Abstract
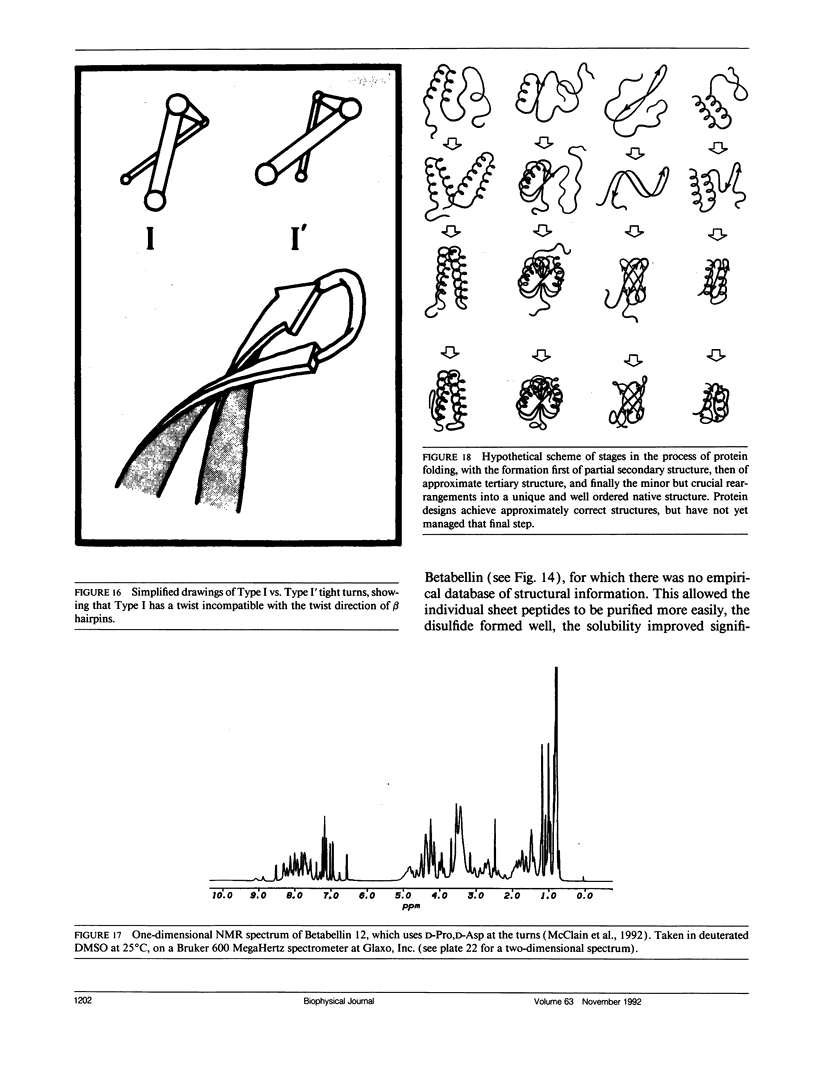
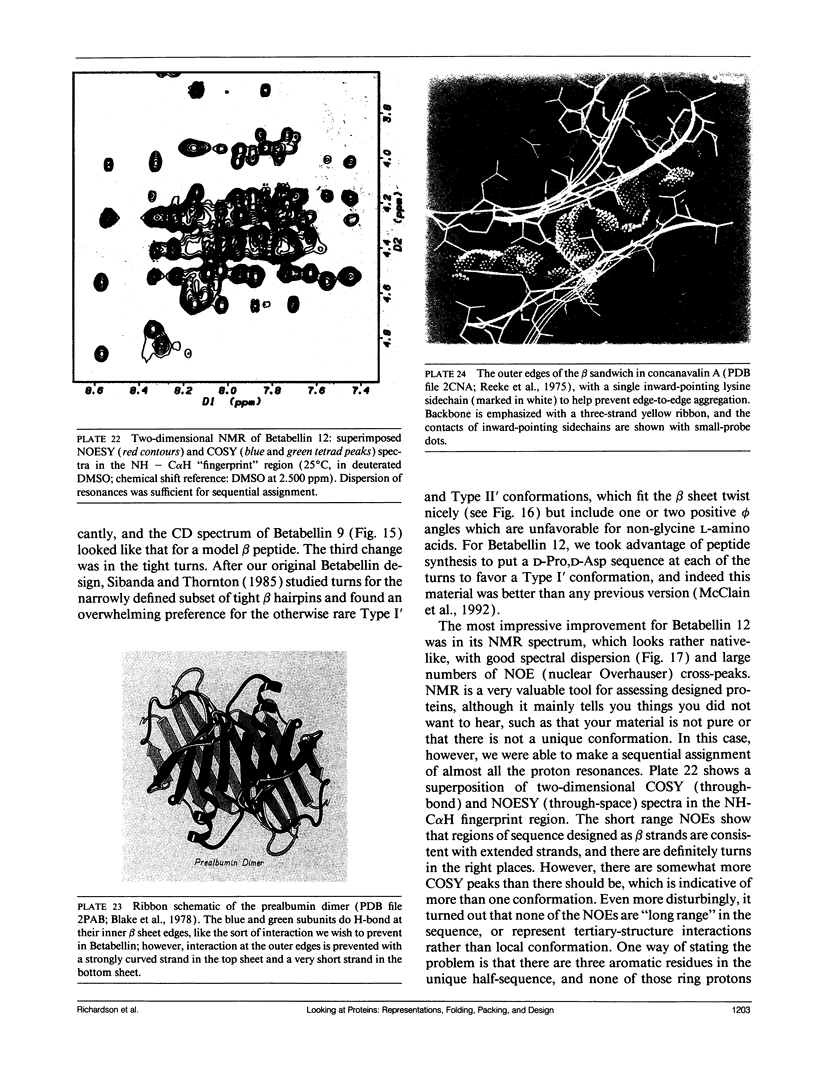
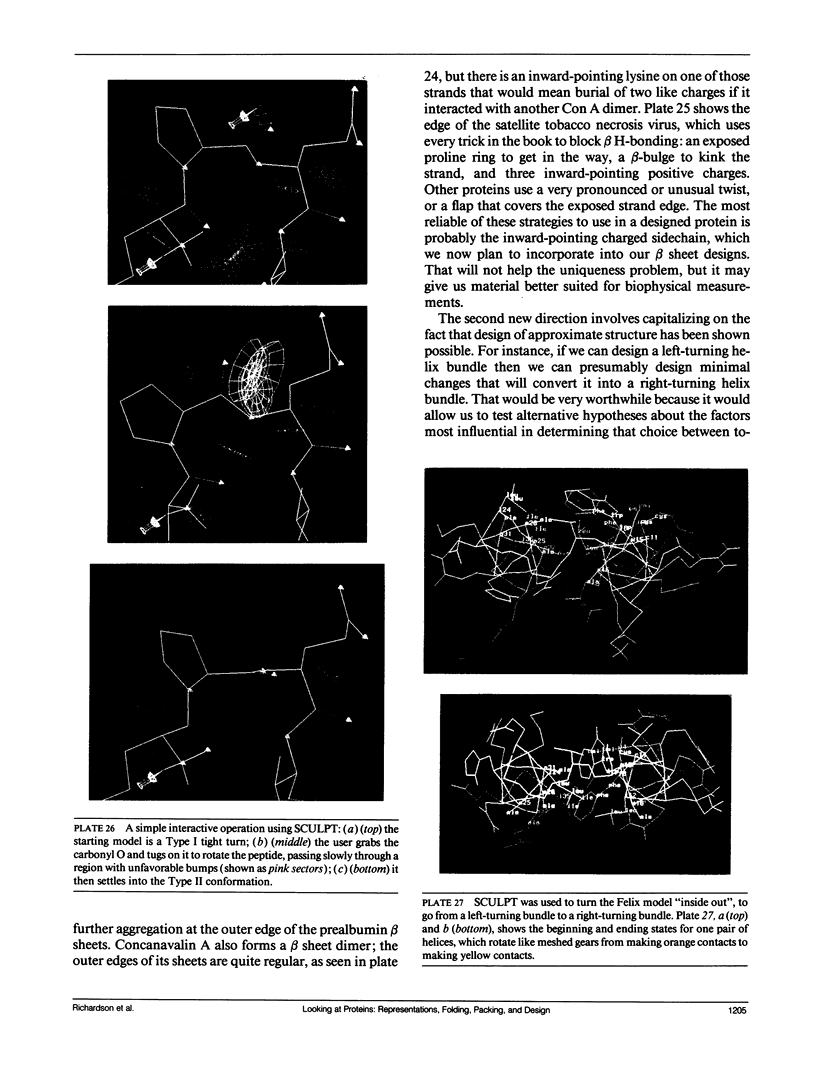
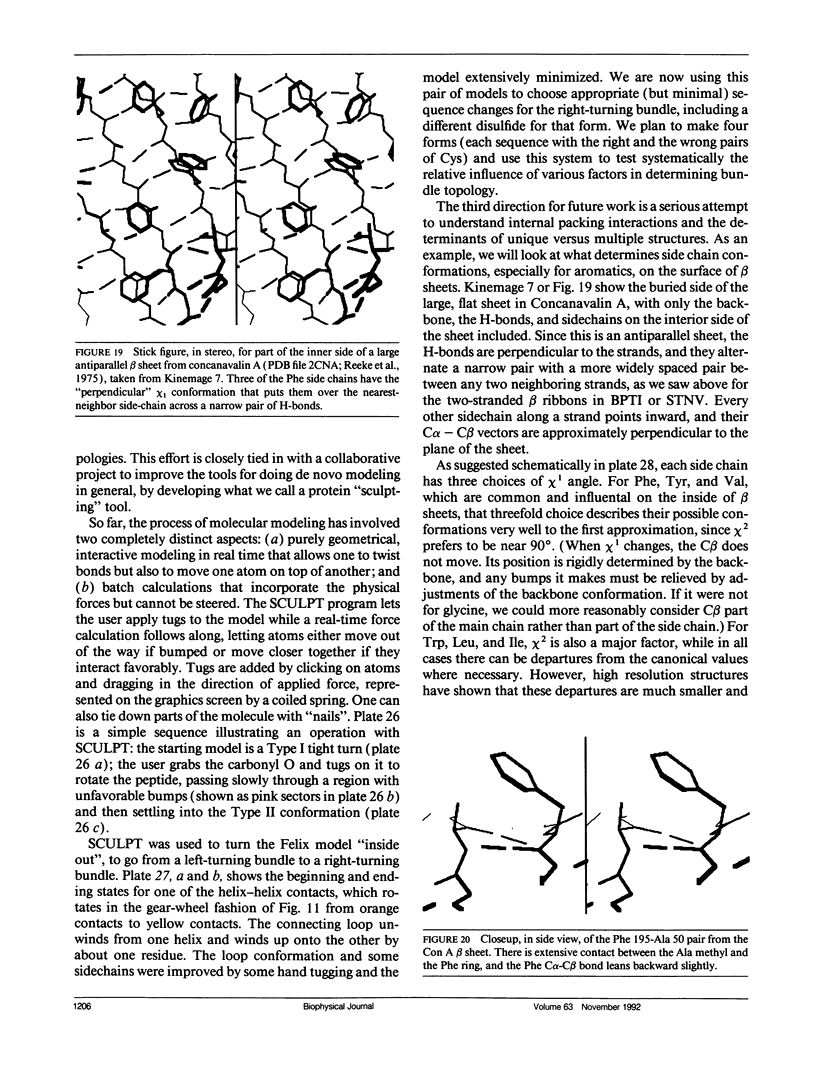
Looking at proteins is an active process of interpretation and selection, emphasizing some features and deleting others. Multiple representations are needed, for such purposes as showing motions or conveying both the chain connectivity and the three-dimensional shape simultaneously. In studying and comparing protein structures, ideas are suggested about the determinants of tertiary structure and of folding (e.g., that Greek key beta barrels may fold up two strands at a time). The design and synthesis of new proteins "from scratch" provides a route toward the experimental testing of such ideas. It has also been a fruitful new perspective from which to look at structures, requiring such things as statistics on very narrowly defined structural categories and explicit attention to "negative design" criteria that actively block unwanted alternatives (e.g., reverse topology of a helix bundle, or edge-to-edge aggregation of beta sheets). Recently, the field of protein design has produced a rather unexpected general result: apparently we do indeed know enough to successfully design proteins that fold into approximately correct structures, but not enough to design unique, native-like structures. The degree of order varies considerably, but even the best designed material shows multiple conformations by NMR, more similar to a "molten globule" folding intermediate than to a well ordered native tertiary structure. In response to this conclusion, we are now working on systems that test useful questions with approximate structures (such as determining which factors most influence the choice of helix-bundle topology) and also analyzing how natural proteins achieve unique core conformations (e.g., for side chains on the interior side of a beta sheet, illustrated in the kinemages).
Full text
PDF












Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
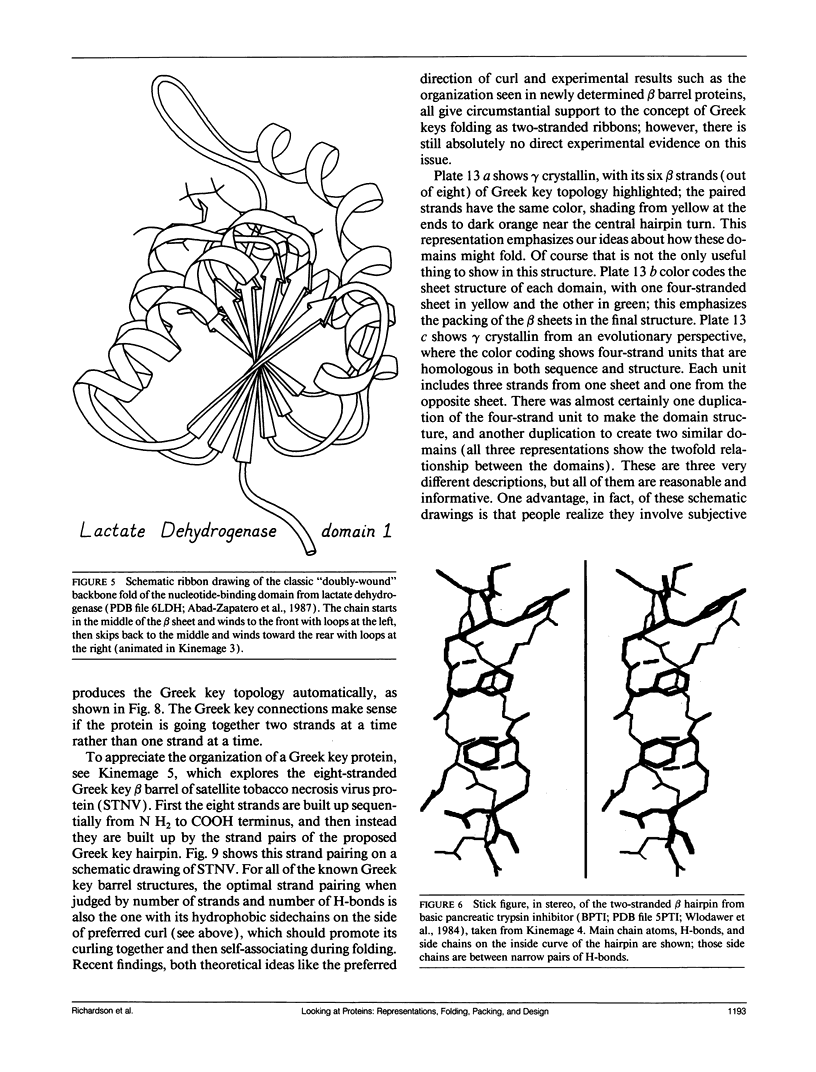
- Abad-Zapatero C., Griffith J. P., Sussman J. L., Rossmann M. G. Refined crystal structure of dogfish M4 apo-lactate dehydrogenase. J Mol Biol. 1987 Dec 5;198(3):445–467. doi: 10.1016/0022-2836(87)90293-2. [DOI] [PubMed] [Google Scholar]
- Arnone A., Bier C. J., Cotton F. A., Day V. W., Hazen E. E., Jr, Richardson D. C., Yonath A., Richardson J. S. A high resolution structure of an inhibitor complex of the extracellular nuclease of Staphylococcus aureus. I. Experimental procedures and chain tracing. J Biol Chem. 1971 Apr 10;246(7):2302–2316. [PubMed] [Google Scholar]
- Banner D. W., Bloomer A. C., Petsko G. A., Phillips D. C., Pogson C. I., Wilson I. A., Corran P. H., Furth A. J., Milman J. D., Offord R. E. Structure of chicken muscle triose phosphate isomerase determined crystallographically at 2.5 angstrom resolution using amino acid sequence data. Nature. 1975 Jun 19;255(5510):609–614. doi: 10.1038/255609a0. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Blake C. C., Geisow M. J., Oatley S. J., Rérat B., Rérat C. Structure of prealbumin: secondary, tertiary and quaternary interactions determined by Fourier refinement at 1.8 A. J Mol Biol. 1978 May 25;121(3):339–356. doi: 10.1016/0022-2836(78)90368-6. [DOI] [PubMed] [Google Scholar]
- Chothia C. Conformation of twisted beta-pleated sheets in proteins. J Mol Biol. 1973 Apr 5;75(2):295–302. doi: 10.1016/0022-2836(73)90022-3. [DOI] [PubMed] [Google Scholar]
- Connolly M. L. Solvent-accessible surfaces of proteins and nucleic acids. Science. 1983 Aug 19;221(4612):709–713. doi: 10.1126/science.6879170. [DOI] [PubMed] [Google Scholar]
- DeGrado W. F., Wasserman Z. R., Lear J. D. Protein design, a minimalist approach. Science. 1989 Feb 3;243(4891):622–628. doi: 10.1126/science.2464850. [DOI] [PubMed] [Google Scholar]
- Hecht M. H., Richardson J. S., Richardson D. C., Ogden R. C. De novo design, expression, and characterization of Felix: a four-helix bundle protein of native-like sequence. Science. 1990 Aug 24;249(4971):884–891. doi: 10.1126/science.2392678. [DOI] [PubMed] [Google Scholar]
- Jacobson R. H., Matsumura M., Faber H. R., Matthews B. W. Structure of a stabilizing disulfide bridge mutant that closes the active-site cleft of T4 lysozyme. Protein Sci. 1992 Jan;1(1):46–57. doi: 10.1002/pro.5560010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
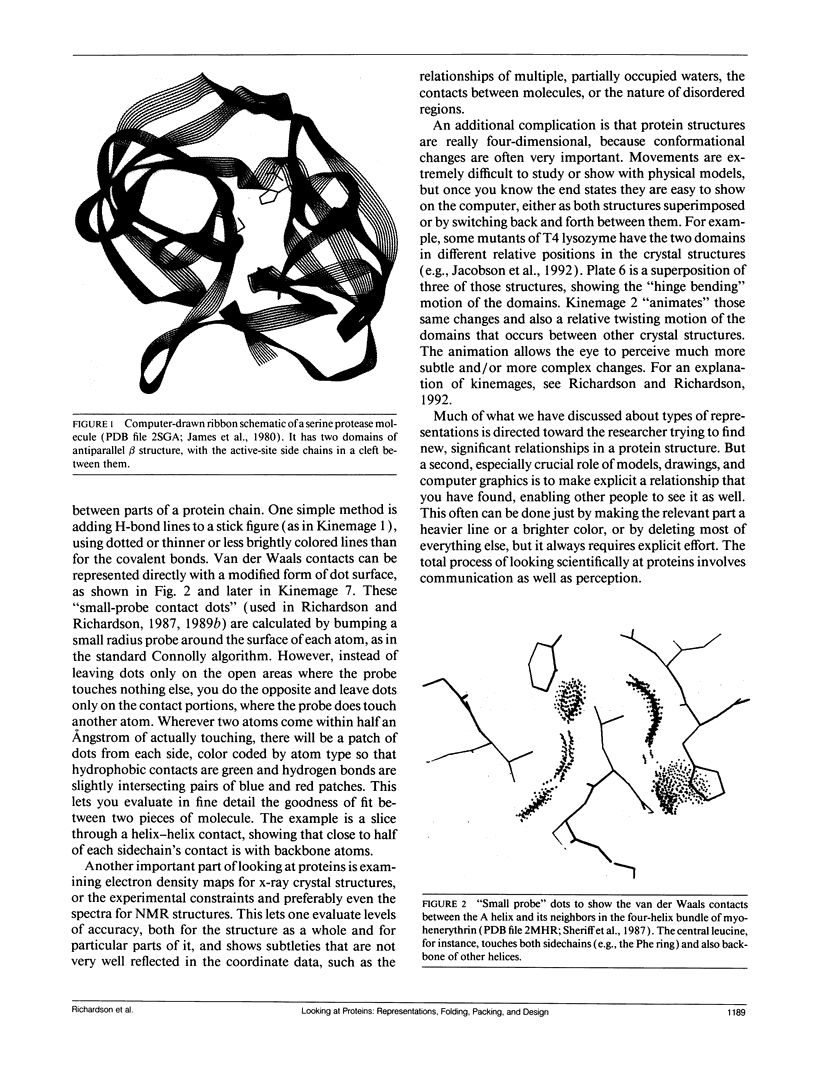
- James M. N., Sielecki A. R., Brayer G. D., Delbaere L. T., Bauer C. A. Structures of product and inhibitor complexes of Streptomyces griseus protease A at 1.8 A resolution. A model for serine protease catalysis. J Mol Biol. 1980 Nov 25;144(1):43–88. doi: 10.1016/0022-2836(80)90214-4. [DOI] [PubMed] [Google Scholar]
- Janin J., Wodak S. Conformation of amino acid side-chains in proteins. J Mol Biol. 1978 Nov 5;125(3):357–386. doi: 10.1016/0022-2836(78)90408-4. [DOI] [PubMed] [Google Scholar]
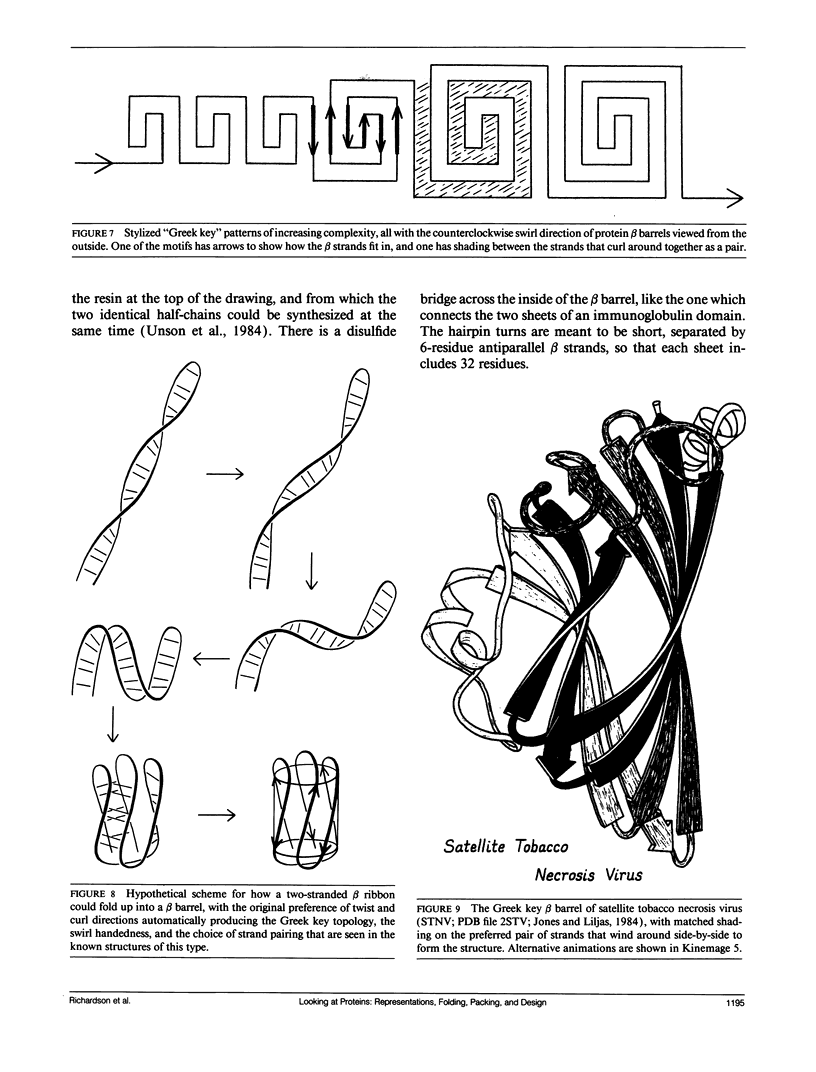
- Jones T. A., Liljas L. Structure of satellite tobacco necrosis virus after crystallographic refinement at 2.5 A resolution. J Mol Biol. 1984 Aug 25;177(4):735–767. doi: 10.1016/0022-2836(84)90047-0. [DOI] [PubMed] [Google Scholar]
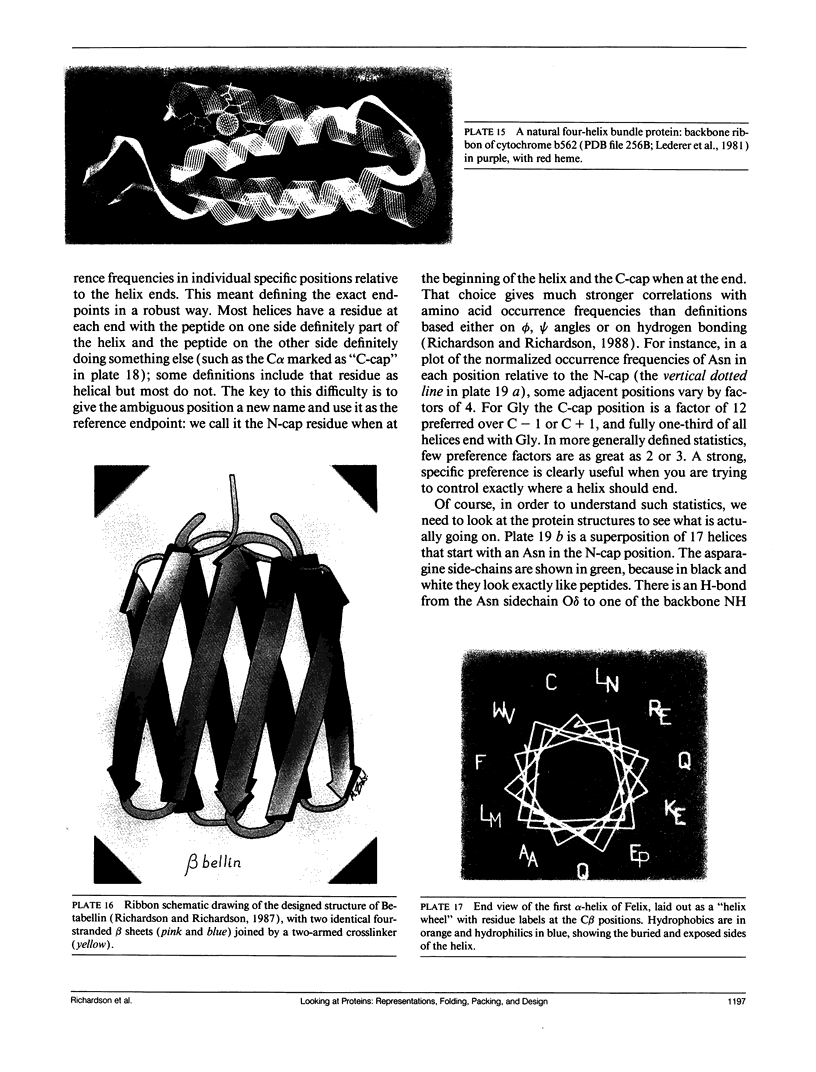
- Lederer F., Glatigny A., Bethge P. H., Bellamy H. D., Matthew F. S. Improvement of the 2.5 A resolution model of cytochrome b562 by redetermining the primary structure and using molecular graphics. J Mol Biol. 1981 Jun 5;148(4):427–448. doi: 10.1016/0022-2836(81)90185-6. [DOI] [PubMed] [Google Scholar]
- Ohgushi M., Wada A. 'Molten-globule state': a compact form of globular proteins with mobile side-chains. FEBS Lett. 1983 Nov 28;164(1):21–24. doi: 10.1016/0014-5793(83)80010-6. [DOI] [PubMed] [Google Scholar]
- Ponder J. W., Richards F. M. Tertiary templates for proteins. Use of packing criteria in the enumeration of allowed sequences for different structural classes. J Mol Biol. 1987 Feb 20;193(4):775–791. doi: 10.1016/0022-2836(87)90358-5. [DOI] [PubMed] [Google Scholar]
- Rao S. T., Rossmann M. G. Comparison of super-secondary structures in proteins. J Mol Biol. 1973 May 15;76(2):241–256. doi: 10.1016/0022-2836(73)90388-4. [DOI] [PubMed] [Google Scholar]
- Reeke G. N., Jr, Becker J. W., Edelman G. M. The covalent and three-dimensional structure of concanavalin A. IV. Atomic coordinates, hydrogen bonding, and quaternary structure. J Biol Chem. 1975 Feb 25;250(4):1525–1547. [PubMed] [Google Scholar]
- Richardson D. C., Richardson J. S. The kinemage: a tool for scientific communication. Protein Sci. 1992 Jan;1(1):3–9. doi: 10.1002/pro.5560010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson J. S. Handedness of crossover connections in beta sheets. Proc Natl Acad Sci U S A. 1976 Aug;73(8):2619–2623. doi: 10.1073/pnas.73.8.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson J. S., Richardson D. C. Amino acid preferences for specific locations at the ends of alpha helices. Science. 1988 Jun 17;240(4859):1648–1652. doi: 10.1126/science.3381086. [DOI] [PubMed] [Google Scholar]
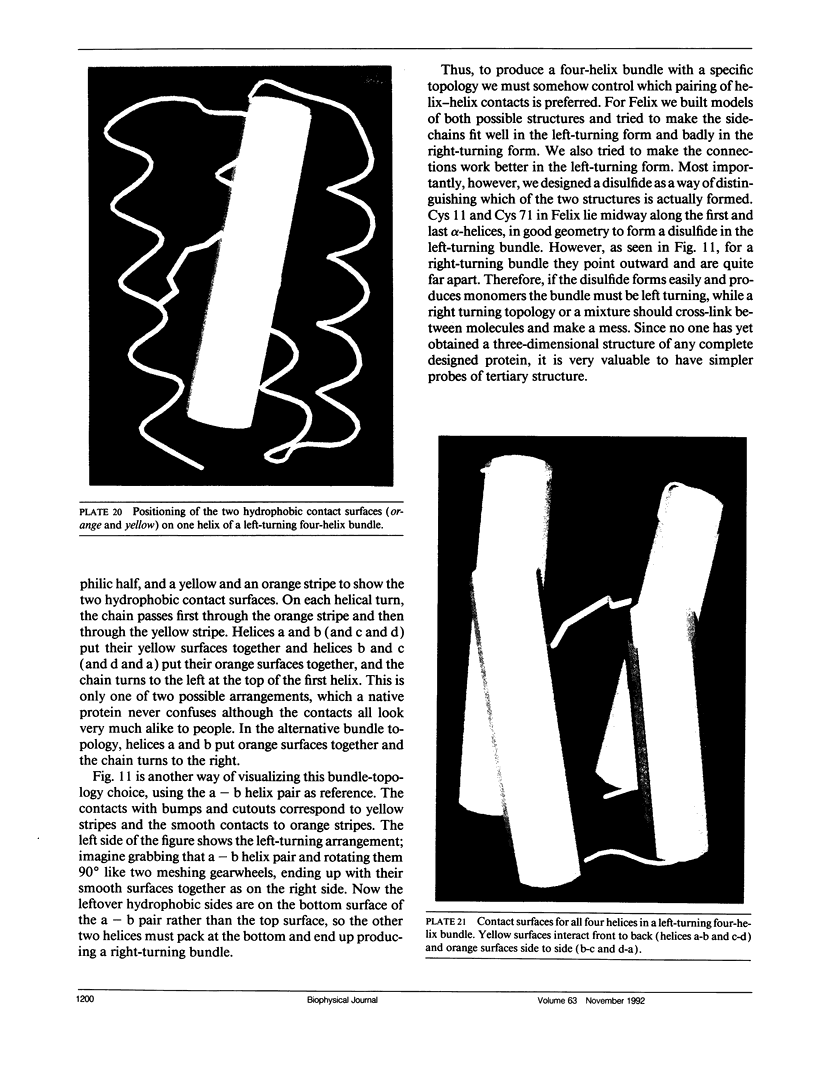
- Richardson J. S., Richardson D. C. The de novo design of protein structures. Trends Biochem Sci. 1989 Jul;14(7):304–309. doi: 10.1016/0968-0004(89)90070-4. [DOI] [PubMed] [Google Scholar]
- Richardson J. S. Schematic drawings of protein structures. Methods Enzymol. 1985;115:359–380. doi: 10.1016/0076-6879(85)15026-3. [DOI] [PubMed] [Google Scholar]
- Richardson J. S. The anatomy and taxonomy of protein structure. Adv Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- Richardson J. S. beta-Sheet topology and the relatedness of proteins. Nature. 1977 Aug 11;268(5620):495–500. doi: 10.1038/268495a0. [DOI] [PubMed] [Google Scholar]
- Rubin B., Richardson J. S. The simple construction of protein alpha-carbon models. Biopolymers. 1972;11(11):2381–2385. doi: 10.1002/bip.1972.360111116. [DOI] [PubMed] [Google Scholar]
- Salemme F. R. Structural properties of protein beta-sheets. Prog Biophys Mol Biol. 1983;42(2-3):95–133. doi: 10.1016/0079-6107(83)90005-6. [DOI] [PubMed] [Google Scholar]
- Shoemaker K. R., Kim P. S., York E. J., Stewart J. M., Baldwin R. L. Tests of the helix dipole model for stabilization of alpha-helices. Nature. 1987 Apr 9;326(6113):563–567. doi: 10.1038/326563a0. [DOI] [PubMed] [Google Scholar]
- Sibanda B. L., Thornton J. M. Beta-hairpin families in globular proteins. Nature. 1985 Jul 11;316(6024):170–174. doi: 10.1038/316170a0. [DOI] [PubMed] [Google Scholar]
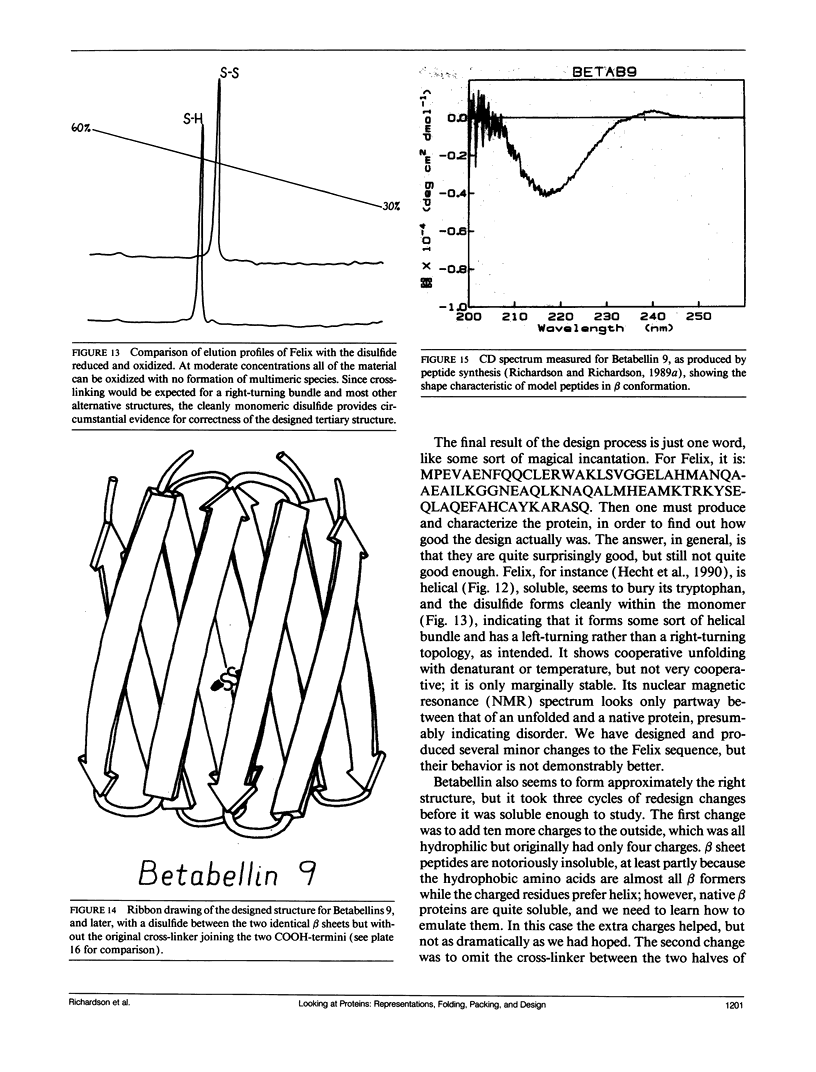
- Tainer J. A., Getzoff E. D., Beem K. M., Richardson J. S., Richardson D. C. Determination and analysis of the 2 A-structure of copper, zinc superoxide dismutase. J Mol Biol. 1982 Sep 15;160(2):181–217. doi: 10.1016/0022-2836(82)90174-7. [DOI] [PubMed] [Google Scholar]
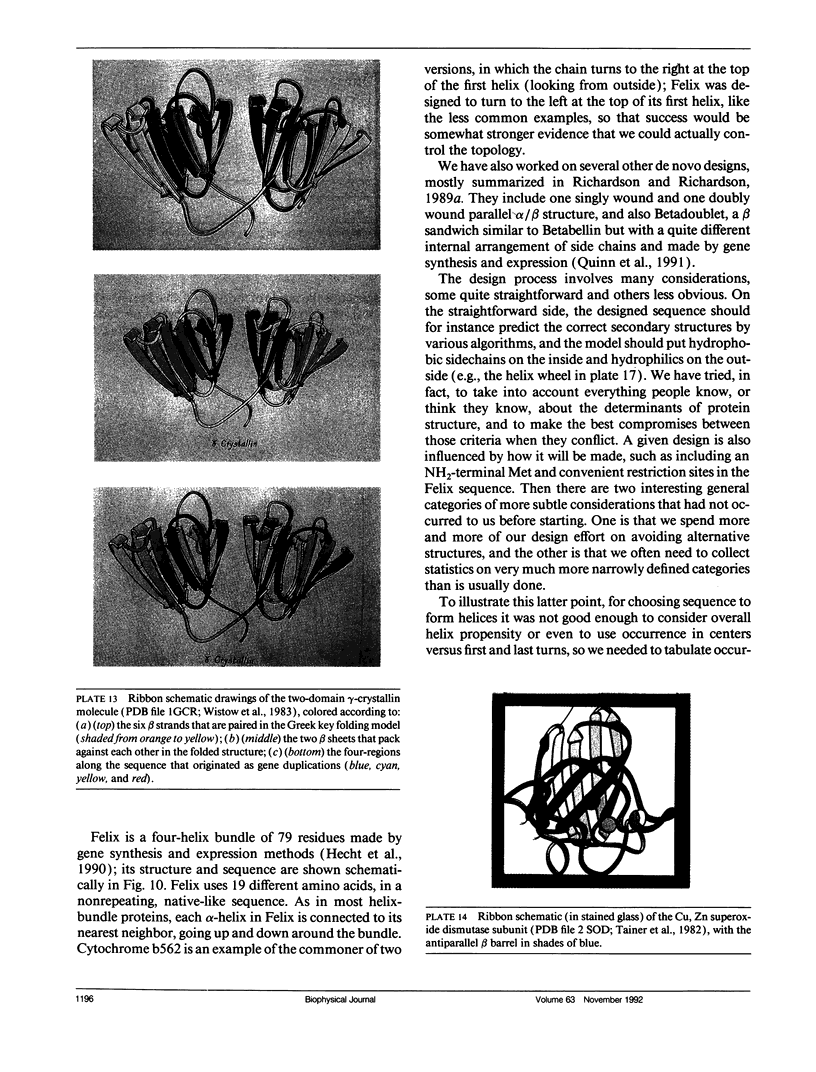
- Wistow G., Turnell B., Summers L., Slingsby C., Moss D., Miller L., Lindley P., Blundell T. X-ray analysis of the eye lens protein gamma-II crystallin at 1.9 A resolution. J Mol Biol. 1983 Oct 15;170(1):175–202. doi: 10.1016/s0022-2836(83)80232-0. [DOI] [PubMed] [Google Scholar]
- Wlodawer A., Sjölin L. Structure of ribonuclease A: results of joint neutron and X-ray refinement at 2.0-A resolution. Biochemistry. 1983 May 24;22(11):2720–2728. doi: 10.1021/bi00280a021. [DOI] [PubMed] [Google Scholar]
- Wlodawer A., Walter J., Huber R., Sjölin L. Structure of bovine pancreatic trypsin inhibitor. Results of joint neutron and X-ray refinement of crystal form II. J Mol Biol. 1984 Dec 5;180(2):301–329. doi: 10.1016/s0022-2836(84)80006-6. [DOI] [PubMed] [Google Scholar]