Abstract
Methylation of the ribose 2′-hydroxyl, the most widespread modification of ribosomal and splicesomal RNAs, is guided by the box C/D class of small nucleolar RNAs (snoRNAs). Box C/D small nucleolar ribonucleoproteins (snoRNPs) contain four core proteins: fibrillarin, Nop56, Nop58 and 15.5 kDa. We constructed U25 snoRNAs containing a single photoactivatable 4-thiouridine at each U position within the conserved box C/D and C′/D′ motifs. Proteins assembled on the snoRNA after injection into Xenopus oocyte nuclei were identified by cross-linking, and reconstituted particles characterized by functional rescue and mutational analyses. Our data argue that box C/D snoRNPs are asymmetric, with the C′ box contacting Nop56 and fibrillarin, the C box interacting with Nop58, and the D and D′ boxes contacting fibrillarin. No cross-link to 15.5 kDa was detected; its binding is disrupted by 4-thiouridine substitution in position 1 of the C box. Repositioning the guide sequence of U25 upstream of box D instead of D′ revealed that both C/D motifs have the potential to function as guide centers, but, surprisingly, there was no alteration in protein cross-linking.
Keywords: box C/D snoRNA/cross-linking/nucleolus/snoRNP/Xenopus
Introduction
The 18S, 5.8S and 28S rRNAs are transcribed as one contiguous unit. Maturation of this transcript requires two critical events: the covalent modification of highly conserved regions of the rRNA, and a series of endo- and exonucleolytic cleavages liberating the mature rRNA products (Sollner-Webb et al., 1996; Venema and Tollervey, 1999). The two predominant covalent modifications are the addition of a methyl group to the 2′-hydroxyl of ribose sugars and the isomerization of uridines to pseudouridines (Maden, 1990). These modifications, as well as many essential nucleolytic cleavages of the pre-rRNA, are directed by small nucleolar ribonucleoproteins (snoRNPs) (reviewed in Venema and Tollervey, 1999; Kiss, 2001; Terns and Terns, 2002).
The snoRNPs involved in rRNA modification can be divided into two classes based on their conserved sequence elements and associated proteins. Both classes select the nucleotide to be modified through specific base pairing between the snoRNA and rRNA substrate. Box C/D snoRNPs guide 2′-O-methylation and are characterized by the conserved terminal sequence elements box C [RUGAUGA] and box D [CUGA], as well as a somewhat less conserved matching set of internal sequence elements referred to as C′ and D′ (Balakin et al., 1996; Tycowski et al., 1996; Kiss-Laszlo et al., 1998). In the extended duplex formed with the substrate, box D or D′ is located precisely five nucleotides from the residue that base-pairs to the rRNA nucleotide to be modified (Cavaille et al., 1996; Kiss-Laszlo et al., 1996; Tycowski et al., 1996). The box H/ACA snoRNPs guide pseudouridylation and are characterized by two conserved sequence elements, the H box [ANANNA] and the 3′-terminal ACA sequence, and form a short duplex with the rRNA on each side of the uracil to be modified (Balakin et al., 1996; Ganot et al., 1997; Ni et al., 1997; Bortolin et al., 1999).
The 5′ and 3′ ends of box C/D snoRNAs form a terminal stem, bringing together the C and D boxes. This box C/D motif (Watkins et al., 2000) has been shown to be essential for snoRNA processing, stability, methylation activity, nucleolar localization and 5′ cap hypermethylation (Filipowicz and Pogacic, 2002; Terns and Terns, 2002). The C′ and D′ boxes are generally found within several nucleotides of one another (3–9 nucleotides) or are predicted to be brought together through the formation of an internal stem–loop (Kiss-Laszlo et al., 1998). The H/ACA snoRNAs likewise fold into a common hairpin– hinge–hairpin–tail structure (Balakin et al., 1996; Ganot et al., 1997), with the conserved sequence elements essential for snoRNA processing, stability, pseudouridylation activity and nucleolar localization (Filipowicz and Pogacic, 2002; Terns and Terns, 2002).
Four core proteins are associated with each snoRNP class. The box C/D class contains fibrillarin (yeast Nop1p, the methyltransferase; Omer et al., 2002), Nop58, Nop56 and the 15.5 kDa protein (yeast Snu13p) (Schimmang et al., 1989; Tyc and Steitz, 1989; Gautier et al., 1997; Wu et al., 1998; Lafontaine and Tollervey, 1999; Lyman et al., 1999; Newman et al., 2000; Watkins et al., 2000). The proteins of the box H/ACA class are dyskerin (yeast Cbf5p, the pseudouridine synthase), Gar1, Nhp2 and Nop10 (Kiss, 2001; Filipowicz and Pogacic, 2002).
A number of box C/D and box H/ACA snoRNAs guide two independent modification reactions making use of two separate antisense regions. This ability together with the symmetry observed for both the box C/D and box H/ACA snoRNAs at the RNA level has suggested that members of both classes of snoRNPs each possess two sets of core proteins (Watkins et al., 1998a; Venema and Tollervey, 1999). In support of this notion, electron micrographic studies of H/ACA snoRNPs revealed a rather symmetric bipartite particle, with an estimated mass consistent with two copies of each protein (Watkins et al., 1998a). We have undertaken site-specific cross-linking analyses to examine in greater detail the RNA–protein interactions of a modification guide box C/D snoRNP, Xenopus U25. In addition to providing a map of the interactions between core proteins and conserved box elements, the data surprisingly reveal that the core box C/D proteins assemble asymmetrically on the U25 snoRNA molecule.
Results
Fibrillarin, Nop56 and Nop58 assemble on and cross-link to the box C/D elements of U25 constructs injected into Xenopus oocytes
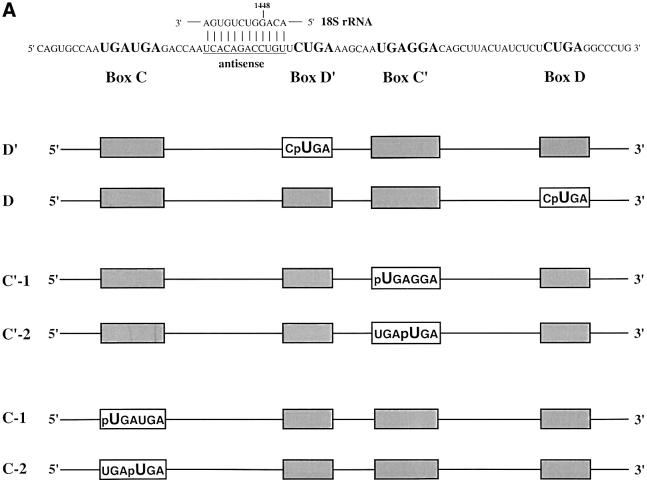
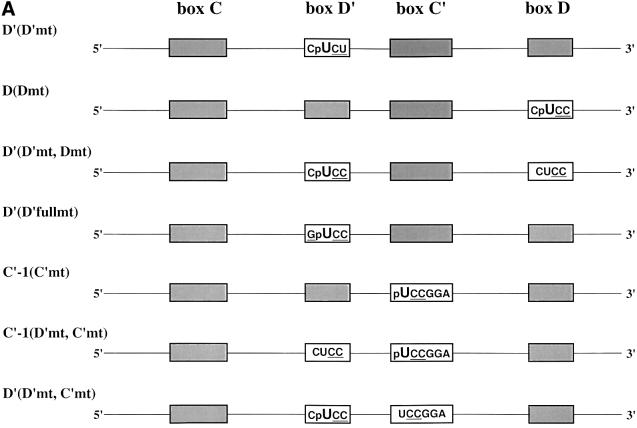
To identify proteins interacting with the conserved box C and D sequences, the photoactivatable cross-linking agent 4-thiouridine (4SU) was inserted site-specifically into the U25 snoRNA by RNA ligation methods (Moore and Sharp, 1992). Constructs were designed such that every uridine position within each of the four conserved sequence elements was replaced individually with the zero-length cross-linker, 4SU (Sontheimer, 1994). The constructs also contained a single radioactive phosphate immediately 5′ of the photoactivatable nucleotide and were named with reference to the site of 4SU incorporation (Figure 1A).
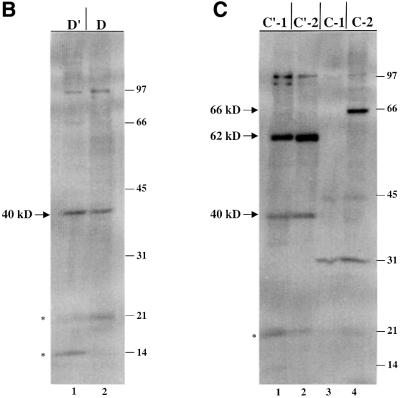
Fig. 1. U25 snoRNA constructs and cross-linking analyses. (A) The Xenopus U25 snoRNA sequence is shown, highlighting in bold the conserved box elements. Three additional nucleotides were added to the 5′ end and two nucleotides to the 3′ end to extend the terminal stem (Tycowski et al., 1996). The antisense region (underlined) is shown base-paired to the Xenopus 18S rRNA sequence it targets. For site- specific cross-linking, a uridine within one of the conserved sequence elements was replaced with a 4SU (U) with a single radioactive label immediately 5′ of the 4SU, denoted by p. Constructs are named based on the site of 4SU incorporation. (B and C) SDS–polyacrylamide gel analyses of proteins cross-linked to the U25 constructs. After assembly of the injected RNAs in oocyte nuclei, photocross-linking and RNase digestion, proteins were resolved by 15% SDS–PAGE. The 40, 62 and 66 kDa cross-linked proteins indicated with an arrow are identified in Figure 2. RNA bands are indicated by an asterisk. The positions of molecular weight markers (kDa) are shown on the right.
In vitro transcribed U25 snoRNA previously had been shown to reconstitute site-specific methylation activity within Xenopus oocytes (Tycowski et al., 1996). In a typical experiment, nuclei isolated from Xenopus oocytes and maintained under paraffin oil were microinjected with a 4SU-containing snoRNA transcript (Lund and Paine, 1990). After a 4 h incubation to allow snoRNP assembly, the nuclei were irradiated at 365 nm, the RNA was digested with RNase T1 and nuclease P1, and 32P-labeled proteins were resolved by SDS–PAGE. The amount of material injected, the assembly time and the extent of irradiation were all optimized to minimize non-specific cross-linking (Sontheimer, 1994; data not shown). Visualization of labeled protein bands was dependent upon irradiation at 365 nm and was sensitive to proteinase K digestion (data not shown).
Figure 1B and C illustrates the cross-linking patterns generated with the six wild-type U25 constructs. Note that, technically, the C′-2 construct is not wild-type, since the C box nucleotide that has been substituted with 4SU is not a U, but rather a G in U25; we refer to it as ‘wild-type’ since the 4SU-containing sequence conforms to the consensus for box C/C′, UGAUGA (Kiss-Laszlo et al., 1998).
Several cross-linked protein bands are seen with these six constructs, notably a protein migrating at ∼40 kDa with the D′ and D constructs (Figure 1B) as well as with the C′-1 and C′-2 constructs (Figure 1C, lanes 1 and 2). Three additional strongly cross-linked proteins were generated with the C′ and C constructs, migrating at ∼66, 62 and 31 kDa (Figure 1C). Surprisingly, the 66 kDa cross-link exhibits a striking specificity for only one of the two substituted positions in the C box (C-2 construct). The 40, 62 and 66 kDa cross-linked bands are of particular interest because their mobilities on SDS–PAGE correspond to those expected for the box C/D-associated proteins fibrillarin (40 kDa; Lapeyre et al., 1990), Nop56 (62–65 kDa; Newman et al., 2000; Watkins et al., 2000) and Nop58 (65–66 kDa; Newman et al., 2000; Watkins et al., 2000), respectively. The radioactivity migrating at ∼21 kDa (and a band occasionally running below this at ∼14 kDa; Figure 1B, lane 1) is RNA; undigested transcript migrates at 21 kDa (data not shown). Despite quite stringent RNase digestion conditions, such bands typically were seen, perhaps because a portion of the injected RNA forms aggregates resistant to nuclease action.
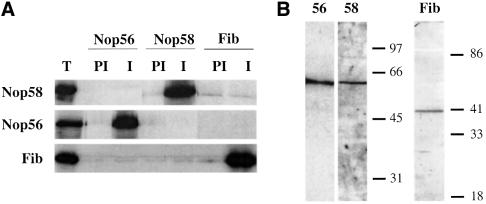
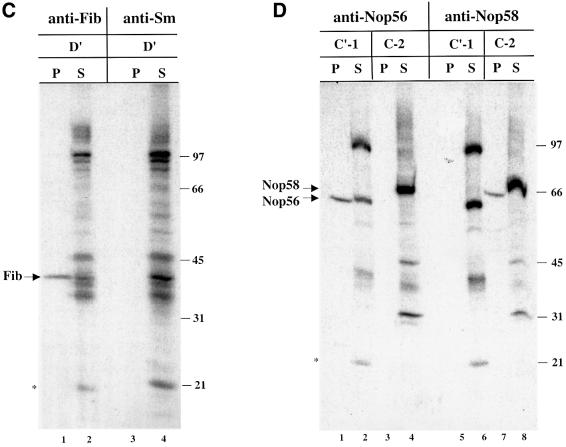
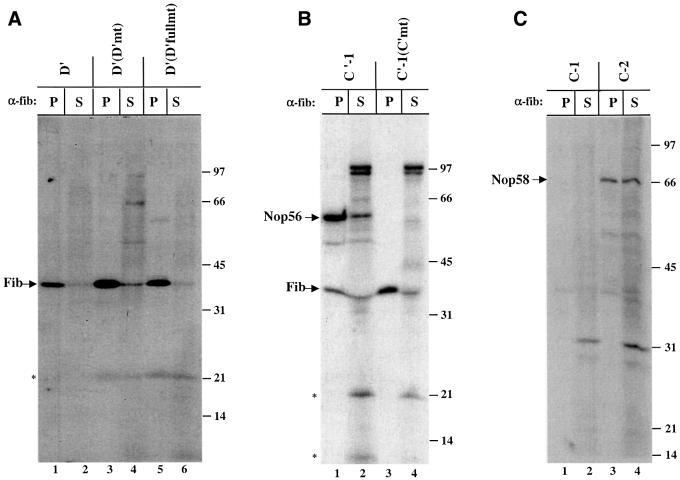
The identities of the 40, 62 and 66 kDa proteins were verified by immunoprecipitation of the RNase-digested, cross-linked samples with antibodies targeting the predicted box C/D snoRNP components (Speckmann et al., 2002). The specificity of these antibodies for Xenopus fibrillarin, Nop56 and Nop58 is confirmed by immunoprecipitation and western blot in Figure 2A and B. The 40 kDa band cross-linked to the D′ element was precipitated selectively by anti-fibrillarin antibodies (Figure 2C, lanes 1 and 2), but not by the control anti-Sm antibodies (Figure 2C, lanes 3 and 4). Although the 62 and 66 kDa cross-linked bands migrate close to one another, precipitation with anti-Nop56 and anti-Nop58 antibodies clearly demonstrated that the 62 kDa protein, cross-linked by the C′-1 construct, is Nop56, while the 66 kDa protein, cross-linked to C-2, is Nop58 (Figure 2D).
Fig. 2. Identification of the 40, 62 and 66 kDa cross-linked proteins. (A) Antibodies generated against Xenopus Nop56, Nop58 and fibrillarin were used for immunoprecipitation of in vitro translated, 35S-labeled Nop56, Nop58 and fibrillarin proteins. Immunoprecipitations with pre-immune and immune sera are shown. Total lanes (T) show 20% of the 35S-labeled protein included in the assays. (B) Immunoblot analysis using oocyte crude nuclear extracts was performed with the Nop56, Nop58 or fibrillarin antisera as indicated. (C and D) After injection of the D′, C′-1 or C-2 construct, the irradiated and RNase-digested nuclear extracts were subjected to immunoprecipitation prior to SDS–PAGE using anti-Sm (Y12) or the antisera characterized in (A) and (B). The resulting pellets (P) and 1/10 of the supernatants (S) were resolved on 12.5% SDS–polyacrylamide gels. The bands identified as fibrillarin (Fib), Nop56 and Nop58 are indicated with arrows. RNA bands are indicated with an asterisk. The positions of protein molecular weight markers are shown on the right.
Mutation of the conserved boxes alters cross-linking to Nop56, but not to fibrillarin
Site-specific U25 constructs with mutations in the four conserved sequence elements were generated to examine their effects on protein association (Figure 3A). It is well known that mutations in the C or D box destabilize box C/D snoRNAs (Venema and Tollervey, 1999; Weinstein and Steitz, 1999), as we confirmed in oocyte injection experiments (see below). In contrast, mutation of the internal C′ or D′ element does not alter the stability of the snoRNA (Kiss-Laszlo et al., 1998), allowing analysis of alterations in protein binding as revealed by cross-linking.
Fig. 3. Proteins cross-linked to mutant U25 constructs. (A) Mutant constructs are named with reference to the site of 4SU incorporation, as in Figure 1A, followed by (in parentheses) the box element that has been mutated. Mutated sequences are underlined. (B–D) The labeling of gels resolving proteins cross-linked to the mutant constructs is as in Figure 2B and C.
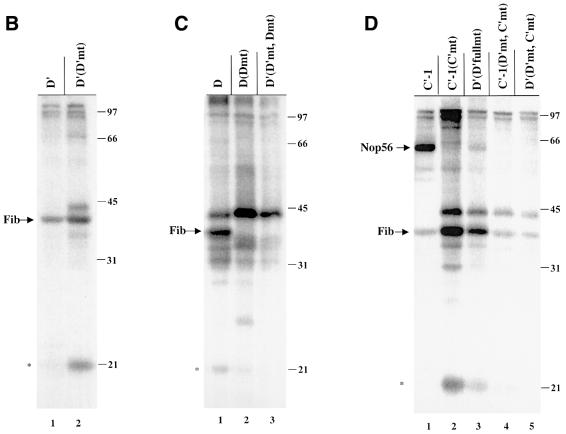
Mutation of the D′ element from C4SUGA to C4SUCU [D′(D′mt)] did not alter cross-linking to fibrillarin (Figure 3B), whereas mutation of the D element from C4SUGA to C4SUCC [D(Dmt)] resulted in the loss of fibrillarin cross-linking (Figure 3C, lane 2). This is most probably a consequence of snoRNA destabilization; during the 4 h incubation, >80% of the box D mutant relative to wild-type constructs was degraded (data not shown). Likewise, the D′(D′mt, Dmt) construct, with 4SU within the internal D′ element and both the D′ and D boxes mutated (Figure 3C, lane 3), generated no cross-linked fibrillarin, but rather greatly enhanced the appearance of a 45 kDa cross-linked protein (a cross-link also occasionally seen with wild-type snoRNAs, particularly at incubation times <4 h; Figures 2C and D, and 3C, and data not shown).
Mutation of the C′ element from 4SUGAGGA to 4SUCCGGA resulted in the loss of cross-linked Nop56, but fibrillarin cross-linking was still observed (Figure 3D, lane 2). Thus, Nop56’s interaction with the C′ box appears to be very sequence specific, whereas fibrillarin interacts with the C′ box in a sequence-independent fashion. More extensive mutation of the D′ element [D′(D′fullmt): C4SUGA to G4SUCC] (Figure 3D, lane 3) or mutation of both the D′ and C′ elements, with the 4SU in either element [C′-1(D′mt, C′mt) and D′(D′mt, C′mt)], likewise revealed no alteration in the cross-linking of fibrillarin (Figure 3D, lanes 4 and 5). The sequence requirements for Nop58 binding were not assessed since mutation of the C box element destabilizes the snoRNA in the oocyte (Caffarelli et al., 1998).
Nop56 and Nop58 cross-linking occurs in fibrillarin-containing snoRNPs
We next asked whether any of the additional cross-linked bands visualized in Figures 1, 2 and 3 might be previously unrecognized, but genuine snoRNP proteins. For instance, there is a major cross-linked protein at 31 kDa, a doublet migrating at ∼97 kDa, and the 45 kDa band that is enhanced with the mutant constructs. To determine whether these proteins are a part of fibrillarin-containing snoRNPs, nuclei were injected with a 4SU-containing U25 snoRNA, incubated, and irradiated as above. Only after immunoprecipitation with anti-fibrillarin antibodies were snoRNPs disassembled by RNase treatment and the cross-linked proteins analyzed.
As expected, fibrillarin itself was strongly labeled in the immunoprecipitate, whether cross-linked to the wild-type D′ construct (Figure 4A, lane 1), either D′ mutant [D′(D′mt) and D′(D′fullmt)] (Figure 4A, lanes 3 and 5) or the C′ wild-type and mutant construct (Figure 4B, lanes 1 and 3). Importantly, both labeled Nop56, generated with the C′-1 construct (Figure 4B, lane 1), and labeled Nop58, generated with the C-2 construct (Figure 4C, lane 3), were precipitated by anti-fibrillarin antibodies even though fibrillarin does not cross-link to the C-1 or C-2 constructs (Figure 4C, lanes 2 and 4, and Figure 1C). In contrast, the strong doublet at 97 kDa generated by either the C′-1 wild-type or mutant construct (Figure 4B, lanes 2 and 4) was not precipitated, indicating that these two proteins interact non-specifically with the U25 snoRNA or perhaps represent assembly factors that are not part of a mature snoRNP. Likewise, the cross-linked 45 kDa protein generated with mutant constructs (Figure 3) was not precipitated with anti-fibrillarin (Figure 4B, lanes 3 and 4, and data not shown) nor was the 31 kDa protein generated by the C-1 or C-2 construct (Figure 4C, lanes 1 and 3). We conclude that the 97, 45 and 31 kDa cross-linked proteins are not components of mature fibrillarin-containing box C/D particles.
Fig. 4. Anti-fibrillarin antibodies co-immunoprecipitate cross-linked fibrillarin, Nop56 and Nop58. After injection of nuclei with the D′, D′(D′mt), D′(D′fullmt), C′-1, C′-1(C′mt), C-1 or C-2 construct, cross-linked samples were immunoprecipitated with the anti-fibrillarin monoclonal antibody (72B9). The pellets (P) and supernatants (S) were then RNase digested and equal amounts were resolved on 12.5% SDS–polyacrylamide gels. Labeling is as in Figures 2 and 3.
The 15.5 kDa protein is not detected by cross-linking, but is present in oocyte assembled snoRNPs
The fourth known box C/D-associated protein, the 15.5 kDa protein, is essential for the stability of a box C/D snoRNP (Watkins et al., 2000). However, it did not appear in any of our cross-linking profiles (Figures 1, 2, 3 and 4; data not shown). This was surprising since structural information predicts that the first nucleotide of the C box, which is replaced with 4SU in the C-1 construct, should closely contact the 15.5 kDa protein, participating in several hydrogen bonds and forming extensive van der Waals interactions (Vidovic et al., 2000). Instead, no cross-links to any bona fide box C/D-associated protein were detected with the C-1 construct; only the 31 kDa non-specifically cross-linked band was seen (Figure 4C, lanes 1 and 2).
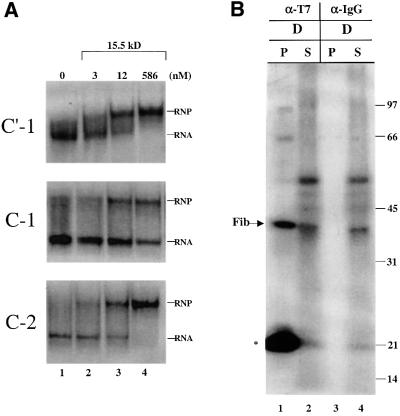
There are two potential explanations. First, the 4SU of the C-1 construct, when buried in the 15.5 kDa protein, may be constrained such that it is unable to adopt a position optimal for cross-linking. Secondly, substitution of the first U within the C box with 4SU may adversely affect the binding of the 15.5 kDa protein to U25. A gel shift assay was therefore employed to compare the binding of recombinant 15.5 kDa protein to the C′-1, C-1 and C-2 constructs (Figure 5A). Whereas all three RNAs formed a shifted complex with the 15.5 kDa protein in the presence of an ∼10-fold excess of protein, both the C′-1 and C-2 constructs were fully shifted compared with only 50% of the C-1 construct. We conclude that the substitution of 4SU in the first position of the C box significantly lowers the RNA’s affinity for the 15.5 kDa protein and could explain the absence of a cross-link. This explanation is supported by the inability of the C-1 construct to rescue methylation activity, as described below.
Fig. 5. The 15.5 kDa protein assembles on U25 constructs in the oocyte. (A) Substitution of 4SU at the C-1 position adversely affects the binding of the 15.5 kDa protein. The C′-1, C-1 or C-2 RNA was incubated either alone (lane 1) or in the presence of recombinant 15.5 kDa protein at the concentrations indicated (see Materials and methods). Complexes were resolved on an 8% non-denaturing polyacrylamide gel, with the RNA alone (RNA) and the shifted RNA complex (RNP) indicated. (B) Approximately 20 nuclei isolated under paraffin oil were co-injected with the D construct (21 fmol/nucleus) and a T7 epitope-tagged recombinant 15.5 kDa protein (12.5 µM; gift of T.Hirose). The nuclei were incubated, irradiated and immunoprecipitated with anti-T7 tag antibody or mouse IgG. Samples were RNase digested, and equal fractions of the pellets (P) and supernatants (S) were resolved on a 12.5% SDS–polyacrylamide gel as in Figures 2–4.
To demonstrate that the 15.5 kDa protein nonetheless was assembling onto the other U25 constructs, recombinant human 15.5 kDa protein containing a T7 epitope tag was co-injected with the D box construct. Cross-linked nuclear extracts, prepared as above, were incubated with a monoclonal antibody that targets the T7 tag followed by RNase digestion. Figure 5B shows that labeled fibrillarin was co-immunoprecipitated by this antibody, demonstrating that the 15.5 kDa protein had indeed assembled onto the injected RNA construct. The large amount of undigested RNA (*) precipitated was most probably due to mixing the protein and RNA prior to injection, leading to aggregates resistant to digestion.
The cross-linking constructs reconstitute methylation activity
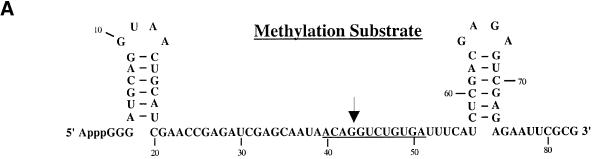
The oocyte system provides the opportunity to assess whether snoRNPs assembled with the 4SU-containing U25 RNAs are functional. Endogenous U25 snoRNA was depleted from a pool of oocytes by injecting an antisense DNA oligonucleotide that elicits selective degradation of U25 by endogenous RNase H and is then itself hydrolyzed (Prives and Foukal, 1991). These oocytes next received a U25 cross-linking construct co-injected with a 32P- labeled artificial methylation substrate (Tycowski et al., 1998; Figure 6A), containing a 12 nucleotide sequence identical to that which is methylated in 18S rRNA (see Figure 1A). Site-specific methylation of the substrate within the sequence complementary to the U25 snoRNA was assayed by exploiting the fact that RNase H does not cleave a site that is 2′-O-methylated when targeted by an RNA–DNA chimera (Yu et al., 1997).
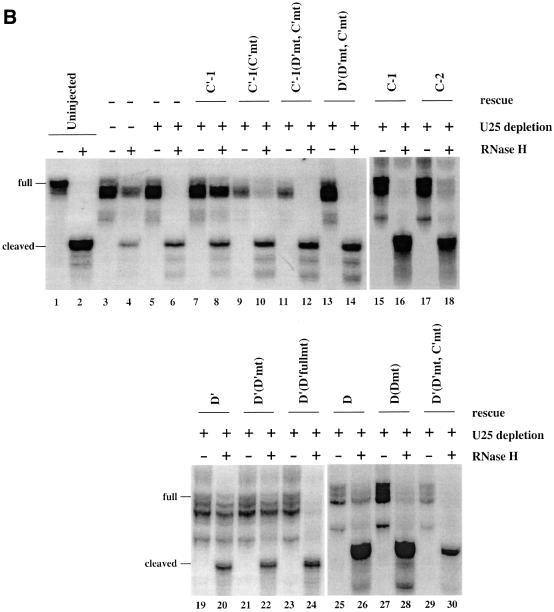
Fig. 6. U25 constructs reconstitute methylation activity in vivo. (A) The substrate used to assay the methylation activity of substituted U25 snoRNPs is shown, with underlining indicating sequences complementary to the U25 snoRNA and the arrow indicating the nucleotide targeted for methylation. (B) The methylation activity of reconstituted U25 snoRNPs was assessed by co-injecting a 4SU-containing U25 snoRNA construct with the methylation substrate into nuclei 18 h after oocytes were injected with an antisense U25 DNA oligonucleotide. Nuclear RNA was isolated 4 h later and incubated with the chimeric oligonucleotide chim-U25 and RNase H as indicated. Lanes 1 and 2 show the uninjected substrate incubated in the absence or presence of RNase H. In lanes 3 and 4, the endogenous U25 snoRNA was not depleted. The amount of residual full-length substrate (full) and the cleaved substrate (cleaved) provides a qualitative assessment of the extent of site-specific methylation. In lanes 19–30, the full-length product was more degraded than in lanes 1–18. The activity of each construct was reproduced in three separate experiments.
Figure 6B shows that uninjected substrate can be fully cleaved in the presence of RNase H and the chimeric oligonucleotide (Figure 6B, lanes 1 and 2). Substrate recovered from undepleted oocytes was methylated by the endogenous U25 snoRNA, as revealed by its partial protection from cleavage by RNase H (Figure 6B, lanes 3 and 4). In oocytes depleted of the U25 snoRNA, the substrate was not methylated (Figure 6B, lanes 5 and 6). When 4SU-containing constructs having the wild-type U25 sequence were injected, methylation activity (appearance of uncleaved full-length substrate) was observed (Figure 6B, lanes 8, 18, 20 and 26). Only the C-1 construct showed no rescue (lane 16), presumably because its binding of the 15.5 kDa protein is perturbed, as shown in Figure 5A. The C-2 construct was active in methylation, although its activity appeared weak in comparison with the other wild-type constructs (lane 18) in this non-quantitative assay.
SnoRNA constructs with mutations in the C, C′, D′ or D boxes exhibited either partial (lane 10) or complete loss (lanes 12, 14, 24, 28 and 30) of methylation activity, with all double mutants showing complete loss. One surprise was the D′(D′mt) construct, whose methylation activity was comparable reproducibly with that of the wild-type D′ construct (lanes 20 and 22). Although the G and A nucleotides are the most conserved nucleotides within the D′ box (Kiss-Laszlo et al., 1998), the activity of this mutant suggests that they may not be required for methylation. We conclude that the incorporation of 4SU within the individual box elements (with the exception of the C-1 position) does not disrupt the proper assembly of a box C/D snoRNP or adversely affect the ability of the synthetic snoRNA to reconstitute site-specific methylation. It therefore seems likely that cross-linking takes place within functional reconstituted snoRNPs.
The position of the guide sequence does not alter the asymmetric cross-linking of Nop56 and Nop58
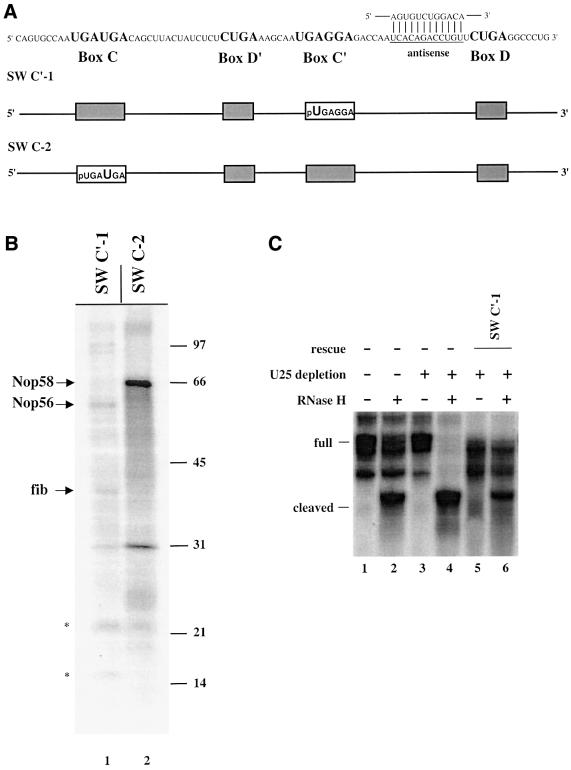
Finally, we asked whether the exclusive cross-linking of Nop56 and Nop58 to boxes C′ and C, respectively, might reflect the position of the guide sequence, which in U25 is located upstream of box D′ (Figure 1A). The antisense element was thus relocated 5′ to the D box (Figure 7A) by switching the 19 nucleotide sequence that lies between the C′ box and D box of U25 with the 15 nucleotide sequence between the C box and D′ box. The C′-1 position and the C-2 position were again substituted with 4SU and the constructs analyzed by cross-linking (Figure 7B). Nop56 cross-linked exclusively to box C′ and Nop58 to box C in the switched (SW) constructs, the same pattern as observed for wild-type U25. Moreover, the construct with the D box guiding methylation was functional in rescuing methylation activity in the oocyte (Figure 7C), showing that U25 possesses two potentially functional guide centers—at the D and D′ boxes.
Fig. 7. The C′ box cross-links to Nop56 and the C box to Nop58, regardless of the location of the guide sequence. (A) U25 constructs were designed with the 15 nucleotide sequence between the C box and D′ box switched for the 19 nucleotide sequence between the C′ box and the D box. The constructs are schematized and named as in Figure 1A, with SW indicating switched. (B) Cross-linking and analyses of labeled proteins were as in Figure 1. (C) Reconstitution of methylation activity with the SW C′-1 construct was assessed as in Figure 6B.
Discussion
The symmetry observed at the RNA level for virtually all methylation guide snoRNAs (currently ∼90 identified in mammals; Huttenhofer et al., 2001) and the ability of some of these to direct two independent modification reactions led to the suggestion that box C/D snoRNPs are highly symmetric, consisting of two sets of core proteins (Kiss-Laszlo et al., 1998; Venema and Tollervey, 1999). We tested this hypothesis by site-specific cross-linking using 4SU inserted into the box C, C′, D and D′ elements of the U25 snoRNA. Since these are the only sequences conserved within this snoRNA class, it seemed likely that observed interactions with the four proteins known to associate specifically with box C/D snoRNAs would be general. Our analyses surprisingly demonstrate that the U25 snoRNP is in fact asymmetric, with each box C/D motif interacting with a distinct protein subset.
The RNA–protein contacts identified by 4SU cross-linking are summarized in Figure 8A. We found no evidence that the conserved sequence boxes within a mature fibrillarin-containing snoRNP contact any proteins other than the four known box C/D components. It is possible that the additional bands we observed are proteins that aid in snoRNP assembly, but are not present in fibrillarin-containing assembled particles (Figure 4). Yet, none of these additional cross-linked proteins exactly correspond to the size of any protein so far implicated in snoRNP biogenesis (Filipowicz and Pogacic, 2002).
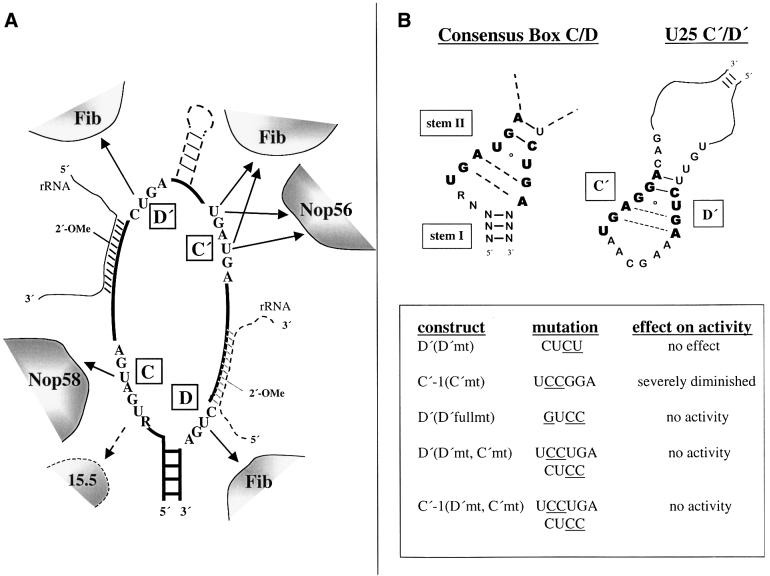
Fig. 8. (A) Summary of the RNA–protein contacts identified by 4SU site-specific cross-linking. The position of the 15.5 kDa protein is indicated with a dashed arrow, since this reported interaction (Watkins et al., 2000) was not observed by cross-linking. The three contacts to fibrillarin do not imply three copies of the fibrillarin molecule; they could reside on one, two or three copies of the protein (see text). Whereas the antisense sequence is 5′ of the D′ box in wild-type U25, the antisense region was switched to be 5′ of the D box in the ‘SW’ constructs, with the rRNA substrate and additional sequences between boxes C′ and D′ found in some snoRNAs indicated by dashed lines. (B) Comparison of the C′/D′ sequences of the U25 snoRNA with those of the consensus box C/D motif (Watkins et al., 2000).
Fibrillarin (recently identified as the methyltransferase; Omer et al., 2002) becomes cross-linked to the D, D′ and (both substituted positions of the) C′ boxes, but does not appear to contact either 4SU position within the C box. In box C/D snoRNAs, the guide sequence is positioned 5′ of the D or D′ box, with the nucleotide to be modified located exactly 5 bp upstream of the conserved sequence. Fibrillarin cross-linking to boxes D and D′ is consistent with its role as the 2′-O-methytransferase. Surprisingly, none of the mutations within the C′ or D′ boxes perturbed the observed cross-linking interaction with fibrillarin.
The asymmetry of the box C/D particle is evident from the distinct cross-linking patterns of Nop56 and Nop58, homologous proteins that exhibit 37% sequence identity in human (Lyman et al., 1999). Cross-linked Nop58 was observed only with 4SU in the second position of the C box, while Nop56 appears to contact both U positions in the consensus C′ box. Our mutational analyses further indicated that Nop56 interacts with the C′ element in a highly sequence-specific manner; a change in the two residues 3′ to the 4SU position completely abrogated cross-linking (Figure 3D). The cross-linking profiles produced by the constructs in which the location of the antisense element was switched (‘SW’ constructs; Figure 7B) demonstrated that the positioning of the guide sequence does not alter the RNA contacts made by Nop56 and Nop58. The Nop56 and Nop58 cross-links generated here are likely to correspond to the Xenopus p62 and p68 cross-links previously characterized (Caffarelli et al., 1998; Fillippini et al., 2000).
The methylation activity assay (Figure 6) verified that incorporation of 4SU within the individual box elements did not adversely affect the assembly and function of a box C/D snoRNP (with the exception of substitution at the C-1 position). The mutant constructs provide further insights into the requirements for site-specific methylation. Kiss-Laszlo et al. (1998) demonstrated that significant mutation of either the D′ (CUGA to AGUC) or C′ (UGAUGA to CCAUGG) sequence resulted in the complete loss of methylation activity, establishing that the C′ element functions in conjunction with the D′ element in the modification reaction. The sequences of these internal boxes can be quite divergent from the consensus box C and D elements and yet they still maintain the necessary sequence information required to direct methylation. What then are the features of these elements that are truly indispensable for site-specific methylation? Mutating the D′ sequence from CUGA to CUCU had little or no effect on methylation (Figure 6B), consistent with the generally loose conservation of the D′ sequence (Kiss-Laszlo et al., 1998), whereas further mutation of the D′ sequence (CUGA to GUCC) abolished activity. In contrast, mutation of just two nucleotides of the C′ element (UGAGGA to UCCGGA) severely diminished the methylation activity (Figure 6B) and resulted in the loss of the Nop56 cross-link (Figure 3D).
The architectural data we have obtained for the U25 snoRNP (Figure 8A) are probably generalizable to all methylation guides regardless of the position of the antisense element. This is supported by the switch experiments (Figure 7), which demonstrate the potential of U25 to utilize either the C/D or the C′/D′ motif to guide methylation without changing its RNA–protein contacts. Additional support for the asymmetric assembly of proteins within a methylation guide RNA comes from studies in yeast. Binding of proteins to the box C/D terminal motif is believed to stabilize the snoRNA by protecting its 5′ and 3′ ends from exonucleolytic digestion (Venema and Tollervey, 1999). The report that Nop56 is not essential for snoRNP stability (Lafontaine and Tollervey, 2000) therefore rationalizes lack of a cross-link between Nop56 and the terminal box C/D motif. Also, the experimental observation (Gautier et al., 1997; Lafontaine and Tollervey, 2000) that yeast Nop56 interacts with box C/D snoRNAs only in association with Nop1p (fibrillarin) is consistent with 4SU in the C′ box cross-linking to both Nop56 and fibrillarin. This protein– protein association may explain further why both Nop56 and Nop58 were affinity purified with a truncated mouse U14 snoRNA lacking internal C′ and D′ elements (Watkins et al., 1998b; Newman et al., 2000).
One limitation of cross-linking approaches is that contacts can be deduced from the appearance of a cross-link, but the absence of a cross-link does not establish lack of interaction. Thus, our inability to cross-link the 15.5 kDa protein provides no information on whether this protein interacts with the C′/D′ boxes as well as with the terminal C/D motif (Watkins et al., 2000). However, interference mapping of a box C/D snoRNP both in vivo and in vitro (L.B.Weinstein Szewczak, S.J.DeGregorio, S.A.Strobel and J.A.Steitz, submitted) provides evidence that the 15.5 kDa protein interacts solely with the C/D elements, consistent with asymmetric protein assembly. The presence of 15.5 kDa in at least one copy in snoRNPs assembled on our U25 constructs was confirmed by co-immunoprecipitation (Figure 5B). Although the stoichiometry of the proteins in the particle is not addressed by cross-linking, we can conclude that the second U of the C box and both substituted positions of the C′ box are in close contact with Nop58 and Nop56, respectively. Thus, unique protein contacts are made with the C′ and C box elements, not only by fibrillarin, but also by Nop56 and Nop58. The switch experiments (Figure 7) and the observation that both boxes D and D′ cross-link fibrillarin strongly argue for the presence of two fibrillarin molecules in box C/D snoRNPs.
The asymmetrical arrangement of proteins within box C/D snoRNPs could result if the internal C′ and D′ elements do not adopt the same RNA fold as the well-characterized stem–internal loop–stem structure of the terminal C/D element (Watkins et al., 2000). Nonetheless, our cross-linking data do hint at common features of the two motifs (Figure 8B). For instance, mutation of the second and third residues of the C′ box [C′-1(C′mt)] would be expected to disrupt the sheared G–A base pairs. Indeed, this construct exhibits altered protein interactions (Figure 3) and severely diminished methylation activity (Figure 6). Yet, in yeast, depletion of neither Nop56 nor Nop58 noticeably alters ribose methylation (Lafontaine and Tollervey, 1999, 2000), suggesting that the C′ mutation produces an RNA structural change that leads to both the loss of methylation activity and perturbation of an RNA–protein contact. When both the D′ and C′ elements are mutated, no methylation activity remains (Figure 6), presumably because all potential for correct orientation of the C′ and D′ elements has been eliminated. The D′ full mutant (GUCC) additionally lacks all methylation activity (Figure 6); this mutation would be predicted to disrupt both the sheared G–A base pairs and the formation of the conserved G–C base pair in stem II. Altering the sequence of stem II of the box C/D motif perturbs the in vitro assembly of the U14 snoRNP (N.J.Watkins, personal communication), supporting the importance of this stem. The loss of activity of the U25 mutants studied therefore highlights the necessity of bringing the C′ and D′ elements together when the guide sequence resides upstream of box D′.
Why two closely related proteins, Nop56 and Nop58, differentially recognize the two box C/D like motifs is not clear. The greatest conservation between mouse Nop56, Nop58 and their archaeal homolog, aNop56 (Omer et al., 2000), lies within the central domain of the proteins (amino acids 78–411; Newman et al., 2000), with the termini diverging in sequence. Disruption of either yeast Nop56 or Nop58 cannot be compensated by the overexpression of the other, emphasizing their uniqueness (Gautier et al., 1997). Thus, it seems likely that the divergent termini of Nop56 and Nop58 make differential contacts with the snoRNA or with other protein components of the snoRNP. In archaea, in contrast, methylation guide RNPs (Gaspin et al., 2000) possess the terminal C/D motif, C′/D′ elements that are more highly conserved than in eukaryotes, homologs to the other associated proteins, but only a single Nop56/58 homolog (aNop56). Thus, it will be most interesting to learn the actual stoichiometry of the individual proteins in both eukaryotic and archaeal box C/D snoRNPs.
Materials and methods
Oligonucleotides for site-specific constructs
All RNA and DNA oligonucleotides were purchased either from the Keck Oligonucleotide Synthesis Facility, Yale University or from Dharmacon Research, Inc. [5′D′(D′fullmt), 5′ and 3′ C-1, 5′ and 3′ C-2, 5′ and 3′ SW C′-1, 5′ 4-thioU and 3′ SW C-2]. Oligonucleotides containing 4-thiouridine purchased from the Keck facility were synthesized with 2′-deoxy-4-thiouridines (4SdU). The site-specific constructs were generated by ligating together two or three pieces of RNA with T4 DNA ligase. A DNA bridging oligonucleotide (Br) spanning the ligation junction(s) mediated each ligation reaction (see Ligations). The RNA and DNA Br oligonucleotides used to generate site-specific cross-linking constructs are listed in the Supplementary data available at The EMBO Journal Online.
Ligations
Ligation reactions were based on Moore and Sharp (1992) with minor modifications. Twenty picomoles of a 4-thioU-containing RNA was 5′-end labeled with 1 µl (3 U) of T4 polynucleotide kinase (USB) and 1 µl (15 µCi) of [γ-32P]ATP (6000 Ci/mmol; New England Nuclear) in 50 mM Tris–HCl pH 8.0, 10 mM MgCl2 and 5 mM dithiothreitol (DTT) in a total volume of 10 µl for 30 min at 37°C and then ethanol precipitated. The phosphorylated fragment (3′ RNA) was mixed with 40 pmol of a 5′ RNA fragment and 20 pmol of the DNA bridge (Br) spanning the ligation junction in a total volume of 6 µl of H2O. Samples were heated to 95°C for 2 min and cooled slowly to room temperature. Then, 3 µl of a 5× reaction buffer, containing 250 mM Tris–HCl pH 7.5, 50 mM MgCl2, 100 mM DTT, 5 mM ATP and 5% polyvinyl alcohol, was added to the annealed mixture in addition to 1 µl (40 U) of RNasin and 5 µl (1 U/µl) of T4 DNA ligase (United States Biochemical). The reaction was incubated at 37°C for 2 h. The ligated products were gel purified on a 10% denaturing polyacrylamide gel and visualized by autoradiography. For the activity assay, the ligations were scaled up 10-fold to produce in the order of 250–500 pmol of RNA trace-labeled with [γ-32P]ATP for purposes of calculating the exact amount of ligated material recovered.
Oocyte microinjection
Oocytes for microinjection were isolated manually from the ovary in OR3 medium (Romero et al., 1998). For cross-linking analyses, nuclei were isolated from oocytes and prepared for injection as detailed by Lund and Paine (1990). Typically 9 nl of a 4SU-containing RNA (600 000 c.p.m./µl H2O) were injected per nucleus, with seven nuclei injected per construct. For immunoprecipitation experiments, larger numbers of nuclei were injected, as detailed below.
U25-depleted oocytes were generated as described (Tycowski et al., 1996). For ‘rescue’ experiments, after the overnight incubation with DNA at 18°C, the germinal vesicles of the depleted oocytes were injected with 27 nl of the activity assay substrate (2 × 106 c.p.m./µl or ∼90 fmol/µl) with or without 4.7 fmol of a 4-thioU-containing construct to be assayed for activity. After 4 h incubation at 18°C, oocytes were dissected and RNA was isolated from the nuclei (Peculis and Steitz, 1993).
Cross-linking analyses
Seven nuclei were incubated under mineral oil for 4 h post-injection and then irradiated with 365 nm light for 10 min (Sontheimer, 1994) on ice. Nuclei were collected and digested with a mixture of RNase T1 (2.6 U/µl; Calbiochem) and nuclease P1 (0.05 U/µl; Roche) in a final volume of 45 µl for 1 h at 45°C. Samples were brought up in protein sample loading dye (2% SDS, 80 mM Tris–HCl pH 6.8, 10% glycerol, 5% β-mercaptoethanol and Bromophenol Blue marker dye) and resolved by 12.5% (Laemmli, 1970) or 15% (Anderson et al., 1973) SDS–PAGE. Labeled proteins were revealed by autoradiography.
Generation and characterization of antibodies against Xenopus Nop56, Nop58 and fibrillarin
Full-length cDNAs encoding Xenopus laevis Nop56 and Nop58 were obtained by RT–PCR and RACE (Marathon RACE; Clontech) using mRNA from late stage Xenopus oocytes (mRNA separator kit; Clontech) and degenerate oligonucleotides based on sequences available from other organisms. The cDNAs were cloned into the EcoRI site of pET32a (Novagen, WI) and the fusion proteins were expressed in Escherichia coli BL21 (DE3) and isolated from inclusion bodies by SDS gel purification. Xenopus fibrillarin cDNA (Lapeyre et al., 1990) was used to produce recombinant fibrillarin. Polyclonal antibodies were raised by injecting ∼200 µg of each purified protein into New Zealand white rabbits. Immunoprecipitation was performed as described (Terns et al., 1992) using 10 µl of immune or pre-immune serum and 8 µl of rabbit reticulocyte lysate containing either Xenopus Nop56, Nop58 or fibrillarin prepared by in vitro transcription and translation (TnT; Promega) in the presence of [35S]methionine (Amersham Pharmacia Biotech). Immunoblot analysis was performed using five oocyte nuclei per lane. Primary antibodies were diluted 1:1000 (Nop56), 1:500 (Nop58) or 1:100 (fibrillarin) and detected using either horseradish peroxidase- (Nop56 and Nop58) or alkaline phosphatase- (fibrillarin) conjugated secondary antibodies (1:5000). Immunoreactive bands were visualized with ECL (Amersham) for Nop56 and Nop58 or BCIP/NBT (Sigma) for fibrillarin.
Immunoprecipitations
A 30 µl aliquot of anti-fibrillarin, anti-Nop56 or anti-Nop58 polyclonal serum was coupled to 4 mg of pre-swollen protein A–Sepharose beads (Pharmacia) in NET-2 buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.05% NP-40) for 3 h at 4°C. For each immunoprecipitation, ∼40 nuclei, which had been injected and cross-linked as detailed above, were disrupted in 100 µl of NET-2 buffer, with the NaCl concentration adjusted to 400 mM. After centrifugation for 10 min at 10 000 r.p.m., the cleared extracts were diluted to 150 mM NaCl and subjected to nuclease digestion as detailed above. Following digestion, the total volume was adjusted to 400 µl with NET-2 buffer and incubated with antibody-coupled beads for 2 h at room temperature with constant oscillation. The beads were washed four times with 1 ml of NET-2 buffer, and 10 uninjected nuclei were added to each final Sepharose pellet before boiling in loading buffer for 5 min at 95°C and resolution by SDS–PAGE. The additional nuclei reduced smearing of the labeled protein bands on the gels. Co-immunoprecipitation experiments using monoclonal anti-fibrillarin (72B9) (Reimer et al., 1987) or anti-T7 tag (Novagen) antibodies were carried out as above, except that 20 nuclei were injected for each experiment and the antibodies were used in the following amounts: 200 µl of 72B9 and 10 µl of the anti-T7 tag. Importantly, samples were RNase digested following, rather than preceding, the 2 h incubation with antibody-coupled beads.
Gel retardation assay
Gel shifts were performed essentially as described (Nottrott et al., 1999; Watkins et al., 2000). A GST–15.5 kDa fusion protein was expressed from the pGEX4T2-15.5kDa plasmid (gift of R.Luhrmann), the GST tag was removed by thrombin cleavage and the cleaved 15.5 kDa protein was purified (kindly provided by L.Szewczak). Approximately 20 000 c.p.m. of the C′-1, C-1 or C-2 construct, prepared as described above, was incubated with recombinant 15.5 kDa at a final protein concentration of 0, 3, 12 or 586 nM for 1 h at 4°C as described (Nottrott et al., 1999).
Activity assay
The synthetic RNA methylation substrate was prepared by T7 transcription of a DNA template, generated by annealing oligonucleotides 41 and 42 (see Supplementary data) and filling in the single-stranded overhangs with Klenow fragment (Sambrook et al., 1989).
RNA prepared from nuclei co-injected with the assay substrate and a U25 construct (see ‘Oocyte microinjection’) was subjected to RNase H digestion (Yu et al., 1997). Briefly, 10 ng of a chimeric oligonucleotide (U25 chim: 5′-GAdCdCdTdGUUAUUGCUCGAUC-3′) was annealed to RNA isolated from 8–10 nuclei in a total volume of 3 µl of deionized water. Samples received 2 µl of a 5× buffer (250 mM Tris–HCl pH 7.5, 50 mM MgCl2, 100 mM DTT, 5 mM ATP and 1% polyvinyl alcohol), 4 µl (4 U) of RNase H (Roche) and 1 µl (40 U) of RNase inhibitor (Roche) and were incubated for 2 h at 37°C. RNAs were resolved on 10% denaturing polyacrylamide gels.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Tets Hirose, Robin Lytle, Kazio Tycowski and Lara Szewczak for critical reading of the manuscript. The following provided generous gifts of reagents: Reinhard Luhrmann (pGEX4T2-15.5 kDa plasmid), Lara Weinstein (purified recombinant human 15.5 kDa), Tets Hirose (purified recombinant human his-T7-15.5 kDa protein), Andrew Lukowiak (fibrillarin western blot) and Michele Caizergues-Ferrer (Xenopus fibrillarin cDNA). This work was supported by National Institutes of Health grants GM26154 to J.A.S. and GM54682 to M.P.T. J.A.S. is an investigator of the Howard Hughes Medical Institute.
References
- Anderson C.W., Baum,P.R. and Gesteland,R.F. (1973) Processing of adenovirus 2-induced proteins. J. Virol., 12, 241–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakin A.G., Smith,L. and Fournier,M.J. (1996) The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell, 86, 823–834. [DOI] [PubMed] [Google Scholar]
- Bortolin M.-L., Ganot,P. and Kiss,T. (1999) Elements essential for accumulation and function of small nucleolar RNAs directing site-specific pseudouridylation of ribosomal RNAs. EMBO J., 18, 457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffarelli E., Losito,M., Giorgi,C., Fatica,A. and Bozzoni,I. (1998) In vivo identification of nuclear factors interacting with the conserved elements of box C/D small nucleolar RNAs. Mol. Cell. Biol., 18, 1023–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaillé J., Nicoloso,M. and Bachellerie,J.P. (1996) Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature, 383, 732–735. [DOI] [PubMed] [Google Scholar]
- Filipowicz W. and Pogacic,V. (2002) Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol., 14, 319–327. [DOI] [PubMed] [Google Scholar]
- Fillipini D., Bozzoni,I. and Caffarelli,E. (2000) p62, a novel Xenopus laevis component of box C/D snoRNPs. RNA, 6, 391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganot P., Bortolin,M.-L. and Kiss,T. (1997) Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell, 89, 799–809. [DOI] [PubMed] [Google Scholar]
- Gaspin C., Cavaille,J., Erauso,G. and Bachellerie,J.P. (2000) Archaeal homologs of eukaryotic methylation guide small nucleolar RNAs: lessons from the Pyrococcus genomes. J. Mol. Biol., 297, 895–906. [DOI] [PubMed] [Google Scholar]
- Gautier T., Berges,T., Tollervey,D. and Hurt,E. (1997) Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol. Cell. Biol., 17, 7088–7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhofer A., Kiefmann,M., Meier-Ewert,S., O’Brien,J., Lehrach,H., Bachellerie,J.P. and Brosius,J. (2001) RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J., 20, 2943–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T. (2001) Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J., 20, 3617–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss-László Z., Henry,Y., Bachellerie,J.-P., Caizergues-Ferrer,M. and Kiss,T. (1996) Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell, 85, 1077–1088. [DOI] [PubMed] [Google Scholar]
- Kiss-László Z., Henry,Y. and Kiss,T. (1998) Sequence and structural elements of methylation guide snoRNAs essential for site-specific ribose methylation of pre-rRNA. EMBO J., 17, 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lafontaine D.L. and Tollervey,D. (1999) Nop58p is a common component of the box C+D snoRNPs that is required for snoRNA stability. RNA, 5, 455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine D.L. and Tollervey,D. (2000) Synthesis and assembly of the box C+D small nucleolar RNPs. Mol. Cell. Biol., 20, 2650–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapeyre B., Mariottini,P., Mathieu,C., Ferrer,P., Amaldi,F., Amalric,F. and Caizergues-Ferrer,M. (1990) Molecular cloning of Xenopus fibrillarin, a conserved U3 small nuclear ribonucleoprotein recognized by antisera from humans with autoimmune disease. Mol. Cell. Biol., 10, 430–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E. and Paine,P.L. (1990) Nonaqueous isolation of transcriptionally active nuclei from Xenopus oocytes. Methods Enzymol., 181, 36–43. [DOI] [PubMed] [Google Scholar]
- Lyman S.K., Gerace,L. and Baserga,S.J. (1999) Human Nop5/Nop58 is a component common to the box C/D small nucleolar ribonucleo proteins. RNA, 5, 1597–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden B.E. (1990) The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol., 39, 241–303. [DOI] [PubMed] [Google Scholar]
- Moore M.J. and Sharp,P.A. (1992) Site-specific modification of pre-mRNA: the 2′-hydroxyl groups at the splice sites. Science, 256, 992–997. [DOI] [PubMed] [Google Scholar]
- Newman D.R., Kuhn,J.F., Shanab,G.M. and Maxwell,E.S. (2000) Box C/D snoRNA-associated proteins: two pairs of evolutionarily ancient proteins and possible links to replication and transcription. RNA, 6, 861–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J., Tien,A.L. and Fournier,M.J. (1997) Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell, 89, 565–573. [DOI] [PubMed] [Google Scholar]
- Nottrott S., Hartmuth,K., Fabrizio,P., Urlaub,H., Vidovic,I., Ficner,R. and Luhrmann,R. (1999) Functional interaction of a novel 15.5kD [U4/U6·U5] tri-snRNP protein with the 5′ stem–loop of U4 snRNA. EMBO J., 18, 6119–6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer A.D., Lowe,T.M., Russell,A.G., Ebhardt,H., Eddy,S.R. and Dennis,P.P. (2000) Homologs of small nucleolar RNAs in Archaea. Science, 288, 517–522. [DOI] [PubMed] [Google Scholar]
- Omer A.D., Ziesche,S., Ebhardt,H. and Dennis,P.P. (2002) In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex. Proc. Natl Acad. Sci. USA, 99, 5289–5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peculis B.A. and Steitz,J.A. (1993) Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell, 73, 1233–1245. [DOI] [PubMed] [Google Scholar]
- Prives C. and Foukal,D. (1991) Use of oligonucleotides for antisense experiments in Xenopus laevis oocytes. In Kay,B.K. and Peng,H.B. (eds), Xenopus laevis: Practical Uses in Cell and Molecular Biology, Vol. 36. Academic Press, San Diego, CA, pp. 185–210. [DOI] [PubMed]
- Reimer G., Pollard,K.M., Penning,C.A., Ochs,R.L., Lischwe,M.A., Busch,H. and Tan,E.M. (1987) Monoclonal autoantibody from a (New Zealand black × New Zealand white) F1 mouse and some human scleroderma sera target an Mr 34,000 nucleolar protein of the U3 RNP particle. Arthritis Rheum., 30, 793–800. [DOI] [PubMed] [Google Scholar]
- Romero M.F., Kanai,Y., Gunshin,H. and Hediger,M.A. (1998) Expression cloning using Xenopus laevis oocytes. Methods Enzymol., 296, 17–52. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schimmang T., Tollervey,D., Kern,H., Frank,R. and Hurt,E.C. (1989) A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J., 8, 4015–4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Tycowski,K.T. and Steitz,J.A. (1996) Ribosomal RNA processing in eukaryotes. In Zimmerman,R.A. and Dahlberg,A.E. (eds), Ribosomal RNA: Structure, Evolution, Processing and Function in Protein Biosynthesis. CRC Press, New York, NY, pp. 469–490.
- Sontheimer E.J. (1994) Site-specific RNA crosslinking with 4-thiouridine. Mol. Biol. Rep., 20, 35–44. [DOI] [PubMed] [Google Scholar]
- Speckmann W.A., Li,Z.H., Lowe,T.M., Eddy,S.R., Terns,R.M. and Terns,M.P. (2002) Archaeal guide RNAs function in rRNA modification in the eukaryotic nucleus. Curr. Biol., 12, 199–203. [DOI] [PubMed] [Google Scholar]
- Terns M.P. and Terns,R.M. (2002) Small nucleolar RNAs: versatile trans-acting molecules of ancient evolutionary origin. Gene Expr., 10, 17–39. [PMC free article] [PubMed] [Google Scholar]
- Terns M.P., Lund,E. and Dahlberg,J.E. (1992) 3′-end-dependent formation of U6 small nuclear ribonucleoprotein particles in Xenopus laevis oocyte nuclei. Mol. Cell. Biol., 12, 3032–3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyc K. and Steitz,J.A. (1989) U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J., 8, 3113–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski K.T., Smith,C.M., Shu,M.-D. and Steitz,J.A. (1996) A small nucleolar RNA requirement for site-specific ribose methylation of rRNA in Xenopus. Proc. Natl Acad. Sci. USA, 93, 14480–14485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski K.T., You,Z.-H., Graham,P.J. and Steitz,J.A. (1998) Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol. Cell, 2, 629–638. [DOI] [PubMed] [Google Scholar]
- Venema J. and Tollervey,D. (1999) Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet., 33, 261–311. [DOI] [PubMed] [Google Scholar]
- Vidovic I., Nottrott,S., Hartmuth,K., Luhrmannn,R. and Ficner,R. (2000) Crystal structure of the spliceosomal 15.5kDa protein bound to a U4 snRNA fragment. Mol. Cell, 6, 1331–1342. [DOI] [PubMed] [Google Scholar]
- Watkins N.J., Gottschalk,A., Neubauer,G., Kastner,B., Fabrizio,P., Mann,M. and Lührmann,R. (1998a) Cbf5p, a potential pseudo uridine synthase and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H box/ACA-motif snoRNPs and constitute a common bipartite structure. RNA, 4, 1549–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins N.J., Newman,D.R., Kuhn,J.F. and Maxwell,E.S. (1998b) In vitro assembly of the mouse U14 snoRNP core complex and identification of a 65-kDa box C/D binding protein. RNA, 4, 582–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins N.J. et al. (2000) A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell, 103, 457–466. [DOI] [PubMed] [Google Scholar]
- Weinstein L.B. and Steitz,J.A. (1999) Guided tours: from precursor snoRNA to functional snoRNP. Curr. Opin. Cell Biol., 11, 378–384. [DOI] [PubMed] [Google Scholar]
- Wu P., Brockenbrough,J.S., Metcalfe,A.C., Chen,S. and Aris,J.P. (1998) Nop5p is a small nucleolar ribonucleoprotein component required for pre-18 SrRNA processing in yeast. J. Biol. Chem., 273, 16453–16463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y.T., Shu,M.D. and Steitz,J.A. (1997) A new method for detecting sites of 2′-O-methylation in RNA molecules. RNA, 3, 324–331. [PMC free article] [PubMed] [Google Scholar]