Abstract
The spatial distribution of four subunits specifically associated to the yeast DNA-dependent RNA polymerase I (RNA pol I) was studied by electron microscopy. A structural model of the native enzyme was determined by cryo-electron microscopy from isolated molecules and was compared with the atomic structure of RNA pol II Δ4/7, which lacks the specific polypeptides. The two models were aligned and a difference map revealed four additional protein densities present in RNA pol I, which were characterized by immunolabelling. A protruding protein density named stalk was found to contain the RNA pol I-specific subunits A43 and A14. The docking with the atomic structure showed that the stalk protruded from the structure at the same site as the C-terminal domain (CTD) of the largest subunit of RNA pol II. Subunit A49 was placed on top of the clamp whereas subunit A34.5 bound at the entrance of the DNA binding cleft, where it could contact the downstream DNA. The location of the RNA pol I-specific subunits is correlated with their biological activity.
Keywords: atomic structure docking/cryo-electron microscopy/epitope localization/immunolabelling/yeast RNA polymerase I (A)
Introduction
In all living cells, gene expression is mediated and regulated by DNA-dependent RNA polymerases, a ubiquitous family of multi-subunit nuclear enzymes. Eukaryotes developed three distinct enzymatic forms specialized in the synthesis of rRNA precursors (RNA pol I), pre-messenger RNA (RNA pol II) and small RNAs such as tRNA or 5S RNA (RNA pol III). The α2ββ′ω subunit composition of the unique bacterial core enzyme is conserved in the three eukaryotic RNA polymerases, as demonstrated by primary sequence analysis, functional and structural homologies (Thuriaux and Sentenac, 1992). The largest and the second largest subunit of each eukaryotic enzyme form are highly homologous to the bacterial β′ and β subunits, respectively, and contain the active site taken in a broad sense. However, the two largest subunits are encoded by different genes for each enzyme form and contain amino acid sequences determining their specific assembly and, indirectly, their promoter recognition functions. The homologues of the prokaryotic α2 dimer in Saccharomyces cerevisiae are heterodimers of two distinct α-like subunits, which differ between RNA pol II (Rpb3/Rpb11) and RNA pol I or III (AC40/AC19) (Heyduk et al., 1993). Recently, a functional and structural homology was described between the shared yeast subunit Rpb6, also called ABC23, and the ω subunit of the bacterial enzyme, where it is believed to promote enzyme assembly by constraining the fold of the largest subunit (Minakhin et al., 2001).
In contrast to the bacterial core enzyme, there is no evidence that the five homologous eukaryotic subunits are sufficient to form a functional enzyme. To synthesize RNA processively from a non-specific DNA template, the yeast S.cerevisiae requires a set of four additional subunits shared by all three enzyme forms (Rpb5, Rpb8, Rpb10 and Rpb12) (Krapp et al., 1998; Dumay et al., 1999; Rubbi et al., 1999; Wei et al., 2001). A fifth subunit, although distinct in each enzyme form (A12.2, Rpb9 and C11 for RNA pol I, II and III, respectively), shows important structural and functional homologies (Chedin et al., 1998). In the RNA pol I and II systems, these subunits modulate transcription either through changes in enzyme process ivity or through interactions with transcription factors (Hemming and Edwards, 2000), but are not strictly required for cell viability (Woychik and Young, 1989; Nogi et al., 1993). Subunit C11, on the other hand, is essential for cell growth and for the intrinsic RNA cleavage activity of RNA pol III, therefore playing a role in RNA pol III termination (Chedin et al., 1998). This set of 10 subunits constitutes an extended core RNA polymerase specific for eukaryotic transcription whose structure for the class II enzyme has been studied extensively by electron microscopy (Darst et al., 1991) and culminated recently in the determination of its atomic structure by X-ray diffraction (Cramer et al., 2001).
In addition, each enzyme class contains a distinct set of specific subunits, whose number varies: four for RNA pol I, two for RNA pol II and seven for RNA pol III. The RNA pol I-specific subunits can be divided tentatively into two groups. The first group is composed of subunits A49 and A34.5, which are labile through a high salt treatment resulting in the purification of the A* form of RNA pol I (Huet et al., 1975). Further evidence for a coordinated interaction of these subunits with the core enzyme comes from the observation that a genetic depletion of subunit A34.5 is viable, but results in the loss of subunit A49 upon purification, and vice versa (Gadal et al., 1997). Genetic depletion of A49 or of both A49 and A34.5 subunits results in a cryo-sensitive but viable strain, indicating that these RNA pol I-specific subunits are not strictly required in normal growing conditions (Liljelund et al., 1992; Gadal et al., 1997). The A* form of RNA pol I shows a reduced transcriptional activity at high salt and a higher sensitivity to α-amanitine as compared with the wild-type enzyme, suggesting a role of these subunits in the elongation process (Huet et al., 1975).
The second group of subunits, which are only present in RNA pol I, is composed of A14 and A43. A yeast strain where the Rpa14 gene is disrupted is viable, but the purified RNA pol I was found to be inactive in transcription and depleted of subunits ABC23 (Rpb6), A14 and A43 (Lanzendorfer et al., 1997), suggesting that A14 stabilizes the interaction between A43 and the core enzyme (Smid et al., 1995). Deletion mutants showed that A43 is the only specific subunit essential for cell growth in RNA pol I, although not for catalytic activity (Thuriaux et al., 1995; Lanzendorfer et al., 1997). A43 appears to participate in the recruitment of the enzyme to the rDNA promoter, since it interacts with the transcription factor Rrn3p, which bridges RNA pol I to the core factor (CF) through its interaction with the CF subunit Rrn6p (Peyroche et al., 2000).
Structural information on the interaction of the RNA pol I-specific subunits with the extended core enzyme will shed some light on the function of these polypeptides. The structure of RNA pol I was investigated previously by electron microscopy of negatively stained two-dimensional (2D) crystals (Schultz et al., 1993; Klinger et al., 1996), and an improved model is presented here which is derived from the analysis of unstained, non-crystallized RNA pol I molecules preserved in vitreous water. This study exploits the complementarities between cryo- electron microscopy and X-ray crystallography to dock the atomic structure of the homologous RNA pol II Δ4/7 enzyme (Cramer et al., 2001) into the wild-type RNA pol I, and has mapped additional RNA pol I densities on the corresponding atomic model. Using subunit-specific antibodies, these densities could be characterized and attrib uted to RNA pol I-specific features. Altogether our results reveal that A14 and A43 co-localize in a stalk absent in RNA pol II Δ4/7, that A49 interacts with the clamp region involved in DNA binding and that A34.5 is located in the DNA binding cleft. The position of the subunits with respect to the functional sites of the enzyme confirms that A43 is implicated in the interaction with the initiation factors, and suggests that A49 and A34.5 modulate the DNA binding efficiency of the enzyme.
Results
Three-dimensional model of yeast RNA pol I
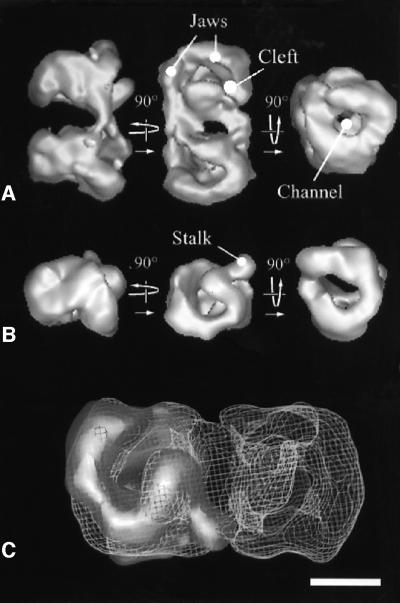
When the purified yeast RNA pol I was adsorbed on a carbon film and negatively stained, dimeric and monomeric enzyme forms could be clearly discriminated by the elongated shape of the dimers (Klinger et al., 1996). A three-dimensional (3D) model of the dimeric form of the enzyme was calculated from 60 different noise-free views (e.g. class averages) obtained upon analysis of 2223 molecular images. The resolution test performed on two independent data sets gave values of 34 and 25 Å for the 0.5 FSC and 3σ criteria, respectively (data not shown). The dimensions of the RNA pol I dimer were 24 × 12 × 14 nm (Figure 1A) and showed an internal 2-fold symmetry. The density threshold of the 3D model represented a volume of 1176 kDa, corresponding to twice the mass of the RNA pol I molecule. When the symmetry axis was in plane (Figure 1A, side view, left panel), each monomer had an elongated wedge shape and the dimerization contacts occurred mainly at the top of the monomers, whereas the narrow apical regions did not interact. When viewed down the 2-fold axis (Figure 1A, top view, middle panel), the top surface showed a deep and curved cleft separating the monomer structure into two parts. The cleft was open near the dimerization interface and changed into a channel closed by two jaws, each formed by a cleft wall. The protein contacts appeared on both sides of the dimerization interface and the extent of each monomer cannot be determined in this region. When viewed down the long dimer axis (Figure 1A, right panel), the whole structure appeared to be traversed by a channel opening into a funnel at each end.
Fig. 1. A 3D model of a negatively stained RNA pol I dimer and monomer. (A) Surface representation of the yeast RNA pol I dimers at 3.4 nm resolution. The 2-fold symmetry axis of the dimer is horizontal in the left panel, perpendicular to the plane in the middle panel and vertical in the right panel. Characteristic structural features described in the text, such as the cleft, the channel and the jaws, are indicated. (B) 3D model of the RNA pol I monomer represented in the same orientations as the lower molecule in the dimer shown in (A). The stalk substructure protruding from the model is indicated. (C) Docking of the monomer into the envelope of the dimer, showing that the interface is constituted mainly of the stalk interacting with the end of the cleft. The position of the stalk may be slightly different in the monomer than in the dimer. Bar represents 10 nm in (A) and (B), and 5.5 nm in (C).
In order to reveal in more detail the structure of the dimerization interface, a 3D model of RNA pol I monomers was determined from 89 different noise-free views obtained upon analysis of 2794 particles. The resolution test performed on two independent data sets gave values of 34 and 29 Å for the 0.5 FSC and 3σ criteria, respectively (data not shown). The monomer model revealed a 4-nm-long and 3-nm-wide protein density, hereafter identified as the stalk, which was protruding out of the structure and originated from one of the cleft walls near the dimerization interface (Figure 1B). When docked into the envelope of the dimer, the stalk of one monomer interacted with the end of the cleft of the second monomer (Figure 1C).
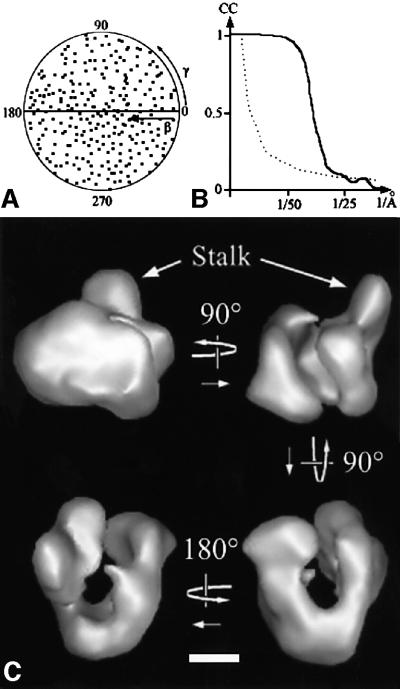
A dataset consisting of 7147 molecular images of unsupported and unstained, frozen hydrated RNA pol I monomers was recorded and analysed. A total of 266 different noise-free views of the molecule were used to calculate the 3D model of the enzyme (Figure 2A). The resolution tests gave values of 32 or 25 Å for the 0.5 FSC and 3σ criteria, respectively (Figure 2B). The overall shape of the unstained, frozen hydrated RNA pol I monomer was similar to that of the negatively stained model (Figure 2C). In the hydrated model, the separation between the two jaws was clearly marked and the stalk appeared larger.
Fig. 2. 3D model of frozen hydrated yeast RNA pol I. (A) Diagram showing the orientations of the 266 class averages used in the final reconstruction. Each class average is represented by a point in a (β,γ) coordinate system. (B) Diagram representing the Fourier shell correlation function between two independent reconstructions (continuous line) and the 3σ threshold curve (dotted line). The value of the Fourier shell correlation coefficient is indicated compared with the resolution in 1/Å. (C) Surface representation of the 3D reconstruction of the yeast RNA pol I at a resolution of 3.2 nm. Bar = 5 nm.
Docking of the atomic model of yeast RNA pol II into the 3D model of the native RNA pol I determined by cryo-electron microcopy
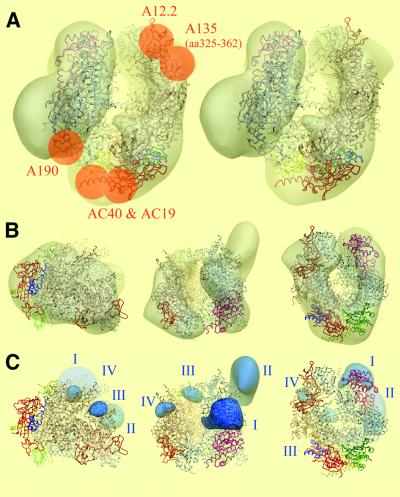
A comparison of the cryo-electron microscopy model of the native RNA pol I monomer with the atomic structure of the RNA pol II Δ4/7 (Cramer et al., 2001) was performed in order to reveal differences due to the presence of the four subunits A49, A34.5, A43 and A14. The Situs program (Wriggers et al., 1999) found two docking solutions with correlation coefficients of 0.628 and 0.497, respectively, corresponding to 180° rotations around the internal pseudo-2-fold axis of the molecule. The solution with the highest correlation was found to be correct according to our previously reported immunomapping experiments (Figure 3A and B). A monoclonal antibody directed against residues 325–362 of A135 labelled the jaw facing the wall from which the stalk protrudes (Klinger et al., 1996) and the homologous Rpb2 sequence was superimposed onto the labelled region in the correct docking solution. With this alignment of the two models, Rpb9 is superimposed to its homologue A12.2, whose position was determined by immunolabelling (Chedin et al., 1998). In this orientation, the positions of AC40 and AC19 in the apical part of the enzyme fitted with the positions of Rpb3 and Rpb11, their RNA pol II homologues, whereas an A190-specific antibody mapped a region occupied by Rpb1 (Klinger et al., 1996). The immunolocalization of the RNA pol I subunits was thus in excellent agreement with the proposed alignment of the atomic model.
Fig. 3. Comparison of the cryo-electron microscopic model of RNA pol I with the atomic model of RNA pol II Δ4/7. (A) Stereoviews of the atomic model of RNA pol II Δ4/7 (ribbon diagram) docked into the cryo-electron microscopy envelope of RNA pol I (transparent shell). The 10 common or homologous subunits are colour coded as follows: Rpb1, white; Rpb2, gold; Rpb3, red; Rpb5, pink; Rpb6, light blue; Rpb8, dark green; Rpb9, orange; Rpb10, dark blue; Rpb11, yellow; Rpb12, light green. The docking was confirmed by previous immunolabelling experiments, represented as orange dots on the left panel. (B) The docking is represented along the three characteristic orientations shown in Figure 1B. (C) Density difference map between RNA pol I and RNA pol II, thresholded at 3σ. Additional densities present in RNA pol I are represented in blue on the atomic model of RNA pol II Δ4/7 (ribbon diagram), and are labelled I, II, III and IV.
This docking allowed the positioning of characteristic structural features described for the atomic model of RNA pol II Δ4/7 in our cryo-electron microscopy model. A deep cleft, which was shown to bind the downstream DNA (Gnatt et al., 2001), separates two distinct masses formed essentially by the two large subunits A190 and A135. Two jaws are apparent at the tips of the cleft; the A190 jaw is formed by A190 and Rpb5 (ABC27), and the A135 jaw contains part of A135 and A12.2 (the homologue of Rpb9). The funnel is also apparent as a depression at the enzyme surface, which communicates with the cleft through a hole. The position of the clamp is inferred from the docking at the top of the A190 cleft wall and is located above the stalk, which participates in the dimer interface. This region also contains the Rpb6 (ABC23) subunit (light blue in Figure 3A).
The 3D density difference map between the cryo-electron microscopy model and the atomic structure revealed four additional densities present in RNA pol I (Figure 3C). Density I is saddle shaped and is placed on top of the clamp head domain of Rpb1 in proximity to the Zn6 binding site (Table I) and close to subunit Rpb5. The corresponding sequence of the A190 subunit maps to a weakly conserved region that shows a poor alignment between Rpb1 and Rpa190. The A190 subunit contains an insertion of 74 residues within the Zn6 binding site, whose structure is Cx2Cx127Cx2C, and an additional insertion of 43 residues after the Zn6 binding site and before conserved region B. These insertions are present in Schizosaccharo myces pombe and also in higher eukaryotes where they are smaller (42 and 23 residues, respectively), but no RNA pol I-specific motif could be detected in this region. The most prominent density II is located in the stalk, which contacts the enzyme in the region where the N- and the resolved C-termini of Rpb1, as well as the C-terminus of Rpb2, are grouped (Table I). Density II also contacts the Rpb1 dock domain and the assembly domain of Rpb6. Two RNA pol I-specific insertions are present in the N-termini of A190; the first one is five residues long and is located between the two conserved Zn binding motifs. The second one is 36 amino acids long and is located two residues after the second Cx2C motif. Both insertions are found in S.pombe, whereas only the first one is present in higher eukaryotes. The smaller density III is detected at the entrance of the cleft, where it contacts distinct loops of the Rpb2 wall domain. Finally, density IV contacts Rpb2 in the protrusion and lobe domains (Table I).
Table I. Structural domains and sequences contacting the additional densities present in RNA pol I.
| Additional density | Structural domain in RNA pol II | Secondary elements in RNA pol II (residue numbers) | Corresponding residues in RNA pol I |
|---|---|---|---|
| I | Rpb1 clamp head | Zn6 binding site: α2 and α3 (aa 105–118), β3–α4 loop (aa 145–170) | A190 (aa 100–120, aa 206–239) |
| β4-β5 loop (aa 183–197) | A190 (aa 255–304) | ||
| α5 and α6 (aa 200–223) | A190 (aa 313–336) | ||
| II | Rpb1 clamp core | N-terminus (aa 4–10, aa 60–75) | A190 (aa 4–10, aa 56–70) |
| Rpb1 linker | Start of CTD | No equivalence | |
| Rpb1 dock | β11–β12 (aa 400–425) | A190 (aa 531–560) | |
| Rpb2 clamp | Zn7 binding site: β43–β44 (aa 1165–1182) | A135 (aa 1107–1128) | |
| Rpb6 assembly | Loop between α1 and α2 (aa 101–118) | Same subunit | |
| III | Rpb2 wall | β28–β33 (aa 855–875, aa 910–935, aa 955–965) | A135 (aa 803–826, aa 857–870, aa 888–902) |
| IV | Rpb2 protrusion | α2 (aa 58–64), β1 (aa 89–98), β2 (aa 125–135), α11 (aa 410–430) | A135 (aa 59–65, aa96–105, aa134–144, aa 406–428) |
| Rpb2 lobe | β8–β9 loop (aa 240–250) | A135 (aa 236–246) |
CTD, C-terminal domain.
Immunolabelling of the RNA pol I-specific subunits
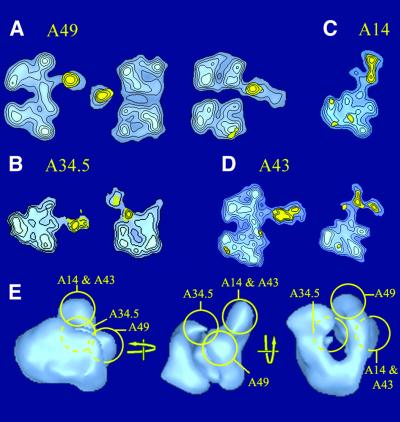
In order to identify the source of the additional protein densities present in the 3D model of RNA pol I, a series of immunolabelling experiments was conducted using subunit-specific antibodies. A data set of 2046 RNA pol I dimers labelled with the A49-specific antibody revealed three characteristic labelled views (Figure 4A). In side views, the A49-specific antibody bound in the vicinity of the DNA binding cleft (left panel in Figure 4A). When viewed down the 2-fold axis, the labelled molecules revealed that the antibody interacted outside of the cleft. When the cleft was oriented away from the supporting carbon foil, the antibody density appeared detached from RNA pol I, probably due to stain accumulation or partial embedding of the immunoglobulin G (IgG) molecule (Figure 4A, top view, central panel). In the reversed orientation, the IgG molecule clearly contacted the stain-exclusion volume of the enzyme (Figure 4A, right panel). The labelled site overlapped with the previously identified density I, indicating that subunit A49 contributed to this density difference between RNA pol I and RNA pol II Δ4/7.
Fig. 4. Immunolabelling of the RNA pol I-specific subunits. Purified RNA pol I was incubated with subunit-specific antibodies, and molecular images of immune complexes were analysed to obtain characteristic noise-free images of the labelled enzyme. The statistically significant difference with the corresponding unlabelled view is shown in yellow and is overlaid on the labelled view, represented as blue isodensity contour levels. (A) Characteristic views of RNA pol I dimers labelled with a subunit A49-specific antibody. Left: side view; middle: top view; right: bottom view. (B) Side view (left) and top view (right) of RNA pol I monomers labelled with a subunit A34.5-specific antibody, showing that A34.5 is located at the entrance to the cleft. (C) Side view of a labelled RNA pol I monomer showing that the HA-tagged N-terminus of subunit A14 is placed at the tip of the stalk. (D) Side views of labelled RNA pol I dimers (left) and monomers (right) showing the HA-tagged N-terminus of subunit A43 in the stalk. (E) Representation of the subunit-specific antibody binding sites on the surface of the 3D model of RNA pol I, oriented as in Figure 1B.
When incubated with the A34.5-specific antibody, RNA pol I dimers were found to be dissociated and a data set of 766 labelled monomers was analysed. The labelled side view (Figure 4B, left panel) showed that A34.5 was also located in proximity to the cleft, but in contrast to the A49 subunit, the top view (right panel in Figure 4B) revealed that the antibody interacted with the entry of the cleft. This location of A34.5 was consistent with the observed interference of antibody binding with dimerization since the stalk of monomer 2 covered this region of monomer 1 to form the dimer (Figure 1C). The antibody binding site superposed to density III (Figure 3B), indicating that subunit A34.5 participated in this density difference.
RNA pol I was purified from yeast strains expressing a HA-tagged version of A43 and A14, which proved to increase significantly the amount of immunolabelled RNA pol I monomers and dimers. The analysis of 527 labelled RNA pol I dimers HA tagged on subunit A43 showed that the side views were labelled in the vicinity of the cleft (Figure 4D, left panel), but failed to reveal additional labelled views, probably because the antibody binding oriented the immune complexes. The analysis of 506 labelled monomers showed that the antibody bound to the stalk, indicating that the HA-tagged N-terminus of subunit A43 was located in that structure (Figure 4D, right panel). When the tag was placed on subunit A14, the analysis of 1803 labelled monomers showed that the specific antibody bound to the stalk, indicating that the N-terminus of A14 was located at the tip of the protrusion (Figure 4C). Both subunits A43 and A14 contribute to the stalk structure and overlap with density difference II, but the immunolabelling experiments were not precise enough to determine the relative positions of the two subunits.
Immunolabelling experiments thus characterized the three major additional densities I, II and III, present in RNA pol I, as corresponding respectively to subunits A49, A14 and A43, and A34.5 (Figure 4E). Density IV was not labelled by any of the antibodies used and was, therefore, not due to the presence of an RNA pol I-specific subunit.
Discussion
In this study, the structure of yeast RNA pol I, derived from the analysis of cryo-electron microscopy images of frozen hydrated molecules, was compared with the atomic structure of the homologous yeast RNA pol II deprived of subunits Rpb4 and Rpb7. The cryo-model obtained from unstained molecules fitted better with the crystallographic data than our previous reconstruction, which was derived from negatively stained 2D crystals, even though the sample composition was identical (see Supplementary data available at The EMBO Journal Online) (Schultz et al., 1993). This improvement is mainly due to a more isotropic sampling of the projection space and to the absence of dehydration-induced flattening. To a resolution of ∼32 Å, the overall shape and dimensions of the enzyme were preserved, and structural features such as the cleft, the channel and the funnel were clearly resolved. This structural information was sufficient to perform an accurate docking of the atomic structure into the cryo-model and was confirmed by previously reported immunomapping of RNA pol I subunits. Once docked, a difference map was calculated between the two models to reveal the distribution of protein densities present only in RNA pol I, whereas an immunolabelling approach was used to characterize those differences due to, or at least involving, specific subunits. Finally, the X-ray model was used to map precisely the density differences on the atomic structure of RNA pol II Δ4/7, which, assuming a similar fold of the largest subunits, identified secondary structures possibly involved in subunit interfaces (Figure 5).
Fig. 5. Mapping the domains of A190 and A135 interacting with the RNA pol I-specific subunits. Schematic alignment of the largest subunit Rpb1 with A190 (A), and of the second largest subunit Rpb2 with A135 (B). Structural domains are indicated on the RNA pol II sequences and are named as in Cramer et al. (2001). Insertions larger than five residues are mapped on top of the RNA pol I sequences if present in the RNA pol II genes, and on the bottom of the RNA pol I sequences if present in the RNA pol I genes. The size of the insertions are indicated. Conserved regions are represented by black boxes. The regions contacting the additional densities present in RNA pol I are indicated by black bars.
Our results show that the four RNA pol I-specific subunits are located on the same side of the enzyme, close to the major cleft, which was shown to bind DNA (Gnatt et al., 2001). Subunits A43 and A14 are co-localized in the stalk, a protruding protein density revealed only in native RNA pol I; the A49 subunit is placed on top of the clamp formed by the largest subunit, whereas subunit A34.5 interacts with the end of the DNA binding cleft. All additional densities present in RNA pol I could be attributed to an enzyme-specific subunit, except for a small density IV, which mapped in the Rpb2-protrusion and lobe domains. This additional protein density could be due to several unresolved loops in this region of Rpb2; this is, however, unlikely since insertions of up to 89 residues are present in Rpa190 as compared with Rpb1 and were not detected. This density difference could also reflect a conformational change between the crystallized and the isolated structure. Alternatively, since this domain is located closest to A34.5, it cannot be excluded that densities III and IV represent two domains of A34.5. To answer these questions, a higher resolution cryo-electron microscopy map is needed in order to recognize and position structural domains.
Localization of subunits A14 and A43 in a pol I-specific substructure
The stalk, a protein density protruding out of the RNA pol I structure, was absent in previously reported 3D models of RNA pol II Δ4/7 obtained by electron microscopy (Darst et al., 1991) or by X-ray diffraction (Cramer et al., 2001) and thus appeared specific to the class I enzyme. This structure contains the additional density II and harbours subunits A43 and A14, as demonstrated by immunolabelling experiments. The co-localization of these subunits is consistent with the subunit composition of an RNA pol I variant purified from the Rpa14-Δ strain, in which the gene encoding the A14 subunit was disrupted. This enzyme variant was found to lack not only A14, but also A43 and Rpb6 (ABC23), suggesting that these three subunits interact (Smid et al., 1995). Electron microscope observations showed further that the stalk was missing in the Rpa14-Δ enzyme and that this structure was not restored upon reassociation of recombinant Rpb6, thus indicating that A14 and A43 contributed to the stalk structure (Lanzendorfer et al., 1997). It could not, however, be excluded from these experiments that the stalk folds back in the absence of A43 and A14; this can be ruled out by the present difference map. The docking of the atomic model of RNA pol II into the electron density map of RNA pol I confirmed that the stalk is close to the C-terminal assembly region of Rpb6, which could explain the cross-talk between the three subunits missing in the Rpa14-Δ enzyme. Subunit ω, the prokaryotic homologue of Rpb6, was shown to be required for the proper assembly of the enzyme by latching the C- and N-termini of the largest subunit (Minakhin et al., 2001). The role of Rpb6 is more complex in eukaryotes because the properly assembled Rpa14-Δ enzyme was shown to be inactive in non-specific transcription, whereas the addition of recombinant Rpb6 restored the transcriptional activity of the Rpa14-Δ enzyme (Lanzendorfer et al., 1997). It was shown by electron microscopy that the binding of Rpb6 is accompanied by a local and reversible conformational change, which, according to our docking, corresponds to a movement of the clamp domain. Consequently, the association of Rpb6 may be required for a correct position of the clamp in addition to a role in latching the largest subunit.
Among the RNA pol I-specific subunits, A43 is the only essential one and homologues have been found in fission yeast and in higher eukaryotes, which indicates that its function has been conserved throughout evolution. Several lines of evidence indicate that A43 is implicated in the recruitment of the enzyme on the rDNA promoter by interacting through transcription factor Rrn3p with the core factor, which binds directly to the rDNA core promoter element (Keys et al., 1994; Peyroche et al., 2000). A43 and Rrn3p form a stable complex when co-expressed in Escherichia coli, and Rrn3p is believed to bridge RNA pol I with the C-terminal domain (CTD) of the Rrn6 core factor subunit (Peyroche et al., 2000). Immunoelectron microscopy data showed that Rrn3p binds to the stalk, which identifies this structure as an interface for assembly of the initiation factors. In this respect, it is interesting to note that the stalk structure is located at the site where the CTD of Rpb1 exits the RNA pol II structure (Cramer et al., 2001) (Figure 5). This CTD is composed of heptapeptide repeats and plays a role in the initiation phase by interacting with the basal transcription factors (Thompson et al., 1993; Myers et al., 1998) and in the elongation phase by binding RNA processing factors (Proudfoot, 2000). The CTD repeats are absent in A190, and our results show that a dedicated structure containing A43 is positioned at the same place and might fulfil similar functions, at least by binding the core transcription factor. The role of subunit A14 is more elusive; it is not essential for growth and the only documented role of A14 is to stabilize the interaction of subunits A43 and Rpb6 within the enzyme.
Localization and possible role of subunits A49 and A34.5
Antibody labelling revealed two distinct interaction sites for subunits A34.5 and A49 on the core enzyme at the entrance of the cleft and on the clamp head region, respectively. The subunit A49-specific density I maps near a 56-residue insertion in A190, which is only found in RNA pol I genes and is conserved, although with different lengths, from yeast to humans. Although no conserved motif could be detected in this insertion, it constitutes a good candidate to form an RNA pol I-specific interface for the interaction of the A49 subunit. Subunit A34.5 interacts mainly with the second largest subunit and the sequence elements in contact with density III involve structure elements of the wall domain (Figure 5). The corresponding sequences in A135 are highly variable and do not show any obvious RNA pol I-specific sequences. In keeping with the absence of sequence conservation is the observation that the orthologues of A34.5 remain to be discovered in other eukaryotes, since no clear sequences homologous to Rpa34.5 could be found in the relevant databases.
The distant localization of A49 and A34.5 is in contrast to the expected direct interaction inferred from the observation that RNA pol I purified from a Rpa34-Δ strain lacks both A34.5 and A49 (Gadal et al., 1997), and that the two subunits dissociate from the enzyme at high ionic strength (Huet et al., 1975). Since no complex of the two RNA pol I-specific subunits has been reported so far, it is possible that the dissociation of one subunit causes a conformational change in the core enzyme, which weakens the interaction of the second subunit. On the other hand, our results cannot rule out a direct interaction between the two subunits inside the cleft and over the Rpb1 wall, because of the uncertainty with respect to the exact position of A49 due to the flexibility of the enzyme. The clamp domain was shown to adopt distinct positions in different crystal forms (Cramer et al., 2001) and an outward movement of the clamp, resulting in an opening of the cleft, is conceivable upon A49 binding. In such a position, the clamp contributes to the observed density difference, which would then not outline the exact position of A49. As a result, A49 could reside on the side of the clamp, either inside or outside the DNA binding groove. A higher resolution cryo-electron microscopy map or a more precise mapping of A49 is needed to address this question.
The role of these subunits remains uncertain as they are not necessary for growth in normal conditions. The localization of A34.5 at the end of the cleft is consistent with a role in contacting the upstream DNA (Poglitsch et al., 1999; Gnatt et al., 2001). The C-terminal 44 residues of A34.5 are highly charged and contain 50% lysines, which yields a very hydrophilic and positively charged domain that could stabilize the interaction of the template with the core enzyme. This role may be dispensable in normal conditions, but could become essential in particular genetic contexts (Gadal et al., 1997). The described co-lethality of DNA topoisomerase I and A34.5 suggests that both polypeptides could collaborate to remove the topological stress that accumulates as negative supercoils upstream of the moving elongation complex (Liu and Wang, 1987; Masse and Drolet, 1999).
Several lines of evidence suggest that A49 could influence transcription by affecting the elongation process. The slow-growing and cryo-sensitive Rpa49-Δ strain shows a markedly reduced synthesis of rRNA (Liljelund et al., 1992) and early experiments on the A* form of RNA pol I indicated that the depletion of A49 and A34.5 had an ionic strength-dependent effect on transcriptional elongation (Huet et al., 1975). Consistent with a role of A49 in facilitating RNA pol I processivity, the nuclear amount of PAF53, the mouse orthologue of A49, was shown to be related to the proportion of active transcription units (Hanada et al., 1996; Hannan et al., 2000). In the context of an elongation complex, crystallographic studies showed that the clamp moves inwards in order to hold the downstream DNA more firmly and to allow processivity of the enzyme (Gnatt et al., 2001). In its position on the clamp head domain, A49 could influence the conformation of the clamp or interact directly with DNA in order to modify its association with the enzyme.
The paradox of the specific subunits
With the exception of A43, all RNA pol I-specific subunits, including subunit A12.2, are dispensable for growth in normal conditions. Any double gene disruptions of A12.2, A34.5 and A49, and even the inactivation of all three genes, are viable. Paradoxically, a double disruption of any of these three subunits with A14 is lethal through a defect in rDNA transcription (Gadal et al., 1997). A possible explanation for this co-lethality is that the inactivation of two independent non-essential functions may destabilize the enzymatic reaction and create a lethal phenotype. Accordingly, two groups of subunits, A14 on one side and A49/A34.5/A12.2 on the other side, cooperate to perform an essential function. We propose that the latter group contributes to the stability of the elongating RNA pol I by acting through different mechanisms on the association of the upstream and downstream DNA with the cleft. The depletion of these subunits would result in a less processive, but still functional enzyme. Subunit A14 could stabilize the interaction between Rpb6 and the largest subunit, which is essential for the correct operation of the clamp. A weaker association of Rpb6 could still result in a large enough fraction of active enzyme and permit cell viability. By combining a less processive and a reduced fraction of active enzyme, the cells could reach a threshold where rRNA cannot be synthesized to a sufficient level to allow viability.
Materials and methods
RNA polymerases and antibody purification
Saccharomyces cerevisiae cells were grown in YPD to an OD600 of 4. Starting with 400 g of cells, the RNA pol I was purified to homogeneity as described previously by loading the B2000 fraction onto a Superose 6 column (Milkereit and Tschochner, 1998). The purified enzyme was stored at –20°C in buffer P300 (20 mM HEPES pH 7.8, 300 mM potassium acetate, 2 mM MgCl2, 0.02 mM EDTA, 20% glycerol) at a concentration of 0.3 mg/ml. We used the yeast strains CPy2 and Rpa14-HA, where the N-termini of the A43 and A14 subunits, respectively, were tagged with a haemagglutinin (HA) epitope (Peyroche et al., 2000), to purify RNA pol I, in which a specific subunit is HA tagged. Polyclonal antibodies directed against subunits A49 and A34.5 were raised in rabbits upon injection of gel-purified subunits. The antibodies were purified by ammonium sulfate precipitation and DEAE–cellulose chromatography, and assayed for their ability to immunoprecipitate the native enzyme (Buhler et al., 1980).
Electron microscopy
For negatively stained preparations, 10 µl of purified RNA pol I fractions diluted to a concentration of 20 µg/ml in 20 mM Tris–HCl pH 7.4, 150 mM NaCl and 20% glycerol were placed on a 10-nm-thick carbon film, previously treated by a glow discharge in air. After 2 min of adsorption, the grid was negatively stained with a 2% (w/v) uranyl acetate solution. Immune complexes were formed and observed as described previously (Klinger et al., 1996). Briefly, a 3- to 5-fold molar excess of antibodies was incubated with purified RNA pol I at a final protein concentration of 20 µg/ml for 1 h at 20°C, and processed for electron microscopy as described above. The putatively labelled complexes were identified by a stain-excluding domain protruding out of the RNA pol I molecule that had a size and a shape consistent with that of an IgG molecule. Areas covered with individual molecules were recorded under low-dose conditions (<20 electrons/Å2) at a nominal magnification of ×45 000 on SO163 photographic plates.
For cryo-electron microscopy, a 5 µl droplet of purified RNA pol I at a concentration of 0.3 mg/ml was applied on a glow discharged holey carbon foil, washed twice with distilled water, blotted for 5–10 s with filter paper and plunged into an ethane slush cooled with liquid nitrogen. The frozen specimen was transferred to liquid nitrogen and mounted onto a pre-cooled cryo-specimen holder (Model 626; Gatan, Pleasanton, CA). The holder was inserted in the cryo-electron microscope (Model CM120; Philips, Eindhoven, The Netherlands), which was equipped with a liquid nitrogen-cooled anti-contaminator (Homo et al., 1984) and was operating at 100 kV with a LaB6 filament. Images were recorded under low-dose conditions at a magnification of ×31 000 on SO163 photographic plates.
Image processing
The micrographs were checked by optical diffraction for absence of astigmatism and optimal contrast transfer function. The selected micrographs of negatively stained samples were digitized at 18 µm raster size, resulting in a pixel spacing of 0.4 nm on the object. Cryo-electron micrographs were scanned at a 14 µm raster size (0.45 nm at the object level) using a flat bed microdensitometer (SCAI; Z/I imaging, Oberkochen, Germany).
The image analysis was performed using the IMAGIC software package (van Heel et al., 1996). In order to investigate the 3D organization of the enzyme, a multistep approach was used to assign the viewing directions of the noise-free molecular projections, as described previously (Schultz et al., 2000). The untilted RNA pol I dimer images were iteratively aligned and analysed by multivariate statistical methods (van Heel and Frank, 1981) to identify preferential views, four of which, representing ∼60% of the total data set, were analysed further. For each of these orientations, an independent 3D model was calculated using a conical tilt approach (Radermacher et al., 1986) and the resulting four reconstructions were averaged to reduce the flattening effect. For further refinement, the untilted data set was aligned against reprojections of the averaged model, clustered into classes and the angular assignment of each class average was performed using sinogram correlation functions (van Heel, 1987b). This refinement cycle was iterated three times until the model did not evolve further. The RNA pol I monomer data set was analysed in a similar manner, except that the first angular assignment of the class averages used the dimer model, interactively masked to generate a series of monomer projections. The frozen hydrated data set was extracted from 19 images recorded at defocus values ranging from 1.2 to 3 µm. The images were partially corrected for the contrast transfer function of the microscope (CTF) by phase inversion between the minima. The viewing directions of the class averages were assigned by comparison with the negatively stained monomer model, and the cycles described above were used to refine the model. The resolution of the final reconstructions was estimated by the Fourier shell correlation function obtained by comparing two independent reconstructions generated by randomly splitting the data set in half, each being independently aligned and clustered. The resolution is indicated according to two criteria: the 0.5 cut-off Fourier shell correlation curve (0.5 FSC criterion) and the intersection point of the 3σ curve with the FSC curve (3σ criterion) (van Heel, 1987a).
In order to align in translation, rotation and magnification the cryo-electron microscopy model of RNA pol I with respect to the atomic model of RNA pol II Δ4/7, the atomic coordinates of the 10 subunit complex [Protein Data Bank (PDB) code 1I50, chain identification A, B, C, E, F, H, I, J, K and L] were transformed into a density map, low pass filtered to 1/2.5 nm–1, and compared with the RNA pol I electron density map. The density maps were quantified by vectors using the Situs program (Wriggers et al., 1999), followed by an exhaustive search of all possible docking solutions. The highest correlation (0.628) was obtained when the density maps were described by four codebook vectors and, in these conditions, the 2-fold symmetric alignment due to a partial internal symmetry of the monomer showed a value of 0.497. The correct docking solution between the two symmetric orientations was validated by previously reported immunolabelling experiments of key subunits, and a density difference map was calculated after normalization of the densities.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank M.Schatz for assistance with the IMAGIC software and for customizing the package, E.Orlova for help in angular assignment, the structural department at EMBL for providing access to the microdensitometer, and P.Cramer and R.D.Kornberg for providing the atomic coordinates of RNA pol II Δ4/7 before publication. This work was supported by the Institut National de la Santé et de la Recherche Médicale, the Centre National pour la Recherche Scientifique, the Hôpital Universitaire de Strasbourg (HUS), the Association pour la Recherche sur le Cancer and the Human Frontier Science Program Organization (grant No. RG 336/99). N.B. was supported by a grant from La Ligue Contre le Cancer.
References
- Buhler J.M., Huet,J., Davies,K.E., Sentenac,A. and Fromageot,P. (1980) Immunological studies of yeast nuclear RNA polymerases at the subunit level. J. Biol. Chem., 255, 9949–9954. [PubMed] [Google Scholar]
- Chedin S., Riva,M., Schultz,P., Sentenac,A. and Carles,C. (1998) The RNA cleavage activity of RNA polymerase III is mediated by an essential TFIIS-like subunit and is important for transcription termination. Genes Dev., 12, 3857–3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P., Bushnell,D.A. and Kornberg,R.D. (2001) Structural basis of transcription: RNA polymerase II at 2.8 Å resolution. Science, 292, 1863–1876. [DOI] [PubMed] [Google Scholar]
- Darst S.A., Edwards,A.M., Kubalek,E.W. and Kornberg,R.D. (1991) Three-dimensional structure of yeast RNA polymerase II at 16 Å resolution. Cell, 66, 121–128. [DOI] [PubMed] [Google Scholar]
- Dumay H., Rubbi,L., Sentenac,A. and Marck,C. (1999) Interaction between yeast RNA polymerase III and transcription factor TFIIIC via ABC10α and τ131 subunits. J. Biol. Chem., 274, 33462–33468. [DOI] [PubMed] [Google Scholar]
- Gadal O., Mariotte-Labarre,S., Chedin,S., Quemeneur,E., Carles,C., Sentenac,A. and Thuriaux,P. (1997) A34.5, a nonessential component of yeast RNA polymerase I, cooperates with subunit A14 and DNA topoisomerase I to produce a functional rRNA synthesis machine. Mol. Cell. Biol., 17, 1787–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnatt A.L., Cramer,P., Fu,J., Bushnell,D.A. and Kornberg,R.D. (2001) Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 Å resolution. Science, 292, 1876–1882. [DOI] [PubMed] [Google Scholar]
- Hanada K., Song,C.Z., Yamamoto,K., Yano,K., Maeda,Y., Yamaguchi,K. and Muramatsu,M. (1996) RNA polymerase I associated factor 53 binds to the nucleolar transcription factor UBF and functions in specific rDNA transcription. EMBO J., 15, 2217–2226. [PMC free article] [PubMed] [Google Scholar]
- Hannan K.M., Kennedy,B.K., Cavanaugh,A.H., Hannan,R.D., Hirschler-Laszkiewicz,I., Jefferson,L.S. and Rothblum,L.I. (2000) RNA polymerase I transcription in confluent cells: Rb downregulates rDNA transcription during confluence-induced cell cycle arrest. Oncogene, 19, 3487–3497. [DOI] [PubMed] [Google Scholar]
- Hemming S.A. and Edwards,A.M. (2000) Yeast RNA polymerase II subunit RPB9. Mapping of domains required for transcription elongation. J. Biol. Chem., 275, 2288–2294. [DOI] [PubMed] [Google Scholar]
- Heyduk T., Lee,J.C., Ebright,Y.W., Blatter,E.E., Zhou,Y. and Ebright,R.H. (1993) CAP interacts with RNA polymerase in solution in the absence of promoter DNA. Nature, 364, 548–549. [DOI] [PubMed] [Google Scholar]
- Homo J.C., Booy,F.P., Labouesse,P., Lepault,J. and Dubochet,J. (1984) Improved anticontaminator for cryo-electron microscopy with Philips EM400. J. Microsc., 136, 337–340. [Google Scholar]
- Huet J., Buhler,J.M., Sentenac,A. and Fromageot,P. (1975) Dissociation of two polypeptide chains from yeast RNA polymerase A. Proc. Natl Acad. Sci. USA, 72, 3034–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keys D.A., Vu,L., Steffan,J.S., Dodd,J.A., Yamamoto,R.T., Nogi,Y. and Nomura,M. (1994) RRN6 and RRN7 encode subunits of a multiprotein complex essential for the initiation of rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae. Genes Dev., 8, 2349–2362. [DOI] [PubMed] [Google Scholar]
- Klinger C., Huet,J., Song,D., Petersen,G., Riva,M., Bautz,E.K., Sentenac,A., Oudet,P. and Schultz,P. (1996) Localization of yeast RNA polymerase I core subunits by immunoelectron microscopy. EMBO J., 15, 4643–4653. [PMC free article] [PubMed] [Google Scholar]
- Krapp S., Kelly,G., Reischl,J., Weinzierl,R.O. and Matthews,S. (1998) Eukaryotic RNA polymerase subunit RPB8 is a new relative of the OB family. Nat. Struct. Biol., 5, 110–114. [DOI] [PubMed] [Google Scholar]
- Lanzendorfer M., Smid,A., Klinger,C., Schultz,P., Sentenac,A., Carles,C. and Riva,M. (1997) A shared subunit belongs to the eukaryotic core RNA polymerase. Genes Dev., 11, 1037–1047. [DOI] [PubMed] [Google Scholar]
- Liljelund P., Mariotte,S., Buhler,J.M. and Sentenac,A. (1992) Characterization and mutagenesis of the gene encoding the A49 subunit of RNA polymerase A in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 89, 9302–9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.F. and Wang,J.C. (1987) Supercoiling of the DNA template during transcription. Proc. Natl Acad. Sci. USA, 84, 7024–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse E. and Drolet,M. (1999) Relaxation of transcription-induced negative supercoiling is an essential function of Escherichia coli DNA topoisomerase I. J. Biol. Chem., 274, 16654–16658. [DOI] [PubMed] [Google Scholar]
- Milkereit P. and Tschochner,H. (1998) A specialized form of RNA polymerase I, essential for initiation and growth-dependent regulation of rRNA synthesis, is disrupted during transcription. EMBO J., 17, 3692–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minakhin L., Bhagat,S., Brunning,A., Campbell,E.A., Darst,S.A., Ebright,R.H. and Severinov,K. (2001) Bacterial RNA polymerase subunit ω and eukaryotic RNA polymerase subunit RPB6 are sequence, structural, and functional homologs and promote RNA polymerase assembly. Proc. Natl Acad. Sci. USA, 98, 892–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers L.C., Gustafsson,C.M., Bushnell,D.A., Lui,M., Erdjument-Bromage,H., Tempst,P. and Kornberg,R.D. (1998) The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev., 12, 45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi Y., Yano,R., Dodd,J., Carles,C. and Nomura,M. (1993) Gene RRN4 in Saccharomyces cerevisiae encodes the A12.2 subunit of RNA polymerase I and is essential only at high temperatures. Mol. Cell. Biol., 13, 114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyroche G., Milkereit,P., Bischler,N., Tschochner,H., Schultz,P., Sentenac,A., Carles,C. and Riva,M. (2000) The recruitment of RNA polymerase I on rDNA is mediated by the interaction of the A43 subunit with Rrn3. EMBO J., 19, 5473–5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poglitsch C.L., Meredith,G.D., Gnatt,A.L., Jensen,G.J., Chang,W.H., Fu,J. and Kornberg,R.D. (1999) Electron crystal structure of an RNA polymerase II transcription elongation complex. Cell, 98, 791–798. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. (2000) Connecting transcription to messenger RNA processing. Trends Biochem. Sci., 25, 290–293. [DOI] [PubMed] [Google Scholar]
- Radermacher M., Wagenknecht,T., Verschoor,A. and Frank,J. (1986) A new 3-D reconstruction scheme applied to the 50S ribosomal subunit of E.coli. J. Microsc., 141, RP1–RP2. [DOI] [PubMed] [Google Scholar]
- Rubbi L., Labarre-Mariotte,S., Chedin,S. and Thuriaux,P. (1999) Functional characterization of ABC10α, an essential polypeptide shared by all three forms of eukaryotic DNA-dependent RNA polymerases. J. Biol. Chem., 274, 31485–31492. [DOI] [PubMed] [Google Scholar]
- Schultz P., Celia,H., Riva,M., Sentenac,A. and Oudet,P. (1993) Three-dimensional model of yeast RNA polymerase I determined by electron microscopy of two-dimensional crystals. EMBO J., 12, 2601–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz P., Fribourg,S., Poterszman,A., Mallouh,V., Moras,D. and Egly,J.M. (2000) Molecular structure of human TFIIH. Cell, 102, 599–607. [DOI] [PubMed] [Google Scholar]
- Smid A., Riva,M., Bouet,F., Sentenac,A. and Carles,C. (1995) The association of three subunits with yeast RNA polymerase is stabilized by A14. J. Biol. Chem., 270, 13534–13540. [DOI] [PubMed] [Google Scholar]
- Thompson C.M., Koleske,A.J., Chao,D.M. and Young,R.A. (1993) A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell, 73, 1361–1375. [DOI] [PubMed] [Google Scholar]
- Thuriaux P. and Sentenac,A. (1992) Yeast nuclear RNA polymerases. In Jones,E.W., Pringle,J.T. and Broach,J.R. (eds), The Molecular Biology of the Yeast Saccharomyces: Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 1–48.
- Thuriaux P., Mariotte,S., Buhler,J.M., Sentenac,A., Vu,L., Lee,B.S. and Nomura,M. (1995) Gene RPA43 in Saccharomyces cerevisiae encodes an essential subunit of RNA polymerase I. J. Biol. Chem., 270, 24252–24257. [DOI] [PubMed] [Google Scholar]
- van Heel M. (1987a) Similarity measures between images. Ultramicroscopy, 21, 95–100. [Google Scholar]
- van Heel M. (1987b) Angular reconstitution: a-posteriori assignment of projection directions for 3D reconstruction. Ultramicroscopy, 21, 111–124. [DOI] [PubMed] [Google Scholar]
- van Heel M. and Frank,J. (1981) Use of multivariate statistics in analysing the images of biological macromolecules. Ultramicroscopy, 6, 187–194. [DOI] [PubMed] [Google Scholar]
- van Heel M., Harauz,G., Orlova,E.V., Schmidt R. and Schatz,M. (1996) A new generation of the IMAGIC image processing system. J. Struct. Biol., 116, 17–24. [DOI] [PubMed] [Google Scholar]
- Wei W., Dorjsuren,D., Lin,Y., Qin,W., Nomura,T., Hayashi,N. and Murakami,S. (2001) Direct interaction between the subunit RAP30 of transcription factor IIF (TFIIF) and RNA polymerase subunit 5, which contributes to the association between TFIIF and RNA polymerase II. J. Biol. Chem., 276, 12266–12273. [DOI] [PubMed] [Google Scholar]
- Woychik N.A. and Young,R.A. (1989) RNA polymerase II subunit RPB4 is essential for high- and low-temperature yeast cell growth. Mol. Cell. Biol., 9, 2854–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wriggers W., Milligan,R.A. and McCammon,J.A. (1999) Situs: A package for docking crystal structures into low-resolution maps from electron microscopy. J. Struct. Biol., 125, 185–195. [DOI] [PubMed] [Google Scholar]