter RARγ2 levels in the absence of RA (Figure 2A, compare lanes 1 and 3), nor β-actin levels (Figure 2A, lower panel). MG132 also abrogated the RA-induced expression of RA target genes (Figure 2B). This lack of induction did not result from a general block in transcription, as MG132 did not affect the basal level of expression of the tested genes and the expression of RA unresponsive genes (RARγ2, 36B4, Nedd4, Pin-1 and many others; Figure 2B; data not shown). Collectively these results indicate that proteasome activity is also required for RA-induced RARγ2-mediated transcriptional activation through endogenous promoters.
Fig. 2. RA-induced RARγ2 degradation and transcription are also reversed by MG132 in F9 cells. (A) F9 WT cells were treated for 48 h with vehicle or 1 × 10–7 M RA. When mentioned, MG132 was added 15 h before harvesting. Whole cell extracts (WCEs) were resolved by SDS–10% PAGE and immunoblotted with RPγ(F), RPRXα(A), or actin antibodies. (B) F9 WT cells were treated for 48 h as in (A), as indicated. Transcripts for collagen IV, laminin B1, Stra4, HNF1β, HNF3α and RARγ2 were analyzed by quantitative RT–PCR. The presented values are the mean ± SD of three individual experiments and correspond to the fold-induction relative to the amount of transcripts present in vehicle-treated cells which was given an arbitrary value of 1. (C) RXRα–/– F9 cells were treated as in (A) and WCEs were immunoblotted with RPγ(F) or actin antibodies. (D) F9 cells that were either WT (lanes 1–4) or expressing RARγΔH12 (lanes 5–8) or RARγS66/68A (lanes 9–12) were treated as in (A) and WCEs were immunoblotted with RPγ(F) or actin antibodies.
Contribution of the activation domains AF-1 and AF-2 to RA-induced RARγ2 degradation
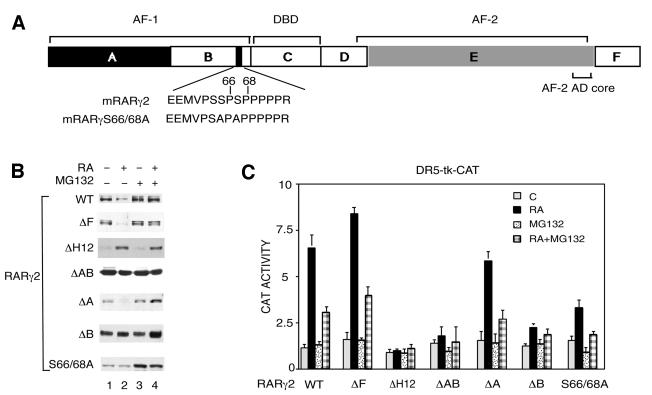
To determine which receptor domain(s) (see Figure 3A) contribute to the proteasome-dependent degradation of RARγ2, expression vectors for mutant forms of the receptor were cotransfected with the DR5-tk-CAT reporter gene in COS-1 cells. After treatment with RA, in the presence or absence of MG132, the degradation (Figure 3B) and the transcriptional activity (Figure 3C) of each mutant were analyzed.
Fig. 3. Both the AF-1 and AF-2 activation domains contribute to the RA-induced degradation of RARγ2. (A) Schematic representation (not to scale) of mRARγ2 with the DBD and the functional AF-1 and AF-2 domains, which lie in the A/B and E regions, respectively. The target sequence for phosphorylation by proline-directed kinases in the B region is shown and the corresponding serine residues, which have been mutated to alanine (S66 and S68), are indicated. (B) COS-1 cells were cotransfected with the DR5-tk-CAT reporter construct and the expression vector for mRARγ2 either WT, ΔF, ΔH12, ΔAB, ΔA, ΔB or S66/68A and treated with vehicle or 1 × 10–6 M RA as indicated. When mentioned, MG132 was added 15 h before harvesting. Equal amounts of WCEs, as estimated by immunoblotting with actin antibodies (data not shown) were resolved by SDS–10% PAGE and immunoblotted with RPγ(F) or Ab5γ(D) in the case of RARγ2ΔF. (C) Cells transfected as in (B) were analyzed for CAT activity. The results are the mean ± SD of three independent experiments.
When compared with the wild-type receptor, deletion of the C-terminal F region (RAR2γΔF) did not affect the RA-induced degradation and transcriptional activity of the receptor, nor the inhibition by MG132. Thus, the F region appears to be dispensible for RA-dependent degradation of RARγ2. The E region contains the transactivation function AF-2 whose activity is dependent on the integrity of the AF-2 AD core, which corresponds to helix 12 of the C-terminal end of the ligand-binding domain. Deletion of helix 12 in RARγ2 (RARγ2ΔH12) resulted in abrogation of both RA-induced RARγ2 transcriptional activity and degradation (Figure 3B and C). Note that RARγ2ΔH12 was in fact stabilized by RA, most likely because the ligand induced a conformational change protecting the receptor from proteolytic digestion (Wijayaratne and McDonnell, 2001).
RARγ2, lacking the entire N-terminal A/B region which contains the ligand-independent transactivation function AF-1 (RARγΔAB), was transcriptionally impaired and was not degraded in response to RA (Figure 3B and C). In contrast, deletion of the A region (RARγΔA) had no effect on RA-induced receptor degradation and transactivation. On the other hand, elimination of the B region (RARγΔB) resulted in a receptor with the same characteristics as RARγΔAB. Interestingly, mutation of the two phosphorylation sites, serines 66 and 68 (see Figure 3A) present in the B region of RARγ2 (RARγS66/68A) (Bastien et al., 2000) decreased RA-induced transactivation and abrogated RA-induced degradation. Note that the addition of MG132 suppressed the residual amount of RA-induced transactivation and further protected this mutant against degradation.
Similar results were obtained in RARγ–/– F9 embryocarcinoma cells expressing RARγ2ΔH12 or RARγ2S66/68A. These cells are known to be deficient for RA-induced expression of RA-target genes (Taneja et al., 1997; Plassat et al., 2000). In both cell lines, RARγ2 levels were not affected by RA addition (Figure 2D, lanes 6 and 10). Thus, taken together, the above results indicate that the integrity of the AF-2 domain and of the phosphorylation sites located in the B region are essential for both RA-induced degradation of, and transactivation by, RARγ2.
Ubiquitylation is required for RA-induced RARγ2 degradation and transactivation
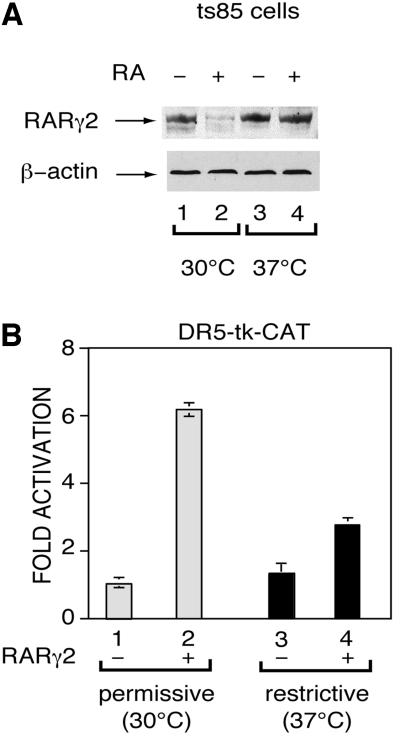
Polyubiquitylation is a prerequisite for the degradation of proteins through the proteasome pathway (Laney and Hochstrasser, 1999). The ts85 mouse mammary carcinoma cell line, which harbors a temperature-sensitive mutation of the ubiquitin activating (UBA) enzyme, is defective for protein ubiquitylation (Finley et al., 1984). These cells transiently transfected with the RARγ2 expression vector and the DR5-tk-CAT reporter gene, were treated with RA and incubated at the permissive (30°C) or the restrictive (37°C) temperatures. Under these conditions, RARγ2 was degraded at the permissive, but not at the restrictive temperature (Figure 4A, compare lanes 2 and 4). Moreover, incubation at the restrictive temperature decreased RA-induced RARγ2-mediated transcription (Figure 4B, compare lanes 2 and 4). Incubation of ts85 cells at the restrictive temperature had no influence on the basal level of CAT activity observed in the absence of RARγ2 and/or in the absence of RA (Figure 4B, lanes 1 and 3; data not shown). Moreover, it did not affect the expression of β-actin (Figure 4A, lower panel) or the levels of β-galactosidase activity expressed from the cotransfected control plasmid pCH110 (data not shown), indicating specificity of the RA-induced degradation of RARγ2 by the ubiquitin–proteasome pathway. Altogether, these results indicate that ubiquitylation is essential for both RA-induced RARγ2 degradation by the proteasome pathway and transactivation.
Fig. 4. Disruption of the ubiquitin-activating enzyme (UBA) function abrogates RA-induced RARγ2 degradation and transactivation. The temperature-sensitive UBA mutant ts85 cell line was transfected with the DR5-tk-CAT reporter gene and the expression vector for mRARγ2, treated with vehicle or 1 × 10–6 M RA and incubated at permissive (30°C) or restrictive (37°C) temperature for 24 h before harvesting. Extracts were immunoblotted with RPγ(F) and actin antibodies (A) and analyzed for CAT activity (B). The results, which correspond to the fold-induction relative to the CAT activity in vehicle-treated cells, are the mean ± SD of three independent experiments.
RA induces phosphorylation of RARγ2 in its AF-1 domain through activation of p38MAPK
Serine 66 and/or serine 68 in the AF-1 domain of RARγ2 are constitutively phosphorylated by cdk7 within TFIIH (Bastien et al., 2000). As these residues are also substrates for MAPKs and SAPKs in vitro (Bastien et al., 2000; our unpublished data), we investigated whether RARγ2 degradation might be linked to phosphorylation of the receptor by MAPKs in response to RA. RARγ2-transfected COS-1 cells were treated with RA and labelled with [32P]orthophosphate. Interestingly, the phosphorylation of RARγ2 was increased after 24 h of RA treatment (Figure 5A, lane 2), thus before its degradation could be seen (data not shown). This increase was not detectable up to 16 h of RA treatment (Bastien et al., 2000; data not shown).
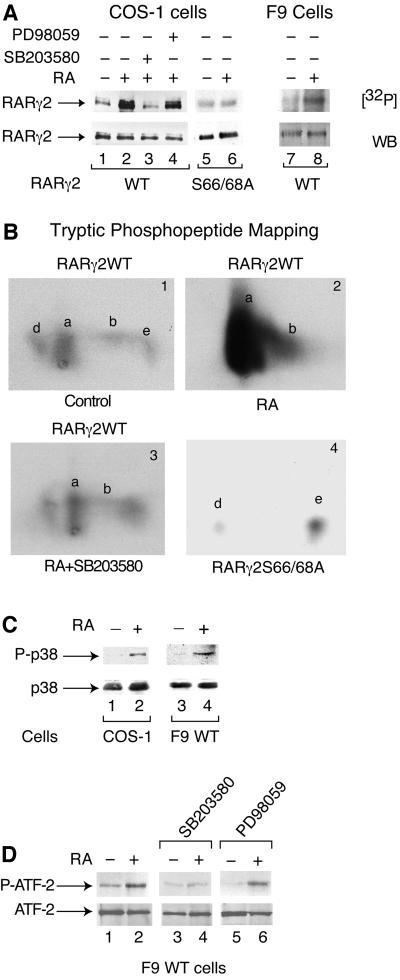
Fig. 5. RA increases the amount of RARγ2 phosphorylated in its AF-1 domain, subsequent to the activation of p38MAPK. (A) COS-1 cells plated in 10 cm Petri dishes and cotransfected with the DR5-tk-CAT reporter construct and the expression vector for mRARγ2 either WT (lanes 1–4) or S66/68A (lanes 5 and 6) were treated with vehicle (lane 1) or 1 × 10–6 M RA (lane 2). In lanes 3 and 4, RA was combined with SB203580 (10 µM) or PD98059 (5 µM), respectively. Lanes 7 and 8 correspond to F9 WT cells, treated or not with RA (1 × 10–7 M). Cells were labelled with [32P]orthophosphate and WCEs were immunoprecipitated with mAb2γ(mF). Immunoprecipitates containing equal amounts of RARγ2 were resolved by SDS–10% PAGE, electrotransferred onto nitrocellulose (NC) filters, autoradiographed [32P] and immunoprobed with RPγ(F) by western blotting (WB). (B) Two-dimensional tryptic phosphopeptide mapping of 32P-labelled immunoprecipitated RARγ2WT (panels 1–3) and RARγ2S66/68A (panel 4). (C) RA activates p38MAPK. Transfected COS-1 cells (lanes 1 and 2) and F9 WT cells (lanes 3 and 4), were treated for 24 h with vehicle or RA as indicated. Then the cells were lysed and immunoprecipitated with a p38MAPK rabbit polyclonal antibody immobilized on Protein A– Sepharose beads. The immunoprecipitates were immunoblotted with antibodies recognizing specifically p38MAPK or its phosphorylated form, P-p38MAPK. (D) Phosphorylation of ATF-2 upon activation of p38MAPK. F9 WT cells were treated for 48 h with vehicle (lane 1), 1 × 10–7 M RA (lane 2), 10 µM SB203580 (lane 3), or 5 µM PD98058 (lane 5). In lanes 4 and 6, RA was combined with SB203580 or PD98058. WCEs were immunoprecipitated with a Phospho-p38MAPK rabbit polyclonal antibody immobilized on Protein A–Sepharose beads, washed and processed for phosphorylation of the 40 kDa ATF-2 fusion protein (5 µg) in kinase buffer (25 mM HEPES, 25 mM MgCl2, 25 mM β-glycerophosphate, 2 mM DTT, 0.1 mM Na3V04 and 20 µM ATP) for 30 min at room temperature. The reaction was terminated upon addition of the SDS sample buffer, and the phospho (P-ATF-2) and non-phospho forms of ATF-2 were detected by immunoblotting with specific antibodies.
In the absence of RA treatment, tryptic phosphopeptide mapping of RARγ2 yields four phosphorylated peptides (Figure 5B, panel 1). Phosphopeptides a and b contain the phosphorylated residues of the AF-1 domain (spot a is diphosphorylated at serines 66 and 68, while spot b is monophosphorylated at serine 68; see Bastien et al., 2000). The other phosphopeptides (d and e) map to region F (Bastien et al., 2000). RA markedly increased the phosphorylation of peptide a, and to a lesser extent that of peptide b (Figure 5B, panel 2). Thus, RA treatment increased selectively the phosphorylation of both serines 66 and 68. Accordingly, in COS-1 cells overexpressing a RARγ2 with the two serine residues mutated into alanine (RARγS66/68A), no increase in the amount of phosphorylated receptor was observed upon RA treatment (Figure 5A, lanes 5 and 6) and spots a and b were absent (Figure 5B, panel 4).
F9 WT cells also responded to RA by an increase in the amount of phosphorylated RARγ2 (Figure 5A, lanes 7 and 8). No such increase could be seen in F9 cells expressing RARγS66/68A in a RARγ-null background (Taneja et al., 1997; data not shown).
To determine whether MAPKs are involved in RA-induced phosphorylation of RARγ2, transfected COS-1 cells were treated with RA in the absence or presence of MAPK inhibitors before labeling with 32P. SB203580 (10 µM), a highly specific inhibitor of p38MAPK (Cohen, 1996), abrogated the RA-dependent increase in phosphorylated RARγ2 (Figure 5A, lane 3) and in phosphopeptides a and b (Figure 5B, panel 3). In contrast, the MEK1 inhibitor PD98059 (5 µM; Cohen, 1996) had no significant effect (Figure 5A, lane 4). Altogether, these results indicate that RA increases the amount of phosphorylated RARγ2, through activation of p38MAPK, but not of Erks.
Thus, we determined the state of phosphorylation and activation of p38MAPK in RA-treated transfected COS-1 cells and F9 WT cells. In both cell systems, the phosphorylation of p38MAPK was induced after 24 h of RA treatment (Figure 5C), and was maximal at 48 h (data not shown), as assessed by western blotting analysis with specific antibodies recognizing the phosphorylated form of the kinase (P-p38). RA had no effect at earlier times (data not shown). Under these conditions, p42/p44 Erks were not activated (data not shown). That RA activates p38MAPK was confirmed by the observation that p38MAPK phosphorylates ATF-2 (a well known substrate of the kinase) more efficiently when immunoprecipitated from RA-treated F9 cells rather than from control untreated cells. (Figure 5D, lane 2). As expected, this phosphorylation of ATF-2 was completely abolished upon incubation of RA-treated F9 cells with the p38MAPK inhibitor SB203580, but not with the MEK1 inhibitor PD98059 (Figure 5D, lanes 4 and 6).
RA-induced phosphorylation of RARγ2 is required for RARγ2 degradation and transactivation
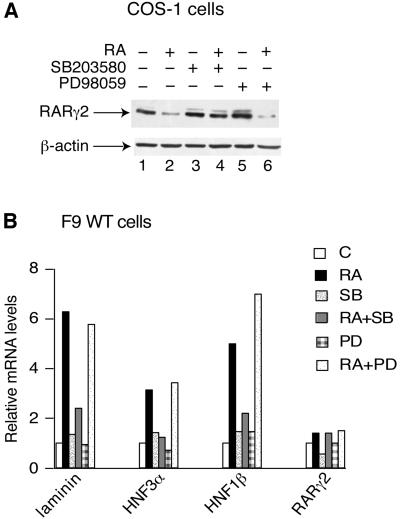
As RARγS66/68A is resistant to RA-induced degradation (Figure 3A and Figure 2D, lanes 9–12) and impaired in transactivation (Rochette-Egly et al., 1997; Taneja et al., 1997; Figure 3C), we investigated whether the RA-induced phosphorylation of RARγ2 affects its degradation and transactivation activity. To this end we inhibited the RA-dependent RARγ2 phosphorylation. SB203580 treatment of transfected COS-1 cells abrogated RA-induced degradation of RARγ2 (Figure 6A, lanes 3 and 4), whereas PD98059 was ineffective (Figure 6A, lanes 5 and 6). SB203580 also decreased RA-induced degradation of RARγ2 in F9 cells (data not shown) and blocked the induced expression of laminin, HNF3α and HNF1β, which was observed after 48 h of RA treatment (Figure 6B), while PD98059 had no effect (Figure 6B).
Fig. 6. Phosphorylation by p38MAPK is required for RA-induced RARγ2 degradation and transactivation. (A) COS-1 cells cotransfected with the DR5-tk-CAT reporter construct and the expression vector for mRARγ2 were treated for 48 h with vehicle, RA, SB203580 or PD98058, either alone or in combination, as indicated. WCEs were resolved by SDS–10% PAGE and immunoblotted with RPγ(F) (upper panels) or actin antibodies (lower panels). (B) F9 WT cells were treated for 48 h with vehicle, 1 × 10–7 M RA, 10 µM SB203580 or 5 µM PD98059 either alone or in association as indicated. Transcripts for laminin B1, HNF3α, HNF1β and RARγ2 were analyzed by quantitative RT–PCR. The presented results are an average of at least three independent experiments which agreed within ±15%. The values correspond to the fold-induction relative to the amount of transcripts present in vehicle-treated cells.
Collectively, these results indicate that the RA-induced phosphorylation of RARγ2 by p38 MAPK is required for both RARγ2 degradation and RARγ2-mediated activation of transcription.
Overexpression of SUG-1, a subunit of the 26S proteasome complex, reverses RA-induced degradation of RARγ2
As the proteasome activity is required for RA-induced degradation of RARγ2, we investigated whether SUG-1 could be involved in this process. SUG-1 is a nuclear receptor-interacting protein which belongs to the 19S regulatory complex of the 26S proteasome (Rubin et al., 1996; Glickman et al., 1998; DeMartino and Slaughter, 1999). The RA-dependent interaction of SUG-1 with RARs requires the integrity of the receptor AF2-AD core (helix 12) (vom Baur et al., 1996) and of the SUG-1 AAA module (ATPases Associated with a variety of cellular Activities), which contains a putative ATP binding site consensus motif (Fraser et al., 1997).
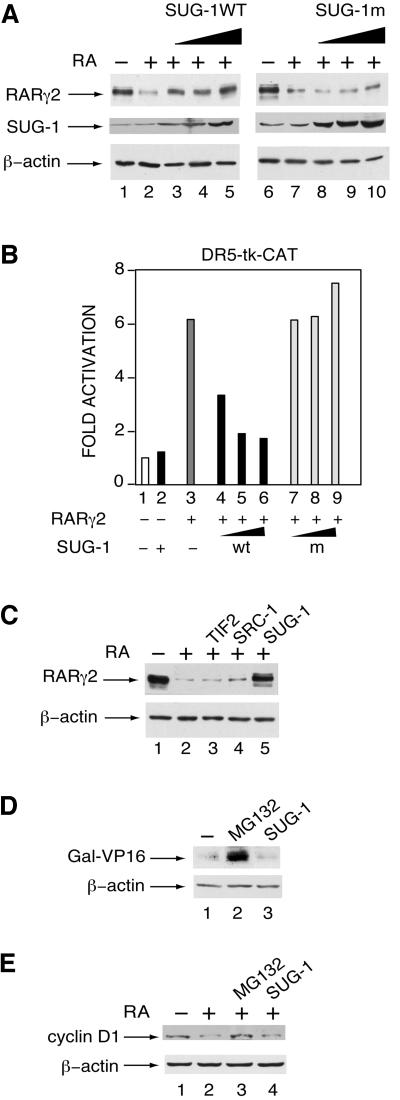
To investigate whether proteasomal SUG-1 is recruited by liganded RARγ2 to selectively specify its degradation, competition experiments were performed by transfecting COS-1 cells with increasing concentrations of an expression vector for SUG-1, either WT or mutated at its AAA module (K196H mutation in SUG-1m) (Fraser et al., 1997), along with RARγ2 and the DR5-tk-CAT reporter gene. Interestingly, overexpressed SUG-1WT abrogated the RA-induced degradation of RARγ2 (Figure 7A, lanes 3–5) and inhibited its transactivation activity (Figure 7B, lanes 4–6). In contrast, the SUG-1 K196H mutant which cannot bind nuclear receptors (vom Baur et al., 1996) did not affect RARγ2 degradation (Figure 7A, lanes 8–10) and transactivation activity (Figure 7B, lanes 7–9). These effects of SUG-1 were specific for RARγ2, as β-actin levels (Figure 7A, lower panels) and the basal CAT activity observed in the absence of RARγ2 (Figure 7B, lanes 1 and 2) were unaffected. In contrast, overexpression of the coactivators TIF2 and SRC-1 did not exert any significant effect on the RA-induced degradation of RARγ2 (Figure 7C, compare lanes 1–4). Note that overexpressed SUG-1, TIF2 and SRC-1 did not undergo any degradation upon addition of RA (Figure 7A; data not shown). Importantly, SUG-1 overexpression has no effect on the turnover of the chimeric transcriptional activator Gal-VP16 (Figure 7D, lane 3), which is also targeted by the ubiquitin–proteasome pathway (Molinari et al., 1999; Thomas and Tyers, 2000; Tansey, 2001) as shown by the accumulation of the protein in cells treated with the proteasome inhibitor MG132 (Figure 7D, lane 2). It did not reverse either the degradation of cyclin D1 that is induced by RA and also involves the proteasome pathway (Boyle et al., 1999) (Figure 7E).
Fig. 7. Overexpression of SUG-1 interferes with RA-induced RARγ2 degradation and transactivation. (A) COS-1 cells were cotransfected with the DR5-tk-CAT reporter construct and the expression vector for mRARγ2, without (lanes 1 and 2) or with increasing amounts (0.1, 0.2 and 0.5 µg) of an expression vector for SUG-1 [either WT (lanes 3–5), or mutated (lanes 8–10)] and treated for 48 h with vehicle (lanes 1 and 6) or 1 × 10–6 M RA (lanes 2–5 and 7–10). Equal amounts of WCEs were immunoblotted with RPγ(F), SUG-1 antibodies or actin antibodies. (B) COS-1 cells were cotransfected and RA-treated as in (A). After 48 h, extracts were analyzed for CAT activity. Lanes 1 and 2 correspond to the activity of the reporter gene in the absence of cotransfected RARγ2. The results correspond to the fold-induction relative to the CAT activity in the absence of RA. They are the mean ± SD of three independent experiments. (C) COS-1 cells were cotransfected with the DR5-tk-CAT reporter construct and the expression vector for mRARγ2 with or without the vectors (0.5 µg) for SUG-1WT, TIF2 and SRC-1 as indicated. Cells were treated for 48 h with vehicle or 1 × 10–6 M RA and processed as in (A). (D) COS-1 cells were cotransfected with the (17mer)×5-TATA-CAT reporter construct and the expression vector for Gal-VP16 with or without the SUG-1 vector. WCEs were immunoblotted with antibodies recognizing the DNA binding domain of GAL4 or β-actin. In lane 2, cells were treated with MG132, 15 h before harvesting. (E) COS-1 cells were treated for 48 h with vehicle or with RA. In lane 2, cells were treated with MG132, 15 h before harvesting. In lane 4, cells were transfected with the SUG-1 expression vector and RA-treated. WCEs were immunoblotted with antibodies recognizing cyclin D1 or β-actin.
That overexpressed SUG-1 competes with proteasomal SUG-1 indicates that RARγ2 degradation involves the RA-dependent recruitment of the proteasome through SUG-1, and is in agreement with our observation that RARγ2ΔH12 cannot be degraded in response to RA (see Figures 2D and 3B). Such a mechanism would be specific for liganded RARγ2, as it does not appear to be involved in the turnover of other substrates of the proteasome.
Discussion
It is now well established that eukaryotic transcription factors as well as steroid hormone receptors, including retinoic acid receptors, are degraded by the ubiquitin– proteasome pathway. As there is increasing evidence that the degradation of transactivators can be associated with their transcriptional activation function (Molinari et al., 1999; Thomas and Tyers, 2000; Tansey, 2001) we have investigated whether this is also the case for RARs. We demonstrate that RARγ2 is degraded only in its liganded transcriptionally active state when engaged in transcription of a RA-responsive gene. Additionally, inhibition of RARγ2 degradation subsequent to inhibition of the 26S proteasome activity or to disruption of the UBA enzyme, severely impairs RA-induced transcriptional activity mediated by RARγ2. Thus, proteasome-mediated degradation of the receptor and RARγ2-mediated transcription appear to be intimately linked. This association between the two processes is illustrated by the overlap of the AF-1 and AF-2 domains involved in transcription activation with the domains signaling the degradation of the receptor. The AF-1 domain signals degradation through a marked increase of its phosphorylation that is secondary to a RA-induced activation of p38MAPK, while the AF-2 domain acts through a RA-dependent recruitment of the proteasomal SUG-1 subunit.
Role of AF-1 phosphorylation in RA-induced degradation of RARγ2
We had previously shown that part of the cellular unliganded RARγ2 pool is constitutively (i.e. in the absence of RA) phosphorylated at Ser 66 and/or Ser 68 located in the AF-1 domain and that this phosphorylation is necessary, but not sufficient for transactivation that occurs only in the presence of ligand (Bastien et al., 2000). This constitutive RAR phosphorylation is carried out by the kinase (cdk7) subunit of the general transcription factor TFIIH and very likely occurs during the formation of the initiation complex (Keriel et al., 2002).
The novelty of the present study is that the amount of RARγ2 phosphorylated at these sites is markedly increased in response to RA, through activation of p38MAPK. The other novelty is that this RA-induced phosphorylation of RARγ2 is important for both the degradation of the receptor by the ubiquitin–proteasome pathway and the transcription of RA-responsive genes. Indeed, blocking the RA-induced activation of p38MAPK inhibits not only the RA-induced phosphorylation of RARγ2, but also the degradation and the transactivation activity of the receptor. Thus, we have uncovered a novel link between RA-induced, RARγ2-mediated transcription and RARγ2 degradation through phosphorylation of the receptor by p38MAPK. Such an overlap of two activities, transcriptional activation and targeting for degradation, is not unique to the AF-1 domain of RARγ2, as it has been also reported for the transcriptional activation domains of the progesterone receptor (Lange et al., 2000; Shen et al., 2001) and of a number of transcription factors such as VP16, Myc, ATF-2, E2F-1 and others (Molinari et al., 1999; Salghetti et al., 1999, 2000; Vandel and Kouzarides, 1999; Fuchs et al., 2000) which contain either acidic or phosphorylated residues.
It is noteworthy that the phosphorylation sites located in the AF-1 domain of RARγ2 are located within a PEST-rich sequence (Kopf et al., 2000), known as a potential signal for binding of ubiquitin ligases and subsequent degradation. In addition, phosphorylation of these sites is required for the increase in RARγ2 ubiquitylation that occurs in response to RA (Kopf et al., 2000). As phosphorylation has been reported to be a signal for ubiquitylation (Orford et al., 1997; Laney and Hochstrasser, 1999; Mitsui and Sharp, 1999) this raises the interesting possibility that the marked p38MAPK-induced phosphorylation of RARγ2 in its AF-1 domain could be a positive signal for ubiquitylation and subsequent targeting of the receptor to the proteasome.
Role of SUG-1 in proteasome recruitment by liganded RARγ2
We have shown here that RARγ helix 12, whose integrity is known to be crucial for the agonist-dependent binding of coactivators, and therefore to be required for the AF-2 function of nuclear receptors, is also essential for RA-induced RARγ2 degradation. Our data strongly suggest that the ligand-dependent binding of the receptor-interacting protein SUG-1 which also requires the integrity of helix 12 (vom Baur et al., 1996) is involved in the degradation of RARγ2. In this respect, we note that SUG-1 has recently been shown to be also involved in the proteasome-dependent degradation of the vitamin D receptor (Masuyama and MacDonald, 1998). Interestingly, SUG-1 is one of the ATP-dependent subunits belonging to the 19S regulatory component of the 26S proteasome (Fraser et al., 1997; DeMartino and Slaughter, 1999). In that context, SUG-1 is thought to be involved in proteasome-dependent degradation through binding polyubiquitylated polypeptides, unwinding them and feeding the resulting unstructured chain into the 20S catalytic component of the 26S proteasome (Rubin et al., 1996; Fraser et al., 1997; DeMartino and Slaughter, 1999). Therefore it is likely that RARγ2 degradation involves the proteasomal SUG-1 subunit bound to the AF-2 domain, once the receptor has been ubiquitylated, subsequent to increased phosphorylation of its AF-1 domain by p38MAPK.
It has been proposed that upon ligand binding, initiation of transcription occurs through temporally ordered exchanges of regulatory protein complexes that bind to a common interacting surface in which helix 12 is involved (for review see Dilworth and Chambon, 2001; Naar et al., 2001). Thus, SUG-1 within the 19S regulatory component of the proteasome may displace coactivators through its ATP-dependent chaperone activity (Hochstrasser, 1995). This exchange may funnel RARγ2 into the 20S subunit of the proteasome and subsequently result in its degradation. Such a scenario is in agreement with the competition effect observed upon overexpression of exogenous SUG-1. Indeed, an excess of free SUG-1 would make RARγ2 inaccessible to coactivators (e.g. SRC-1, TIF2) and proteasome-associated SUG1, resulting in inhibition of RA-dependent transactivation and proteasome-mediated degradation. Such a mechanism, which reflects the ability of RARs to bind SUG-1, is not valid for other substrates of the ubiquitin–proteasome pathway.
Relationship between RARγ2 phosphorylation, transactivation and degradation
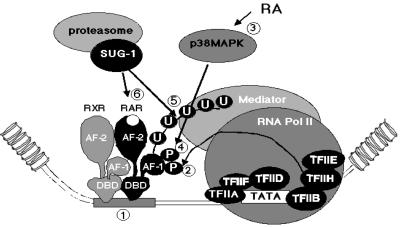
Our present data and previous studies (Rochette-Egly et al., 1997; Bastien et al., 2000; Kopf et al., 2000; Keriel et al., 2002) suggest the following model (see Figure 8). Upon ligand binding, the fraction of RARγ2 that is bound to cognate response elements as heterodimers with RXR and constitutively phosphorylated by the cdk7 subunit of TFIIH, activates transcription initiation. Subsequently, the RA-induced increased activity of p38MAPK would lead to further phosphorylation of the AF-1 domain. This marked increase in phosphorylation may act as a permissive signal paving the way to RARγ2 degradation through an increase in its ubiquitylation and subsequent recognition by the proteasomal SUG-1 subunit bound to the AF-2 domain. Whether an interaction of SUG-1 with TFIIH (Weeda et al., 1997) which also interacts with RARγ2 (Bastien et al., 2000) may play a role in this process, requires further investigation. In any event, hyperphosphorylation of the AF-1 domain would be a signal for receptor degradation, and SUG-1 would belong to a surveillance machinery that may prevent a single RAR/RXR heterodimer bound to a response element to perform endless rounds of transcription of the cognate RA target genes, thus allowing rapid reprogramming of transcriptional patterns (Tansey, 2001). Alternatively or concomitantly, the receptor proteasomal degradation may contribute to gene transcription by disrupting the preinitiation complex, allowing elongation to proceed (Thomas and Tyers, 2000).
Fig. 8. RA-induced degradation of RARγ2 by the ubiquitin–proteasome pathway. Upon ligand binding, RARγ2/RXR heterodimers bound to a RAR response element (step 1) recruit coactivators that decompact chromatin and allow the recruitment of the general transcription machinery at the promoter. Then RARγ2 becomes phosphorylated at Ser66 or Ser68 by the cdk7 subunit of TFIIH (step 2). Assembly of the transcription initiation complex leads to transcription initiation. After 24 h of RA treatment, p38MAPK is activated (step 3), resulting in the increase of the phosphorylation of both serine residues located in the AF-1 domain (step 4). This acts as a signal for ubiquitylation (step 5) and subsequent recognition and degradation (step 6) by proteasomal SUG-1 bound to the AF-2 domain.
Materials and methods
Plasmids and chemicals
The pSG5-based expression vectors for mRXRα, mRARγ2, mRARγΔAB, mRARγΔA, mRARγΔB, mRARγΔC, mRARγΔ399–407 (RARγΔH12), mRARγΔF and mRARγS66/66A were previously described (Nagpal et al., 1992, 1993; Bastien et al., 2000; Plassat et al., 2000), as well as pSG5-TIF2 (Voegel et al., 1998), pSG5-SUG-1WT, pSG5-SUG-1K196H (vom Baur et al., 1996), and pSG5-Gal4-VP16 (Tora et al., 1989). The SRC-1 cDNA (a gift from B.O’Malley) was cloned into pBKCMV (Stratagene). PD98059, SB203580 and MG-132 and lactacystin were from Calbiochem. All-trans retinoic acid was from Sigma-Aldrich.
Antibodies
Rabbit polyclonal antibodies against the F region of RARγ, RPγ(F) and mouse monoclonal antibodies against the D and F regions of RARγ, Ab5γ(D) and Ab2γ(mF), respectively, were as described (Rochette-Egly et al., 1991; Bastien et al., 2000) as well as rabbit polyclonal antibodies against RXRα, RPRXα(A) (Rochette-Egly et al., 1994) and mouse monoclonal antibodies against mSUG-1 (MAb 2SU1B8) (vom Baur et al., 1996) and the Gal4-DNA binding domain (MAb 3GV2) (White et al., 1992). Rabbit polyclonal antibodies against SRC-1 and Cyclin D1, goat polyclonal antibodies against β-actin, rabbit polyclonal antibodies against ATF-2 (N-96) and mouse monoclonal antibodies against the phosphorylated form of ATF-2 (F-1) were from Santa Cruz Biotechnology Inc. Rabbit polyclonal antibodies raised against a TIF2 epitope within residues 624 and 869 were a gift from H.Gronemeyer. Rabbit polyclonal antibodies against p38MAPK and its active phosphorylated form, P-p38MAPK (Thr180/Tyr182) were from Cell Signaling Technology, Inc. (USA).
Cells, transfections, CAT assays and immunoblotting
F9 cells ablated for RXRα (RXRα–/– cells) and F9 cells re-expressing RARγS66/68A and RARγΔH12 (RARγΔAF-2) were as described (Clifford et al., 1996; Taneja et al., 1997; Plassat et al., 2000) and cultured as monolayers on gelatinized surfaces. The UBA (E1) temperature-sensitive cell line ts85, which is derived from the FM3A mouse mammary carcinoma cell line, was maintained at either the restrictive (37°C) or permissive temperature (30°C) (Finley et al., 1984).
COS-1 cells were grown and transiently transfected in six-well plates, using the DMRIE-C reagent, according to the manufacturer’s protocol (Gibco-BRL-Life Technologies). In addition to the D5-tk-CAT reporter gene (1 µg/well) and the mRARγ2 pSG5-based expression vector (0.05 µg/well), all transfections contained Bluescript as a carrier and 0.5 µg of the β-galactosidase expression vector pCH110 to correct for variations in transfection efficiency. After a 16 h incubation with the DNA, the cells were washed, and maintained for a further 48 h in the appropriate medium with or without RA (1 × 10–6 M). When mentioned, MG132 (40 µM, France CalBiochem) was added 15 h before harvesting. CAT assays were performed using the ELISA method (CAT ELISA, Roche Molecular Biochemicals). All assays were normalized to equal β-galactosidase activity and the results were expressed as pg CAT/unit of β-galactosidase.
Whole cell extracts (WCEs) were prepared from F9 or transfected COS-1 cells as described (Rochette-Egly et al., 1991). For the detection of active phosphorylated forms of p38MAPK, WCEs were prepared in phosphorylation lysis buffer (PBL) (Alsayed et al., 2001). Proteins (40 µg) were resolved by SDS–10% polyacrylamide gel electrophoresis, electrotransferred onto nitrocellulose membranes (NC), and detected by immunoblotting and chemiluminescence according to the manufacturer’s protocol (Amersham Pharmacia Biotech).
In vivo phosphorylation and tryptic phosphopeptide mapping
F9 WT cells or COS-1 cells, cotransfected with the mRARγ2 expression vector and the DR5-TK-CAT reporter gene in 10 cm Petri dishes, were starved in phosphate-deficient medium, incubated for 4 h in the same medium containing 250 µCi [32P]orthophosphate, and lysed in RIPA buffer (Rochette-Egly et al., 1995). RA was added 24 h before harvesting the cells. Extracts were immunoprecipitated with mAb2γ(mF). Immunoprecipitates containing equal amounts of RARγ2 were resolved by SDS–10% PAGE, electrotransferred onto NC filters, autoradiographed and immunoprobed with RPγ(F). Two-dimensional tryptic phosphopeptide mapping was carried out on thin layer cellulose plates using the HTLE system (Rochette-Egly et al., 1995).
RNA isolation and real-time RT–PCR
Total RNAs were isolated using the guanidinium thiocyanate method and aliquots (500 ng) were subjected to real-time quantitative RT–PCR by using the SYBR Green Light-Cycler Detection System (Roche, Idaho Technologies). Transcript levels were normalized according to 36B4 transcripts which are unresponsive to retinoids treatment. The oligonucleotides for 36B4, laminin B1, collagen IV, HNF3α and HNF1β were as described (Taneja et al., 1997) as well as those for RARγ2 (Kopf et al., 2000). The RT–PCR primers for Stra4 were as follows: 5′-CCCATGTGGAGCTAACGAGT-3′ and 5′-GAGTAGATAGCAGCGCACGA-3′.
Acknowledgments
Acknowledgements
We thank R.Losson for the SUG-1 expression vectors and antibodies and H.Gronemeyer for the TIF2 expression vectors and antibodies. J.Bastien and A.Tarrade are thanked for critically reading the manuscript. We also thank Jean-Luc Plassat for helpful advice on quantitative RT–PCR as well as members of the cell culture and oligonucleotides facilities for help. We are grateful to Dr Finley for the generous gift of ts85 cells. This work was supported by funds from the Centre National de la Recherche Scientifique (CNRS), the Institut National de la Recherche Médicale (INSERM), the Collège de France, the Hôpital Universitaire de Strasbourg, the Association pour la Recherche sur le Cancer, and Bristol-Myers Squibb. M.G. was supported by short term fellowships from Human Frontier Science Program, FIRC (Fondazione Italiana per la Ricerca sul Cancro) and the Association pour la Recherche sur le Cancer.
References
- Alsayed Y., Uddin,S., Mahmud,N., Lekmine,F., Kalvakolanu,D.V., Minucci,S., Bokoch,G. and Platanias,L.C. (2001) Activation of Rac1 and the p38 mitogen-activated protein kinase pathway in response to all-trans-retinoic acid. J. Biol. Chem., 276, 4012–4019. [DOI] [PubMed] [Google Scholar]
- Bastien J., Adam-Stitah,S., Riedl,T., Egly,J.M., Chambon,P. and Rochette-Egly,C. (2000) TFIIH interacts with the retinoic acid receptor γ and phosphorylates its AF-1-activating domain through cdk7. J. Biol. Chem., 275, 21896–21904. [DOI] [PubMed] [Google Scholar]
- Boyle J.O., Langenfeld,J., Lonardo,F., Sekula,D., Reczek,P., Rusch,V., Dawson,M.I., Dmitrovsky,E. (1999) Cyclin D1 proteolysis: a retinoid chemoprevention signal in normal, immortalized and transformed human bronchial epithelial cells. J. Natl Cancer Inst., 91, 373–379. [DOI] [PubMed] [Google Scholar]
- Chambon P. (1996) A decade of molecular biology on retinoic acid receptors. FASEB J., 10, 940–954. [PubMed] [Google Scholar]
- Chen J.D. (2000) Steroid/nuclear receptor coactivators. Vitam. Horm., 58, 391–448. [DOI] [PubMed] [Google Scholar]
- Chiba H., Clifford,J., Metzger,D. and Chambon,P. (1997) Distinct retinoid X receptor–retinoic acid receptor heterodimers are differentially involved in the control of expression of retinoid target genes in F9 embryonal carcinoma cells. Mol. Cell. Biol., 17, 3013–3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford J., Chiba,H., Sobieszczuk,D., Metzger,D. and Chambon,P. (1996) RXRα-null F9 embryonal carcinoma cells are resistant to the differentiation, anti-proliferative and apoptotic effects of retinoids. EMBO J., 15, 4142–4155. [PMC free article] [PubMed] [Google Scholar]
- Cohen P. (1996) Dissection of protein kinase cascades that mediate cellular response to cytokines and cellular stress. Adv. Pharmacol., 36, 15–27. [DOI] [PubMed] [Google Scholar]
- DeMartino G.N. and Slaughter,C.A. (1999) The proteasome, a novel protease regulated by multiple mechanisms. J. Biol. Chem., 274, 22123–22126. [DOI] [PubMed] [Google Scholar]
- Dilworth F.J. and Chambon,P. (2001) Nuclear receptors coordinate the activities of chromatin remodeling complexes and coactivators to facilitate initiation of transcription. Oncogene, 20, 3047–3054. [DOI] [PubMed] [Google Scholar]
- Egly J.M. (2001) The 14th Datta Lecture. TFIIH: from transcription to clinic. FEBS Lett., 498, 124–128. [DOI] [PubMed] [Google Scholar]
- Finley D., Ciechanover,A. and Varshavsky,A. (1984) Thermolability of ubiquitin-activating enzyme from the mammalian cell cycle mutant ts85. Cell, 37, 43–55. [DOI] [PubMed] [Google Scholar]
- Fraser R.A., Rossignol,M., Heard,D.J., Egly,J.M. and Chambon,P. (1997) SUG1, a putative transcriptional mediator and subunit of the PA700 proteasome regulatory complex, is a DNA helicase. J. Biol. Chem., 272, 7122–7126. [DOI] [PubMed] [Google Scholar]
- Fuchs S.Y., Tappin,I. and Ronai,Z. (2000) Stability of the ATF2 transcription factor is regulated by phosphorylation and dephosphorylation. J. Biol. Chem., 275, 12560–12564. [DOI] [PubMed] [Google Scholar]
- Glass C.K. and Rosenfeld,M.G. (2000) The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev., 14, 121–141. [PubMed] [Google Scholar]
- Glickman M.H., Rubin,D.M., Fried,V.A. and Finley,D. (1998) The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol. Cell. Biol., 18, 3149–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser S., Adelmant,G., Sarraf,P., Wright,H.M., Mueller,E. and Spiegelman,B.M. (2000) Degradation of the peroxisome proliferator-activated receptor gamma is linked to ligand-dependent activation. J. Biol. Chem., 275, 18527–18533. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. (1995) Ubiquitin, proteasomes and the regulation of intracellular protein degradation. Curr. Opin. Cell Biol., 7, 215–223. [DOI] [PubMed] [Google Scholar]
- Kastner P., Mark,M. and Chambon,P. (1995) Nonsteroid nuclear receptors: what are genetic studies telling us about their role in real life? Cell, 83, 859–869. [DOI] [PubMed] [Google Scholar]
- Keriel A., Stary,A., Sarasin,A., Rochette-Egly,C. and Egly,J.M. (2002) XPD mtation prevents TFIIH-dependent phosphorylation of nuclear receptors and transactivation. Cell, 109, 125–135. [DOI] [PubMed] [Google Scholar]
- Kopf E., Plassat,J.L., Vivat,V., de The,H., Chambon,P. and Rochette-Egly,C. (2000) Dimerization with retinoid X receptors and phosphorylation modulate the retinoic acid-induced degradation of retinoic acid receptors alpha and gamma through the ubiquitin–proteasome pathway. J. Biol. Chem., 275, 33280–33288. [DOI] [PubMed] [Google Scholar]
- Laney J.D. and Hochstrasser,M. (1999) Substrate targeting in the ubiquitin system. Cell, 97, 427–430. [DOI] [PubMed] [Google Scholar]
- Lange C.A., Shen,T. and Horwitz,K.B. (2000) Phosphorylation of human progesterone receptors at serine-294 by mitogen-activated protein kinase signals their degradation by the 26S proteasome. Proc. Natl Acad. Sci. USA, 97, 1032–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonard D.M., Nawaz,Z., Smith,C.L. and O’Malley,B.W. (2000) The 26S proteasome is required for estrogen receptor-α and coactivator turnover and for efficient estrogen receptor-α transactivation. Mol. Cell., 5, 939–948. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf D.J. and Evans,R.M. (1995) The RXR heterodimers and orphan receptors. Cell, 83, 841–850. [DOI] [PubMed] [Google Scholar]
- Mark M., Ghyselinck,N.B., Wendling,O., Dupe,V., Mascrez,B., Kastner,P. and Chambon,P. (1999) A genetic dissection of the retinoid signalling pathway in the mouse. Proc. Nutr. Soc., 58, 609–613. [DOI] [PubMed] [Google Scholar]
- Masuyama H. and MacDonald,P.N. (1998) Proteasome-mediated degradation of the vitamin D receptor (VDR) and a putative role for SUG1 interaction with the AF-2 domain of VDR. J. Cell. Biochem., 71, 429–440. [PubMed] [Google Scholar]
- Mitsui A. and Sharp,P.A. (1999) Ubiquitylation of RNA polymerase II large subunit signaled by phosphorylation of C-terminal domain. Proc. Natl Acad. Sci. USA, 96, 6054–6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari E., Gilman,M. and Natesan,S. (1999) Proteasome-mediated degradation of transcriptional activators correlates with activation domain potency in vivo. EMBO J., 18, 6439–6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moras D. and Gronemeyer,H. (1998) The nuclear receptor ligand-binding domain: structure and function. Curr. Opin. Cell Biol., 10, 384–391. [DOI] [PubMed] [Google Scholar]
- Morgan D.O. (1997) Cyclin-dependent kinases: engines, clocks and microprocessors. Annu. Rev. Cell Dev. Biol., 13, 261–291. [DOI] [PubMed] [Google Scholar]
- Naar A.M., Lemon,B.D. and Tjian,R. (2001) Transcriptional coactivator complexes. Annu. Rev. Biochem., 70, 475–501. [DOI] [PubMed] [Google Scholar]
- Nagpal S., Saunders,M., Kastner,P., Durand,B., Nakshatri,H. and Chambon,P. (1992) Promoter context- and response element-dependent specificity of the transcriptional activation and modulating functions of retinoic acid receptors. Cell, 70, 1007–1019. [DOI] [PubMed] [Google Scholar]
- Nagpal S., Friant,S., Nakshatri,H. and Chambon,P. (1993) RARs and RXRs: evidence for two autonomous transactivation functions (AF-1 and AF-2) and heterodimerization in vivo. EMBO J., 12, 2349–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orford K., Crockett,C., Jensen,J.P., Weissman,A.M. and Byers,S.W. (1997) Serine phosphorylation-regulated ubiquitination and degradation of β-catenin. J. Biol. Chem., 272, 24735–24738. [DOI] [PubMed] [Google Scholar]
- Pearson G., Robinson,F., Beers Gibson,T., Xu,B.E., Karandikar,M., Berman,K. and Cobb,M.H. (2001) Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr. Rev., 22, 153–183. [DOI] [PubMed] [Google Scholar]
- Plassat J., Penna,L., Chambon,P. and Rochette-Egly,C. (2000) The conserved amphipatic α-helical core motif of RARγ and RARα activating domains is indispensable for RA-induced differentiation of F9 cells. J. Cell Sci., 113, 2887–2895. [DOI] [PubMed] [Google Scholar]
- Rochette-Egly C. and Chambon,P. (2001) F9 embryocarcinoma cells: a cell autonomous model to study the functional selectivity of RARs and RXRs in retinoid signaling. Histol. Histopathol., 16, 909–922. [DOI] [PubMed] [Google Scholar]
- Rochette-Egly C., Lutz,Y., Saunders,M., Scheuer,I., Gaub,M.P. and Chambon,P. (1991) Retinoic acid receptor gamma: specific immunodetection and phosphorylation. J. Cell Biol., 115, 535–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochette-Egly C., Lutz,Y., Pfister,V., Heyberger,S., Scheuer,I., Chambon,P. and Gaub,M.P. (1994) Detection of retinoid X receptors using specific monoclonal and polyclonal antibodies. Biochem. Biophys. Res. Commun., 204, 525–536. [DOI] [PubMed] [Google Scholar]
- Rochette-Egly C., Oulad-Abdelghani,M., Staub,A., Pfister,V., Scheuer,I., Chambon,P. and Gaub,M.P. (1995) Phosphorylation of the retinoic acid receptor-α by protein kinase A. Mol. Endocrinol., 9, 860–871. [DOI] [PubMed] [Google Scholar]
- Rochette-Egly C., Adam,S., Rossignol,M., Egly,J.M. and Chambon,P. (1997) Stimulation of RARα activation function AF-1 through binding to the general transcription factor TFIIH and phosphorylation by CDK7. Cell, 90, 97–107. [DOI] [PubMed] [Google Scholar]
- Rubin D.M., Coux,O., Wefes,I., Hengartner,C., Young,R.A., Goldberg,A.L. and Finley,D. (1996) Identification of the gal4 suppressor Sug1 as a subunit of the yeast 26S proteasome. Nature, 379, 655–657. [DOI] [PubMed] [Google Scholar]
- Salghetti S.E., Kim,S.Y. and Tansey,W.P. (1999) Destruction of Myc by ubiquitin-mediated proteolysis: cancer-associated and transforming mutations stabilize Myc. EMBO J., 18, 717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salghetti S.E., Muratani,M., Wijnen,H., Futcher,B. and Tansey,W.P. (2000) Functional overlap of sequences that activate transcription and signal ubiquitin-mediated proteolysis. Proc. Natl Acad. Sci. USA, 97, 3118–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen T., Horwitz,K.B. and Lange,C.A. (2001) Transcriptional hyperactivity of human progesterone receptors is coupled to their ligand-dependent down-regulation by mitogen-activated protein kinase-dependent phosphorylation of serine 294. Mol. Cell. Biol., 21, 6122–6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneja R., Rochette-Egly,C., Plassat,J.L., Penna,L., Gaub,M.P. and Chambon,P. (1997) Phosphorylation of activation functions AF-1 and AF-2 of RARα and RARγ is indispensable for differentiation of F9 cells upon retinoic acid and cAMP treatment. EMBO J., 16, 6452–6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tansey W.P. (2001) Transcriptional activation: risky business. Genes Dev., 15, 1045–1050. [DOI] [PubMed] [Google Scholar]
- Thomas D. and Tyers,M. (2000) Transcriptional regulation: Kamikaze activators. Curr. Biol., 10, R341–R343. [DOI] [PubMed] [Google Scholar]
- Tora L., White,J., Brou,C., Tasset,D., Webster,N., Scheer,E. and Chambon,P. (1989) The human estrogen receptor has two independent nonacidic transcriptional activation functions. Cell, 59, 477–487. [DOI] [PubMed] [Google Scholar]
- Vandel L. and Kouzarides,T. (1999) Residues phosphorylated by TFIIH are required for E2F-1 degradation during S-phase. EMBO J., 18, 4280–4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voegel J.J., Heine,M.J., Tini,M., Vivat,V., Chambon,P. and Gronemeyer,H. (1998) The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J., 17, 507–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- vom Baur E. et al. (1996) Differential ligand-dependent interactions between the AF-2 activating domain of nuclear receptors and the putative transcriptional intermediary factors mSUG1 and TIF1. EMBO J., 15, 110–124. [PMC free article] [PubMed] [Google Scholar]
- Weeda G., Rossignol,M., Fraser,R.A., Winkler,G.S., Vermeulen,W., van’t Veer,L.J., Ma,L., Hoeijmakers,J.H.J. and Egly,J.M. (1997) The XPB subunit of repair/transcription factor TFIIH directly interacts with SUG-1, a subunit of the 26S proteasome and putative transcription factor. Nucleic Acids Res., 25, 2274–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J., Brou,C., Wu,J., Lutz,Y., Moncollin,V. and Chambon,P. (1992) The acidic transcriptional activator Gal-VP16 acts on preformed template-commited complexes. EMBO J., 11, 2229–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijayaratne A.L. and McDonnell,D.P. (2001) The human estrogen receptor-α is a ubiquitinated protein whose stability is affected differentially by agonists, antagonists and selective estrogen receptor modulators. J. Biol. Chem., 276, 35684–35692. [DOI] [PubMed] [Google Scholar]