Abstract
The cis-acting sequences required for chromosome segregation are poorly understood in most organisms, including bacteria. Sporulating cells of Bacillus subtilis undergo an unusual asymmetric cell division during which the origin of DNA replication (oriC) region of the chromosome migrates to an extreme polar position. We have now characterized the sequences required for this migration. We show that the previously characterized soj–spo0J chromosome segregation system is not essential for chromosome movement to the cell pole, so this must be driven by an additional segregation mechanism. Observations on a large set of precisely engineered chromosomal inversions and translocations have identified a polar localization region (PLR), which lies ∼150–300 kbp to the left of oriC. Surprisingly, oriC itself has no involvement in this chromosome segregation system. Dissection of the PLR showed that it has internal functional redundancy, reminiscent of the large diffuse centromeres of most eukaryotic cells.
Keywords: Bacillus subtilis/centromere/chromosome segregation/spo0J gene/sporulation
Introduction
Centromeres were originally identified cytologically in certain eukaryotic cells as specialized heterochromatic regions where chromosomes become attached to the mitotic spindle. The term is now used more generally to describe the cis-acting DNA sequences responsible for accurate chromosome segregation. In eukaryotes, centromeres generally comprise huge regions containing many tens or hundreds of kilobase pairs of DNA. The sequences seem to be poorly conserved, as are the proteins that recognize them (reviewed by Karpen and Allshire, 1997; Pidoux and Allshire, 2000).
In bacteria, almost nothing is known about cis-acting sequences needed for chromosome segregation (Gordon and Wright, 2000; Hiraga, 2000). It has long been thought that oriC, the site where bi-directional chromosome replication usually begins, also acts as the target for the segregation machinery (Jacob et al., 1963), but there is little direct evidence to support this notion. Work on the oriC region of Bacillus subtilis has shown that soon after a new round of replication initiates, the two daughter oriC regions separate and abruptly move apart towards opposite poles of the cell, suggesting the existence of an active segregation apparatus (Glaser et al., 1997; Lewis and Errington, 1997; Webb et al., 1997, 1998; Sharpe and Errington, 1998). Similar events occur in other bacterial cells (Gordon et al., 1997; Mohl and Gober, 1997; Niki and Hiraga, 1998), although the timing of events and precise positioning of oriC show some phylogenetic variation (Sharpe and Errington, 1999; Gordon and Wright, 2000; Hiraga, 2000). In general, the experiments described above show that the oriC region moves actively during chromosome segregation, but they do not distinguish whether the mechanism responsible for the movement acts at oriC or some other site on the chromosome.
Several candidates for genes involved in chromosome segregation have been identified. However, in most cases, a direct role in chromosome segregation is difficult to distinguish from an effect on nucleoid replication, condensation or organization (Wake and Errington, 1995; Hiraga, 2000). One of the best known systems comprises the soj and spo0J genes of B.subtilis. Mutations in spo0J have a weak segregation phenotype in B.subtilis (Ireton et al., 1994). Spo0J binds to a series of sites around the oriC region of B.subtilis (Lin and Grossman, 1998) and forms tight, highly condensed foci that co-localize with oriC throughout the cell cycle in wild-type cells (Glaser et al., 1997; Lewis and Errington, 1997; Lin et al., 1997). Soj protein has a clear role as a negative regulator of transcription during sporulation (Ireton et al., 1994; Cervin et al., 1998; Marston and Errington, 1999; Quisel et al., 1999; Quisel and Grossman, 2000). There is no gross effect on chromosome segregation in the absence of Soj, but recent work has shown that Spo0J foci are fragmented and dispersed in soj mutants, suggesting that Soj has a role in some aspect of chromosome organization (Marston and Errington, 1999; Autret et al., 2001). Genes closely related to soj and spo0J are found in many diverse bacteria, generally in the vicinity of oriC, though curiously, they are absent from Escherichia coli and its close relatives. There is good evidence that they have a direct role in segregation of plasmids (Gordon and Wright, 2000; Hiraga, 2000), but the underlying mechanism remains obscure.
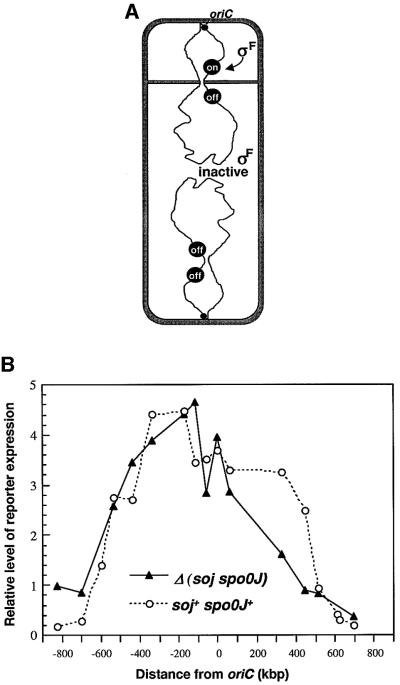
The asymmetric cell division that occurs early in spore formation of B.subtilis provides a unique opportunity for studying chromosome organization and orientation in bacteria. Soon after the onset of sporulation, the two sister chromosomes elongate along the long axis of the cell to form a structure known as the axial filament (Bylund et al., 1993). When the cell divides near to the cell pole, the division septum initially closes around one of the two nucleoids, trapping only one-third of a chromosome in the small (prespore) compartment. The remainder of the chromosome is then transported into the prespore by an unusual DNA translocation process requiring a protein called SpoIIIE, which is probably the transporter, to complete segregation (Wu and Errington, 1994; Wu et al., 1995; Bath et al., 2000). Translocation can therefore be blocked by use of a mutation in the spoIIIE gene, allowing the segment trapped in the prespore compartment to be analysed. A convenient way to detect DNA sequences present in the prespore is to take advantage of a transcription factor, σF, which is activated specifically in the small compartment. This activation occurs even in the absence of DNA transfer in a particular class of spoIIIE mutants (Wu and Errington, 1994; Figure 1A). If a σF-dependent reporter gene is located in the segment of DNA that is trapped in the prespore, it will be turned on in these spoIIIE mutants. Consequently, the level of reporter enzyme activity gives an indication of the frequency of trapping of the reporter gene in the prespore. We previously used this phenomenon to map the segment of DNA that is trapped in the prespore compartment and therefore the part of the chromosome that migrates to the cell pole early in sporulation. We showed that it comprises a fairly specific part of the chromosome, centred approximately on oriC, the origin of bi-directional chromosome replication (Wu and Errington, 1998).
Fig. 1. Effect of deletion of the soj–spo0J locus on chromosome trapping in the prespore compartment. (A) Schematic representation of the reporter trapping system. Soon after the onset of sporulation an asymmetrically positioned division septum closes around one of the two chromosomes of the cell to form the prespore compartment. SpoIIIE protein then normally drives transfer of the remainder of the chromosome into the small compartment but in spoIIIE mutants the chromosome remains in this bisected state. The prespore chromosome has a relatively fixed orientation with the oriC region invariably located within the prespore compartment and oriC-distal sequences located outside in the mother cell. A prespore-specific transcription factor, σF, becomes active in the small compartment and can drive expression of a σF-dependent reporter gene if located in the segment of the chromosome that is trapped in the prespore (‘on’) but not if it is located in the oriC-distal part of the chromosome (‘off’). (B) Effect of soj–spo0J deletion on the accumulation of β-galactosidase expressed from the σF-dependent gpr–lacZ reporter at different chromosomal positions (in a spoIIIE36 background). Strains were induced to sporulate and assayed at intervals for β-galactosidase activity. Open circles show results obtained previously with soj+ spo0J+ cells (Wu and Errington, 1998). Filled triangles show the data for the soj–spo0J [Δ(yyaA–yyaC)::kan] deletion derivatives (average of two to eight experiments, in samples taken after 3 h of sporulation, representative of the full time courses). The values given have been normalized to the activities obtained with the equivalent spoIIIE+ strains (set at 1), induced to sporulate and sampled in parallel (see Wu and Errington, 1998).
We have now used the reporter trapping system and other methods to identify the DNA sequences required for orientation of the chromosome at the onset of sporulation and investigated the nature of the interaction between this and the soj–spo0J system.
Results
Deletion of the soj–spo0J locus has only a minor effect on polar localization of the chromosome during sporulation
Previous application of the reporter trapping method (Figure 1A) showed that the segment of DNA trapped in the prespore compartment of spoIIIE mutant cells is a fairly specific one, of ∼1 Mbp, roughly centred on oriC, the site of initiation of bi-directional chromosome replication (Wu and Errington, 1998). It was also known that in the absence of the soj–spo0J system, the trapping of the amyE locus on the right arm of the chromosome was reduced, while that of the DNA near the terminus was slightly increased, suggesting that Spo0J is needed to specify the precise orientation of the chromosome at the time of polar division (Sharpe and Errington, 1996). On the other hand, Fransden et al. (1999) have described an elegant experiment demonstrating that sequences located at –421 kbp (positions on the 4.2 Mbp circular chromosome, here and after, are given in kilobase pairs relative to oriC; ‘+’ and ‘–’ indicate the right and left arms of the chromosome, respectively) on the chromosome are always trapped in the prespore in a soj–spo0J deletion mutant. To further understand the role of the soj–spo0J system in chromosome orientation during sporulation, we used the reporter trapping approach to map the segment of DNA trapped in the prespore compartment of a soj–spo0J spoIIIE mutant. (We actually used a larger deletion removing the adjacent, spo0J-like yyaA gene and the yyaC gene—the phenotypic effect of this larger deletion was not discernibly different from that of a soj–spo0J deletion; results not shown.) On the basis of the β-galactosidase activities obtained with insertions of the σF-dependent reporter (gpr–lacZ) at different sites, it appeared, surprisingly, that the chromosome still had a fairly specific orientation (Figure 1B). Moreover, the position of the peak activity overlapped substantially with that obtained in soj+ spo0J+ (spoIIIE) cells. As in the previous experiments, β-galactosidase activities to the left of oriC were generally higher than to the right. This is probably due, in part, to the presence of the rsfA gene, which enhances the transcription of some σF-dependent genes when present in the prespore (Wu and Errington, 2000).
The pattern did differ from that of soj+ spo0J+ cells in several minor ways (and these differences were borne out qualitatively by experiments with all of the strains patched out on X-Gal plates; data not shown). First, a large region of the right arm of the chromosome (from about +100 to +500) was trapped at a substantially lower frequency in the mutant background than in the wild type. This was surprising because most reported Spo0J binding sites lie to the left of oriC (Lin and Grossman, 1998). This right arm effect changed the overall shape of the distribution from a plateau with fairly steep edges in the wild type, to that of a peak in the –100 to –300 region, bounded on each side by more gentle gradients. These gradients probably reflect variability in positioning of the chromosome at the time of septation (Wu and Errington, 1998). Finally, at the outside edges of the gradients, significant activity could be detected at several positions where trapping is virtually absent in the wild type, for example –703, –826 and +698. This was consistent with our previous report of being able to detect significant expression of markers at very distant sites such as –1580 (gpr) and –1769 (dacF) in the soj–spo0J background (Sharpe and Errington, 1996).
These results showed that in B.subtilis cells devoid of the soj–spo0J system, there is still a mechanism imposing a distinct orientation on the chromosome during sporulation. They also suggested that the region of chromosome responsible for this Soj–Spo0J-independent orientation, the polar localization region (PLR), might lie some distance away from, and to the left of, oriC.
Use of directed chromosome inversions to define the PLR
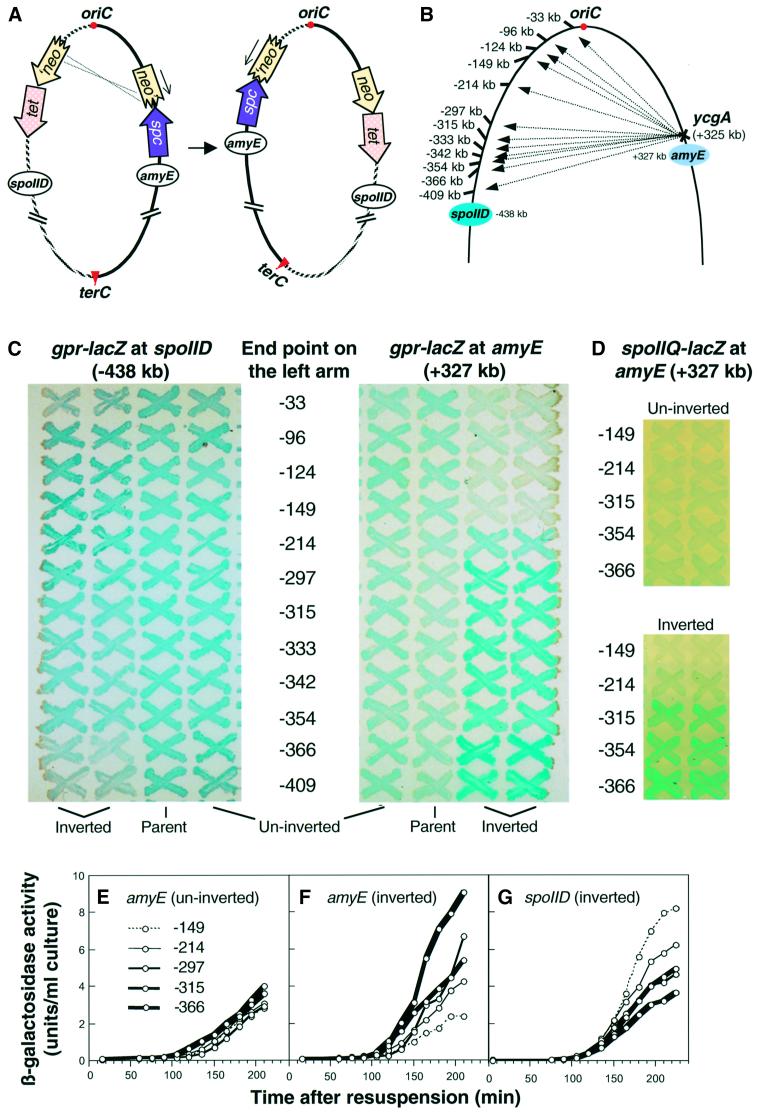
Extensive attempts to isolate regions of DNA of up to 10 kbp or so that could direct reorientation of the chromosome failed (results not shown), leading us to believe that no single short sequence or cluster of sequences was sufficient for this function. If an extended chromosomal region was responsible for chromosome migration to the cell pole during sporulation, it should be possible to map its position by making defined rearrangements in chromosome structure and using the reporter trapping method to examine the effects on chromosome capture by the prespore. We therefore constructed a large set of defined chromosome inversions, using methods pioneered by Itaya and colleagues (Toda et al., 1996; Figure 2A). To avoid the contribution of the Soj–Spo0J system to chromosome orientation, a spoIIIE36 soj–spo0J mutant was used for the analysis. We took advantage of the relatively poor trapping of the σF-dependent reporter gene at the amyE locus (+327) in the absence of soj–spo0J (Figure 1B). A common site for inversion was constructed just to the oriC-proximal side of amyE (at ycgA; +325) (Figure 2B). After generation of an inversion, the σF-dependent reporter at amyE would be placed adjacent to sequences near oriC on the other arm of the chromosome. All of the exchanges would be reciprocal, so no information would be lost from the chromosome. Also, the orientation of genes relative to DNA replication forks emanating from oriC would be preserved. Finally, the inversion points were carefully chosen to be positioned in intergenic regions, or in known non-essential genes. Having made the inversion, we could determine what effect this had on reporter trapping efficiency. If the inversion brought the cis-acting sequences required for chromosome orientation closer to the reporter, it should enhance the efficiency of trapping in the prespore and β-galactosidase synthesis by the population of cells should increase.
Fig. 2. Effect of defined chromosome inversions on the segment of the chromosome localized in the prespore. (A) Schematic of the method used to construct defined chromosome inversions. The left and right arms of the circular chromosome are represented in hatched and solid lines, respectively. These are bounded at oriC (the origin of bi-directional chromosome replication) and terC (the terminus of replication). Various inserted resistance genes are indicated as follows: spc, spectinomycin; tet, tetracycline; neo, neomycin. ‘neo, neo’ and ‘neo’ represent non-functional derivatives of the neo gene truncated at the 5′, 3′, or 5′ and 3′ ends, respectively. Homologous recombination between the truncated neo genes in the left and right chromosome arms results in a chromosome inversion in which the upper part of the chromosome, containing oriC, is switched around relative to the lower part. Note that this does not change the direction of DNA replication through the chromosome arms. (B) Locations of the inversion end points (numbered) and the σF-dependent gpr–lacZ reporters (blue ovals). (C) β-galactosidase accumulation in different inversion strains. Two independently isolated strains with inversions from ycgA (+325) to various locations on the left arm of the chromosome (‘Inverted’) were patched onto agar plates containing X-Gal, together with the un-inverted strains (‘Un-inverted’) containing the inversion elements and the parent strain (‘Parent’) 1265 [spoIIIE36 Δ(yyaA–yyaC)::erm amyE::gpr–lacZ] or 1266 [spoIIIE36 Δ(yyaA–yyaC)::erm spoIID::gpr–lacZ], and grown at 37°C for 3 days. The left panel shows the strains with the σF-dependent gpr–lacZ fusion at the spoIID locus, and the right panel shows strains with the fusion at amyE. The locations of the inversion end points on the left chromosome arm are shown in the middle. (D) β-galactosidase accumulation in a subset of inversion strains bearing a spoIIQ–lacZ fusion at the amyE locus. Inversion end points labelled as for (C). (E–G) Quantitative analysis of reporter expression during sporulation in liquid culture. The strains containing gpr–lacZ at amyE [(E), before inversion; (F), after inversion] or at spoIID [(G), after inversion] were induced to sporulate at time zero, and samples were taken at intervals for assay of β-galactosidase activity. A single experiment representative of several repeats is shown.
Inversions were generated by selection for kanamycin resistance (neo; Figure 2A) and the structure of each insertion was verified by long-range PCR. Each inversion was also checked in a wild-type background (i.e. soj+ spo0J+ spoIIIE+) to ensure that there were no deleterious effects on growth or sporulation. Only one inversion, the most asymmetric of the set with respect to oriC (to –30), had an overt phenotypic effect; the growth rate and sporulation frequency were both mildly reduced (data not shown). It is possible that this effect is due to the extreme asymmetry imposed on the chromosome by this inversion.
The effects of different inversions were most readily compared by examining the strains patched on agar plates containing the β-galactosidase indicator, X-Gal (Figure 2C). As expected, uniform activities were obtained with each of the strains in the uninverted state, and the activities were similar to those of the parent strains without the inversion generating elements. The inverted strains, however, showed a wide range of activities. Remarkably, the results suggested that the PLR was not localized at, or even near, oriC. Inversions that placed the amyE reporter much closer to oriC than it normally is, at –96, –124 or –149, gave levels of β-galactosidase, if anything, less than at their normal, more distant position (+327). Thus, the sequences required for delivery of the chromosome to the cell pole do not include the oriC region but must lie at least 150 kbp away from oriC down the left arm of the chromosome. With inversion end points located further from oriC, an increase in the expression of the reporter at amyE began to be seen, indicating that in these cells the amyE locus had become associated with sequences that promote more efficient trapping in the prespore. Step-wise increases in expression were evident as the left inversion end point reached –297 and then –366. (The last of these increases but not earlier ones, was at least partly due to the proximity of the regulatory gene, rsfA at –355; see above.)
As a control, a complete set of inversion strains were constructed with the same gpr–lacZ reporter in the left arm of the chromosome, at spoIID (–438). Expression at this site was generally stronger, as expected. Overall, the results with these strains were essentially the reciprocal of those at amyE, excluding the possibility that the gradient of expression patterns obtained were due to non-specific effects of the inversions on growth or sporulation.
Quantitative measurements of β-galactosidase activity in cultures induced to sporulate in liquid medium were entirely consistent with the above results (Figure 2E–G). Figure 2E shows accumulation of β-galactosidase during sporulation in a set of uninverted strains. Although there were small differences in the time of onset of β-galactosidase accumulation, the overall levels of expression were uniform in each of the strains. In contrast, with the inverted strains, expression levels varied considerably from strain to strain (Figure 2F and G). There was a clear reciprocal relationship between the levels of expression at amyE versus spoIID, and there was a good correlation with the results obtained with the plate cultures. Thus, the –149 inversion gave the lowest level of expression for fusions at amyE and the highest levels for those at spoIID, and the inversion at –366 had a completely opposite effect. Inversions located between these extremes gave results that reflected their positions on the chromosome. These experiments also confirmed that the intensity of the blue colour on plates was a useful qualitative indicator of enzyme activity (and thus trapping efficiency; Wu and Errington, 1998; also see below).
To confirm that similar effects would be seen with other promoters, we reconstructed some of the key inversion strains with the highly specific σF-dependent promoter spoIIQ fused to lacZ. The results obtained were almost indistinguishable from those of the equivalent gpr–lacZ-containing strains (Figure 2D).
Functioning of the PLR at large distances from oriC
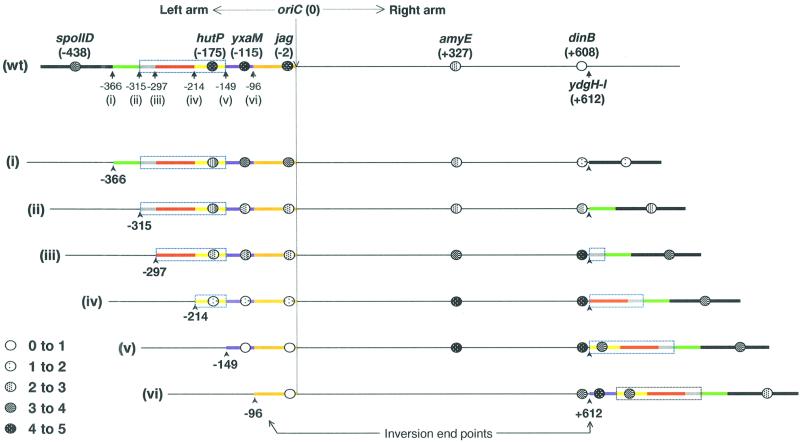
To test whether the putative PLR would function at a greater distance from oriC, a series of inversions were made with right-arm inversion end points at +612 (between the ydgH and ydgI genes), where reporter gene expression is normally virtually undetectable because of its distance from the PLR (Figure 1B). Six left-arm inversion end-points from the above experiments were used, and for each inversion, chromosome orientation was assessed by measuring reporter activity at a range of well-spaced reference points (Figure 3). In general, these inversion strains grew and sporulated less well and less reliably than the less extreme inversions to the +325 end-point. However, again, no significant difference in viability or general sporulation efficiency between the various inverted strains was evident.
Fig. 3. Summary of the effect of inversions at a distant location (+612) on chromosome trapping as measured by the accumulation of β-galactosidase from the σF-dependent gpr–lacZ reporter located at six different chromosomal positions. Strains were patched to and grown on agar plates containing X-Gal as described in the legend to Figure 2. The activities of the reporter in a series of experiments were scored and depicted as circles with different degrees of shading at the corresponding locations. The top line (labelled wt) shows the oriC region of the wild-type chromosome. The right arm of the chromosome is depicted with a thin black line. The left arm is depicted with a thick line, with different segments, demarcated by the different inversion sites used, shown in different colours. The positions of the different gpr–lacZ reporter insertions are shown as circles, with the corresponding level of β-galactosidase activity indicated by the shading (scored on a scale of 1 to 5 as indicated). Lines labelled (i)–(vi) below show the chromosomal structures of the different inversion strains tested, with the inversion end points indicated by black arrowheads. The location of the putative PLR is indicted by the boxed region (though some polar localizing activity clearly lies outside this region).
The lacZ fusion located at dinB (+608) is normally virtually excluded from the prespore, so little or no β-galactosidase activity was detected in the uninverted strains (wild type). However, inversions that brought parts of the –149 to –315 region adjacent to dinB (inversions iii–vi) allowed strong expression and thus efficient trapping of this site in the prespore. Similar results were obtained for the reporter when placed on the other side of the PLR, at spoIID. Again, this fusion was only expressed poorly if none of the region from –366 was nearby (i.e. inversion i). In contrast, an almost exactly reciprocal effect was seen when the lacZ fusion was placed on the oriC-proximal side of the PLR (jag at –2, or yxaM at –115). With these reporter positions expression was strong in the uninverted state (wild type) and with an inversion distant from oriC (at –366; inversion i), but it fell away to negligible levels as inversions removed from their proximity increasing segments of the –366 to –149 region, especially from –297 to –149. Note that jag is only 2 kbp from oriC and thus in strains with left arm inversion end-points at –96 (vi) or –149 (v), the chromosome appears to have been completely reoriented in such a way that oriC is almost always excluded from the prespore. Furthermore, in the same inversion strains spoIID was trapped efficiently despite being located almost 1 Mbp (i.e. about one-quarter of the chromosome) away from oriC. On the basis of these results, we conclude that the region comprising approximately –149 to –366 contains sequences that can direct chromosome trapping in the prespore compartment.
Inversions that bisected the PLR allowed us to determine whether subdivisions could have independent activity, as is sometimes the case with diffuse eukaryotic centromeres (Dobie et al., 1999). With the inversions at –297 (iii), for example, it did appear that partial activity was retained by both PLR segments. Here the fusion at hutP at one extreme retained substantial trapping, as did the most distant one, >900 kbp away, at spoIID. Thus, when the PLR is divided into two parts, both parts appear to retain substantial activity and can influence the orientation of the chromosome.
In further experiments we constructed double inversion strains to specifically translocate full-length or truncated segments of the PLR from the left chromosome arm to the right arm. The resultant strains again grew and sporulated normally, and again the results were consistent with the active region lying between –149 and –315 (data not shown).
It was important to exclude the possibility that some of the effects we were seeing were due to repositioning of an as yet undetected σF regulatory gene (in addition to rsfA) lying in the oriC region. For this, a σF-dependent spoIIQ– gfp fusion was introduced into various translocation strains and the proportion of prespores producing green fluorescent protein (GFP) was determined by direct counting from fluorescence microscopic images of sporulating cultures. Control experiments showed that even weak expression of the spoIIQ–gfp fusion was readily detected by this method. Again the results were consistent with the sequences responsible for polar localization lying mainly in the –149 to –315 region (data not shown).
Visualizing chromosome orientation with LacI–GFP
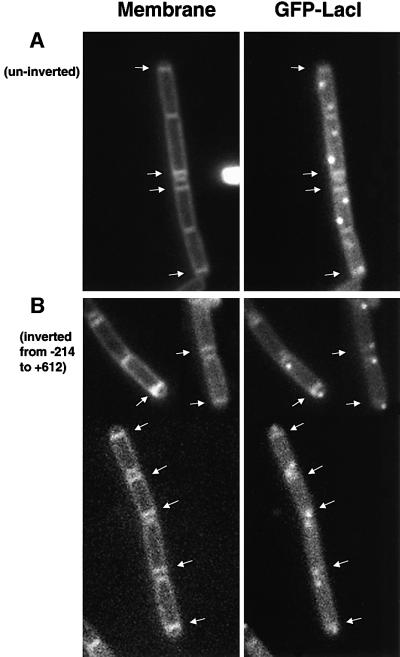
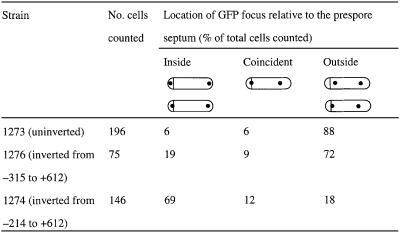
As a final test of the effects of the PLR on chromosome orientation we used the lacO/GFP–LacI system of Webb et al. (1997) to examine chromosome position directly. The strains containing the lacO–lacI system seemed to sporulate less efficiently (Webb et al., 1997), but we were nevertheless able to detect many cells with an asymmetric septum and two clear GFP spots (representing the tags on the prespore and mother cell chromosomes; Figure 4). The lacO array was placed at the dinB locus at a position (+608) in which trapping in the prespore of a soj–spo0J spoIIIE mutant was thought to be rare (see above). Accordingly, for this strain the majority (88%) of the sporulating cells had GFP spots located outside the pre-spore compartment (strain 1273, Table I; Figure 4A). However, when the fragment from –214 to +612 was inverted (strain 1274), bringing dinB right next to the putative cis-acting region (–214 to –315), at least 69% of the GFP spots were located inside the prespore compartment (Figure 4B), consistent with the reporter trapping results (see Figure 3). Note also the striking difference in the spacing of the GFP spots between the two strains: in the inverted strain, most GFP spots were closely apposed to the cell poles, whereas in the uninverted strain, the spots tended to be much closer together and away from the cell poles, at approximately one-third and two-thirds positions in the cell.
Fig. 4. Fluorescence microscopy showing sporulating cells with a lacO array at dinB (+608) before [(A), strain 1273] and after [(B), strain 1274] chromosome inversion from –214 to +612 kbp. The strains were induced to sporulate, stained with the membrane dye FM5-95 and viewed under fluorescence microscope as described in Materials and methods. Left panels show the cell outline and the asymmetric septa as revealed by the membrane dye. Right panels are GFP signals showing the cellular location of the lacO. It also shows faint staining of the membrane because the signals from the dye crossed into the GFP channel. Arrows point to the positions of asymmetric septa.
Table I. Effect of chromosome inversion on localization of the lacO cassette at +608 kbp in sporulating cells.
On the basis of the experiments shown in Figures 2 and 3, if the inversion end-point was moved a little further from oriC, and hence to the other side of the PLR, polar localization of the GFP tag should be reduced. As shown in Table I, with this inversion (–315 to +612, strain 1276), the frequency of trapping of the GFP tag was greatly reduced, to a level only a little higher than trapping in the uninverted state.
Discussion
A prespore chromosome segregation system independent of Soj–Spo0J
We used the reporter trapping method to test the role of the Soj–Spo0J system in orientation of the chromosome at the onset of sporulation. As expected based on previous work (Sharpe and Errington, 1996), there was an increase in trapping frequency for markers normally excluded from the prespore (e.g. at –826; gerA; see Figure 1B). There was also a substantial fall in trapping efficiency to the right of oriC, over a region from approximately +100 to +500. However, the equivalent region in the left arm of the chromosome was trapped just as efficiently as in the Soj+ Spo0J+ background. In the soj–spo0J background, a marked peak in trapping efficiency became evident, centred at about –200 kbp. Two important conclusions follow from these observations. First, the peak in trapping efficiency revealed the likely location of cis-acting site for prespore chromosome segregation, the PLR (see below). Secondly, migration of the PLR to the cell pole is not dependent on the soj–spo0J system. Nevertheless, the soj–spo0J system clearly does have a role in defining the precise configuration of the chromosome, because inversions that moved the intact PLR and therefore substantially affected chromosome orientation in a soj–spo0J deletion had much less effect when examined in the presence of a functional Soj–Spo0J system (data not shown).
The new data agree quantitatively with the previous results of Sharpe and Errington (1996), although in the previous work positions to the left of oriC were not examined, so the efficient trapping of this part of the chromosome was not detected. In contrast, Fransden et al. (1999) concluded that the precision of chromosome orientation was retained in a soj–spo0J background, and our new data suggest that this is because their test location (–421) lies close to the PLR.
Spo0J protein forms large protein–DNA complexes that are associated with the oriC region of the chromosome throughout growth, sporulation and germination (Glaser et al., 1997; Lewis and Errington, 1997; Lin et al., 1997). Chromatin immunoprecipitation experiments have revealed a set of preferred DNA binding sites for Spo0J protein (Lin and Grossman, 1998). All were located in the oriC-proximal part of the chromosome, with strong binding sites at +48, –10, –49, –63 and –68 (Figure 5A), and weaker sites at +185, +471 and –305. We recently showed that in soj mutants, Spo0J complexes associated with these various sites around the oriC region fail to condense into the bright discrete foci normally observed in vegetative and sporulating cells (Marston and Errington, 1999; Autret et al., 2001). Thus, Soj presumably acts to bring together Spo0J molecules bound at dispersed sites on the chromosome, so as to condense or otherwise impose order on the oriC region of the chromosome. Interestingly, most of the strong binding sites for Spo0J lie in the left arm of the chromosome between oriC and the PLR (Figure 5A). On the basis of these observations the Soj–Spo0J system could condense the oriC region so that it moves as a discrete unit to the cell pole, directed by the PLR (Figure 5B). Alternatively, or additionally, as indicated by the arrows in Figure 5B, Spo0J may be attracted to the cell pole independently of the PLR. In either case, in the absence of the soj–spo0J system the left arm of the chromosome is still efficiently positioned close to the cell pole via the PLR, but sequences to the right of the PLR are no longer as efficiently attracted to the cell pole (Figure 5C), giving the reduced reporter trapping efficiencies seen in Figure 1B.
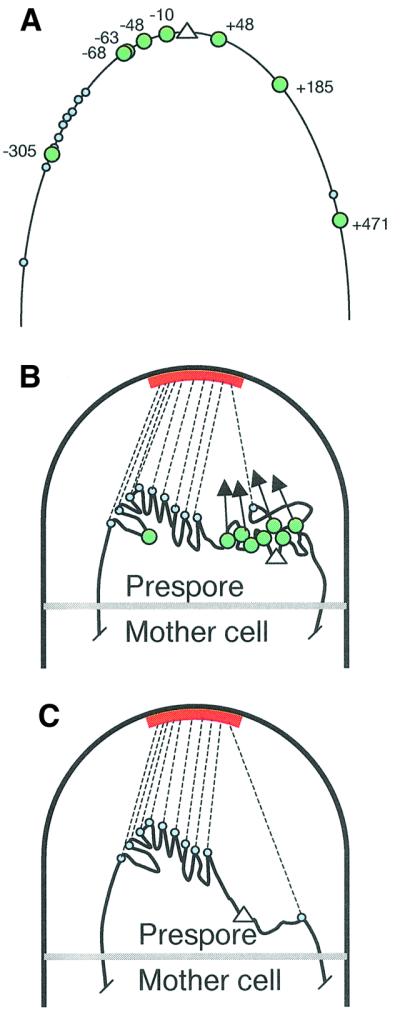
Fig. 5. A model for prespore chromosome segregation and possible roles for the Soj–Spo0J system. (A) Layout of known Spo0J-binding sites (labelled large green circles; as defined by Lin and Grossman, 1998) and putative PLR sites (small blue circles) in the oriC (white triangle) region of the chromosome. The segment of DNA depicted (thin black line) shows approximately the segment of DNA that is normally trapped in the prespore compartment of spoIIIE mutant cells (soj+ spo0J+) during sporulation (approximately +600 to –700; see Figure 1B). On the basis of our results, the hypothetical PLR sites mainly lie in the –149 to –315 region. (B) Association of proteins bound to sites in the PLR with a recognition site or anchor (red bar) in the vicinity of the cell pole during sporulation results in dragging of the PLR towards the cell pole (dotted black lines). oriC itself is efficiently trapped in the prespore because Spo0J protein forms a complex that condenses the oriC region in close juxtaposition to the PLR. Alternatively or additionally, Spo0J may be directly attracted towards the cell pole, as indicated by the arrows. (C) Effect of disruption of the soj–spo0J system on trapping of DNA in the prespore compartment.
Localization of the PLR
The existence of the PLR was revealed by the relatively efficient orientation of the chromosome in the absence of the Soj–Spo0J system. To locate the cis-acting sequences or region responsible for PLR activity we took a soj–spo0J mutant and constructed a series of defined chromosomal inversions. The effects of each inversion on trapping of σF reporter genes was measured with reporters at different locations. The clearest results were obtained when one inversion point was placed a large distance from oriC (+612; Figures 3 and 4), but similar conclusions could be drawn from both the inversions anchored at ycgA (+325; Figure 2) and from translocation experiments (not shown). These results unambiguously demonstrated that oriC itself has no role in the PLR function. Thus, with certain inversions (e.g. –96 to +608; Figure 3) a reporter gene located next to oriC (jag; +2) was not detectably expressed. Efficient trapping of the reporter at this position only occurred when the left arm inversion end point was such that ∼315 kbp of oriC-proximal DNA was retained in its normal position. [The further increase in activity as the inversion end point was moved out from –315 to –366 was predominantly or exclusively due to the regulatory gene, rsfA, at –355 (Wu and Errington, 2000), and was not seen when cells expressing a gfp fusion were counted directly (data not shown).] This provides a limit for the oriC-distal border of the PLR, at about –315. A limit for the oriC-proximal border could be placed at –149 on the basis of inversions, giving minimal expression of the reporter near oriC. The intervening region of ∼150 kbp contained sequences both necessary and sufficient for orientation of the chromosome.
The most striking demonstration of the effect of the PLR on chromosome orientation came from the experiment with the lacO array placed at +608 and tagged with GFP–LacI (Figure 4; Table I). In this experiment, the GFP spots were clearly localized well away from the cell poles in the uninverted strain or in an inversion that maintained the putative PLR at a large distance from the lacO array. However, with an inversion that brought the lacO array close to the PLR, the spots were dramatically repositioned close to the cell poles. This experiment also suggested that the putative attachment site for the PLR is close to or at the cell pole.
The nature of the PLR
When the 150 kbp region was bisected, such as when the right arm inversion end point lay at –214 or –297, significant levels of gene expression were detected with reporters lying at extremely distant positions (e.g. 928 kbp apart for hutP and spoIID in inversion iii; Figure 3). This was not consistent with the PLR comprising a small discrete sequence. Two other kinds of result also suggested a relatively diffuse structure. First, the inversions that brought oriC very close to the reporter at +327 (Figure 2B), which gave a readily visible reduction in lacZ expression, even though they did not change the total distance from +327 to the 150 kbp region. A simple explanation for this effect would be that a site or sites with polar localization activity lies just oriC-proximal to the inversion end point at +325 (Figure 5A), so that the inversions take the sites further away from the reporter. Secondly, expression of the reporter at dinB (+608) increased, albeit slightly, when placed next to sequences distal to –366 (inversion i in Figure 3). This suggests that there is weak chromosome orientating activity beyond –366. On the basis of these observations we suppose that the PLR comprises a diffuse zone with many relatively weak cis-acting sites, mainly, but not exclusively, located between –149 and –315, and that the sites act with some co-operativity (Figure 5).
Our favoured explanation for the intermediate expression profiles obtained with strains in which the PLR was bisected is that in these strains there is cell-to-cell variation in the specific region that becomes localized at the pole. In other words, there could be competition between the two partial PLR segments for binding to a putative receptor at the cell pole. Alternatively, there may simply be some general loss of specificity in chromosome orientation.
In any event, the PLR we have discovered resembles the diffuse centromeres of eukaryotic cells (Karpen and Allshire, 1997) in a number of respects. It occupies a large zone covering many tens or hundreds of kilobase-pairs. It appears to be internally redundant and when bisected, the separated segments retain at least partial activity. The B.subtilis system differs from higher eukaryotic centromeres in containing many genes (though active genes are beginning to be found in the centromeres of some higher organisms, notably Arabidopsis; Copenhaver et al., 1999) and in the relative absence of repetitive DNA.
Chromosome segregation in non-sporulating cells
The results described herein may explain why it has not been possible to directly isolate cis-acting sequences required for chromosome stability in bacteria so far. If we assume that chromosome segregation sequences in vegetative cells of B.subtilis and other bacteria have some functional similarity to the system defined here, they may contain considerable functional redundancy and be spread over relatively large segments of DNA. Bacterial cells are extremely small and their DNA is relatively diffuse, so it is difficult to characterize chromosome position and orientation directly. We have overcome this problem by using the asymmetric septum of sporulating cells (of spoIIIE mutants) as a fixed marker in the cell against which chromosome position can be precisely measured (Wu and Errington, 1994, 1998). We have tried several experimental approaches to test whether the region we have identified is also involved in vegetative chromosome segregation. So far these have been inconclusive because of the technical difficulties of measuring chromosome orientation mentioned above, and because the nature of the large region with many essential genes makes it difficult to manipulate (data not shown). However, it seems likely that the system we have uncovered is at least partly a specialized adaptation for the asymmetrical cell division that occurs during sporulation, in which the chromosome needs to move an extreme distance to the cell pole. Nevertheless, it is possible that it is used in a more general capacity by related bacteria. We recently discovered that the divIVA gene of B.subtilis has a role in chromosome segregation during sporulation (Thomaides et al., 2001). DivIVA protein is targeted to and retained at the cell poles (Edwards and Errington, 1997), and in its absence, chromosome trapping in the small prespore compartment frequently fails, producing prespores devoid of DNA (Thomaides et al., 2001). Since DivIVA is localized at the extreme cell pole, and this is where the sporulation PLR appears to localize (when tagged with GFP–LacI), DivIVA is a good candidate for a factor involved directly or indirectly in PLR binding (red bars in Figure 5). This latter function for DivIVA is completely separable from its function in division-site selection (Thomaides et al., 2001). Furthermore, clear DivIVA homologues are present in Gram-positive cocci, such as Staphylococcus and Streptococcus, which do not have a MinCD system, so it seems possible that this important group of pathogenic organisms may use a sporulation-like chromosome segregation system in the course of their normal growth. A preliminary report of the phenotype of a divIVA mutant of Streptococcus appears to support this notion (Massidda et al., 1998).
Materials and methods
Bacterial strains and plasmids
These are listed in Table II. Details of strain constructions are given in the Supplementary data available at The EMBO Journal Online.
Table II. Bacillus subtilis strains and plasmids.
| Strain/plasmid |
Relevant genotypea |
Construction, source or referenceb |
|---|---|---|
| Bacillus subtilis | ||
| 168 | trpC2 | Laboratory stock |
| SG38 | trpC2 amyE | Errington and Mandelstam (1986) |
| 36.3 | trpC2 spoIIIE36 | Piggot (1973) |
| 1249 | trpC2 amyE::gpr-lacZ cat | Wu and Errington (2000) |
| 1221 | trpC2 amyE Δ(yyaA-yyaC)::kan | See Supplementary data |
| 1222 | trpC2 amyE Δ(yyaA-yyaC)::erm | See Supplementary data |
| 1223 | trpC2 amyE Δ(soj-yyaC)::spc | See Supplementary data |
| 1258 | trpC2 ycgA::(neo′-spc) | See Supplementary data |
| 1259 | trpC2 ydgH-ydgI::(neo′-spc) | See Supplementary data |
| 1260 | trpC2 yexD-yxeC::(′neo-tet) | See Supplementary data |
| 1261 | trpC2 spoIIIE36 ycgA::(neo′-spc) yexD-yxeC::(′neo-tet) | See Supplementary data |
| 1263 | trpC2 spoIIIE36 | 1222 (chr)→36.3 |
| Δ(yyaA-yyaC)::erm | ||
| 1264 | trpC2 spoIIIE36 | 1221 (chr)→36.3 |
| Δ(yyaA-yyaC)::kan | ||
| 1265 | trpC2 spoIIIE36 | 1249 (chr)→1263 |
| Δ(yyaA-yyaC)::erm | ||
| amyE::gpr-lacZ cat | ||
| 1266 | trpC2 spoIIIE36 | 36::pSG862 (chr)→1263 |
| Δ(yyaA-yyaC)::erm | ||
| spoIID::gpr-lacZ cat | ||
| 1269 | trpC2 spoIIIE36 | See Supplementary data |
| Δ(yyaA-yyaC)::erm dinB::pSG4920 (cat lacOx256 PspoVG-gfp-lacI) Δ(yxeD-yxjF)::(neo-tet) ycgA::(yxeD-yxjF ‘neo’-spc) | ||
| 1270 | trpC2 spoIIIE36 | See Supplementary data |
| Δ(yyaA-yyaC)::erm dinB::pSG4920 (cat lacOx256 PspoVG-gfp-lacI) Δ(yxeD-ywdA)::(neo-tet) ycgA::(yxeD-ywdA ‘neo’-spc) | ||
| 1273 | trpC2 spoIIIE36 | See Supplementary data |
| Δ(yyaA-yyaC)::erm dinB::pSG4920 (cat lacOx256 PspoVG-gfp-lacI) ydgH-I:: (neo′-spc) yxjG-F::(′neo-tet) | ||
| 1274 | trpC2 spoIIIE36 | See Supplementary data |
| Δ(yyaA-yyaC)::erm dinB::pSG4920 (cat lacOx256 PspoVG-gfp-lacI) ydgH-I:: (neo′-spc) yxjG-F::(′neo-tet) Inv (+612/-214) (NeoR) | ||
| 1275 | trpC2 spoIIIE36 | See Supplementary data |
| Δ(yyaA-yyaC)::erm dinB::pSG4920 (cat lacOx256 PspoVG-gfp-lacI) ydgH-I:: (neo′-spc) thiD-ywdA::(′neo-tet) | ||
| 1276 | trpC2 spoIIIE36 | See Supplementary data |
| |
Δ(yyaA-yyaC)::erm dinB::pSG4920 (cat lacOx256 PspoVG-gfp-lacI) ydgH-I:: (neo′-spc) thiD-ywdA::(′neo-tet) Inv (+612/–315) (NeoR) |
|
| Plasmids |
|
|
| pAT12 | bla cat lacOx256 | Webb et al. (1997) |
| pBEST502 | bla neo | Itaya et al. (1989) |
| pIC156 | bla spc | Steinmetz and Richter (1994) |
| pSG122 | bla aphA-3 | Daniel and Errington (1993) |
| pSG250 | bla erm | Errington et al. (1992) |
| pSG840 | bla erm | pSG250 (XbaI + SstI), religated |
| pSG862 | bla cat gpr-lacZ ′spoIID | Wu and Errington (1998) |
| pSG1189 | bla amyE′ spc Pxyl-gfp-lacI ′amyE | P.Lewis, unpublished |
| pSG4906 | bla ′neo tet | Wu and Errington (2000) |
| pSG4913 | bla spc | pIC156 (XbaI) religated |
| pSG4914 | bla neo′ spc | spc from pSG4913 (BamHI + SstI) into pBEST501 (BglII + SstI) |
| pSG4918 | bla cat lacOx256 ‘dinB ydgF’ | ‘dinB ydgF’(PCR, HindIII) into pAT12 (HindIII) |
| pSG4919 | bla cat lacOx256 ‘dinB ydgF’ Pxyl-gfp-lacI | Pxyl-gfp-lacI (PCR, SstI + XhoI) into pSG4918 (SstI + XhoI) |
| pSG4920 | bla cat lacOx256 ‘dinB ydgF’ PspoVG-gfp-lacI | PspoVG (PCR, SstI + KpnI) into pSG4919 (SstI + KpnI) |
a′X or X′, the 5′-end or the 3′-end of the gene X has been truncated; aphA3, kanamycin resistance gene; cat, chloramphenicol resistance gene; tet, tetracycline resistance gene; neo, neomycin resistance gene; spc, spectinomycin resistance; erm, erythromycin resistance; bla, ampicillin resistance. Inv(+612/–214), the strain contains an inversion with end points at +612 and –214.
bFor strains constructed by transformation, the source of the DNA used in the transformation is given first and the recipient strain is indicated after the arrow. Restriction enzymes used for digesting DNA are given in parentheses.
General methods
Bacillus subtilis cells were made competent for transformation with DNA either by the method of Kunst et al. (1995), or by the method of Anagnostopoulos and Spizizen (1961) as modified by Jenkinson (1983). DNA manipulations and E.coli transformations were carried out using standard methods (Sambrook et al., 1989). Solid medium used for growing B.subtilis was nutrient agar (Oxoid) and liquid medium was PAB (Oxoid Antibiotic Medium no. 3). Chloramphenicol (5 µg/ml), kanamycin (5 µg/ml), tetracycline (12 µg/ml), erythromycin (1 µg/ml) and lincomycin (25 µg/ml), or 0.012% X-gal, were added as required. Media used for growing E.coli were 2xTY (Sambrook et al., 1989) and nutrient agar supplemented with ampicillin (100 µg/ml) as required.
Induction of sporulation and assay for β-galactosidase activity
Bacillus subtilis cells grown in hydrolysed casein growth media at 37°C were induced to sporulate by the resuspension method of Sterlini and Mandelstam (1969), as modified by Partridge and Errington (1993). Times (in minutes) after resuspension of cells in the starvation medium were denoted t0, t30, etc.
β-galactosidase activity was measured by the method of Errington and Mandelstam (1986). One unit of β-galactosidase catalyses the production of 1 nmol 4-methylumbelliferone per min under the standard conditions.
PCR primers
Owing to the length limitation, information on the primers will not be listed here but will be available on request.
Fluorescence microscopy
Cells containing GFP–LacI constructs were grown at 30°C after induction of sporulation, and membrane dye FM5-95 (Molecular Probes) was added to the culture at 110 min at a final concentration of 22 µg/ml. The concentration of the dye was limited to minimize the FM5-95 signal crossing over into the GFP channel. Cells were then viewed on agarose slides and images were obtained as described previously (Marston and Errington, 1999).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank P.Lewis for plasmid pSG1189, and Meriem El Karoui and Sabine Autret for helpful comments on the manuscript. This work was funded by grants from the Biotechnology and Biological Sciences Research Council. J.E. acknowledges receipt of a Senior Research Fellowship of the BBSRC.
References
- Anagnostopoulos C. and Spizizen,J. (1961) Requirements for transformation in Bacillus subtilis. J. Bacteriol., 81, 741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autret S., Nair,R. and Errington,J. (2001) Genetic analysis of the chromosome segregation protein Spo0J of Bacillus subtilis: evidence for separate domains involved in DNA binding and interactions with Soj protein. Mol. Microbiol., 41, 743–755. [DOI] [PubMed] [Google Scholar]
- Bath J., Wu,L.J., Errington,J. and Wang,J.C. (2000) Role of Bacillus subtilis SpoIIIE in DNA transport across the mother cell–prespore division septum. Science, 290, 995–997. [DOI] [PubMed] [Google Scholar]
- Bylund J.E., Haines,M.A., Piggot,P.J. and Higgins,M.L. (1993) Axial filament formation in Bacillus subtilis: induction of nucleoids of increasing length after addition of chloramphenicol to exponential-phase cultures approaching stationary phase. J. Bacteriol., 175, 1886–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervin M.A., Spiegelman,G.B., Raether,B., Ohlsen,K., Perego,M. and Hoch,J.A. (1998) A negative regulator linking chromosome segregation to developmental transcription in Bacillus subtilis. Mol. Microbiol., 29, 85–95. [DOI] [PubMed] [Google Scholar]
- Copenhaver G.P. et al. (1999) Genetic definition and sequence analysis of Arabidopsis centromeres. Science, 286, 2468–2474. [DOI] [PubMed] [Google Scholar]
- Daniel R.A. and Errington,J. (1993) Cloning, DNA sequence, functional analysis and transcriptional regulation of the genes encoding dipicolinic acid synthesis required for sporulation in Bacillus subtilis. J. Mol. Biol., 232, 468–483. [DOI] [PubMed] [Google Scholar]
- Dobie K.W., Hari,K.L., Maggert,K.A. and Karpen,G.H. (1999) Centromere proteins and chromosome inheritance: a complex affair. Curr. Opin. Genet. Dev., 9, 206–217. [DOI] [PubMed] [Google Scholar]
- Edwards D.H. and Errington,J. (1997) The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division. Mol. Microbiol., 24, 905–915. [DOI] [PubMed] [Google Scholar]
- Errington J. and Mandelstam,J. (1986) Use of a lacZ gene fusion to determine the dependence pattern of sporulation operon spoIIA in spo mutants of Bacillus subtilis. J. Gen. Microbiol., 132, 2967–2976. [DOI] [PubMed] [Google Scholar]
- Errington J., Appleby,L., Daniel,R.A., Goodfellow,H., Partridge,S.R. and Yudkin,M.D. (1992) Structure and function of the spoIIIJ gene of Bacillus subtilis: a vegetatively expressed gene that is essential for σG activity at an intermediate stage of sporulation. J. Gen. Microbiol., 138, 2609–2618. [DOI] [PubMed] [Google Scholar]
- Fransden N., Barák,I., Karmazyn-Campelli,C. and Stragier,P. (1999) Transient gene asymmetry during sporulation and establishment of cell specificity in Bacillus subtilis. Genes Dev., 13, 394–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser P., Sharpe,M.E., Raether,B., Perego,M., Ohlsen,K. and Errington,J. (1997) Dynamic, mitotic-like behaviour of a bacterial protein required for accurate chromosome partitioning. Genes Dev., 11, 1160–1168. [DOI] [PubMed] [Google Scholar]
- Gordon G.S. and Wright,A. (2000) DNA segregation in bacteria. Annu. Rev. Microbiol., 54, 681–708. [DOI] [PubMed] [Google Scholar]
- Gordon G.S., Sitnikov,D., Webb,C.D., Teleman,A., Straight,A., Losick,R., Murray,A.W. and Wright,A. (1997) Chromosome and low copy plasmid segregation in E. coli: visual evidence for distinct mechanisms. Cell, 90, 1113–1121. [DOI] [PubMed] [Google Scholar]
- Hiraga S. (2000) Dynamic localization of bacterial and plasmid chromosomes. Annu. Rev. Genet., 34, 21–59. [DOI] [PubMed] [Google Scholar]
- Ireton K., Gunther,N.W.I. and Grossman,A.D. (1994) spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J. Bacteriol., 176, 5320–5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itaya M., Kondo,K. and Tanaka,T. (1989) A neomycin resistance gene cassette selectable in a single copy state in the Bacillus subtilis chromosome. Nucleic Acids Res., 17, 4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F., Brenner,S. and Cuzin,F. (1963) On the regulation of DNA replication in bacteria. Cold Spring Harbor Symp. Quant. Biol., 28, 329–348. [Google Scholar]
- Jenkinson H.F. (1983) Altered arrangement of proteins in the spore coat of a germination mutant of Bacillus subtilis. J. Gen. Microbiol., 129, 1945–1958. [DOI] [PubMed] [Google Scholar]
- Karpen G.H. and Allshire,R.C. (1997) The case for epigenetic effects on centromere identity and function. Trends Genet., 13, 489–496. [DOI] [PubMed] [Google Scholar]
- Kunst F. and Rapoport,G. (1995) Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J. Bacteriol., 177, 2403–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis P.J. and Errington,J. (1997) Direct evidence for active segregation of oriC regions of the Bacillus subtilis chromosome and co-localization with the Spo0J partitioning protein. Mol. Microbiol., 25, 945–954. [DOI] [PubMed] [Google Scholar]
- Lin D.C.-H. and Grossman,A.D. (1998) Identification and characterization of a bacterial chromosome partitioning site. Cell, 92, 675–685. [DOI] [PubMed] [Google Scholar]
- Lin D.C.-H., Levin,P.A. and Grossman,A.D. (1997) Bipolar localization of a chromosome partition protein in Bacillus subtilis. Proc. Natl Acad. Sci. USA, 94, 4721–4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston A.L. and Errington,J. (1999) Dynamic movement of the ParA-like Soj protein of B. subtilis and its dual role in nucleoid organization and developmental regulation. Mol. Cell, 4, 673–682. [DOI] [PubMed] [Google Scholar]
- Massidda O., Anderluzzi,D., Friedli,L. and Feger,G. (1998) Unconventional organization of the division and cell wall gene cluster of Streptococcus pneumoniae. Microbiology, 144, 3069–3078. [DOI] [PubMed] [Google Scholar]
- Mohl D.A. and Gober,J.W. (1997) Cell cycle-dependent polar localization of chromosome partitioning proteins in Caulobacter crescentus. Cell, 88, 675–684. [DOI] [PubMed] [Google Scholar]
- Niki H. and Hiraga,S. (1998) Polar localization of the replication origin and terminus in Escherichia coli nucleoids during chromosome partitioning. Genes Dev., 12, 1036–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge S.R. and Errington,J. (1993) The importance of morphological events and intercellular interactions in the regulation of prespore-specific gene expression during sporulation in Bacillus subtilis. Mol. Microbiol., 8, 945–955. [DOI] [PubMed] [Google Scholar]
- Pidoux A.L. and Allshire,R.C. (2000) Centromeres: getting a grip of chromosomes. Curr. Opin. Cell Biol., 12, 308–319. [DOI] [PubMed] [Google Scholar]
- Piggot P.J. (1973) Mapping of asporogenous mutations of Bacillus subtilis: a minimum estimate of the number of sporulation operons. J. Bacteriol., 114, 1241–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quisel J.D. and Grossman,A.D. (2000) Control of sporulation gene expression in Bacillus subtilis by the chromosome partitioning proteins Soj (ParA) and Spo0J (ParB). J. Bacteriol., 182, 3446–3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quisel J.D., Lin,D.C.-H. and Grossman,A.D. (1999) Control of development by altered localization of a transcription factor in B. subtilis. Mol. Cell, 4, 665–672. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sharpe M.E. and Errington,J. (1996) The Bacillus subtilis soj–spo0J locus is required for a centromere-like function involved in prespore chromosome partitioning. Mol. Microbiol., 21, 501–509. [DOI] [PubMed] [Google Scholar]
- Sharpe M.E. and Errington,J. (1998) A fixed distance for separation of newly replicated copies of oriC in Bacillus subtilis: implications for co-ordination of chromosome segregation and cell division. Mol. Microbiol., 28, 981–990. [DOI] [PubMed] [Google Scholar]
- Sharpe M.E. and Errington,J. (1999) Upheaval in the bacterial nucleoid: an active chromosome segregation mechanism. Trends Genet., 15, 70–74. [DOI] [PubMed] [Google Scholar]
- Steinmetz M. and Richter,R. (1994) Plasmids designed to alter the antibiotic resistance expressed by insertion mutations in Bacillus subtilis, through in vivo recombination. Gene, 142, 79–83. [DOI] [PubMed] [Google Scholar]
- Sterlini J.M. and Mandelstam,J. (1969) Committment to sporulation in Bacillus subtilis and its relationship to the development of actinomycin resistance. Biochem. J., 113, 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomaides H.B., Freeman,M., El Karoui,M. and Errington,J. (2001) Division-site-selection protein DivIVA of Bacillus subtilis has a second distinct function in chromosome segregation during sporulation. Genes Dev., 15, 1662–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Tanaka,T. and Itaya,M. (1996) A method to invert DNA segments of the Bacillus subtilis 168 genome by recombination between two homologous sequences. Biosci. Biotechnol. Biochem., 60, 773–778. [DOI] [PubMed] [Google Scholar]
- Wake R.G. and Errington,J. (1995) Chromosome partitioning in bacteria. Annu. Rev. Genet., 29, 41–67. [DOI] [PubMed] [Google Scholar]
- Webb C.D., Teleman,A., Gordon,S., Straight,A., Belmont,A., Lin, D.C.-H., Grossman,A.D., Wright,A. and Losick,R. (1997) Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell, 88, 667–674. [DOI] [PubMed] [Google Scholar]
- Webb C.D., Graumann,P.L., Kahana,J.A., Teleman,A.A., Silver,P.A. and Losick,R. (1998) Use of time-lapse microscopy to visualize rapid movement of the replication origin region of the chromosome during the cell cycle in Bacillus subtilis. Mol. Microbiol., 28, 883–892. [DOI] [PubMed] [Google Scholar]
- Wu L.J. and Errington,J. (1994) Bacillus subtilis SpoIIIE protein required for DNA segregation during asymmetric cell division. Science, 264, 572–575. [DOI] [PubMed] [Google Scholar]
- Wu L.J. and Errington,J. (1998) Use of asymmetric cell division and spoIIIE mutants to probe chromosome orientation and organization in Bacillus subtilis. Mol. Microbiol., 27, 777–786. [DOI] [PubMed] [Google Scholar]
- Wu L.J. and Errington,J. (2000) Identification and characterization of a new prespore-specific regulatory gene, rsfA, of Bacillus subtilis. J. Bacteriol., 182, 418–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L.J., Lewis,P.J., Allmansberger,R., Hauser,P.M. and Errington,J. (1995) A conjugation-like mechanism for prespore chromosome partitioning during sporulation in Bacillus subtilis. Genes Dev., 9, 1316–1326. [DOI] [PubMed] [Google Scholar]