Abstract
The BRCT (BRCA1 C-terminus) is an evolutionary conserved protein–protein interacting module found as single, tandem or multiple repeats in a diverse range of proteins known to play roles in the DNA-damage response. The BRCT domains of 53BP1 bind to the tumour suppressor p53. To investigate the nature of this interaction, we have determined the crystal structure of the 53BP1 BRCT tandem repeat in complex with the DNA-binding domain of p53. The structure of the 53BP1–p53 complex shows that the BRCT tandem repeats pack together through a conserved interface that also involves the inter-domain linker. A comparison of the structure of the BRCT region of 53BP1 with the BRCA1 BRCT tandem repeat reveals that the interdomain interface and linker regions are remarkably well conserved. 53BP1 binds to p53 through contacts with the N-terminal BRCT repeat and the inter-BRCT linker. The p53 residues involved in this binding are mutated in cancer and are also important for DNA binding. We propose that BRCT domains bind to cellular target proteins through a conserved structural element termed the ‘BRCT recognition motif’.
Keywords: 53BP1/BRCA1/BRCT/DNA repair/p53
Introduction
DNA damage triggers a variety of cellular responses in eukaryotic cells, including the induction of a regulatory signalling network that activates DNA-repair pathways. DNA-repair proteins are highly diverse structurally, but many have been shown to contain a conserved globular domain, first identified in the breast cancer tumour suppressor protein BRCA1 and thus designated BRCT (BRCA1 C-terminus) (Bork et al., 1997; Callebaut and Mornon, 1997). The BRCT domain consists of ∼95 amino acid residues and has been observed as single, tandem or multiple repeats in a multitude of proteins of different apparent functions. The structures of the single BRCT domains of XRCC1 (Zhang et al., 1998) and ligase III (Krishnan et al., 2001) and the tandem repeat of BRCA1 (Williams et al., 2001) have been reported. The BRCT domain consists of a four-stranded parallel β-sheet flanked by three α-helices. BRCT-containing proteins are known to play roles (directly or indirectly) in DNA-damage response, including cell-cycle control and checkpoint-mediated repair. This includes proteins such as BRCA1, 53BP1, RAD9, Rad4, ECT2, XRCC1, RAP1, terminal transferases, poly(ADP-ribose) polymerases and DNA ligases III and IV (Bork et al., 1997; Callebaut and Mornon, 1997). Despite the overall functional diversity of these BRCT-containing proteins, participation in the DNA-damage response appears to be a unifying theme. Furthermore, BRCT domains are likely to perform critical functions in the cell-cycle control of organisms from bacteria to humans. It is believed that BRCT modules can interact in various ways forming either homo- or hetero- BRCT dimers and complexes with non-BRCT proteins (Huyton et al., 2000), as well as complexes with DNA (Yamane and Tsuruo, 1999). It has been suggested that BRCT domains may act as signal transducers that transmit the signals from DNA-damage detectors to other components of the DNA-repair machinery via specific protein–protein interactions (Bork et al., 1997). The BRCT domain of mammalian DNA ligase III forms a specific complex with the C-terminal BRCT domain of XRCC1 (Nash et al., 1997), and DNA ligase IV, shown to be important in double-strand break repair, interacts with XRCC4 via a BRCT tandem repeat (Critchlow et al., 1997; Grawunder et al., 1997). These examples provide direct evidence that BRCT domains physically mediate contacts between components of the DNA-repair machinery. The C-terminal BRCT domain of BRCA1 corresponds precisely to the minimal transcription activation domain of this protein (Chapman and Verma, 1996), indicating another possible function for BRCT domains.
A two-hybrid screen has shown that the BRCT-containing protein, p53-binding protein 1 (53BP1) binds to wild-type p53 but fails to bind to mutant p53, suggesting that it may be involved in the ability of wild-type p53 to suppress transformation (Iwabuchi et al., 1994). The tumour suppressor protein p53 plays a critical role in many key cellular processes (Lane, 1992; Levine, 1993), acting as a transcriptional activator for a number of DNA damage and growth arrest genes, as well as negatively regulating the transcription of genes that have TATA box-initiated promoters, and is thought to be involved directly in checking both viral and eukaryotic DNA replication (Horikoshi et al., 1995). p53 binds as a tetramer to response elements through a sequence-specific DNA-binding core domain extending from amino acid residues 96–308 (Pavletich et al., 1993). Studies of tumour-derived p53 mutants have shown that they are defective in sequence-specific DNA-binding and, consequently, cannot activate transcription (Hollstein et al., 1991). These studies strongly suggest that sequence-specific DNA binding and transactivation are the key biochemical activities responsible for much of the biological function of p53. Mutations in the p53 protein have been associated with human cancers and the vast majority of these mutations are located in the DNA-binding core domain (Pavletich et al., 1993) and affect its sequence-specific DNA-binding activity (Cho et al., 1994). This suggests that p53 function is mediated by its DNA-binding and transactivation properties (Bargonetti et al., 1993; Pavletich et al., 1993). The co-crystal structure of the p53 nucleoprotein complex has provided valuable insights into the binding specificity of p53 by identifying specific binding contacts (Cho et al., 1994).
It has been reported that 53BP1 binds to the DNA-binding domain of p53 and enhances p53-mediated transcriptional activation (Iwabuchi et al., 1994). The C-terminal region of 53BP1 interacts with p53 and sequence analysis has shown that this region contains a BRCT tandem repeat (Iwabuchi et al., 1998). It has been reported recently that after DNA damage, 53BP1 re-localizes to discrete nuclear foci that are thought to represent sites of DNA lesions and becomes hyperphosphorylated (Schultz et al., 2000; Anderson et al., 2001; Rappold et al., 2001; Xia et al., 2001). 53BP1 is readily phosphorylated in vitro whilst ataxia-telangiectasia mutated kinase (ATM)-deficient cells show no 53BP1 hyperphosphorylation and reduced 53BP1 foci formation in response to γ-radiation compared with cells expressing wild-type ATM. These results suggest that 53BP1 is a target substrate for ATM and is involved early in the DNA-damage signalling pathways in mammalian cells. A recent report suggests that 53BP1 may also have a distinct role in checkpoint signalling during mitosis (Jullien et al., 2002). It has been shown that the C-terminal BRCT domain of BRCA1 also interacts with the p53 DNA-binding domain both in vivo and in vitro, and stimulates transcription (Chai et al., 1999). This suggests possible parallels between the functioning of BRCA1 and 53BP1.
To investigate the nature of the molecular interactions between 53BP1 and p53, we have determined the 2.6 Å crystal structure of the DNA-binding domain of p53 in complex with the predicted BRCT tandem repeat of 53BP1. In this report, we have elucidated the structural basis for the interactions of BRCT domains with p53. A comparison of the structure of the 53BP1 BRCT domains with other BRCTs in the Protein Data Bank (PDB), including the human BRCA1 tandem repeat, shows that they have remarkably similar structures and provides important insights into the molecular basis for protein– protein contacts mediated by BRCT domains.
Results and discussion
Iwabuchi et al. (1998) reported that 53BP1 interacts with the DNA-binding domain of p53 via two C-terminal BRCT domains. We have defined the minimal region of 53BP1, containing a tandem repeat of BRCT domains, that interacts directly with the core domain of p53 (Derbyshire et al., 2002). We produced a series of N-terminal truncated forms of 53BP1 to define the most stable interacting domain of 53BP1. These truncated proteins were expressed in Escherichia coli and purified to homogeneity. We incubated the proteins with the recombinantly expressed DNA-binding domain of p53 (residues 96–308) and analysed the resulting complexes by gel filtration (Derbyshire et al., 2002). The region comprising residues 1722–1971 was found to form a stable complex with p53 suitable for crystallographic investigation. This region encompasses both predicted BRCT domains. The p53– BP1 complex has been crystallized and its three- dimensional structure determined, in part, by molecular replacement from the co-ordinates of the p53 core domain (Cho et al., 1994). Finally, the structure determination was completed by using a molecular replacement solution obtained using the co-ordinates of the p53–BP1 complex as a search model (PDB ID, 1KZY; Joo et al., 2002) and refined at 2.6 Å resolution (Table I; PDB ID, 1GZH).
Table I. Data collection and structure statistics.
| X-ray source | CuKα |
| Wavelength (Å) | 1.541 |
| Cell dimensions: a; b; c (Å), P212121 | 71.52; 94.57; 136.20 |
| Resolution range (Å) | 55.0–2.6 |
| % Completeness | 99.1 (96.9) |
| Multiplicity | 7.7 (3.70) |
| Rsym | 10.4 (35.8) |
| I/σ(I) | 6.2 (1.40) |
| R-factor/correlationa | 51.1/41.02 |
| R-factor/correlationb | 42.2/66.01 |
|
VM/% solvent |
2.13/41.76 |
| Resolution range (Å) | 500–2.6 |
| No. reflections (working) | 51 284 (93.4% of theoretical) |
| No. reflections (free) | 2835 (5.20% of theoretical) |
| No. non-hydrogen atoms | 6510 |
| No. waters | 110 |
| No. SO4 ions | 2 |
| R.m.s.ds from ideality: | |
| Bond lengths (Å) | 0.00782 |
| Bond angles (°) | 1.34850 |
| Average temperature factors: | |
| Protein | 37.30 |
| Water | 31.06 |
| SO4 ions | 48.76 |
| R-factor/Rfree | 23.81/28.83 |
| Ramachandran plot: | |
| % Most favoured regions | 85.9 |
| % Additionally allowed regions | 14.1 |
| Overall G-factor | 0.24 |
ap53 partial structure from AMoRe, using p53 core co-ordinates.
bp53–Bp1 complex from AMoRe, using p53 and Bp1 co-ordinates.
Rsym = ΣhΣi|Ih,I – <Ih>|/ΣhΣiIh,I where <Ih> is the mean intensity of the ith observation of the reflection h.
R-factor = Σ||Fobs| – |Fcalc||/Σ|Fobs|.
Rfree is calculated from data excluded from refinement.
G-factor is the overall measure of structure quality from Procheck (Laskowski et al., 1993).
R.m.s.ds, root mean square deviations.
Overall structure of the complex
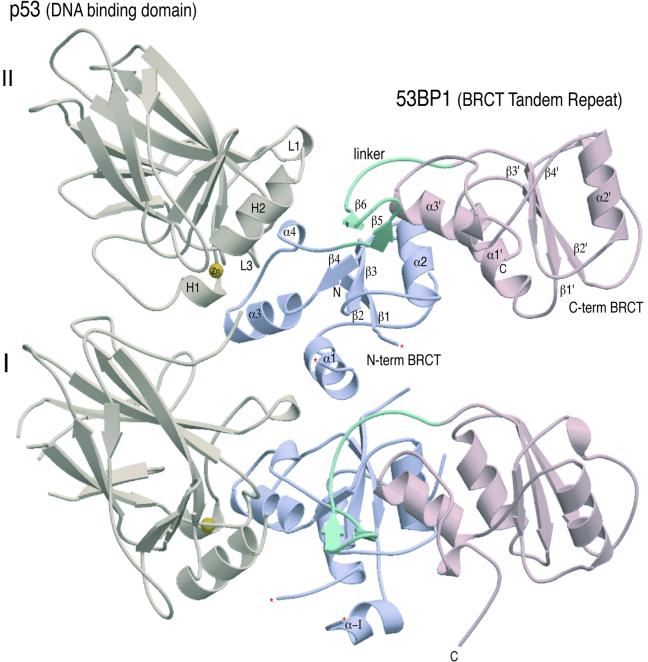
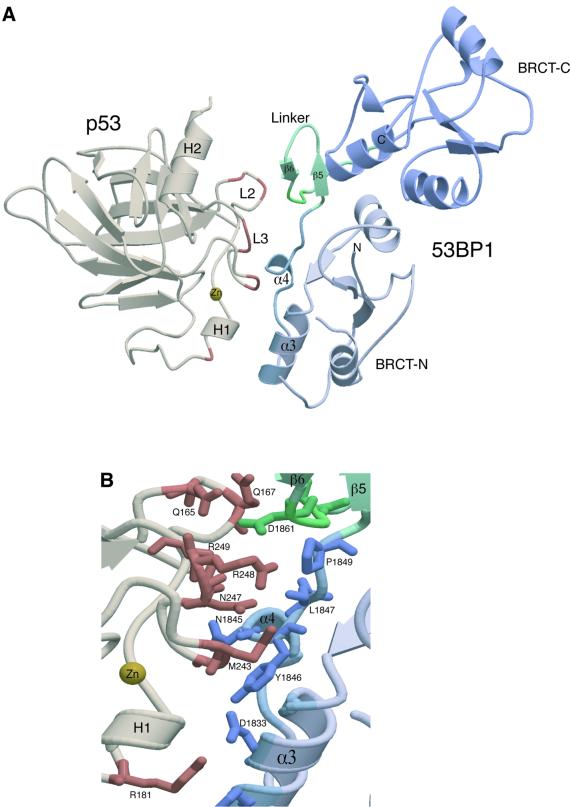
Crystals of the complex display orthorhombic symmetry with two heterodimers (I and II) of p53 in complex with the C-terminal region of 53BP1(BP1) in the asymmetric unit (Figure 1). The heterodimers predominantly pack together via contacts between the N-terminal BRCT domains. There are few molecular contacts between the p53 domains despite their proximity to one another. Overall, clear electron density was interpretable throughout the structures of both p53–BP1 heterodimers, with slight variations in the structure of the loop regions involved in close crystal contacts. The first molecule of p53 extends from residue S95 to K292 with breaks in the electron density between residues 183–188 and 224–227, and at residue 200. The second copy of p53 has no such breaks, but only stretches from S95 to L289. Electron density for the first copy of BP1 was interpretable from L1724 to S1971, and in the second BP1 molecule from N1725 to Y1969. With the exception of breaks between residues 1793–1798 (first copy) and 1794–1796 (second copy), the only region of the binding protein not clearly visible in our structure corresponds to a unique insertion found in BP1 between residues 1740 and 1768, although there is some limited density for residues 1741–1750 (α-I) in the first copy of BP1 (Figure 1).
Fig. 1. Crystal structure of the 53BP1–p53 complex. Structure of the asymmetric unit containing four molecules, two heterodimers (I and II) of p53 DNA-binding domain (brown) in complex with the BRCT tandem repeat of 53BP1 (blue and pink). The heterodimers pack together mainly through contacts made between the N-terminal region of 53BP1. There are no contacts made between the adjacent p53 molecules. The single zinc atom bound to p53 is shown in yellow and the 53BP1 inter-domain linker is shown in green. ‘Missing’ residues in the structure are denoted by an asterisk.
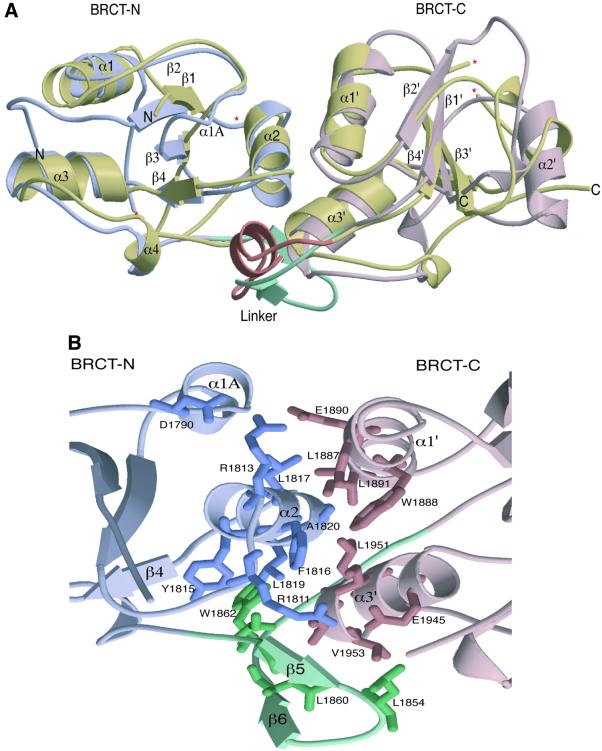
The structure of the heterodimer reveals that the DNA-binding domain of p53 interacts specifically with the N-terminal BRCT domain of 53BP1 (Figure 1). The overall morphology of the complex is roughly L-shaped (∼60 Å long in both directions and ∼30 Å in thickness), with p53 binding to a structural motif extending from α3 of the N-terminal BRCT domain and including the linker region that extends between the two BRCT domains. The C-terminal BRCT domain extends away from the p53 interaction surface. The overall fold of the 53BP1 BRCT tandem repeat is similar to the BRCT domains of human BRCA1 (Figure 2A). The fold of the BRCT domains is characterized by a central four-stranded β-sheet, with a pair of α-helices (α1 and α3) packed against one face and a single helix (α2) packed against the opposite face of the sheet (Figure 2A). The β-sheet forms the core of the structure, with helix α1 forming hydrophobic interactions with residues from β1 and β2. Helix 2 (α2) interacts with β4, also through hydrophobic interactions (Figure 2B). This is further stabilized by hydrogen bonding, curiously only in the N-terminal domain, between the base of α2 to the α2/β4 loop (R1813 and D1790) (Figure 2B). In the N-terminal domain, α1 and α3 form a two-helical bundle, solely through hydrophobic interactions. Helix 3 (α3) contains the highly conserved tryptophan residue, which interacts with other conserved ‘core’ residues from β1, β3 and β4 as well as the segment involved in p53 binding (Figure 2B).
Fig. 2. Structural features of the 53BP1 tandem BRCT repeat. (A) A secondary structure representation of the superimposition of the BRCT tandem repeats from human BRCA1 and 53BP1. The N- and C-terminal BRCT domains of 53BP1 are coloured in light blue and pink, respectively, whilst both BRCT domains of human BRCA1 are coloured gold. The linker regions of each tandem repeat are also highlighted; BRCA1 (a loop–helix–loop structure) in orange and 53BP1 (a β-ribbon-like motif) in green. ‘Missing’ residues in the structure are denoted by an asterisk. (B) Intra-molecular interactions at the inter-BRCT repeat interface of 53BP1. The side chains from the first BRCT repeat are shown in blue, those from the linker are in green and those from the second BRCT repeat are pink. The α2 helix of the first repeat together with α1′ and α3′ helices of the second repeat form a three-helical bundle that is stabilized by α1A and the β-hairpin linker (β5–β6). (C) An amino acid sequence alignment of the regions of BRCA1, 53BP1 and RAD9 that are predicted to form BRCT–BRCT interfaces. Residues that constitute this interface in human 53BP1, as well as conserved residues in human BRCA1 and Sacchromyces cerevisiae RAD9, are coloured red.
The 53BP1 inter-BRCT repeat interface contains both core BRCT structural elements and also the inter-domain linker (Figure 2B). The interface buries ∼2500 Å2 of surface area, with one third of this area contributed by the inter-domain linker. The α1′ and α3′ helices pack against α2, forming a three-helical bundle structure. The interface is stabilized by hydrophobic residues contributed by the bundle, with extra stability being provided by two arginine residues; R1813 at the base of α2, which interacts directly with E1890 on α1′ and D1790 on α1A, and R1811, which forms a salt bridge with E1945 of α3′ (Figure 2B). The inter-domain linker adopts a β-hairpin structure (β5–β6; residues 1851–1860) followed by an extended structure (residues 1861–1867). The linker packs extensively with both BRCT repeats at the inter-BRCT interface (Figure 2B). This packing is stabilized by hydrophobic residues from one face of the β-hairpin structure that stacks against residues from the α2 and α3′ helices of the three-helical bundle.
The structure of the p53 core domain in this complex is similar to the crystal structures of free and DNA-bound forms of p53 (Figure 1; Cho et al., 1994). p53 domain consists of a β-sandwich with a ‘greek key’ topology that serves as a scaffold for two large loops, L2 (163–195) and L3 (236–251), and a loop–sheet–helix motif, H2 (278–286). The two loops bind a single zinc atom, which is tetrahedrally co-ordinated by C176 and H179 of L2, and C238 and C242 of L3 (Figure 1). This region, together with the H2 helix, forms a continuous surface at one end of the β-sandwich, which is known to be critical for p53’s function and its inactivation in tumours. This area is one of the most conserved regions of p53 to which most of the tumorigenic mutations map (Cho et al., 1994). Interestingly, this conserved region of p53 represents the sole 53BP1-binding interface. In this complex, ∼1400 Å2 of mostly polar surface area is buried in the p53–BP1 interface. In the BP1–p53 complex, the C-terminal portion of the first BRCT repeat and the linker region of 53BP1 forms a continuous surface that binds part of the DNA-binding surface of the p53 core domain, mostly through contacts to the L3 loop, but a smaller number of contacts are made via the L2 loop (Figure 1). A surprising feature of the complex is that the C-terminal BRCT domain of 53BP1 is positioned distantly from the p53 molecule and makes no contacts with the core domain (Figure 1). The significance of this architecture will be discussed later.
Comparison of 53BP1 with other BRCT structures
The overall structure of the individual 53BP1 BRCT domains is similar to that observed in the C-terminal repeat of BRCA1 (Figure 2; Williams et al., 2001), as well as in the individual BRCT domains of XRCC1 and ligase III (Zhang et al., 1998; Krishnan et al., 2001). A structural alignment of the BRCT repeats of 53BP1 with the BRCA1, XRCC1 and ligase III BRCT domains reveals that the relative arrangement of the central β-sheet and helices (α1 and α3) is conserved in all the four repeats (data not shown). However, the conformations of the β1–α1, β2–β3 and β3–α2 connecting loops, as well as the orientation of α2 relative to the central β-sheet, are much less conserved. The conservation of the α1–α3–β-sheet structure is maintained by the packing of a limited number of key conserved hydrophobic residues in the core of the BRCT fold. The structure of the 53BP1 BRCT region closely resembles that of human BRCA1, especially in the N-terminal domain. However, the structures of the C-terminal BRCTs are considerable different. BRCA1 lacks the α2′ helix, which in p53BP1 is presented on the surface of this BRCT domain. Furthermore, the adjacent loop (β2′–α3′) has a different conformation between these two proteins. The N-terminal BRCT domain of BP1 con tains a unique large insertion of 29 residues (1741–1769) rich in polar and charged residues between β1 and α1 that are not found in BRCA1 or other BRCTs. In our structure, this region is very disordered, probably indicating that it is highly flexible. However, in the first copy of BP1, some broken electron density is interpretable for residues 1741–1750 and there is good indication from model building that these residues form an α-helical structure (α-I) that extends from the surface of the N-terminal BRCT domain into the solvent (Figure 1).
The entire tandem BRCT structures of 53BP1 and BRCA1 can be superimposed with a root mean square deviation (r.m.s.d.) of 6.27 Å (Figure 2A). This value improves to 1.85 Å when matching only 135 Cα positions, including all the central β-strands and α-helices (Figure 2A). The most notable structural conservation between these two structures exists at the inter-BRCT repeat interface. As discussed above, two inter-BRCT repeats of 53BP1 interact through intramolecular head-to-tail contacts, burying a large amount of surface area in this interface (Figure 2B). The core of this interface is formed by the interaction of three α-helices: α2 of the N-terminal repeat, and α1′ and α3′ from the C-terminal repeat (Figure 2B). The residues in these helices that contribute to the interface are almost all hydrophobic and pack tightly in a knobs-in-holes manner. In the crystal structure of human BRCA1, the two inter-BRCT repeats also pack via a triple helical interface similar to that seen in the 53BP1 structure (Williams et al., 2001; Figure 2B). Multiple BRCT repeats are common in many of the BRCT-containing proteins such as BRCA1, RAD9 and DNA ligase IV. An amino acid sequence alignment of the two 53BP1 BRCT repeats with those in BRCA1 and Rad9 shows that the residues occupying the interface between the two repeats in α2, α1′ and α3′ of 53BP1 are highly conserved in BRCA1 (Figure 2C; Williams et al., 2001), and it is likely that this mode of intra-molecular packing will be common within the repeats of the BRCT family. The inter- repeat linker of 53BP1 occupies a similar position as that of human BRCA1, packing on one side of the three-helical bundle and stabilizing the inter-BRCT repeat interface (Figure 2B). There is continuous electron density for the linker connecting the two BP1 BRCTs. In contrast, the structure of the linker connecting the human BRCA1 BRCT repeats is poorly ordered, possibly indicating flexibility. The central portion of the BRCA1 linker, however, is clearly α-helical. In the BP1 linker, this region forms a distinct β-ribbon structure (β5–β6; Figure 2B).
Many BRCT domains have been shown to interact with specific partners through either homo (e.g. Rad9) or hetero (e.g. XRCC1–ligase III) BRCT–BRCT interactions (Huyton et al., 2000). In our crystals, we noted that the N-terminal BRCT domains from neighbouring 53BP1 molecules in the unit cell form non-crystallographic interactions. The interface is composed of interactions between α1′ and an extended loop region between β1 and α1 (Figure 3). A closer examination of this interface showed that there are no hydrogen bonds or electrostatic contacts, suggesting that this interaction is unlikely to represent a specific BRCT–BRCT interface.
Fig. 3. Contacts between 53BP1 BRCT–BRCT domains. The N-terminal BRCT domains form non-crystallographic interactions. The interface is composed of non-specific interactions between residues in α1 and an extended loop region, shown in shaded colour.
Structure of the 53BP1–p53 interface
A striking feature of the complex is that 53BP1 interacts with p53 through a very limited number of specific contacts localized around the inter-domain linker. The p53-binding region of 53BP1 forms an unusual extended structure, which extends from the C-terminal region of the first BRCT (α3 helix) into the central helical region (α4) of the inter-domain linker (approximately residues 1843–1867). The linker, a β-ribbon structure (β5–β6; Figure 4A), then proceeds into the next domain as strand β1′ in a fashion not dissimilar to the extended region leading into β1 of the N-terminal domain. This may represent a BRCT–BRCT linkage more suited to multiple BRCT repeats. Interestingly, this linker region is distinct from that of the human BRCA1 tandem repeat, which forms a loop–helix–loop structure (Figure 2A). All the 53BP1 contacts are with highly conserved residues on the L2 and L3 loops of p53. The L2 and L3 loops form rigid hairpin structures stabilized by a single co-ordinating zinc atom and also by multiple intra-molecular hydrogen bonds. The COOH-terminal of the L3 hairpin (247–249) is juxtaposed to the extended linker region at the C-terminal region of the first BRCT of 53BP1 (Figure 4A). Arg 248 from this loop hydrogen bonds with Asp 1861 from the β-hairpin of the inter-repeat linker and also hydrogen bonds to the backbone carbonyl group of Leu 1847 at the beginning of the linker (Figure 4B). The conformation of this arginine residue is stabilized by stacking interaction with the ring of Pro 1849 (Figure 4B). Asn 247 also forms a main-chain hydrogen bond with Leu 1847, and Arg 249 hydrogen bonds with Arg 1844 and Asn 1845 from α4 (Figure 4B). The Met 243 side-chain of p53 makes van der Waals contacts with Val 1829 and Tyr 1846 (Figure 4B). The L2 loop (163–195) of p53 also makes major contributions to the 53BP1-binding surface (Figure 4A). Arg 181 forms a charge-stabilized hydrogen bond with Asp 1833 and also forms a stacking interaction with the aromatic ring of His 1836 (Figure 4B). Glu 177 hydrogen bonds to Asn 1842 and additional hydrogen bonds are also made in the complex (Gln 165–Asp 1861, Gln 167–Gln 1863, Glu 171–Asn 1842).
Fig. 4. Structure of the 53BP1–p53 interface. (A) The heterodimeric complex of 53BP bound to p53. The p53 binding region of 53BP1 forms an extended structure from the N-terminal region of the first BRCT (shown in light blue) to the central helical region of the inter-domain linker (α4). The 53BP1 contacts (shown in brown) are with highly conserved residues on the L2 and L3 loops of p53. The L2 and L3 loops form rigid hairpin structures that are stabilized by a single co-ordinating zinc atom (yellow). (B) Specific amino acid contacts at the complex interface between 53BP1 (blue) and p53 (brown). The inter-domain β-turn (β5-β6) is shown in green.
Effects of site-specific and tumorigenic p53 mutations
It has been reported that the binding of 53BP1 to p53 inhibits binding to DNA and 53BP2 in vitro (Iwabuchi et al., 1994). Our structure reveals the molecular basis for this inhibition. The p53-binding site of 53BP1 directly overlaps with the DNA- and 53BP2-binding sites, and therefore sterically blocks the docking of these ligands (Figure 4A; Cho et al., 1994; Gorina and Pavletich, 1996). More specifically, a comparison of the 53BP1–p53 structure with both the p53–DNA and p53–53BP2 complexes showed that many common residues, including those on the highly conserved loops L2 and L3, provide many of the important contacts in these complexes. Alanine scanning mutagenesis has confirmed this and has shown that a number of these conserved residues are involved in the binding of all three ligands (Thukral et al., 1994). The significance of many of these conserved interactions is highlighted by the residue Arg 248 (Figure 4B). In vivo, this residue is the most frequently mutated amino acid of p53. The replacement of this residue by alanine abolishes 53BP1, DNA and 53BP2 binding, confirming its pivotal role in p53 ligand interactions (Figure 4B; Thukral et al., 1994). The failure of many tumour-derived p53 mutants to bind to 53BP1 also closely correlates with their failure to bind to DNA or 53BP2. This is not unexpected, since we have demonstrated that the 53BP1-, DNA- and 53BP2-binding surfaces all appear to overlap considerably. Is it possible that the disruption of 53BP1 binding by tumorigenic mutations may be secondary to their disruption of DNA binding, and the loss of 53BP1 binding may have no functional consequence for tumorigenesis? Pavletich and colleagues noted previously that among the p53 surface residues with no apparent structure-stabilizing roles, those that contact either 53BP1 or 53BP2 but not DNA (Gorina and Pavletich, 1996) are also found mutated, although at lower frequencies and, conversely, residues that contact only the DNA but not 53BP1 or 53BP2 also show similarly low mutational frequencies. However, the highest mutational frequencies occur at residues involved in 53BP1, DNA and 53BP2 binding (e.g. Arg 248). It is also notable that Met 243, an important 53BP1 contact but has no apparent role in structure stabilization or DNA recognition, is also mutated in cancer.
A conserved protein–protein binding motif in BRCT domains
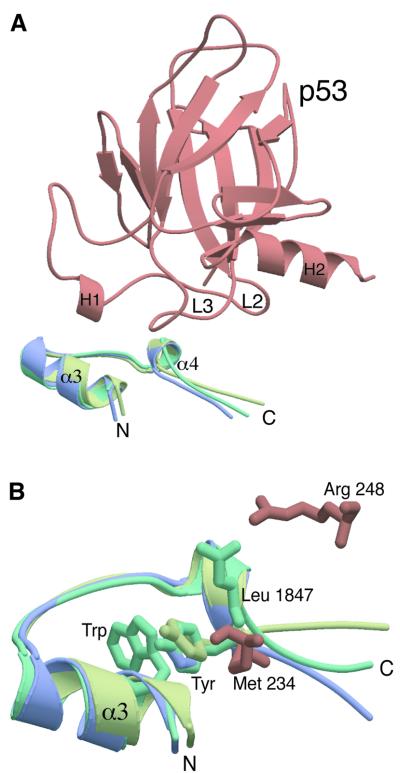
It is now evident that BRCT domains physically mediate interactions between different proteins (Huyton et al., 2000), suggesting that one specific role for these domains is to promote specific protein–protein interactions in vivo. The importance of the inter-BRCT linker in mediating protein–protein interactions was first highlighted by biochemical studies (Grawunder et al., 1998; Taylor et al., 1998) and subsequently supported by the structure of the XRCC4–ligase IV linker complex (Sibanda et al., 2001). A notable feature of the 53BP1–p53 complex is also the very localized nature of the interactions, around the linker, between the BP1 BRCTs and p53. We examined the local structure of BP1 at the BRCT–p53 interface and compared this with the uncomplexed BRCT domains of BRCA1, XRCC1 and ligase III. Figure 5 shows a superimposition of the p53-binding interface of BP1 with the equivalent regions of BRCA1 and XRCC1. The Cα backbone of these regions overlap with an r.m.s.d. of <1 Å, confirming that they form almost identical structural motifs. This structure has a highly conserved helix (α3)–loop–helix (α4)–loop conformation in all three structures (Figure 5A). Does this structural conservation also reflect a conserved functional role for this motif? In the 53BP1–p53 complex, the majority of the contacts are made with residues in the short α helix (α4; NYRNYLL, residues 1842–1848). In the BRCA1 and XRCC1 BRCT domains, many of these residues are also highly conserved (N-x-H/R-N/D-Y/F-x-L/V). The position of the side chains of these conserved Asn, Asn/Asp and Tyr/Phe are absolutely conserved in the short helix (α4) and, more importantly, these residues make important interactions with p53 (Figure 5B). Tyr 1846 makes van der Waals contact with Met 243, Asn 1842 hydrogen bonds to Glu 177 and Asn 1845 hydrogen bonds with Arg 249. Based on these findings, it seems likely that this conserved ‘BRCT recognition motif’ may be important for the binding of target proteins by BRCT domains. We also noted that the highly conserved tryptophan residue (Trp 1830 in BP1), found in most BRCT domains on α3, is positioned in an identical position in all three structures (Figure 5B). This tryptophan residue stacks tightly against the conserved tyrosine residue (Tyr 1846 in BP1) in all the BRCT structures (Figure 5B), suggesting that this conserved interaction may be important for stabilizing the conformation of the conserved BRCT recognition motif.
Fig. 5. A conserved protein binding motif in BRCT domains. (A) Superimposition of the conserved p53-binding motif. This shows a superimposition of the p53-binding interface of 53BP1 (green; residues 1829–1849) with the comparable regions of the BRCT domains of BRCA1 (blue; 1716–1738) and XRCC1 (yellow; 72–94). The Cα backbone of these regions overlap with an r.m.s.d. of <1 Å. The position of this motif in relation to p53 binding is also shown (p53 core domain is shown in brown). (B) The highly conserved Trp residue on α3 is positioned in an identical position in all three structures (for clarity only a single Trp is shown) and stacks against the conserved Tyr residue (the superimposed Tyr shown). Two conserved 53BP1–p53 interactions are shown, Arg 248–Leu 1847 and Met 243–Tyr1846.
Conclusions
Since it was first reported that the breast cancer susceptibility gene contained two copies of a conserved BRCT domain at its C-terminus, and found subsequently in a variety of proteins involved in DNA repair, recombination and cell-cycle control, there has been a lot of speculation about the physiological role of this highly conserved structural motif. Recent biochemical data has indicated that the BRCT domain functions as a protein–protein interaction module. In this report, we describe the crystal structure of a tandem repeat of BRCT domains (53BP1) in complex with an intra-cellular target protein (p53) allowing us to directly visualize the molecular contacts in a BRCT–protein complex. The structure of this complex highlights the importance of the BRCT inter-domain linker and interface in protein–protein interactions. The α4 helix of the N-terminal BRCT domain and the inter-domain linker region form what we have termed the ‘BRCT recognition motif’. This recognition motif stacks against the inter-repeat interface, which is highly conserved in other tandem BRCT repeats, including BRCA1. This structural motif is responsible for all the major interactions with p53 and is conserved in the structures of other BRCT domains, probably reflecting a conserved functional role in protein recognition. BRCT domains are often found in pairs (Bork et al., 1997; Callebaut and Mornon, 1997), and it is probable that the conserved interface between BRCT tandem repeats is required to stabilize and present the correct conformation of the recognition motif, allowing this region to bind to protein partners. This mode of binding may represent only one of many distinct modes of protein recognition utilized by BRCT domains. Strikingly, in the 53BP1–p53 complex, the position of the C-terminal BRCT domain is distant from the p53-binding site and, consequently, it plays no role in p53 binding. It is tempting to speculate that BRCTs may also interact with other ligands via contacts with the central globular core of the BRCT motif. This promiscuity would allow BRCTs, such as the C-terminal BRCT of BP1, to simultaneously mediate interactions with multiple protein partners. However, we must first elucidate the structures of many more BRCT–protein complexes before we can delineate the determinants that specify the full range of BRCT–protein interactions.
After submission of this manuscript, the structure of the 53BP1 BRCT region bound to p53 was reported (Joo et al., 2002). This structure is in good agreement with the findings reported in this paper.
Materials and methods
Purification and crystallization of the p53–53BP1 complex
p53 (residues 94–312) and 53BP1 (residues 1722–1972) were overexpressed separately in the E.coli strain B834(DE3) plysS (Novagen) as described previously (Derbyshire et al., 2002). The p53 DNA-binding domain was purified by SP and heparin–Sepharose column chromatography. The 53BP1 BRCT tandem repeat protein was purified by Ni-NTA agarose and Hi-Trap Q Sepharose column chromatography (Derbyshire et al., 2002). The p53–BP1 complex was formed by dialysis and the heterodimer purified by size exclusion chromatrography using a prepara tive Superdex-75 column. For optimal crystallization, the p53–BP1 complex was concentrated to ∼15 mg/ml. Crystals were grown by the micro-batch technique under mineral oil by mixing protein with an equal volume of a precipitant solution containing 50 mM Tris–HCl pH 7.4, NH3SO4 and 20–25% polyethylene glycol 4 kDa (PEG 4k; Derbyshire et al., 2002).
Structure determination
A native data set was measured to a resolution of 2.6 Å from a crystal of the complex cooled to 100 K. Indexing was performed using the MOSFLM software (Leslie, 1992). The data were scaled and reduced using the CCP4 suite (CCP4, 1994). Initial work on the structure of the p53–BP1 complex proceeded via a partial molecular replacement solution obtained using the co-ordinates of the DNA-binding domain of p53 alone. Two p53 molecules were found in the asymmetric unit by molecular replacement using AMoRe (Navaza, 1994) and the co-ordinates of the p53 DNA-binding domain (PDB ID, 1TSR; Cho et al., 1994). Through the use of multi-domain non-crystallographic symmetry averaging (DM in CCP4, 1994), as well as cross-crystal averaging (DMmulti in CCP4, 1994), both being combined with solvent flattening and extensive phase extension, a gradual improvement in electron density quality and interpretability was observed (Derbyshire et al., 2002). Finally, the structure was completed using a molecular replacement solution obtained using the co-ordinates of the p53–BP1 complex (PDB ID, 1KZY; Joo et al., 2002). Rigid body refinement by Refmac (CCP4, 1994) was conducted with four independent bodies, two molecules each of p53 and BP1, giving a final correlation coefficient of 47.6%. Once again, density modification procedures were adopted to improve the electron density prior to molecular modelling. Model building with the Xfit program of XTALVIEW (McRee, 1999) was alternated with crystallographic refinement using CNS (Brünger et al., 1998). After the addition of 110 water molecules and two SO4 ions, the crystallographic R-factor and Rfree factor were 23.81 and 28.83, respectively. The stereochemistry of the structure was assessed with PROCHECK (Laskowski et al., 1993) and 100% of the residues were in the favourable regions of the Ramachandran plot. Details and statistics of the final refined model are presented in Table I. The atomic co-ordinates have been deposited in the Protein Data Bank (PDB ID, 1GZH). Figures were generated with several combined uses of MOLSCRIPT (Kraulis, 1991) and Raster3D (Merritt and Bacon, 1997).
Acknowledgments
Acknowledgements
We are indebted to Dr N.Pavletich for providing co-ordinates of the p53–53BP1 complex prior to publication. We would like to thank Professor R.Read and Drs R.Pannu, Y.Muller, G.Spraggon and M.Noble for invaluable crystallographic advice. We would also like to thank Drs L.Itzhaki, F.Rousseau and J.Schymkowitz for their support in the early stages of this work. A.J.D. is Royal Society University Research Fellow, and work in the A.J.D. laboratory is supported by grants from Cancer Research UK, Leukaemia Research Fund, BBSRC, the Association for International Cancer Research and the Royal Society. K.I. is supported by project research grants (H99-1, H00-1 and H01-1) from the High-Technology Center of Kanazawa Medical University, Japan.
References
- Anderson L., Henderson,C. and Adachi,Y. (2001) Phosphorylation and rapid relocalization of 53BP1 to nuclear foci upon DNA damage. Mol. Cell. Biol., 21, 1719–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargonetti J., Manfredi,J.J., Chen,X., Marshak,D.R. and Prives,C. (1993) A proteolytic fragment from the central region of p53 has marked sequence-specific DNA-binding activity when generated from wild-type but not from oncogenic mutant p53 protein. Genes Dev., 7, 2565–2574. [DOI] [PubMed] [Google Scholar]
- Bork P., Hofmann,K., Bucher,P., Neuwald,A.F., Altschul,S.F. and Koonin,E.V. (1997) A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J., 11, 68–76. [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr., 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Callebaut I. and Mornon,J.P. (1997) From BRCA1 to RAP1: a widespread BRCT module closely associated with DNA repair. FEBS Lett., 400, 25–30. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr., 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Chai Y.L., Cui,J., Shao,N., Shyam,E., Reddy,P. and Rao,V.N. (1999) The second BRCT domain of BRCA1 proteins interacts with p53 and stimulates transcription from the p21WAF1/CIP1 promoter. Oncogene, 18, 263–268. [DOI] [PubMed] [Google Scholar]
- Chapman M.S. and Verma,I.M. (1996) Transcriptional activation by BRCA1. Nature, 382, 678–679. [DOI] [PubMed] [Google Scholar]
- Cho Y.J., Gorina,S., Jeffrey,P.D. and Pavletich,N.P. (1994) Crystal structure of a p53 tumor suppressor–DNA complex: understanding tumorigenic mutations. Science, 265, 346–355. [DOI] [PubMed] [Google Scholar]
- Critchlow S.E., Bowater,R.P. and Jackson,S.P. (1997) Mammalian DNA double-strand break repair protein XRCC4 interacts with DNA ligase IV. Curr. Biol., 7, 588–598. [DOI] [PubMed] [Google Scholar]
- Derbyshire D.J., Basu,B.P., Iwabuchi,K. and Doherty,A.J. (2002) Purification, crystallization and preliminary X-ray analysis of a complex of the BRCT domains of human 53BP1 bound to the p53 tumour suppressor. Acta Crystallogr. D Biol. Crystallogr., in press. [DOI] [PubMed] [Google Scholar]
- Gorina S. and Pavletich,N.P. (1996) Structure of the p53 tumor suppressor bound to the ankyrin and SH3 domains of 53BP2. Science, 274, 1001–1005. [DOI] [PubMed] [Google Scholar]
- Grawunder U., Wilm,M., Wu,X., Kulesza,P., Wilson,T.E., Mann,M. and Lieber,M.R. (1997) Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature, 388, 492–495. [DOI] [PubMed] [Google Scholar]
- Grawunder U., Zimmer,D. and Leiber,M.R. (1998) DNA ligase IV binds to XRCC4 via a motif located between rather than within its BRCT domains. Curr. Biol., 8, 873–876. [DOI] [PubMed] [Google Scholar]
- Hollstein M., Sidransky,D., Vogelstein,B. and Harris,C.C. (1991) p53 mutations in human cancers. Science, 253, 49–53. [DOI] [PubMed] [Google Scholar]
- Horikoshi N., Usheva,A., Chen,J.D., Levine,A.J., Weinmann,R. and Shenk,T. (1995) Two domains of p53 interact with the TATA-binding protein and the adenovirus 13S E1A protein disrupts the association, relieving p53-mediated transcriptional repression. Mol. Cell. Biol., 15, 227–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyton T., Bates,P.A., Zhang,X., Sternberg,M.J. and Freemont,P.S. (2000) The BRCA1 C-terminal domain: structure and function. Mutat. Res., 460, 319–332. [DOI] [PubMed] [Google Scholar]
- Iwabuchi K., Bartel,P.L., Li,B., Marraccino,R. and Fields,S. (1994) Two cellular proteins that bind to wild-type but not mutant p53. Proc. Natl Acad. Sci. USA., 91, 6098–6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwabuchi K., Li,B., Massa,H.F., Trask,B.J., Date,T. and Fields,S. (1998) Stimulation of p53-mediated transcriptional activation by the p53-binding proteins, 53BP1 and53BP2. J. Biol. Chem., 273, 26061–26068. [DOI] [PubMed] [Google Scholar]
- Joo W.S., Jeffrey,P.D., Cantor,S.B., Finnin,M.S., Livingston,D.M. and Pavletich,N.P. (2002) Structure of the 53BP1 BRCT region bound to p53 and its comparison to the Brca1 BRCT structure. Genes Dev., 16, 583–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien D., Vagnarelli,P., Earnshaw,W.C. and Adachi,Y. (2002) Kinetochore localisation of the DNA damage response component 53BP1 during mitosis. J. Cell Sci., 115, 71–79. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Krishnan V.V., Thornton,K.H., Thelen,M.P. and Cosman,M. (2001) Solution structure and backbone dynamics of the human DNA ligase III α BRCT domain. Biochemistry, 40, 13158–13166. [DOI] [PubMed] [Google Scholar]
- Lane D.P. (1992) Cancer. p53, guardian of the genome. Nature, 358, 15–16. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Leslie A.G.W. (1992) Molecular data processing. In Moras,D., Podjarny,A.D. and Thierry,J.C. (eds), Crystallographic Computing 5: From Chemistry to Biology. Oxford University Press, Oxford, UK, pp. 50–61.
- Levine A.J. (1993) The tumor suppressor genes. Annu. Rev. Biochem., 62, 623–651. [DOI] [PubMed] [Google Scholar]
- McRee D.E., (1999) XtalView/Xfi—a versatile program for manipulating atomic co-ordinates and electron density. J. Struct. Biol., 125, 156–165. [DOI] [PubMed] [Google Scholar]
- Merritt E.A. and Bacon,D.J. (1997) Raster3D photorealistic molecular graphics. Methods Enzymol., 277, 505–524. [DOI] [PubMed] [Google Scholar]
- Nash R.A., Caldecott,K.W., Barnes,D.E. and Lindahl,T. (1997) XRCC1 protein interacts with one of two distinct forms of DNA ligase III. Biochemistry, 36, 5207–5211. [DOI] [PubMed] [Google Scholar]
- Navaza J. (1994) AMoRe: an automated package for molecular replacement. Acta Crystallogr. A, 50, 157–163. [Google Scholar]
- Pavletich N.P., Chambers,K.A. and Pabo,C.O. (1993) The DNA-binding domain of p53 contains the four conserved regions and the major mutation hot spots. Genes Dev., 7, 2556–2564. [DOI] [PubMed] [Google Scholar]
- Rappold I., Iwabuchi,K., Date,T. and Chen,J. (2001) Tumor suppressor p53 binding protein 1 (53BP1) is involved in DNA damage-signaling pathways. J. Cell Biol., 153, 613–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz L.B., Chehab,N.H., Malikzay,A. and Halazonetis,T.D. (2000) p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol., 151, 1381–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibanda B.L., Critchlow,S.E., Begun,J., Pei,X.Y., Jackson,S.P., Blundell,T.L. and Pellegrini,L. (2001) Crystal structure of an XRCC4–DNA ligase IV complex. Nat. Struct. Biol., 8, 1015–1019. [DOI] [PubMed] [Google Scholar]
- Taylor R.M., Wickstead,B., Cronin,S. and Caldecott,K.W. (1998) Role of a BRCT domain in the interaction of DNA ligase III-α with the DNA repair protein XRCC1. Curr. Biol., 8, 877–880. [DOI] [PubMed] [Google Scholar]
- Thukral S.K., Blain,G.C., Chang,K.K. and Fields S. (1994) Distinct residues of human p53 implicated in binding to DNA, simian virus 40 large T-antigen, 53BP1 and 53BP2. Mol. Cell. Biol., 14, 8315–8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R.S., Green,R. and Glover,J.N. (2001) Crystal structure of the BRCT repeat region from the breast cancer-associated protein BRCA1. Nat. Struct. Biol., 8, 838–842. [DOI] [PubMed] [Google Scholar]
- Xia Z., Morales,J.C., Dunphy,W.G. and Carpenter,P.B. (2001) Negative cell cycle regulation and DNA damage-inducible phosphorylation of the BRCT protein 53BP1. J. Biol. Chem., 276, 2708–2718. [DOI] [PubMed] [Google Scholar]
- Yamane K. and Tsuruo,T. (1999) Conserved BRCT regions of TopBP1 and of the tumor suppressor BRCA1 bind strand breaks and termini of DNA. Oncogene, 18, 5194–5203. [DOI] [PubMed] [Google Scholar]
- Zhang X. et al. (1998) Structure of an XRCC1 BRCT domain: a new protein–protein interaction module. EMBO J., 17, 6404–6411. [DOI] [PMC free article] [PubMed] [Google Scholar]