Abstract
Recombination of gene segments at the immunoglobulin and T-cell receptor loci requires that the RAG1 and RAG2 proteins bring together DNA signal sequences (RSSs) with 12- and 23-bp spacers into a synaptic complex and cleave the DNA. A RAG1/2 multimer that can cleave both signals is shown to assemble on an isolated RSS, and the complementary RSS enters this complex as naked DNA. When RAG1/2 is allowed to bind 12 and 23 RSSs separately prior to their mixing, synaptic complex assembly and cleavage activity are greatly reduced, indicating that only a complex initially assembled on a single RSS leads to productive cleavage. RAG1/2 complexes assembled on 12 RSSs will only incorporate 23 partners, while complexes assembled on 23 RSSs show a 5- to 6-fold preference for 12 partners. Thus, initial assembly on a 12 RSS most accurately reflects the strict 12/23 coupled cleavage observed in the cell. Additional cellular factors such as chromatin may ensure that RAG1/2 first assembles on a 12 RSS, and then a free 23 RSS enters to activate cleavage.
Keywords: DNA recombination/nucleoprotein complex/RAG1/RAG2/V(D)J recombination
Introduction
Unlike almost all known proteins, the variable regions of antigen receptor proteins are not encoded by intact germline genes (Tonegawa, 1983). Mature coding regions must be assembled from among multiple variable (V), joining (J) and sometimes diversity (D) gene segments. Rearrangement of these segments occurs through the site-specific V(D)J recombination reaction that is unique to jawed vertebrates. This process requires specific protein and DNA components as well as general DNA repair factors (reviewed in Gellert, 2002).
In the germline, coding segments are flanked by recombination signal sequences (RSSs) comprising conserved heptamer and nonamer sequence motifs, which are separated by spacers of either 12 or 23 bp (called 12 and 23 RSSs). Segments to be recombined are flanked by RSSs with dissimilar spacer lengths (Tonegawa, 1983). For example, at the Igκ locus all V segments are flanked by 12 RSSs, while J segments are flanked by 23 RSSs. During recombination, specific DNA cleavage events introduce double-stranded breaks at one 12 and one 23 RSS (Roth et al., 1992a,b,c, 1993; Schlissel et al., 1993). The coding segments are fused to create coding joints, and the RSSs are then fused to one another to create signal joints. Depending on the germline arrangement, the DNA fragment containing the signal joints may be retained in the chromosome or may be permanently lost from the genome. Coding joints always remain in the genome, and can eventually become mature coding regions. Normally, the vast majority of coding joints in cells that undergo V(D)J recombination are the result of cleavage of a 12/23 RSS pair (Steen et al., 1997), a phenomenon referred to as the 12/23 rule (Tonegawa, 1983). Adherence to this rule is necessary to ensure production of functional joints.
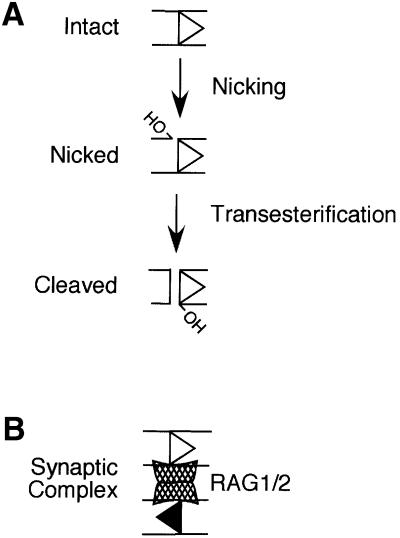
Cleavage is carried out by a recombinase composed of the RAG1 and RAG2 gene products (Oettinger et al., 1990; van Gent et al., 1995). RAG1 and RAG2 form a complex (RAG1/2) that can recognize both 12 and 23 RSSs (Hiom and Gellert, 1997). Binding to the 23 RSS is also facilitated by the high mobility group (HMG) 1 or 2 non-specific DNA binding proteins (van Gent et al., 1997), which are believed to bind within the spacer and bend the DNA (Mo et al., 2000). In the presence of Mn2+ or Mg2+, RAG1/2 introduces a single-stranded nick between the RSS and coding segment (McBlane et al., 1995; Hiom and Gellert, 1997), leaving a hydroxyl group on the 3′ end of the coding flank (Figure 1A). This hydroxyl can then attack the opposite DNA strand in a direct transesterification reaction (van Gent et al., 1996a) that leaves a hairpin on the end of the coding DNA and a blunt-cut RSS end (Figure 1A; McBlane et al., 1995). RAG1 belongs to a family of tranposases and retroviral integrases characterized by similar biochemical mechanisms; in all cases examined cleavage or strand transfer is initiated by nicking events followed by transesterification reactions (reviewed in Rice and Baker, 2001). RAG1 contains the trio of conserved active site residues common to this family (Kim et al., 1999; Landree et al., 1999; Fugmann et al., 2000) that are involved in coordination of divalent metal ion co-factors. RAG1 and RAG2 form a complex in solution (Swanson and Desiderio, 1999). While the role of RAG2 in the complex is not clear, it is known to enhance RAG1 binding to the RSS and is absolutely required for all steps of cleavage (Li et al., 1997; Akamatsu and Oettinger, 1998; Swanson and Desiderio, 1999).
Fig. 1. Steps in RSS cleavage by RAG1/2. In Figures 1–6, 12 RSSs are depicted as open triangles and 23 RSSs are depicted as filled triangles, with both triangles pointing away from the coding flank. (A) RAG1/2 binds to the intact RSS and introduces a nick between the signal and coding DNA. The 3′ hydroxyl (OH) in the nicked species attacks the opposite strand in a direct transesterification reaction to cleave the DNA, leaving a 3′ OH on the blunt-ended RSS. RAG1/2 can also carry out transesterification on pre-nicked substrates. (B) RAG1/2, with the help of HMG1 or HMG2, assembles a synaptic complex including a pair of RSSs (canonically, one 12 RSS and one 23 RSS). The synaptic complex is competent to carry out coupled cleavage of both partners in Mg2+. This cartoon is not meant to reflect the stoichiometry of RAG1 and RAG2 protomers present in the synaptic complex.
When cleavage is carried out in Mg2+, the most likely cellular co-factor, the transesterification step requires the presence of both RSSs (Eastman et al., 1996; van Gent et al., 1996b; West and Lieber, 1998). The core domains of RAG1/2, in recombinant, purified form, can recapitulate the 12/23 rule in that transesterification at a given RSS is strongly stimulated by the presence of the appropriate partner (van Gent et al., 1996b; Hiom and Gellert, 1998; West and Lieber, 1998). Transesterification takes place in a synaptic complex including the RSS pair (Figure 1B; Hiom and Gellert, 1998). RAG1/2, with the help of HMG proteins, has been shown to assemble such a complex in vitro from RSSs supplied on oligonucleotide substrates (Hiom and Gellert, 1998). Evidence suggests that the synaptic complex includes two RAG2 protomers (Mundy et al., 2002) and at least a dimer (Swanson and Desiderio, 1999), or possibly a trimer or tetramer (Landree et al., 2001; Mundy et al., 2002), of RAG1.
There are many potential pathways leading to assembly of the synaptic complex. The observation that RAG1/2 binds to the individual 12 and 23 RSSs and nicks them in Mg2+ (Hiom and Gellert, 1997; van Gent et al., 1997; Kim and Oettinger, 1998; West and Lieber, 1998) implies that these may constitute ‘half complexes’ that come together to form the synaptic complex, activating coupled transesterification of the two partners. However, there is no formal evidence demonstrating that two single-RSS complexes are intermediates in assembly of the synaptic complex. It is possible that a complex capable of binding both partners normally assembles on a single RSS, with the partner RSS being incorporated free of additional protein. Recent work provided evidence that this is one possible route (Mundy et al., 2002), but did not examine whether pre-assembly on a single RSS is absolutely required for building an active complex or whether it is merely one of several possible pathways. Should assembly on a single RSS be the necessary route, it is unknown whether there would be any difference between initial assembly on the 12 or 23 RSS.
We examined the steps leading to assembly of the synaptic complex in an in vitro system that accurately reflects the 12/23 rule. Pre-assembly of RAG1/2 complexes on both the 12 and 23 RSSs prior to their mixing was inhibitory to synaptic complex formation, suggesting that such half complexes are not direct precursors in assembly of the synaptic complex. Complexes pre-assembled on a 12 RSS could incorporate a 23 RSS provided that it was free of additional RAG1/2, and the reciprocal also appeared to be true. However, while complexes assembled first on a 12 RSS adhered strictly to the 12/23 rule, complexes assembled on a 23 RSS showed only a 5- to 6-fold preference for incorporation of a 12 versus a 23 RSS partner. Recent data indicate that on some chromatinized substrates, RAG1/2 is more active on 12 RSSs than on 23 RSSs (Kwon et al., 1998, 2000). In the cell, the presence of nucleosomes at recombining loci may reinforce adherence to the 12/23 rule by helping to ensure that RAG1/2 first assembles on a 12 RSS.
Results
Assembly of RAG1/2 on both RSSs prior to their mixing inhibits cleavage
RAG1/2 can bind to individual 12 or 23 RSSs in the presence of Ca2+, Mg2+ or Mn2+ (Hiom and Gellert, 1997). We examined conditions that promoted assembly of synaptic complexes in which RAG1/2 bound to a pair of RSSs. For the experiments shown in Figures 2–4, cleavage (transesterification) in Mg2+ was used as a measure of synaptic complex assembly. Pre-nicked substrates were used so that transesterification could be specifically examined, as this is the only chemical step that strictly requires synaptic complex assembly (van Gent et al., 1996b; Hiom and Gellert, 1998; Yu and Lieber, 2000).
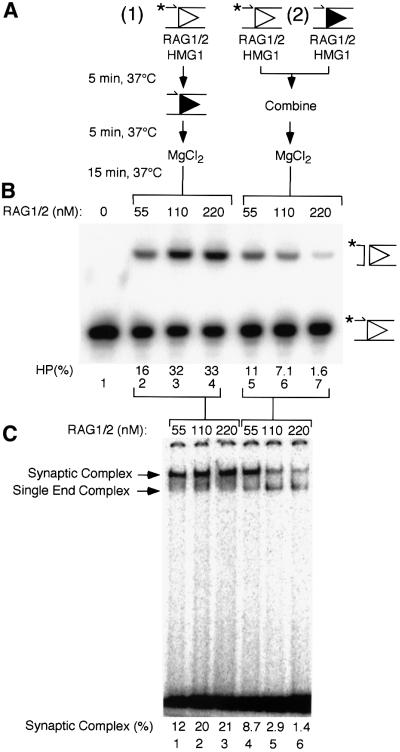
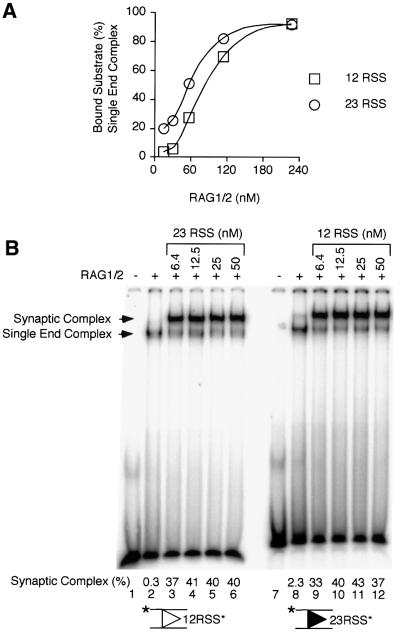
Fig. 2. Cleavage and RAG1/2 binding under two different assembly conditions. (A) Cleavage reactions (10 µl) were assembled in Ca2+ in one of two ways. (1) RAG1/2 at the concentration indicated in (B), HMG1 and pre-nicked 12 RSS substrate (the position of the label is indicated by an asterisk; the OH has been omitted) were first combined (5 min, 37°C), followed by the addition of pre-nicked 23 RSS partner (5 min, 37°C), and finally Mg2+ (15 min, 37°C). (2) Pre-nicked 12 RSS substrate and 23 RSS partner were each individually incubated with HMG1 and RAG1/2 at the concentration indicated in (B) (5 min, 37°C), these mixes were then combined (5 min, 37°C), and finally Mg2+ was added (15 min, 37°C). (B) Reaction products were separated on denaturing polyacrylamide gels as described in Materials and methods. Positions of pre-nicked substrate and hairpin (HP) cleavage products are shown to the right of the gel. Substrate (%) converted to HP product is indicated. (C) Binding reactions using intact RSS substrates were assembled as described in (A), omitting the addition of MgCl2 and the final 15 min incubation. Complexes assembled in these reactions were applied to polyacrylamide gels and separated as described in Materials and methods. The positions of the complex of RAG1/2 with a single RSS end and the synaptic complex are indicated to the left of the gel. Substrate (%) bound in the synaptic complex is indicated.
Fig. 4. Cleavage of intact substrate in the presence of specific competitor without pre-incubation in Ca2+. Cleavage reactions were assembled as indicated with reaction components being added in the order given; HMG1 and buffer components (see Materials and methods) were added prior to intact 12 RSS (lanes 1–6) or 23 RSS (lanes 7–12) substrate (the position of the label is indicated by an asterisk). Reactions did not include Ca2+. Some reactions included specific competitor (intact 12 RSS, lanes 4–6; intact 23 RSS, lanes 10–12) and partner (intact 23 RSS, lanes 3–5; intact 12 RSS, lanes 9–11). Positions of substrates as well as nicked and hairpin (HP) products are shown on either side of the gel. Substrate (%) converted to HP product is indicated.
We determined previously that brief (≤5 min) incubation at 37°C enhanced synaptic complex formation by ∼2-fold (Jones and Gellert, 2001; data not shown), and all incubations were carried out at this temperature; additives such as DMSO had no effect (Melek et al., 2002; data not shown). RAG1/2 was allowed to bind to a pre-nicked 12 RSS in Ca2+ followed by the addition of pre-nicked 23 RSS (Figure 2A, condition 1). RAG1/2–RSS complexes formed in this manner were competent for cleavage after the addition of Mg2+, indicating that synaptic complexes were assembled (Figure 2B, lanes 2–4). Under these assembly conditions, cleavage was enhanced at higher concentrations of RAG1/2 (Figure 2B, lanes 2–4). When RAG1/2 was allowed to bind to isolated 12 and 23 RSSs separately, and the two were then combined (Figure 2A, condition 2), cleavage was greatly reduced relative to pre-assembly on the 12 RSS alone (Figure 2B, lanes 5–7). This effect was most pronounced when the molar ratio of RAG1/2 to total RSS in the reaction was increased to ∼15:1 (Figure 2B, lane 7), conditions under which it is most likely that all the RSSs would be bound by a RAG1/2 complex during the initial incubation. The observation that relatively high concentrations of RAG1/2 did not inhibit cleavage when the proteins were pre-assembled on only one RSS (condition 1; Figure 2B, lanes 2–4) indicates that the inhibitory effect seen in assembly condition 2 could not be explained by factors such as non-specific protein aggregation, excess inactive protein or the presence of maltose-binding protein (MBP) tags fused to the RAG1 and RAG2 constructs. Similar results were obtained when intact (i.e. not pre-nicked) substrates were used (data not shown). These data indicated that at relatively high RAG1/2 concentrations, the complexes that assembled on individual 12 and 23 RSSs could not come together to form an active synaptic complex, and they suggested that the RAG1/2 complex first assembles on a single RSS followed by incorporation of naked partner DNA.
These results were corroborated by band mobility-shift experiments. Conditions have been developed previously in which the synaptic complex can be distinguished from the RAG1/2 complex bound to a single RSS (Hiom and Gellert, 1998; Jones and Gellert, 2001; Mundy et al., 2002). The synaptic complex migrates more slowly than the complex of RAG1/2 with a single RSS (Hiom and Gellert, 1998; Jones and Gellert, 2001; Mundy et al., 2002; see also Figure 2C), and the presence of two RSSs in the synaptic complex has been confirmed using oligonucleotides of different lengths (Jones and Gellert, 2001). The synaptic complex was competent for coupled cleavage after incubation in Mg2+ (Jones and Gellert, 2001; Mundy et al., 2002), so we presume that it represents the active complex formed in solution in the experiments described above, and that it includes RAG1/2 components in the stoichiometry necessary to support coupled cleavage. Binding reactions were staged as described in Figure 2A, with the exception that the final step including Mg2+ was omitted and intact RSS substrates were used. When RAG1/2 was allowed to bind the 12 RSS prior to the addition of 23 RSS partner, robust synaptic complex assembly was observed (Figure 2C, lanes 1–3). With these assembly conditions and this concentration of 23 RSS partner (12.5 nM), nearly all of the bound substrate was found in the synaptic complex (see below and data not shown). However, if RAG1/2 was incubated with both the 12 and 23 RSSs prior to their mixing, synaptic complex assembly was reduced as much as 15-fold (Figure 2C, lanes 4–6). The mobility of the complex of RAG1/2 with the single RSS did not change at relatively high RAG1/2 concentrations (Figure 2C, lanes 4–6), and in assays including a 12 or a 23 RSS alone we observed only one shifted complex, regardless of the concentration of RAG1/2 (see below and data not shown). This suggested that no potentially ‘dead end’ higher order complexes unable to assemble into the synaptic complex were formed at the RAG1/2 concentrations used in these experiments. Although in this assay the RSS substrate did not appear to be saturated even at 220 nM RAG1/2 (Figure 2C, lane 6), we have found that this level is competent to bind 100% of the substrate when binding is conducted at 25°C (see below). Unlike the synaptic complex, the complex of RAG1/2 bound to a single RSS appears to be less stable at 37°C, such that it cannot be captured with 100% efficiency in the gel mobility-shift assay.
RAG1/2 bound to a 12 RSS can incorporate a 23 RSS in the absence of free RAG1/2
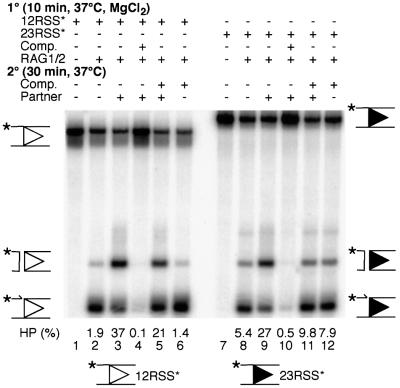
A competition experiment was designed to determine whether the RAG1/2 complex assembled on an isolated, pre-nicked 12 RSS included all of the protein components necessary for incorporation of an intact 23 RSS. Again, cleavage in Mg2+ was used as a measure of synaptic complex formation. Assembly in Ca2+ was performed in stages (Figure 3A), with RAG1/2, HMG1 and labeled 12 RSS always present during the first incubation. In the absence of specific competitor, robust cleavage was observed regardless of whether the 23 RSS partner was added during the first or second stage (Figure 3A, lanes 3 and 6), whereas very little cleavage was observed in the absence of partner (Figure 3A, lane 2). When competitor (100-fold molar excess of unlabeled 12 RSS) was present during the first stage, cleavage was nearly abolished (Figure 3A, lanes 4 and 7), indicating that this level of competitor was sufficient to bind all of the active RAG1/2 present in the reaction. When the addition of competitor was delayed until the second stage, cleavage was observed at 40–50% of the level seen in its absence (Figure 3A, lanes 5 and 8), indicating that this portion of the cleavage seen in the absence of competitor resulted from stable complexes formed during the first stage of incubation. Cleavage was observed at approximately the same level regardless of whether the 23 RSS partner was added during the first or second stage (Figure 3A, lanes 5 and 8). HMG1, which is vital for binding of RAG1/2 to the 23 RSS as well as for synaptic complex formation and cleavage, was present in these reactions at saturating concentrations (data not shown). If the competitor served to bind all of the HMG1 present, we would expect to see no cleavage when the 23 RSS and competitor were added in the second stage, which was not the case. These data indicated that the complex assembled on the 12 RSS during the first stage was sufficient to incorporate the 23 RSS partner even when no free RAG1/2 was present.
Fig. 3. Cleavage under various assembly conditions in the presence of specific competitor. (A) Assembly of cleavage reactions in Ca2+ was staged as indicated with reaction components being added in the order given; HMG1 and buffer components (see Materials and methods) were added prior to pre-nicked 12 RSS substrate (the position of the label is indicated by an asterisk). Competitor was intact 12 RSS, and intact 23 RSS acted as partner. Positions of pre-nicked substrate and hairpin (HP) cleavage products are shown to the right of the gel. Substrate (%) converted to product is indicated. (B) Reactions were staged as per (A), but pre-nicked 23 RSS was used as substrate, intact 23 RSS was used as competitor, and intact 12 RSS acted as partner.
While these experiments do not definitively demonstrate coupled cleavage, we and others have observed that neither partner in a synaptic complex can undergo transesterification before both are nicked, and that transesterification of both partners is usually highly coupled (Eastman et al., 1996; van Gent et al., 1996b; Eastman and Schatz, 1997; Kim and Oettinger, 1998). Therefore, it is very likely that the complex assembled on a single 12 RSS is capable of cleaving both partners once the 23 RSS has been bound.
RAG1/2 complexes assembled on a 23 RSS were also able to incorporate a 12 RSS partner, but this binding was less resistant to a large excess of 23 RSS competitor (Figure 3B). As before, robust cleavage was observed in the absence of competitor regardless of the order of addition (Figure 3B, lanes 3 and 6), and competitor added in the first stage abolished this activity (Figure 3B, lanes 4 and 7). Unlike the results with labeled 12 RSS, low but detectable levels of cleavage occurred in the absence of partner (Figure 3B, lane 2). When 12 RSS partner was present during the first stage and competitor was added in the second, cleavage was restored (Figure 3B, lane 5). However, when both 12 RSS partner and competitor were added in the second stage, no partner-dependent cleavage was observed. This indicates that when a large excess of free 23 RSS was present, the 12 RSS was not able to efficiently enter the complex to activate robust cleavage.
We confirmed that these observations were not an artifact of assembly in Ca2+ or the use of pre-nicked labeled substrates (Figure 4). In an alternative protocol, RAG1/2 was incubated in Mg2+ with labeled, intact 12 or 23 RSS substrate in the absence of partner for 10 min, at which point the nicking reaction is nearly complete (data not shown). Competitor and/or partner was then added, and incubation was continued for an additional 30 min. As before, the addition of complementary partner during the second stage in the absence of competitor strongly stimulated cleavage on both the 12 and 23 RSS substrates (Figure 4, lanes 3 and 9). When competitor was added during the first stage, both nicking and cleavage were abolished (Figure 4, lanes 4 and 10). On the 12 RSS substrate, when competitor was added in the second stage, the addition of 23 RSS partner restored moderate levels of cleavage (Figure 4, lane 5). This indicated that the complex assembled on the 12 RSS could incorporate a partner and carry out both steps of cleavage. Competitor added to the 12 RSS in the absence of partner had no stimulatory effect (Figure 4, lane 6). In contrast, on the 23 RSS substrate, competitor added during the second stage had a small stimulatory effect on cleavage. This was true regardless of whether 12 RSS partner was present (Figure 4, compare lanes 11 and 12 with lane 8). This suggested that under these conditions, excess unlabeled 23 RSS could stimulate cleavage of the labeled 23 RSS to which RAG1/2 was already bound, and that the RAG1/2 complex bound to the 23 RSS did not have as strong a preference for the canonical partner as RAG1/2 bound to a 12 RSS.
RAG1/2 binding to a 12 RSS leads to high affinity binding of a 23 RSS
A RAG1/2 complex assembled on a single RSS always showed some preference for the canonical partner. However, whereas a complex assembled on a 12 RSS appeared to be locked in a conformation that guaranteed incorporation of a 23 RSS, a complex assembled on a 23 RSS showed only a 5- to 6-fold preference for incorporation of a 12 RSS partner over a 23 RSS (see below). Assembly of synaptic complexes with RAG1/2 bound to a pair of RSSs was directly observed by gel mobility-shift experiments. When RAG1/2 and HMG1 were incubated with either a 12 or 23 RSS, one major shifted complex including a single DNA molecule was evident across a range of RAG1/2 concentrations (Figure 5A; data not shown). With HMG1 present, RAG1/2 bound to the individual RSSs with roughly the same affinity to form this complex (Figure 5A). To examine synaptic complex assembly, binding in Ca2+ was staged in a manner similar to cleavage (Figure 5B). RAG1/2 was allowed to bind to labeled 12 or 23 RSSs for 5 min, followed by 5 min incubation in the presence of partner. In the absence of partner, a very small amount of synaptic complex was observed after pre-incubation on the 23 RSS, but not on the 12 RSS (Figure 5B, compare lanes 2 and 8), consistent with the cleavage results obtained in solution. When partner was added, robust synaptic complex assembly was observed across an 8-fold partner concentration range regardless of whether RAG1/2 was pre-assembled on the 12 or 23 RSS (Figure 5B, lanes 3–6 and 9–12). This suggested that complexes assembled on either a 12 or 23 RSS could readily incorporate the appropriate partner, and that there was not a large difference in canonical partner binding affinity between 23 and 12 RSS partners in the concentration range used in our experiments.
Fig. 5. Gel mobility-shift assay for detection of synaptic complex assembly. (A) Binding reactions were assembled in Ca2+ and analyzed as described in Materials and methods. Intact 12 RSS (squares) or 23 RSS (circles) substrate (2 nM) was combined with RAG1/2 at the concentrations indicated and HMG1 (5 min, 25°C). Complexes assembled in these reactions were applied to polyacrylamide gels and separated as described. Substrate (%) bound in the single end complex was determined. (B) Intact 12 RSS (lanes 1–6) or 23 RSS (lanes 7–12) substrate (the position of the label is indicated by an asterisk) was incubated with RAG1/2 (110 nM) and HMG1 (5 min, 37°C), followed by the addition of 23 or 12 RSS partner, respectively, at the concentrations indicated (5 min, 37°C). Complexes assembled in these reactions were applied to polyacrylamide gels and separated as described. The positions of the complex of RAG1/2 with a single RSS end and the synaptic complex are indicated to the left of the gel. Substrate (%) bound in the synaptic complex is indicated.
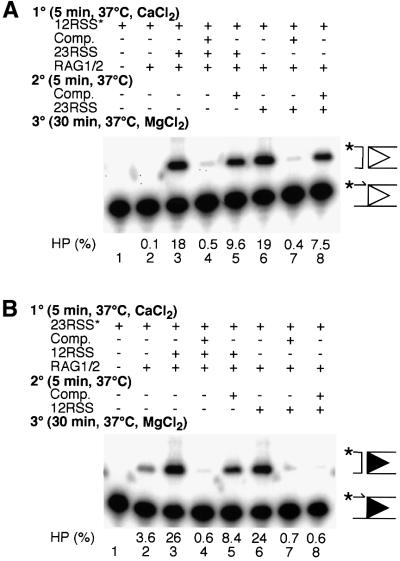
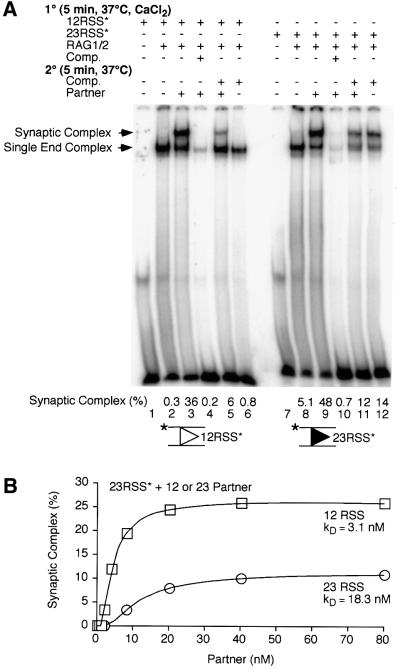
Competition experiments confirmed that RAG1/2 complexes assembled on a 12 RSS bound to the 23 RSS partner with high affinity. Those assembled on a 23 RSS showed a preference for a 12 RSS partner, but could bind a second 23 RSS under certain conditions. Binding was again staged, with partner being added in the second stage, and competitor being added in either the first or second. When RAG1/2 was assembled on a 12 RSS, a synaptic complex could be formed when 23 RSS partner was present (Figure 6A, lane 3), even when excess 12 RSS competitor was also added (Figure 6A, lane 5). No synaptic complex was assembled when the 23 RSS partner was omitted (Figure 6A, lanes 2 and 6). The complex of RAG1/2 bound to a 23 RSS could incorporate a 12 RSS to form a synaptic complex (Figure 6A, lane 9). However, when a large excess of 23 RSS competitor was present, the 12 RSS partner did not stimulate assembly of the synaptic complex beyond what was seen with competitor alone (Figure 6A, lanes 11 and 12). We consistently observed some competitor-dependent reduction in total substrate binding (Figure 6) and cleavage (Figures 3 and 4), even when competitor was added in the second stage, implying that complexes formed in the first stage are not completely resistant to a large excess of competitor.
Fig. 6. Gel mobility-shift assay for detection of synaptic complex assembly in the presence of specific competitor. (A) Binding reactions were assembled as indicated with reaction components being added in the order given; HMG1 and buffer components (see Materials and methods) were added prior to intact 12 RSS (lanes 1–6) or 23 RSS (lanes 7–12) substrate (the position of the label is indicated by an asterisk). Some reactions included specific competitor (intact 12 RSS, lanes 4–6; intact 23 RSS, lanes 10–12) and partner (intact 23 RSS, lanes 3–5; intact 12 RSS, lanes 9–11). The positions of the complex of RAG1/2 with a single RSS end and the synaptic complex are indicated to the left of the gel. Substrate (%) bound in the synaptic complex is indicated. (B) Binding reactions were assembled as in (A) with labeled 23 RSS substrate present in the first stage (5 min, 37°C) and 12 RSS (squares) or 23 RSS (circles) partner, at the concentrations indicated added in the second (5 min, 37°C). Complexes were separated and quantified as described. Substrate (%) bound in the synaptic complex is indicated.
The amounts of synaptic complex formed in the presence of 12 RSS partner versus excess 23 RSS competitor suggested that incorporation of a second 23 RSS was less efficient than incorporation of the 12 RSS partner (Figure 6A, compare lanes 9 and 12). The relative affinity of a complex assembled on a 23 RSS for a 12 or 23 RSS partner was examined using the gel mobility-shift assay. In the first stage, RAG1/2 was allowed to assemble on the labeled 23 RSS substrate in the absence of partner; various concentrations of unlabeled 12 or 23 RSS partner were then added in the second stage (Figure 6B). Partner-stimulated synaptic complex assembly was observed with as little as 2 nM 12 RSS and rose rapidly, peaking at ∼20 nM 12 RSS (Figure 6B, squares), at which point >70% of the shifted species was the synaptic complex (data not shown). Across the entire range of 23 RSS concentrations tested, synaptic complex assembly was lower than that observed with 12 RSS partner (Figure 6B, circles). More robust synaptic complex assembly could not be promoted by the addition of much higher concentrations of 23 RSS (up to 400 nM), and at no concentration was >50% of the shifted species synaptic complex (data not shown). Even with the low level of synaptic complex formation (roughly 25% for 12 RSS partner and 10% for 23 RSS partner), we felt that calculation of the dissociation constant for partner binding would be useful for comparison of the two complexes. After assembly on a 23 RSS, the binding affinity for binding a 12 RSS partner (kD = 3.1 ± 0.5 nM) was five to six times higher than the affinity for binding a second 23 RSS (kD = 18.3 ± 6.5 nM; Figure 6B). These data indicated that under the conditions used in our competition experiments, with 23 RSS competitor present in 16-fold excess over 12 RSS partner, the preference for the canonical partner would not be evident (Figure 6A, lanes 11 and 12). However, the 12 RSS partner would be strongly favored over the 23 RSS, provided that the potential partners were present in roughly equimolar amounts.
Discussion
The complete RAG1/2 complex initially assembles on a single RSS
Our results indicate that a RAG1/2 complex competent for synaptic complex formation first assembles on a single RSS, and that the partner RSS is incorporated as naked DNA. A complex assembled on a single RSS can incorporate a partner RSS in the absence of free RAG1/2 (Figures 3, 4 and 6), and if RAG1/2 is allowed to bind to both RSSs in isolation prior to their mixing, synaptic complex formation is inhibited (Figure 2). In the family of transposases and retroviral integrases to which RAG1 belongs, a synaptic complex between transposon or retroviral ends has commonly been found to be an important intermediate (reviewed in Rice and Baker, 2001). It is not known whether the assembly pathway outlined here for RAG1/2 will have parallels in other members of this family. In the case of transposon Tn5, for example, the transposase binds as a monomer to its DNA binding sites on each transposon end (Weinreich et al., 1994), and interaction between these complexes is hypothesized to produce the dimer which is active for cleavage (Bhasin et al., 2000 and references therein). In fact, binding to single DNA ends has been demonstrated for numerous family members, including RAG1/2, although in some systems intermediates formed during assembly of the synaptic complex are too unstable to be characterized using current techniques. Our observations regarding the ordered assembly of the RAG1/2 synaptic complex suggest that the ability of transposases to assemble on either DNA end in isolation should not necessarily be considered strong evidence that separate, single-end complexes are the direct precursors of synaptic complex assembly.
An assembly pathway analogous to the one we propose for RAG1/2 has been described for integration of bacteriophage λ DNA into the bacterial chromosome, although the λ integrase (Int) protein belongs to a different family of recombinases. The Int protein, with the help of bacterial integration host factor, first assembles on a high affinity attachment site (attP) on the phage genome to form an intasome (Richet et al., 1986). The intasome then captures the low affinity site on the bacterial chromosome (attB) without the requirement for additional protein (Richet et al., 1988; Patsey and Bruist, 1995). In this system, the order of assembly is clearly determined by the large difference in affinity of Int for the attP and attB sites. During V(D)J recombination in the cell, nucleosomes and other factors may influence whether RAG1/2 binds first to a 12 RSS or a 23 RSS (see below).
The results presented herein complement those of Mundy et al. (2002), who used gel mobility-shift assays to examine the stepwise assembly and stoichiometry of the RAG1/2 complex on a 12 RSS, and formation of a synaptic complex upon addition of a 23 RSS. However, our work expands on previous observations in a number of significant ways. First, we demonstrate that synaptic complex assembly probably cannot proceed by an alternative pathway in which RAG1/2 first binds independently to the 12 and 23 RSSs. In addition, we find significant differences between complexes that initially assemble on 12 and 23 RSSs, as is discussed in the next section. While the experiments presented herein were performed using the truncated ‘core’ regions of the RAG1 and RAG2 proteins fused to MBP, those from Mundy et al. employed core RAG1 and RAG2 with and without MBP fusion tags (Mundy et al., 2002). The similarity in behavior between these constructs indicates that the presence of the MBP tag did not significantly affect our findings. However, it is certainly possible that the ‘non-essential’ regions of the proteins will influence assembly. Such issues should be addressed as soon as soluble full-length RAG1 and RAG2 proteins become available.
Binding to a 12 RSS ‘locks’ RAG1/2 conformation
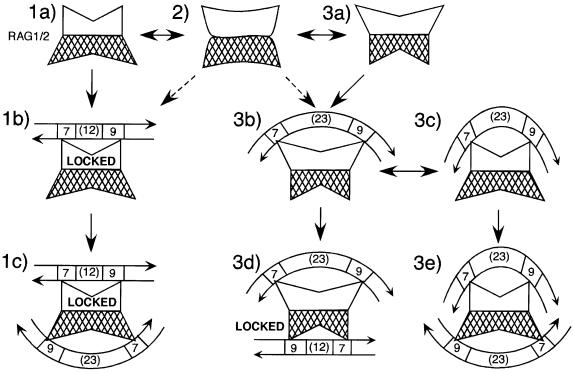
In the system used here, RAG1/2 can bind initially to either the 12 or 23 RSS and incorporate a partner to form a synaptic complex, but initial binding to the 12 RSS results in more faithful adherence to the 12/23 rule. Based on these data, we have developed a hypothetical model for maintenance of the 12/23 rule. The model depicts RAG1/2 in solution as including protein components sufficient to bind two RSSs, but it is also possible that the assembly of multiple components on a single RSS occurs in a stepwise manner. RAG1/2 is both flexible—so that either RSS can initially be bound by either site—and asymmetric (Figure 7, steps 1–3). Such asymmetry can be established by the presence of an uneven number of protomers (e.g. RAG13/RAG22; Mundy et al., 2002) or by complementary, non-identical conformations of the two RSS binding sites transmitted through the multimer interface. In the asymmetric complex, the heptamer and nonamer binding regions within one RSS binding site are closer together than those at the opposite site (Figure 7, step 1a, white versus cross-hatched binding sites). The more narrowly and widely bipartite sites would bind preferentially to 12 and 23 RSSs, respectively (Figure 7, compare step 1b with 3b), with binding of the 23 RSS being further enhanced by HMG1 or HMG2 bound within the 23 bp spacer (van Gent et al., 1997; Mo et al., 2000; HMG proteins are not shown in Figure 7).
Fig. 7. Model for assembly of the synaptic complex. RAG1/2 in solution may include all protein components necessary for binding to two RSSs (1a and 3a), or a bivalent complex could assemble after binding of a monovalent complex to a single RSS (not shown). Heptamer and nonamer binding domains may be contributed by different protomers within the complex (Swanson, 2001). In the bivalent complex, the heptamer and nonamer binding domains within one RSS binding site are optimally arranged to bind a 12 RSS (1a, white binding site), while these domains are farther apart in the second RSS binding site, which can only bind to a 23 RSS (3a, white binding site). These conformations may interchange rapidly in solution (2). Initial binding to a 12 RSS locks that binding site because of the fixed length of the 12 bp spacer (1b); the second site must then be occupied by a 23 RSS (1c). Initial binding to a 23 RSS (3b) does not lock the complex because of the relative flexibility of the 23 bp spacer (3c). The second RSS to enter the complex can be either a 12 (3d) or a 23 (3e). HMG proteins are not shown in this model; they would presumably bind within the 23 bp spacer and increase its bend. Heptamer (7), nonamer (9) and spacer regions are indicated; RSSs are depicted as entering the complex in opposite orientations (DNA arrows).
When a 12 RSS is bound by one of the RAG1/2 RSS binding sites, the complex becomes locked (Figure 7, step 1b) because the 12 bp spacer cannot be expanded to accommodate a 23 bp spacer binding site. This ensures that the second binding site is held in a conformation that can only bind a 23 RSS (Figure 7, step 1c), thus this site will bind a 23 RSS even in the presence of excess free 12 RSS. If a 23 RSS is the first to be bound (Figure 7, step 3b), the complex is not locked. Increased bending within the 23 bp spacer could allow the complex to maintain its flexibility by bringing the heptamer and nonamer binding regions in contact with the 23 RSS into closer proximity (Figure 7, step 3c). The second site could then bind preferentially to a 23 RSS (Figure 7, step 3e). Alternatively, a free 23 RSS could simply bend enough to bind a site that would otherwise bind a 12 RSS (not shown). Once a 12 RSS was bound, the complex would be locked (Figure 7, step 3d).
There is at least a 5- to 6-fold preference for binding to a 12 RSS after initial binding to a 23 RSS (Figure 6B). In addition, after initial binding to a 23 RSS, we were never able to fill the majority of the ‘partner’ binding sites with 23 RSSs, even at very high levels of free 23 RSS; these sites were nearly saturated at much lower levels of free 12 RSS (Figure 5B). It is possible that the 23/23 complex is less stable than the canonical 12/23 complex, and so it may have an increased probability of disassembling prior to activation of cleavage. However, given that the gene segments at loci that undergo V(D)J recombination are arranged so that nearest neighbors have RSSs of like spacer lengths (Tonegawa, 1983) and recombining 12/23 RSS pairs may be separated by distances of 1–2 megabases in the germline, a RAG1/2 complex assembled on a 23 RSS in the cell would have a high probability of encountering a second 23 RSS before a more appropriate 12 RSS partner. With this arrangement, the near invariance of the 12/23 rule during V(D)J recombination in wild-type cells at natural loci would not appear to be consistent with only a 5- to 6-fold preference for the appropriate partner. This suggests that mechanisms may exist to ensure initial binding to a 12 RSS in the cell.
How can initial binding to the 12 RSS be assured during V(D)J recombination at chromosomal loci? While there is not a large difference in the affinity of RAG1/2 for naked 12 and 23 RSSs (Figure 5A), provided that HMG1 or HMG2 is also present (van Gent et al., 1997), there appear to be substantial differences on nucleosomal DNA (Kwon et al., 1998, 2000). Under conditions that support uncoupled cleavage (3 mM Mn2+), 12 RSSs positioned on the nucleosomal dyad can be made cleavage-accessible by either histone acetylation or nucleosome remodeling by the hSWI/SNF complex, while 23 RSSs positioned on the dyad remain refractory to cleavage regardless of nucleosomal modification (Kwon et al., 2000). The complex regulation of accessibility within recombining loci may also play a role. At the TCRβ locus, the Eβ enhancer is absolutely required for both D to J and V to DJ recombination (Bories et al., 1996; Bouvier et al., 1996), but at a TCRδ mini-locus and the IgH locus, the cor responding enhancers are only required for the second stages of rearrangement, VδD to Jδ (Lauzurica and Krangel, 1994; McMurray et al., 1997) and VH to DJH (Chen et al., 1993; Serwe and Sablitzky, 1993), respectively. This has led to the suggestion that promoter and other regulatory regions within the loci can act in a highly localized manner to regulate accessibility. For example, both the DQ52 region in the IgH locus and the T early α element in the TCRα locus have complex and subtle effects on usage of individual gene segments (Villey et al., 1996; Nitschke et al., 2001). It will be interesting to determine whether such intra-locus regulatory mechanisms also promote initial RAG1/2 binding to a 12 RSS during recombination in the cell.
Materials and methods
DNA substrates and proteins
Oligonucleotide substrates have been described previously (McBlane et al., 1995). They were composed of the following oligonucleotides: intact 12 RSS, DAR39 and DAR40; intact 23 RSS, DG61 and DG62; pre-nicked 12 RSS, DAR42, DG10 and DAR40; and pre-nicked 23 RSS, DAR42, DG4 and DG62. All oligonucleotides were purified on denaturing polyacrylamide gels prior to use. Oligonucleotides for each substrate were combined in annealing buffer (20 mM Tris pH 7.5, 100 mM NaCl), heated to 95°C for 5 min, then allowed to cool overnight. Annealed complexes were purified through micro-BioSpin P6 columns (Bio-Rad) in 10 mM Tris pH 7.0 buffer. For radiolabeled substrates, one oligonucleotide in each complex (DAR39, DG61 or DAR42) was phosphorylated using T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (Dupont-NEN), then heated to 80°C for 20 min prior to annealing.
Murine RAG1 (amino acids 384–1008) and RAG2 (amino acids 1–387) proteins tagged on their N-termini with MBP and on their C-termini with polyhistidine and myc epitopes were co-expressed and purified as described previously (McBlane et al., 1995). HMG1 (amino acids 1–163) was expressed from vector pDVG83 (Mo et al., 2000) in RJ1878 bacterial cells (gift of Reid C.Johnson, UCLA Medical Center, Los Angeles, CA), which do not express bacterial HU proteins. Purification was as for HU protein, method A, as described previously (Johnson et al., 1986). Bovine serum albumin (BSA) was purchased from New England BioLabs.
Cleavage reactions
The detailed order of assembly for cleavage reactions is provided for each figure. Cleavage reactions were conducted in volumes of 5 µl unless otherwise indicated. Buffer components (25 mM MOPS pH 7.0, 32 mM KCl, 4 mM dithiothreitol, 0.1 mg/ml BSA, 1% glycerol) and HMG1 (340 nM) were added to reaction mixtures prior to labeled substrate (2 nM), specific competitor (200 nM), partner (12.5 nM) and RAG1/2 (110 nM, based on a probable stoichiometry of RAG14RAG22; Landree et al., 2001; Mundy et al., 2002). The concentration of active RAG1/2 is likely to be lower, as it has been observed that ∼20% of the protein prepared by this method is competent for cleavage (Yu and Lieber, 2000). Unless otherwise indicated, reactions were assembled in 4 mM CaCl2, which was added simultaneously with buffer components. Cleavage was initiated by the addition of 4 mM MgCl2, and the reactions were stopped with one reaction volume of stop buffer (95% formamide, 50 mM EDTA, and 0.05% each Bromophenol Blue and xylene cyanol). Reactions were heated to 95°C for 2 min, then loaded onto warm 15% polyacrylamide gels (cross-linked at a ratio of 20:1) which were electrophoresed in 1× TBE buffer for 1–1.5 h at 20 W. Gels were dried for 3 h at 80°C under a vacuum, then exposed to phosphor-storage autoradiography.
Gel mobility-shift analysis
Reactions were assembled as described for cleavage with detailed order of assembly described for each figure. Binding reactions (5 µl total volume) were assembled in 4 mM CaCl2 and did not include MgCl2. All other buffer, substrate and protein concentrations were as for cleavage unless otherwise indicated. After assembly, 100% glycerol (1 µl) was added and reactions were loaded onto 8% polyacrylamide gels (cross-linked at a ratio of 80:1) in 0.5× TBE buffer. Assembly of binding reactions was staggered such that each reaction was incubated for exactly the time indicated (± 10 s) prior to loading, and gels were electrophoresed at 100 V prior to and throughout loading. After all samples had been applied, gels were electrophoresed at 140 V for 2 h (room temperature). Gels were dried for 1 h at 80°C under a vacuum, and exposed to phosphor-storage autoradiography. Partner binding affinity was determined with KaleidaGraph 3.05 software (Synergy Software, Reading, PA) using the formula Fbound = Fmax/(1 + kD/[RSS]), where Fbound is the fraction of substrate bound in the synaptic complex, Fmax is the maximum amount of synaptic complex formed and [RSS] is the concentration of unlabeled 12 or 23 RSS (potential partner) present in the reaction.
Miscellaneous
All quantification of gel band intensities was by phosphor-storage autoradiography using a Molecular Dynamics Typhoon 8600 and Molecular Dynamics ImageQuant 5.1 software. DNA molar concentrations are given for the whole molecule. All results are representative of at least three independent trials.
Acknowledgments
Acknowledgements
We thank R.C.Johnson for the kind gift of bacterial strain RJ1878. We also thank Eric Green and Miklos Gaszner of the Laboratory of Molecular Biology (NIDDK, NIH) for critical reading of this manuscript and I-hung Shih of this laboratory for assistance in data analysis and many helpful suggestions. J.M.J. is supported by the Damon Runyon Cancer Research Foundation, Fellowship DRG1582.
References
- Akamatsu Y. and Oettinger,M.A. (1998) Distinct roles of RAG1 and RAG2 in binding the V(D)J recombination signal sequences. Mol. Cell. Biol., 18, 4670–4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhasin A., Goryshin,I.Y., Steiniger-White,M., York,D. and Reznikoff,W.S. (2000) Characterization of a Tn5 pre-cleavage synaptic complex. J. Mol. Biol., 302, 49–63. [DOI] [PubMed] [Google Scholar]
- Bories J.C., Demengeot,J., Davidson,L. and Alt,F. (1996) Gene-targeted deletion and replacement mutations of the T-cell receptor β-chain enhancer: the role of enhancer elements in controlling V(D)J recombination accessibility. Proc. Natl Acad. Sci. USA, 93, 7871–7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier G., Watrin,F., Naspetti,M., Verthuy,C., Naquet,P. and Ferrier,P. (1996) Deletion of the mouse T-cell receptor β gene enhancer blocks αβ T-cell development. Proc. Natl Acad. Sci. USA, 93, 7877–7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Young,F., Bottaro,A., Stewart,V., Smith,R.K. and Alt,F. (1993) Mutations of the intronic IgH enhancer and its flanking sequences differentially affect accessibility of the JH locus. EMBO J., 12, 4635–4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman Q.M. and Schatz,D.G. (1997) Nicking is asynchronous and stimulated by synapsis in 12/23 rule-regulated V(D)J cleavage. Nucleic Acids Res., 25, 4370–4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman Q.M., Leu,T.M. and Schatz,D.G. (1996) Initiation of V(D)J recombination in vitro obeying the 12/23 rule. Nature, 380, 85–88. [DOI] [PubMed] [Google Scholar]
- Fugmann S.D., Villey,I.J., Ptaszek,L.M. and Schatz,D.G. (2000) Identification of two catalytic residues in RAG1 that define a single active site within the RAG1/RAG2 protein complex. Mol. Cell, 5, 97–107. [DOI] [PubMed] [Google Scholar]
- Gellert M. (2002) V(D)J recombination: RAG proteins, repair factors and regulation. Annu. Rev. Biochem., 71, 101–132. [DOI] [PubMed] [Google Scholar]
- Hiom K. and Gellert,M. (1997) A stable RAG1–RAG2–DNA complex that is active in V(D)J cleavage. Cell, 88, 65–72. [DOI] [PubMed] [Google Scholar]
- Hiom K. and Gellert,M. (1998) Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol. Cell, 1, 1011–1019. [DOI] [PubMed] [Google Scholar]
- Johnson R.C., Bruist,M.F. and Simon,M.I. (1986) Host protein require ments for in vitro site-specific recombination. Cell, 46, 531–539. [DOI] [PubMed] [Google Scholar]
- Jones J.M. and Gellert,M. (2001) Intermediates in V(D)J recombination: a stable RAG1/2 complex sequesters cleaved RSS ends. Proc. Natl Acad. Sci. USA, 98, 12926–12931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.R. and Oettinger,M.A. (1998) Functional analysis of coordinated cleavage in V(D)J recombination. Mol. Cell. Biol., 18, 4679–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.R., Dai,Y., Mundy,C.L., Yang,W. and Oettinger,M.A. (1999) Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev., 13, 3070–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon J., Imbalzano,A.N., Matthews,A. and Oettinger,M.A. (1998) Accessibility of nucleosomal DNA to V(D)J cleavage is modulated by RSS positioning and HMG1. Mol. Cell, 2, 829–839. [DOI] [PubMed] [Google Scholar]
- Kwon J., Morshead,K.B., Guyon,J.R., Kingston,R.E. and Oettinger,M.A. (2000) Histone acetylation and hSWI/SNF remodeling act in concert to stimulate V(D)J cleavage of nucleosomal DNA. Mol. Cell, 6, 1037–1048. [DOI] [PubMed] [Google Scholar]
- Landree M.A., Wibbenmeyer,J.A. and Roth,D.B. (1999) Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination. Genes Dev., 13, 3059–3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landree M.A., Kale,S.B. and Roth,D.B. (2001) Functional organization of single and paired V(D)J cleavage complexes. Mol. Cell. Biol., 21, 4256–4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauzurica P. and Krangel,M.S. (1994) Enhancer-dependent and -independent steps in the rearrangement of a human T cell receptor δ transgene. J. Exp. Med., 179, 43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Swanson,P. and Desiderio,S. (1997) RAG-1 and RAG-2-dependent assembly of functional complexes with V(D)J recombination substrates in solution. Mol. Cell. Biol., 17, 6932–6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBlane J.F., van Gent,D.C., Ramsden,D.A., Romeo,C., Cuomo,C.A., Gellert,M. and Oettinger,M.A. (1995) Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell, 83, 387–395. [DOI] [PubMed] [Google Scholar]
- McMurray M.T., Hernandez-Munain,C., Lauzurica,P. and Krangel,M.S. (1997) Enhancer control of local accessibility to V(D)J recombinase. Mol. Cell. Biol., 17, 4553–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melek M., Jones,J.M., O’Dea,M.H., Pais,G., Burke,T.B., Pommier,Y., Neamati,N. and Gellert,M. (2002) Effect of HIV integrase inhibitors on the RAG1/2 recombinase. Proc. Natl Acad. Sci. USA, 99, 134–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo X., Bailin,T., Noggle,S. and Sadofsky,M.J. (2000) A highly ordered structure in V(D)J recombination cleavage complexes is facilitated by HMG1. Nucleic Acids Res., 28, 1228–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy C.L., Patenge,N., Matthews,A.G. and Oettinger,M.A. (2002) Assembly of the RAG1/RAG2 synaptic complex. Mol. Cell. Biol., 22, 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitschke L., Kestler,J., Tallone,T., Pelkonen,S. and Pelkonen,J. (2001) Deletion of the DQ52 element within the Ig heavy chain locus leads to a selective reduction in VDJ recombination and altered D gene usage. J. Immunol., 166, 2540–2552. [DOI] [PubMed] [Google Scholar]
- Oettinger M.A., Schatz,D.G., Gorka,C. and Baltimore,D. (1990) RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science, 248, 1517–1523. [DOI] [PubMed] [Google Scholar]
- Patsey R.L. and Bruist,M.F. (1995) Characterization of the interaction between the lambda intasome and attB. J. Mol. Biol., 252, 47–58. [DOI] [PubMed] [Google Scholar]
- Rice P.A. and Baker,T.A. (2001) Comparative architecture of transposase and integrase complexes. Nat. Struct. Biol., 8, 302–307. [DOI] [PubMed] [Google Scholar]
- Richet E., Abcarian,P. and Nash,H. (1986) The interaction of recombination proteins with supercoiled DNA: defining the role of supercoiling in lambda integrative recombination. Cell, 46, 1011–1021. [DOI] [PubMed] [Google Scholar]
- Richet E., Abcarian,P. and Nash,H. (1988) Synapsis of attachment sites during lambda integrative recombination involves capture of a naked DNA by a protein–DNA complex. Cell, 52, 9–17. [DOI] [PubMed] [Google Scholar]
- Roth D.B., Menetski,J.P., Nakajima,P.B., Bosma,M.J. and Gellert,M. (1992a) V(D)J recombination: broken DNA molecules with covalently sealed (hairpin) coding ends in scid mouse thymocytes. Cell, 70, 983–991. [DOI] [PubMed] [Google Scholar]
- Roth D.B., Nakajima,P.B., Menetski,J.P., Bosma,M.J. and Gellert,M. (1992b) Double-strand breaks associated with V(D)J recombination at the TCR delta locus in murine thymocytes. Curr. Top. Microbiol. Immunol., 182, 115–124. [DOI] [PubMed] [Google Scholar]
- Roth D.B., Nakajima,P.B., Menetski,J.P., Bosma,M.J. and Gellert,M. (1992c) V(D)J recombination in mouse thymocytes: double-strand breaks near T cell receptor delta rearrangement signals. Cell, 69, 41–53. [DOI] [PubMed] [Google Scholar]
- Roth D.B., Zhu,C. and Gellert,M. (1993) Characterization of broken DNA molecules associated with V(D)J recombination. Proc. Natl Acad. Sci. USA, 90, 10788–10792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlissel M., Constantinescu,A., Morrow,T., Baxter,M. and Peng,A. (1993) Double-strand signal sequence breaks in V(D)J recombination are blunt, 5′-phosphorylated, RAG-dependent and cell cycle regulated. Genes Dev., 7, 2520–2532. [DOI] [PubMed] [Google Scholar]
- Serwe M. and Sablitzky,F. (1993) V(D)J recombination in B cells is impired but not blocked by targeted deletion of the immunoglobulin heavy chain intron enhancer. EMBO J., 12, 2321–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen S.B., Gomelsky,L., Speidel,S.L. and Roth,D.B. (1997) Initiation of V(D)J recombination in vivo: role of recombination signal sequences in formation of single and paired double-strand breaks. EMBO J., 16, 2656–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson P.C. (2001) The DDE motif in RAG-1 is contributed in trans to a single active site that catalyzes the nicking and transesterification steps of V(D)J recombination. Mol. Cell. Biol., 21, 449–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson P.C. and Desiderio,S. (1999) RAG-2 promotes heptamer occupancy by RAG-1 in the assembly of a V(D)J initiation complex. Mol. Cell. Biol., 19, 3674–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonegawa S. (1983) Somatic generation of antibody diversity. Nature, 302, 575–581. [DOI] [PubMed] [Google Scholar]
- van Gent D.C., McBlane,J.F., Ramsden,D.A., Sadofsky,M.J., Hesse,J.E. and Gellert,M. (1995) Initiation of V(D)J recombination in a cell-free system. Cell, 81, 925–934. [DOI] [PubMed] [Google Scholar]
- van Gent D.C., Mizuuchi,K. and Gellert,M. (1996a) Similarities between initiation of V(D)J recombination and retroviral integration. Science, 271, 1592–1594. [DOI] [PubMed] [Google Scholar]
- van Gent D.C., Ramsden,D.A. and Gellert,M. (1996b) The RAG1 and RAG2 proteins establish the 12/23 rule in V(D)J recombination. Cell, 85, 107–113. [DOI] [PubMed] [Google Scholar]
- van Gent D.C., Hiom,K., Paull,T.T. and Gellert,M. (1997) Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J., 16, 2665–2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villey I.J., Caillol,D., Selz,F., Ferrier,P. and de Villartay,J.P. (1996) Defect in rearrangement of the most 5′ TCR-J α following targeted deletion of T early α (TEA): implication for TCR α locus accessibility. Immunity, 5, 331–342. [DOI] [PubMed] [Google Scholar]
- Weinreich M.D., Mahnke-Braam,L. and Reznikoff,W.S. (1994) A functional analysis of the Tn5 transposase. Identification of domains required for DNA binding and multimerization. J. Mol. Biol., 241, 166–177. [DOI] [PubMed] [Google Scholar]
- West R.B. and Lieber,M.R. (1998) The RAG–HMG1 complex enforces the 12/23 rule of V(D)J recombination specifically at the double-hairpin formation step. Mol. Cell. Biol., 18, 6408–6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K. and Lieber,M.R. (2000) The nicking step in V(D)J recombination is independent of synapsis: implications for the immune repertoire. Mol. Cell. Biol., 20, 7914–7921. [DOI] [PMC free article] [PubMed] [Google Scholar]