Abstract
Background
Laboratory diagnosis of Chlamydophila psittaci, an important turkey respiratory pathogen, is difficult. To facilitate the diagnosis, a nested PCR-enzyme immunoassay (PCR-EIA) was developed to detect the Cp. psittaci outer membrane protein A (ompA) gene in pharyngeal swabs.
Methods
The fluorescein-biotin labelled PCR products were immobilized on streptavidin-coated microtiter plates and detected with anti-fluorescein peroxidase conjugate and a colorimetric substrate. An internal inhibition control was included to rule out the presence of inhibitors of DNA amplification. The diagnostic value of the ompA nested PCR-EIA in comparison to cell culture and a 16S-rRNA based nested PCR was assessed in pharyngeal turkey swabs from 10 different farms experiencing respiratory disease.
Results
The sensitivity of the nested PCR-EIA was established at 0.1 infection forming units (IFU). Specificity was 100%. The ompA nested PCR-EIA was more sensitive than the 16S-rRNA based nested PCR and isolation, revealing 105 out of 200 (52.5%) positives against 13 and 74 for the latter two tests, respectively. Twenty-nine (23.8%) out of 122 ompA PCR-EIA negatives showed the presence of inhibitors of DNA amplification, although 27 of them became positive after diluting (1/10) the specimens in PCR buffer or after phenol-chloroform extraction and subsequent ethanol precipitation.
Conclusion
The present study stresses the need for an internal control to confirm PCR true-negatives and demonstrates the high prevalence of chlamydiosis in Belgian turkeys and its potential zoonotic risk. The ompA nested PCR-EIA described here is a rapid, highly sensitive and specific diagnostic assay and will help to facilitate the diagnosis of Cp. psittaci infections in both poultry and man.
Background
Avian chlamydiosis is caused by the obligate intracellular Gram-negative bacterium Chlamydophila psittaci (formerly Chlamydia psittaci). Currently, seven genotypes of Cp. psittaci are known to infect birds [1-3]. Avian chlamydiosis in birds is usually systemic and occasionally fatal. The clinical signs vary greatly in severity and depend on the species, age of the bird and the strain of Cp. psittaci. Avian chlamydiosis can produce lethargy, hyperthermia, abnormal excretions, nasal and eye discharges, and reduced egg production. Mortality rates range up to 30% [4]. Avian chlamydiosis occurs worldwide, with the incidence and distribution varying greatly with the species of bird and the serotype of the chlamydial organism. In the past, chlamydiosis in turkeys was thought to be limited to the United States and to free-ranging flocks. Most outbreaks in US turkeys were explosive, involving one or more flocks [5-10]. Nowadays, the increase in confinement-rearing of turkeys and the prevention of wild birds flying in and out the turkey houses seems to contribute to a decrease of severe outbreaks. Probably, the situation is comparable to the one in Europe where, at present, Cp. psittaci is nearly endemic in Belgian, German and probably French turkeys [11-13]. However, devastating, explosive outbreaks with high mortality rates occur occasionally, whereas present outbreaks are mostly characterized by respiratory signs without mortality [4]. Nevertheless, Cp. psittaci causes important economical losses as a primary pathogen and trough it's pathogenic interaction with other respiratory pathogens like the avian pneumovirus (APV) and Ornithobacterium rhinotracheale (ORT) [13]. Cp. psittaci is also a threat to public health as this zoonotic agent can infect humans and precautions should be taken when handling infected birds or contaminated materials [14-17]. Human infections are common following handling or processing of infected turkeys or ducks [2,7,8,18]. Most infections are through inhalation of infectious aerosols and subsequently processing plant employees, farm workers, veterinarians and poultry inspectors are at risk. However, personnel who were employed to further process turkey meat could also become infected [19].
Thus, diagnosis is essential. In contrast to cell culture and serology, antigen detection methods like micro-immunofluorescence and PCR provide a more rapid, specific and sensitive alternative for identification of Cp. psittaci infection. However, currently described PCR assays for birds use either labour intensive and/or insensitive post PCR detection methods. A PCR-enzyme immunoassay (PCR-EIA) would circumvent this problem. At the moment, we are not aware of a nested PCR- enzyme immunoassay (PCR-EIA) for demonstrating Cp. psittaci infection, although the method has been successfully used for C. pneumoniae detection in human respiratory specimens [20,21].
The objective of the present study was to develop and evaluate a rapid and simple EIA for semi-quantitative detection of the amplified Cp. psittaci outer membrane protein A (ompA) gene, included with an internal inhibition control to eliminate possible false positive results during field sample analysis.
Methods
Specimens
In the fall of 2001, 200 fattening turkeys from 10 different farms in Belgium (8 farms) or in Northern France (2 farms) were examined at slaughter for the presence of Cp. psittaci. All turkeys had been vaccinated against Newcastle disease (NCD) (Nobilis® ND LaSota; Intervet International, Boxmeer, The Netherlands) and in 7 out of 10 farms turkeys had also been vaccinated against APV (Nobilis® RTV; Intervet International). Farmers provided information about clinical symptoms throughout the rearing period. All farms had experienced one or more periods of respiratory disease.
Pharyngeal swabs were collected from 20 ad randomly selected turkeys on each turkey farm. Of each turkey there was taken 1 sample by using cotton tipped aluminium shafted swabs (Fiers, Kuurne, Belgium) in Cp. psittaci transport medium [22] consisting of: 0.2 M sucrose (VWR International, Haasrode, Belgium); 0.015 M Na2HPO4 (VWR International), 0.01 M NaH2PO4 (VWR International) and 20% inactivated foetal calf serum (Integro, Leuvenheim, The Netherlands). Swabs were shaken vigorously for 1 hour and centrifuged (10 min, 2790 × g, 4°C). One millilitre of supernatant was provided with 1% streptomycin sulphate (10 mg/ml; Invitrogen), 2% vancomycin (5 mg/ml; Glaxo Smith Kline) and 1.6% fungizone (250μl/ml; Invitrogen) and subsequently used for Cp. psittaci isolation.
Generation of the internal control
The internal inhibition control was constructed starting from the pcDNA1 vector in which the ompA gene of a Cp. psittaci serovar D strain 92/1293 was inserted (Fig 1 &2) [23]. First, a fragment of 231 bp of the ompA gene was amplified using primers ML-BbrpI-F01 and ML-Bbrp1-R01 (table 1), which provided a BbrpI restriction site at their 5' end for subsequent cloning. The PCR reaction was performed using Pfu DNA polymerase in 50 μl reactions containing dNTP's (0.2 μM final concentration), Pfu buffer (10x), ML-BbrpI-F01 and ML-Bbrp1-R01 (0.5 μM final concentration), DMSO (7.5 %), Pfu DNA Polymerase (2.5 U, Stratagene, La Jolla, USA) and 2 μg plasmid DNA. After an initial denaturation at 95°C for 1 minute, 30 cycles of 30 seconds at 95°C, 30 seconds at 58°C and 1 minute at 72°C, followed by a final elongation at 72°C for 10 min, were performed. PCR products were subjected to electrophoresis on a 1.2% agarose gel stained with ethidium bromide and photographed under UV illumination. Size was determined using Smart Ladder (Eurogentec, Seraing, Belgium). PCR products were purified using the Qiaquick PCR Purification kit (Qiagen) and ligated into the pPCR-script™ Amp SK (+) vector (Stratagene, La Jolla, USA), as described by the manufacturer. Next, the vector was transformed into Epicurian coli XL10-Gold Kan ultracompetente cells (Stratagene) using heat shock. Clones were selected on Luria-Bertoni (LB) medium containing ampicilin (100 μg/ml) and grown in microtiter plates for 2 hours. The presence of the insert was confirmed by PCR clone analysis. Therefore, 5 μl of each clone was subjected to PCR in a 50 μl reaction mixture containing 50 mM KCl, 20 mM Tris-HCl (pH 8.3), 2 mM MgCl2, 0.1% Tween20, 200 μM each dNTP, 1.25 μM of each inner ompA primer (table 1) and 0.1 U SuperTaq polymerase (5 U/μl). After an initial denaturation at 95°C for 5 minutes, 20 cycles of one min at 95°C, two minutes at 55°C and three minutes at 72°C, with a final extension at 72°C for 5 min, were performed. To ensure PCR accuracy, the construct was sequenced using the ABI PRISM Bigdye™ Terminator Cycle Sequencing Ready Reaction Kit (ABI, Foster City, USA), following the manufacturers' manual. Sequencing samples were analyzed on the ABI PRISM 377 DNA sequencer (Perking Elmer).
Figure 1.
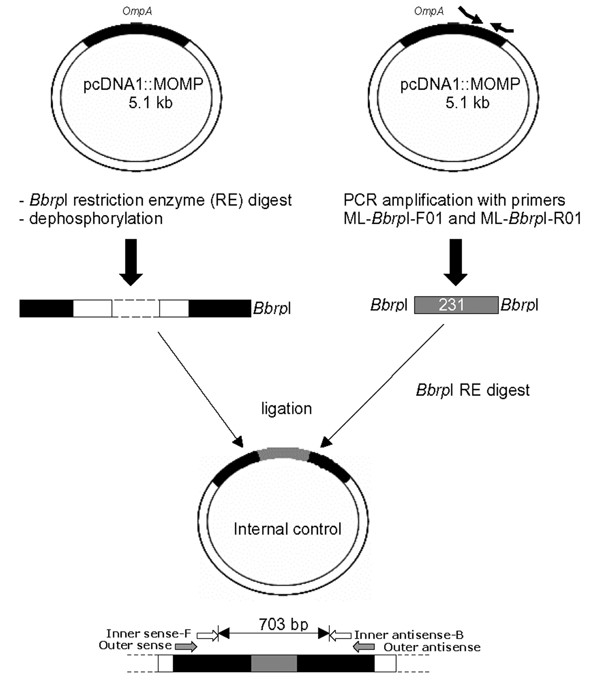
Generation of the internal control using primers ML-Bbrp1-F01 and ML-Bbrp1-R01.
Figure 2.
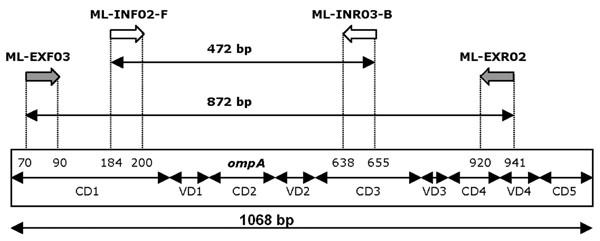
Location of the outer and inner primers in the ompA gene.Numbering according to ompA sequences in Genebank
Table 1.
Oligonucleotides used in this study
| Oligonucleotide | Length (bp) | Sequence (5'-3') |
| Sense outer 16 rRNA | 18 | ACG GAA TAA TGA CTT CGG |
| Anti-sense outer 16S rRNA | 18 | TAC CTG GTA CGC TCA ATT |
| Sense inner 16S rRNA | 21 | ATA ATG ACT TCG GTT GTT ATT |
| Anti-sense inner 16S rRNA | 20 | TGT TTT AGA TGC CTA AAC AT |
| ML-BbrpI-F01 | 29 | GCC ACG TGC GTC TGC AAC ACT CAA ATA TC |
| ML-BbrpI-R01 | 28 | GGC ACG TGC AGT TGT AAG AAG TCA GAG T |
| Sense outer ompA | 21 | CCT GTA GGG AAC CCA GCT GAA |
| Anti-sense outer ompA | 22 | GGT TGA GCA ATG CGG ATA GTA T |
| Fluorescein-sense inner ompA | 17 | GCA GGA TAC TAC GGA GA |
| Biotin-antisense inner ompA | 18 | GGA ACT CAG CTC CTA AAG |
Positive clones were grown overnight in 4 ml LB medium containing ampicilin (100 μg/ml) and subjected to the Qiaprep Spin Miniprep kit (Qiagen) to obtain purified plasmid. Using BbrpI, the 231 bp fragment was cut out of the pPCR-script™ Amp SK (+) vector and ligated into the dephosphorylated Bbrp1 site, situated within the ompA gene of the pcDNA1::MOMP vector [23]. Thus, the Bbrp1 restriction site is located within the target sequence of the inner primers and subsequently, nested PCR amplification of the inhibition control resulted in a PCR product of 703 bp (fig. 1). E. coli strain MC1061/P3 was transfected by electroporation (Gene Pulser, Bio-Rad). Again, selected clones were subjected to PCR clone analysis to asses the presence of the insert and its sequence was determined by the dideoxy chain terminating method, as described above.
Preparation of positive control DNA
Cp. psittaci strains genotype A to F strains were propagated in cycloheximide-treated BGM cells, as described elsewhere [24]. Bacteria were harvested at approximately 72 hours by disrupting the cells by subsequent freezing and thawing, followed by sonication and differential centrifugation (Urografin 76%). Purified elementary bodies were pelleted, washed, resuspended in sucrose-phosphate-glutamate buffer, and stored in aliquots at -70°C. For determination of bacterial titres, BGM monolayers grown on glass cover slips (Chlamydia Trac Bottles, International Medical) were infected and stained by the IMAGEN™ direct immunofluorescence assay [24]. Inclusion forming units (IFU) were determined by counting the numbers of inclusions cultured in duplicate in Chlamydia Trac Bottles using 10-fold serial dilutions of purified EBs. Cp. psittaci titres were expressed as IFU per millilitre and IFU quantitated in this manner were used as the positive-control DNA in PCR assays.
OmpA nested PCR-EIA
Clinical specimens included pharyngeal swabs from turkeys taken at slaughter. These specimens as well as positive control DNA were prepared for ompA nested PCR-EIA by the STD DNA extraction method, performed as followed: Cp. psittaci suspensions or turkey specimens were pelleted at 13,000 x g, resuspended in 198 μl STD buffer (0.01 M Tris-HCl [pH 8.3], 0.05 M KCl, 0.0025 M MgCl2.6H20, 0.5% Tween20) and 2 μl proteinase K (20 mg/ml stock solution; Sigma Chemical Co.). The specimens were incubated at 56°C for one hour and subsequently heated at 100°C for 10 min. Samples for both PCR's were prepared in a class II laminar flow hood, and amplification and analysis of PCR products were performed in separate locations.
The nested ompA PCR-EIA was developed using external and internal primers (table 1) generating a biotin-fluorescein dual labelled internal PCR product of 472 bp. Both inner and outer sense primers are located within the first conserved domain (CD1) of ompA, whereas the inner anti-sense primer is located in CD3 and the outer anti-sense primers overlaps CD4 and variable domain 4 (fig. 2). First round PCR occurred in 50 mM KCl, 20 mM Tris-HCl (pH 8.3), 2 mM MgCl2, 0.1% Tween20, 200 μM each dNTP, 1.25 μM each external primer (table 1) and 0.1 U SuperTaq polymerase (5 U/μl). After an initial denaturation at 95°C for 5 minutes, 20 cycles of one min at 95°C, two min at 59°C and three min at 72°C, with a final extension at 72°C for 5 min were performed. Second round amplification was performed under similar conditions, using labelled internal primers (each 10 μM; table 1), adapted annealing temperature (47°C) and adapted number of cycles (25). Subsequently, nested PCR generated a fluorescein/biotin dual-labelled product of 472 bp. To minimize false-positive results, each step of the nested PCR was performed in physically separated places.
To allow colorimetric detection of the ompA PCR products, 50 μl of the PCR product diluted 1/10 in PBS supplemented with 3% BSA was transferred in duplicate to streptavidin-coated microtiter plates (2 μg/well for 3 hours at 37°C) and incubated at 37°C for one hour. Non-specific binding places were blocked overnight (4°C) with 5% BSA in PBS. Subsequently, the plates were washed twice with PBS and incubated (1 hour, 37°C) with a horseradish peroxidase labelled anti-fluorescein antibody (Invitrogen), diluted 1/1000 in PBS supplemented with 3% BSA. Following incubating and washing with PBS, the ABTS substrate solution (2,2' azino-di-3-ethylbenzothiazoline sulphonate, KPL) was added to the wells. Absorbencies were read at 450 nm after incubating for 30 minutes at 37°C (TiterTek MultiskanR Plus, MKII, TechGen Internatonal). Three positive controls consisting of serial 10-fold dilutions of PCR products generated from 5 IFU of Cp. psittaci and five negative controls (water) were included in each assay. Results were positive if the absorbance exceeded the cut off value of the mean of negative controls plus three times the standard deviation. Nested PCR-EIA negative samples were re-tested after adding 10 ng internal inhibition control and visualized by gel electrophoresis to assess possible inhibition.
The sensitivity of the PCR was evaluated by testing 10-fold serial dilutions of DNA extracted from purified elementary bodies of Cp. psittaci strains 92/1293 [2]. Next, diagnosis of six reference strains of Cp. psittaci serovars A-F was assessed (table 2). The specificity was determined by testing DNA extracted from other bacterial species commonly found in the avian respiratory tract and avian respiratory tract tissue, originating from Cp. psittaci negative specific pathogen free turkeys (CNEVA, Ploufragan, France). The following micro-organisms were tested for cross-reactivity in the PCR assay with both the first- and second-step PCR primers: Acinetobacter species, Aspergillus flavus, Candida albicans, Enterococcus faecelis, Escherichia coli, Klebsiella species, Mycobacterium avium, Mycoplasma gallisepticum, Mycoplasma meleagridis, Ornithobacterium rhinotracheale, Pasteurella species, Proteus mirabilis, Pseudomonas species, Salmonella enteritidis, Salmonella gallinarum, Salmonella pullorum, Staphylococcus species, Streptococcus species and Xanthomonas maltophila. A Cp. psittaci positive control of 1 IFU was included in every test to verify that the PCR was working.
Table 2.
Cp. psittaci reference strains
| Strain | Year | Country | Host | Serovar | Reference |
| VS 1 | 1985 | USA, Georgia | Amazona sp. | A | [34] |
| CP3 | 1957 | USA, Calofornia | Columba livia | B | [35] |
| GD | 1960 | Germany | Anas platyrhynchos | C | [36] |
| NJ1 | 1954 | USA, New Jersey | Meleagris gallopavo | D | [37] |
| MN | 1934 | USA, California | Homo sapiensa | E | [38] |
| VS225 | 1991 | USA, Texas | Parakeet | F | [39] |
a Probably originated from birds, isolated in ferrets from human [39].
16S-rRNA nested PCR – gel electrophoresis
The performance of the ompA nested PCR-EIA was compared to those of isolation and another nested PCR, targeting the 16S rRNA gene [25]. Cp. psittaci suspensions or turkey specimens were prepared for PCR by the QiaAmp Blood kit (Qiagen Inc., Chatworth, Califormia) adapted by [25]. Genus-specific first-step primers and species-specific second step primers generated PCR products of 436 bp and 127 bp, respectively (table 1). Amplification products were visualized by gel electrophoresis (1.5% Nusieve GTG agarose, FMC Bioproducts, Rockland, Maine). PCR negatives were spiked with 5 IFU of Cp. psittaci to control for the presence of inhibitors. The limit of detection of this 16S-rRNA-based PCR was 5 IFU as previously described [25]. Samples for both PCR's were prepared in a class II laminar flow hood, and amplification and analysis of PCR products were each performed in separate locations.
Comparison to Cp. psittaci isolation
Pharyngeal swabs were examined for the presence of viable Cp. psittaci by isolation in cycloheximide-treated Buffalo Green Monkey (BGM) cells. Swabs were shaken at 4°C for 1 hour and centrifuged (10 minutes, 2790 × g, 4°C). The supernatant was used for Cp. psittaci isolation in BGM cells and subsequent identification using the IMAGEN™ direct immunofluorescence assay (DakoCytomation, Denmark), as previously described [24]. All inoculated monolayers were stained at 6 days post inoculation. Inclusion-negative cultures were passaged once. After adding an equal volume of sucrose phosphate glutamate (SPG; [25]) and freezing at -80°C, cultures were thawed, cell suspensions were sonicated and centrifuged once (2000 x g). Supernatant was inoculated in duplicate onto new BGM monolayers as described elsewhere [24]. Staining was performed at 3, and if negative at 6 days post inoculation.
Validation
A specimen was considered positive if culture positive. In addition, a culture-negative, but 16S rRNA-based PCR positive specimen was considered to be a true positive only if it could be verified by ompA-based PCR.
Results
Development of the nested PCR-EIA
Optimizing PCR conditions was performed using STD extracted DNA of Cp. psittaci serovar D strain 92/1293. Initial PCR with temperature gradients were performed with either inner or outer primer sets separately to determine optimal annealing temperature for both primer sets. The optimal annealing temperature for outer and inner primer sets was determined at 59°C and 47°C, respectively (data not shown). Next, optimal primer dilutions were tested to obtain a single band as nested PCR product, after visualization on 1.2% agarose gel (fig. 3). Hereto, external primers were used at 0.625 μM and internal primers at 10 μM. First and second round PCR amplification with the outer and inner primers resulted in PCR products of 872 bp and 472 bp, respectively. After amplification biotinylated PCR products were immobilized to streptavidin-coated microtiter wells and detected with anti-fluorescein peroxidase conjugate and a colorimetric substrate. Next, optimal enzyme immunoassay conditions were realised, among others by diluting the dual labelled PCR product 1/10 in dilution buffer (PBS + 3% BSA + 2% IgG free horse serum). All incubation steps and reaction components of this EIA were optimized prior to use with pharyngeal swabs.
Figure 3.
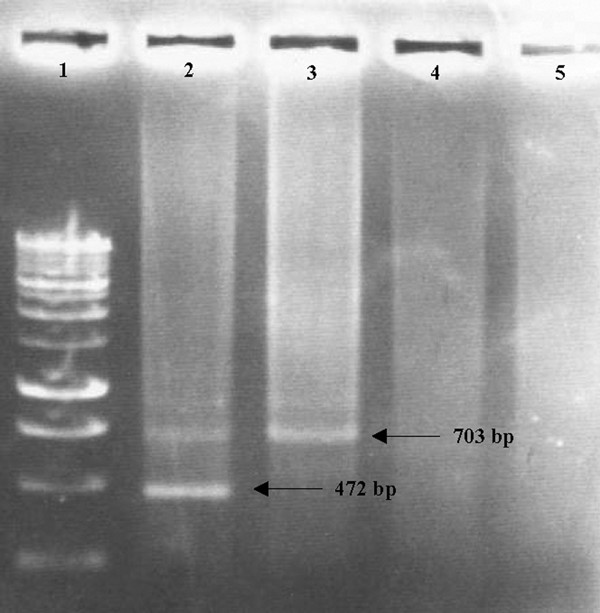
Different conditions observed in the nested PCR-EIA analysis of Cp. psittaci in turkey field samples. Lane 1: molecular marker (BenchTop 1 kb DNA ladder; Promega); lane 2: Cp. psittaci positive sample, showing one band (472 bp) diagnostic for Cp. psittaci and a band (703 bp) for the internal inhibition control; lane 3: Cp. psittaci negative sample, showing only the internal inhibition control band; lane 4: a sample with inhibitory substances lacking both the Cp. psittaci-specific band and the internal inhibition control band; lane 5: negative control, free from Cp. psittaci DNA and internal control DNA.
Sensitivity
Following definition of optimal reagent and reaction conditions, sensitivity and specificity of the ompA nested PCR was determined. The STD DNA-extraction was performed on 108 IFU and tenfold dilutions of the purified DNA were subjected to the nested PCR and visualised on a 1.2% agarose gel. This resulted in a final nested PCR product of 472 bp and a detection limit of 10-2 IFU (fig. 4A). Subjecting the dual-labelled nested PCR product to the EIA, resulted also in a detection limit of 10-2 IFU (fig. 4B) and there was a linear relationship between the measured absorbance and the tenfold dilution series. However, when the tenfold dilution series of Cp. psittaci elementary bodies was made prior to the STD DNA-extraction, sensitivity decreased one log to 10-1 IFU. Similarly, when field samples, which tested negative by nested PCR, were spiked with the serial tenfold dilutions, sensitivity decreased to 10-1 IFU.
Figure 4.
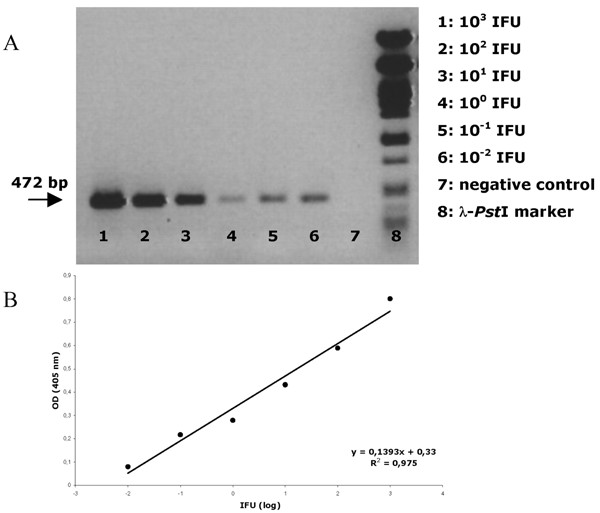
(A) Nested PCR-EIA analysis on a tenfold serial dilution of Cp. psittaci strain 92/1293. (B) Visualisation of the tenfold serial dilution on a 1.2% agarose gel following nested PCR. 103 IFU (lane 1) until 10-2 IFU (lane 6). Lane 7 shows the PCR results on a negative control. Lane 8: Phage lambda PstI fragments as size marker.
Amplification of chlamydial DNA and the internal inhibition control was achieved with the ompA inner and outer primer sets, as the additional DNA fragment for the inhibition control was inserted within the target sequence of the ompA inner primer set. Therefore, the nested PCR amplification of Cp. psittaci cultures or inhibitory substance-free clinical specimens, which were positive for Cp. psittaci exhibited two bands on ethidium-stained agarose gel electrophoresis: one band (472 bp) diagnostic for Cp. psittaci and a control band (703 bp) for the internal inhibition control (fig 3). Inhibitory substance-free clinical specimens negative for Cp. psittaci contained only the control band, indicating that no detectable inhibitors were present and that biochemical conditions were optimal for PCR amplifications. When inhibitory substances were present in field samples, no bands were detected on ethidium-stained agarose gel. Adding 10 ng of inhibition control to the nested PCR mixture was determined as the optimal condition to assess inhibition in field samples (fig 3).
Specificity
The nested PCR-EIA was able to detect al 6 tested Cp. psittaci reference strains (table 2), whereas strains of Chlamydia trachomatis, Chlamydophila pneumoniae, Chlamydophila abortus and Chlamydophila felis remained undetected. Furthermore, avian respiratory tract tissue originating from Cp. psittaci negative specific-pathogen-free turkeys (CNEVA, Ploufragan, France) and a wide range of non-chlamydial bacteria were tested and showed no cross-reactivity with: Acinetobacter species, Aspergillus flavus, Candida albicans, Enterococcus faecelis, Escherichia coli, Klebsiella species, Mycobacterium avium, Mycoplasma gallisepticum, Mycoplasma meleagridis, Ornithobacterium rhinotracheale, Pasteurella species, Proteus mirabilis, Pseudomonas species, Salmonella enteritidis, Salmonella gallinarum, Salmonella pullorum, Staphylococcus species, Streptococcus species and Xanthomonas maltophila. A Cp. psittaci positive control of 1 IFU was included in every test to verify that the PCR was working.
Analysis of pharyngeal specimens
Two hundred turkeys from 10 different farms in Belgium (8 farms) or in Northern France (2 farms) were examined at slaughter for the presence of Cp. psittaci. All farms had experienced one or more periods of respiratory disease. All samples have been analysed for the presence of Cp. psittaci by isolation in BGM cells, nested PCR-EIA and 16S rRNA nested PCR (table 3). The pharyngeal swabs were inoculated onto cycloheximide-treated BGM cells. Cp. psittaci was isolated from 54 specimens (27%) after the first inoculation and from 20 additional samples (10%) following one passage. Thus, 74 out of 200 (37%) specimens revealed to be culture positive. They were all confirmed as culture positive by ompA nested PCR-EIA analysis of the infected BGM monolayers. One hundred and twenty six samples remained negative, notwithstanding an additional 6 days passage on BGM cells. OmpA nested PCR-EIA was able to detect chlamydial DNA successfully in 105 on 200 (52.5%) pharyngeal swabs. However, 29 out of 122 (23.8%) PCR-EIA negatives clearly demonstrated inhibition showing no internal control band on the agarose gel. Seventeen out of 29 samples containing inhibitors became positive after prior 1/10 dilution in PCR buffer. Specimens that still showed inhibition were subjected to phenol-chloroform extraction and ethanol precipitation to further purify the DNA and were retested. Ten tested positive for Cp. psittaci, but 2 of 29 specimens continued to show inhibition. Thus, finally 105 out of 200 (52.5%) pharyngeal swabs tested positive by the ompA nested PCR-EIA. Surprisingly, the 16S rRNA-based PCR could only confirm 13 out of 105 ompA PCR positives revealing a total of 6.5% positive turkeys. Nine of these 13 positives could be confirmed by isolation while the remaining 4 were negative by isolation but positive by ompA PCR, indicating that they were true positives.
Table 3.
Results of nested PCR's on turkey pharyngeal specimens compared to isolation as a reference test
| Isolation | N | OmpA PCR | 16S rRNA PCR | ||
| positive | negative | positive | negative | ||
| Negatives | 126 | 31 | 95 | 4 | 122 |
| Positives | 74 | 74 | 0 | 9 | 65 |
| Total | 200 | 105 | 95 | 13 | 187 |
Referring to isolation as a reference, all 10 examined farms were Cp. psittaci positive at slaughter. The same was true when looking at the ompA nested PCR results. Although apparently less sensitive, the 16S rRNA PCR detected Cp. psittaci in 50% of the examined farms.
Discussion
A nested PCR-EIA based on the detection of the ompA gene was developed and evaluated for the diagnosis of chlamydiosis in turkeys. Nested PCR resulted in a 5' fluorescein and 3' biotin labelled ompA fragment of 472 bp which was subsequently detected in an enzyme immunoassay. Although the designed inner and outer anti-sense primers showed few nucleotide mismatches as compared to the ompA sequences of Cp. psittaci genotype C, D and F reference strains, amplification of Cp. psittaci genotypes A to F strains consistently resulted in the anticipated nested PCR-EIA product. The nested PCR-EIA was 100% specific as all Cp. psittaci genotypes were detected, but no C. trachomatis, C. pneumoniae, C. abortus or C. felis DNA. Additionally, no cross-reactivity was observed with other bacterial respiratory pathogens commonly found in the avian respiratory tract or with turkey respiratory tract DNA.
Nested PCR was chosen in order to obtain high sensitivity and specificity. Amplification of internal control DNA helped us in confirming true-negative PCR results by ruling out the presence of inhibitors of DNA amplification. Adding 10 ng of the internal inhibition control did not comprise sensitivity, as 10-2 IFU of all Cp. psittaci genotype reference strains was detected. However, when field samples, which tested negative in the nested PCR-EIA were spiked with the tenfold DNA dilutions, sensitivity decreased to 10-1 IFU, probably due to the higher amount of background DNA.
In the present study, 200 commercial turkeys, originating from 10 different farms in Belgium (8 farms) or Northern France (2 farms) were sampled at slaughter to examine for the presence of Cp. psittaci. Isolation in BGM cells revealed 74 (37%) positives. Application of the ompA nested PCR-EIA on pharyngeal DNA could confirm all culture positive results. Moreover, the ompA nested PCR-EIA detected 31 additional positives, resulting in a total of 105 (52.5%) Cp. psittaci positive turkeys. However, 29 (23.8%) of the 105 PCR-EIA positives were initially negative by the EIA and during retesting, when the internal control was added to the PCR mix, they demonstrated inhibition lacking the internal control band on an EtBr stained agarose gel. Yet, 17 samples became positive after prior 1/10 dilution of the specimen in PCR buffer. Moreover, after phenol-chloroform extraction and ethanol precipitation all but two of the sample became positive. Those two samples could not be diagnosed by the ompA nested PCR-EIA, as inhibitory substances could not be removed. Culture and 16S rRNA nested PCR for those 2 samples were also negative. However as the latter two tests have shown to be less sensitive and consequently, the presence or absence of Cp. psittaci in these 2 samples cannot be conclude. The present results clearly demonstrate the importance of using an internal control to help identify true-negatives when examining turkey pharyngeal swabs, as inhibition of DNA amplification seem to occur rather frequently in these specimens. Notwithstanding the presence of polymerase inhibitors, pharyngeal swabs still remain the first choice for sampling live birds. Pharyngeal specimens are preferred as cloacal shedding of Cp. psittaci is intermittent and, in contrast, the respiratory tract appears to be the last system to be cleared of infection. Furthermore, pathogenesis of Cp. psittaci revealed that lateral nasal glands can be infected for a extended period [26]. Secretions of these glands function to keep the mucosa moist and drainage of infected secretions into the pharyngeal cavity can serve as source for Cp. psittaci. Also secretions from the lung are expelled into the pharyngeal area [27].
Surprisingly, the 16S rRNA-based PCR could only confirm 13 out of 105 ompA nested PCR-EIA positives revealing only 6.5% positive turkeys. Discrepant results were probably not due to different extraction methods, as 30 16S-rRNA negative samples remained negative even after using the STD extraction method as for the ompA - based nested PCR.
The PCR-EIA turned out to be more sensitive than isolation in cell culture and more sensitive than the 16S rRNA-based nested PCR. The 16S-rRNA PCR primers have already been shown to be sensitive (5 IFU) and specific [25]. The ompA nested PCR-EIA is approximately 50 times more sensitive than the 16S-rRNA based PCR. The sensitivity of the nested PCR-EIA was also superior to isolation of Cp. psittaci in cell culture, which is in agreement with other reports [28-30]. Moreover, the nested PCR-EIA is easy, rapid, and less labour-intensive than isolation and non-viable Chlamydiaceae can be detected, due to the relative high stability of DNA. This allows less stringent demands on collection, transportation and storage of the samples, making the nested PCR-EIA an ideal diagnostic method for monitoring turkey flocks during processing.
Referring to isolation as a reference, all 10 examined farms were Cp. psittaci positive at slaughter. The same was true when looking at the ompA nested PCR results. Although apparently less sensitive, the 16S rRNA PCR still detected Cp. psittaci in 50% of the examined farms. Results are in concordance with previous reports, demonstrating the high prevalence of Cp. psittaci in Belgian turkeys [13]. Public health is here of concern, as poultry workers, veterinary surgeons and slaughterhouse employees are at risk of becoming infected by this zoonotic agent [9,15,16,19,31-33]. In the present study, 37% of the turkeys were still shedding infectious Cp. psittaci, when transported to the slaughterhouse posing a threat to human health. Thus, the presented ompA nested PCR-EIA will help to facilitate the diagnosis of Cp. psittaci infections in both poultry and man.
Competing interests
The author(s)declare that they have no competing interests
Authors' contributions
Marnix Van Loock and Kristel Verminnen have made substantial contributions to conception, design, acquisition of data, analysis and interpretation of data. Trudy Messmer made substantial contributions to acquisition of data, analysis and interpretation. Guido Volckaert, Bruno Goddeeris and Daisy Vanrompay have been involved in revising the manuscript critically for important intellectual content.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Acknowledgments
Acknowledgements
This work was supported by project S6037-section 2 grant from the Belgian Ministry of Public Health and Intervet International N.V. (Boxmeer, The Netherlands).
R. Pensaert, A. Doop, Ph. Deloddere and D. Gilliaert are acknowledged for their assistance in the slaughterhouse (Volys-Star, Lendelede, Belgium). N. Boon and W. Verstraete (Department of Biochemical and Microbial Technology, Ghent University) and E. de Graef and F. Haesebrouck (Department of Veterinary Bacteriology and Mycology, Ghent University) for the bacterial strains used for specificity testing.
Contributor Information
Marnix Van Loock, Email: marnix.van.loock@pandora.be.
Kristel Verminnen, Email: Kristel.Verminnen@UGent.be.
Trudy O Messmer, Email: TMessmer@cdc.gov.
Guido Volckaert, Email: Guido.Volckaert@biw.kuleuven.be.
Bruno M Goddeeris, Email: Bruno.Goddeeris@biw.kuleuven.be.
Daisy Vanrompay, Email: Daisy.Vanrompay@UGent.be.
References
- Andersen AA. Serotyping of Chlamydia psittaci isolates using serovar-specific monoclonal antibodies with the microimmunofluorescence test. J Clin Microbiol. 1991;29:707–711. doi: 10.1128/jcm.29.4.707-711.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanrompay D, Ducatelle F, Haesebrouck F, Hendrickx W. Primary pathogenicity of an European isolate of Chlamydia psittaci from turkey poults. Vet Microbiol. 1993;38:103–113. doi: 10.1016/0378-1135(93)90078-L. [DOI] [PubMed] [Google Scholar]
- Geens T, Van Loock M, Kaleta E, Magnino S, Andersen AA, Vanrompay D. Serological and molecular characterization of Chlamydophila psittaci strains using serovar-specific monoclonal antibodies, OmpA RFLP analysis and OmpA sequencing. In: Deák J, editor. Proceedings of the 5th meeting of the European society for Chlamydia research: Hungary. Budapest: University of Szeged; 2004. p. 302. 1–4 September 2004. [Google Scholar]
- Andersen AA, Vanrompay D. Chlamydiosis. In: Saif YM, editor. Diseases of Poultry. 11. Iowa: Iowa State University Press; 2003. pp. 863–879. [Google Scholar]
- Hall CF, Glass SE, Grimes JE, Moore RW. An epidemic of ornithosis in Texas turkeys in 1974. Southwest Vet. 1975;28:19–21. [Google Scholar]
- Page LA, Derieux WT, Cutlip RC. An epornitic of fatal chlamydiosis (ornithosis) in South Carolina turkeys. J Am Vet Med Ass. 1975;166:175–178. [PubMed] [Google Scholar]
- Anderson DC, Stoesz PA, Kaufmann AF. Psittacosis outbreak in employees of a turkey-processing plant. Am J Epidmiol. 1978;107:140–148. doi: 10.1093/oxfordjournals.aje.a112516. [DOI] [PubMed] [Google Scholar]
- Chalmers ZSK. Duck ornithosis and related zoonosis in the UK. In: Oriel D, Schachter J, Taylor-Robinson D, Ward ME, editor. Chlamydial infections. Cambridge: Cambridge University Press; 1986. pp. 345–348. [Google Scholar]
- Newman JA. Chlamydia spp. Infection in turkey flocks in Minnesota. J Am Vet Med Assoc. 1989;195:1528–1530. [PubMed] [Google Scholar]
- Grimes JE, Wyrick PB. Chlamydiosis (ornithosis) In: Calnek BW, Barnes HJ, Beard CW, Reid M, Yoder HW Jr, editor. Diseases of poultry. 9. Ames: Iowa State University Press; 1991. pp. 311–325. [Google Scholar]
- Vanrompay D, Butaye P, Sayada C, Ducatelle R, Haesebrouck F. Characterization of avian Chlamydia psittaci strains using omp1 restriction mapping and serovar-specific monoclonal antibodies. Res Microbiol. 1997;148:327–333. doi: 10.1016/S0923-2508(97)81588-4. [DOI] [PubMed] [Google Scholar]
- Hafez HM, Sting R, Jodas S, Stadler A. Chlamydia psittaci infections in meat turkey: investigations on the interaction with other avian infectious agents. In: Hafez HM, Mazaheri A, editor. Proceedings of the 1st international symposium on turkey diseases. Giessen, Germany: Verlag der DVG Service GmbH; 1998. pp. 208–217. [Google Scholar]
- Van Loock M, Geens T, De Smit L, Nauwynck H, Van Empel P, Naylor C, Hafez HM, Goddeeris BM, Vanrompay D. Key role of Chlamydophila psittaci on Belgian turkey farms in association with other respiratory pathogens. Vet Microbiol. 2005;107:91–101. doi: 10.1016/j.vetmic.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Moroney JF, Guevara R, Iverson C, Chen FM, Skelton SK, Messmer TO, Plikaytis B, Williams PO, Blake P, Butler JC. Detection of chlamydiosis in a shipment of pet birds, leading to recognition of an autbreak of clinically mild psittacosis in humans. Clin Infect Dis. 1998;26:1425–1429. doi: 10.1086/516368. [DOI] [PubMed] [Google Scholar]
- Soni R, Seale JP, Young IH. Fulminant psittacosis requiring mechanical ventilation and demonstrating serological cross-reactivity between Legionella longbeachae and Chlamydia psittaci. Respirology. 1999;4:203–205. doi: 10.1046/j.1440-1843.1999.00176.x. [DOI] [PubMed] [Google Scholar]
- Gosbell IB, Ross AD, Turner IB. Chlamydia psittaci infection and reinfection in a veterinarian. Aust Vet. 1999;7:511–513. doi: 10.1111/j.1751-0813.1999.tb12121.x. [DOI] [PubMed] [Google Scholar]
- Everett KDE. Chlamydia and Chlamydiales: more than meets the eye. Vet Microbiol. 2000;75:109–126. doi: 10.1016/S0378-1135(00)00213-3. [DOI] [PubMed] [Google Scholar]
- Hinton DG, Shipley A, Galvin JW, Harkin JT, Brunton RA. Chlamydiosis in workers at duck farm and processing plant. Aust Vet J. 1993;70:174–176. doi: 10.1111/j.1751-0813.1993.tb06123.x. [DOI] [PubMed] [Google Scholar]
- Hedberg K, White KE, Forfang JC, Korlath JA, Friendshuh KA, Hedberg CW, MacDonald KL, Osterholm MT. An outbreak of psittacosis in Minnesota turkey industry workers: implications for modes of transmission and control. Am J Epidemiol. 1989;130:569–577. doi: 10.1093/oxfordjournals.aje.a115371. [DOI] [PubMed] [Google Scholar]
- Wilson PA, Phipps J, Samuel D, Saunders NA. Development of a simplified polymerase chain reaction-enzyme immunoassay for detection of Chlamydia pneumoniae. J Appl Bacteriol. 1996;80:431–438. doi: 10.1111/j.1365-2672.1996.tb03239.x. [DOI] [PubMed] [Google Scholar]
- Jantos CA, Roggendorf R, Wuppermann FN, Hegemann JH. Rapid detection of Chlamydia pneumoniae by PCR-enzyme immunoassay. J Clin Microbiol. 1998;36:1890–1894. doi: 10.1128/jcm.36.7.1890-1894.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer WN, Johnson FW. Simple transport medium for the isolation of Chlamydia psittaci from clinical material. Vet Rec. 1983;113:535–536. [PubMed] [Google Scholar]
- Vanrompay D, Cox E, Mast J, Goddeeris B, Volckaert G. High-level expression of Chlamydia psittaci major outer membrane protein in COS cells and in skeletal muscles of turkeys. Infect Immun. 1998;66:5494–5500. doi: 10.1128/iai.66.11.5494-5500.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanrompay D, Van Nerom A, Ducatelle R, Haesebroeck F. Evaluation of five immunoassays for detection of Chlamydia psittaci in cloacal and conjunctival specimens from turkeys. J Clin Microbiol. 1994;32:1470–1474. doi: 10.1128/jcm.32.6.1470-1474.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messmer TO, Skelton SK, Moroney JF, Daugharty H, Fields BS. Application of a nested, multiplex PCR to psittacosis outbreaks. J Clin Microbiol. 1997;35:2043–2046. doi: 10.1128/jcm.35.8.2043-2046.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arizmendi F, Grimes JE. Comparison of the Gimenez staining method and antigen detection ELISA with culture for detecting Chlamydiae in birds. J Vet Diag Invest. 1995;7:400–401. doi: 10.1177/104063879500700320. [DOI] [PubMed] [Google Scholar]
- Andersen AA. Comparison of pharyngeal, fecal and cloacal samples for the isolation of Chlamydia psittaci from experimentally infected cockatiels and turkeys. J Vet Diagn Invest. 1996;8:448–450. doi: 10.1177/104063879600800407. [DOI] [PubMed] [Google Scholar]
- Tong CYW, Donnelly C, Harvey G, Sillis M. Multiplex polymerase chain reaction for the simultaneous detection of Mycoplasma pneumoniae and Chlamydia psittaci in respiratory samples. J Clin Path. 1999;52:257–263. doi: 10.1136/jcp.52.4.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy N, Markey B, Gleeson M, Quinn PJ. Differentiation of Chlamydia psittaci and Chlamydia pecorum strains by species-specific PCR. J Clin Microbiol. 1996;34:3175–3179. doi: 10.1128/jcm.34.12.3175-3179.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewinson RG, Griffiths PC, Bevan BJ, Kirwan SES, Field ME, Woodward MJ, Dawson M. Detection of Chlamydia psittaci DNA in avian clinical samples by polymerase chain reaction. Vet Microbiol. 1997;54:155–166. doi: 10.1016/S0378-1135(96)01268-0. [DOI] [PubMed] [Google Scholar]
- Harris JW. Zoonotic human chlamydiosis of avian origin: a review with particular reference to epidemiology and control. Wild Poultry Sci J. 1983;39:5–23. doi: 10.1079/WPS19830001. [DOI] [Google Scholar]
- Stanford CF, Connolly JH, Ellis WA, Smyth ET, Coyle PV, Montgomery WI, Simpson DI. Zoonotic infections in Northern Ireland farmers. Epidemiol Infect. 1990;105:565–570. doi: 10.1017/s0950268800048196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews BE, Major R, Palmer SR. Ornithosis in poultry workers. Lancet. 1981;1:632–634. doi: 10.1016/S0140-6736(81)91552-X. [DOI] [PubMed] [Google Scholar]
- Andersen AA, Van Deusen RA. Production and partial characterization of monoclonal antibodies to four Chlamydia psittaci isolates. Infect Immun. 1988;56:2075–2079. doi: 10.1128/iai.56.8.2075-2079.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page LA, Bankowski RA. Factors affecting the production and detection of ornithosis antibodies in infected turkeys. Am J Vet Res. 1960;21:971–978. [PubMed] [Google Scholar]
- Illner VF. Zur Frage der Uebertragung des Ornithosevirus durch das Ei. Monatsh Veterinärmed. 1960;17:116–117. [Google Scholar]
- Page LA, Baranowski RA. Investigation of a recent ornithosis epornitic in Californian turkeys. Am J Vet Res. 1959;20:941–945. [Google Scholar]
- Francis T, Jr, Magill TP. An unidentified virus proceeding acute meningitis and pneumonitis in experimental animals. J Exp Med. 1938;68:147–163. doi: 10.1084/jem.68.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen AA. Two new serovars of Chlamydia psittaci from North American birds. Diagn Invest. 1997;9:159–164. doi: 10.1177/104063879700900209. [DOI] [PubMed] [Google Scholar]


