Abstract
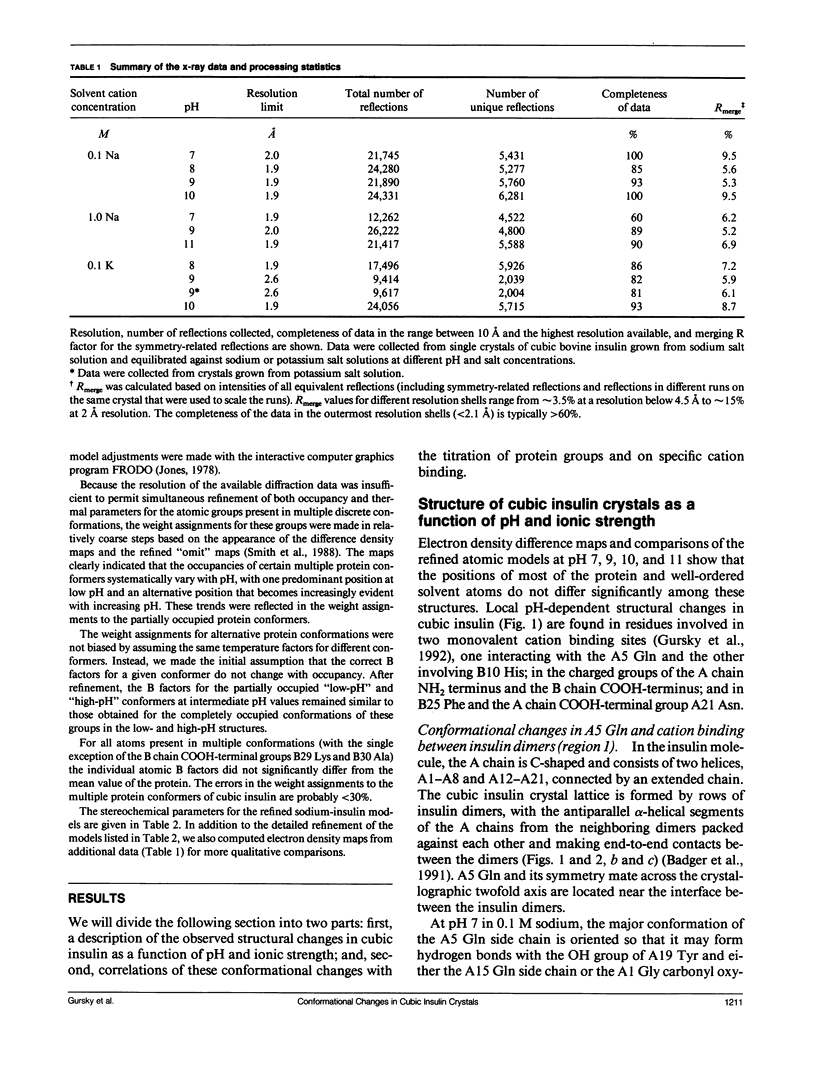
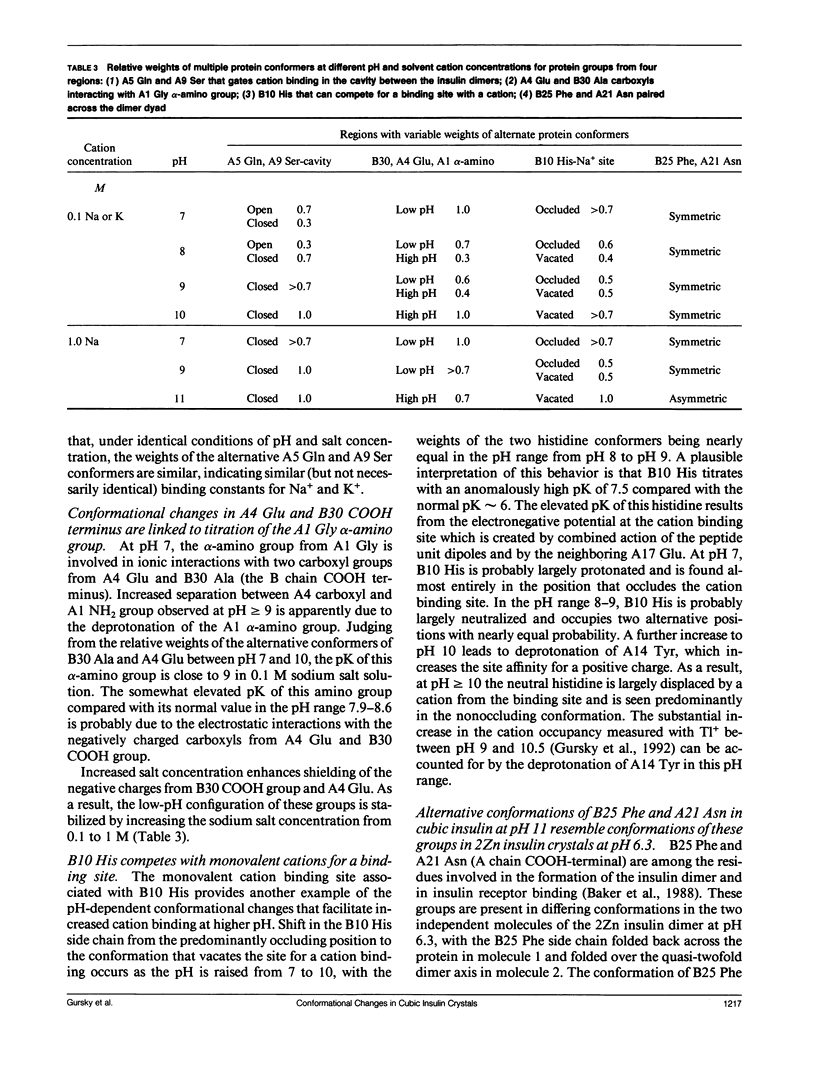
To determine the effect of variations in the charge distribution on the conformation of a protein molecule, we have solved the structures of bovine cubic insulin over a pH range from 7 to 11 in 0.1 M and 1 M sodium salt solutions. The x-ray data were collected beyond 2-A resolution and the R factors for the refined models ranged from 0.16 to 0.20. Whereas the positions of most protein and well-ordered solvent atoms are conserved, about 30% of residues alter their predominant conformation as the pH is changed. Conformational switching of A5 Gln and B10 His correlates with the pH dependence of monovalent cation binding to insulin in cubic crystals. Shifts in the relative positions of the A chain NH2-terminal and B chain COOH-terminal groups are probably due to titration of the A1 alpha-amino group. Two alternative positions of B25 Phe and A21 Asn observed in cubic insulin at pH 11 are similar to those found in two independent molecules of the 2Zn insulin dimer at pH 6.4. The conformational changes of the insulin amino acids appear to be only loosely coupled at distant protein sites. Shifts in the equilibrium between distinct conformational substates as the charge distribution on the protein is altered are analogous to the electrostatically triggered movements that occur in many functional protein reactions.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Badger J., Caspar D. L. Water structure in cubic insulin crystals. Proc Natl Acad Sci U S A. 1991 Jan 15;88(2):622–626. doi: 10.1073/pnas.88.2.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger J. Flexibility in crystalline insulins. Biophys J. 1992 Mar;61(3):816–819. doi: 10.1016/S0006-3495(92)81886-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger J., Harris M. R., Reynolds C. D., Evans A. C., Dodson E. J., Dodson G. G., North A. C. Structure of the pig insulin dimer in the cubic crystal. Acta Crystallogr B. 1991 Feb 1;47(Pt 1):127–136. doi: 10.1107/s0108768190009570. [DOI] [PubMed] [Google Scholar]
- Baker E. N., Blundell T. L., Cutfield J. F., Cutfield S. M., Dodson E. J., Dodson G. G., Hodgkin D. M., Hubbard R. E., Isaacs N. W., Reynolds C. D. The structure of 2Zn pig insulin crystals at 1.5 A resolution. Philos Trans R Soc Lond B Biol Sci. 1988 Jul 6;319(1195):369–456. doi: 10.1098/rstb.1988.0058. [DOI] [PubMed] [Google Scholar]
- Chothia C., Lesk A. M., Dodson G. G., Hodgkin D. C. Transmission of conformational change in insulin. Nature. 1983 Apr 7;302(5908):500–505. doi: 10.1038/302500a0. [DOI] [PubMed] [Google Scholar]
- Derewenda U., Derewenda Z., Dodson E. J., Dodson G. G., Bing X., Markussen J. X-ray analysis of the single chain B29-A1 peptide-linked insulin molecule. A completely inactive analogue. J Mol Biol. 1991 Jul 20;220(2):425–433. doi: 10.1016/0022-2836(91)90022-x. [DOI] [PubMed] [Google Scholar]
- Dixon M. M., Brennan R. G., Matthews B. W. Structure of gamma-chymotrypsin in the range pH 2.0 to pH 10.5 suggests that gamma-chymotrypsin is a covalent acyl-enzyme adduct at low pH. Int J Biol Macromol. 1991 Apr;13(2):89–96. doi: 10.1016/0141-8130(91)90054-x. [DOI] [PubMed] [Google Scholar]
- Dodson E. J., Dodson G. G., Lewitova A., Sabesan M. Zinc-free cubic pig insulin: crystallization and structure determination. J Mol Biol. 1978 Nov 5;125(3):387–396. doi: 10.1016/0022-2836(78)90409-6. [DOI] [PubMed] [Google Scholar]
- Eisenman G., Dani J. A. An introduction to molecular architecture and permeability of ion channels. Annu Rev Biophys Biophys Chem. 1987;16:205–226. doi: 10.1146/annurev.bb.16.060187.001225. [DOI] [PubMed] [Google Scholar]
- Fox R. O., Jr, Richards F. M. A voltage-gated ion channel model inferred from the crystal structure of alamethicin at 1.5-A resolution. Nature. 1982 Nov 25;300(5890):325–330. doi: 10.1038/300325a0. [DOI] [PubMed] [Google Scholar]
- Gursky O., Li Y., Badger J., Caspar D. L. Monovalent cation binding to cubic insulin crystals. Biophys J. 1992 Mar;61(3):604–611. doi: 10.1016/S0006-3495(92)81865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guss J. M., Harrowell P. R., Murata M., Norris V. A., Freeman H. C. Crystal structure analyses of reduced (CuI) poplar plastocyanin at six pH values. J Mol Biol. 1986 Nov 20;192(2):361–387. doi: 10.1016/0022-2836(86)90371-2. [DOI] [PubMed] [Google Scholar]
- Hendrickson W. A. Stereochemically restrained refinement of macromolecular structures. Methods Enzymol. 1985;115:252–270. doi: 10.1016/0076-6879(85)15021-4. [DOI] [PubMed] [Google Scholar]
- Hua Q. X., Shoelson S. E., Kochoyan M., Weiss M. A. Receptor binding redefined by a structural switch in a mutant human insulin. Nature. 1991 Nov 21;354(6350):238–241. doi: 10.1038/354238a0. [DOI] [PubMed] [Google Scholar]
- Karle I. L., Flippen-Anderson J. L., Agarwalla S., Balaram P. Crystal structure of [Leu1]zervamicin, a membrane ion-channel peptide: implications for gating mechanisms. Proc Natl Acad Sci U S A. 1991 Jun 15;88(12):5307–5311. doi: 10.1073/pnas.88.12.5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nar H., Messerschmidt A., Huber R., van de Kamp M., Canters G. W. Crystal structure analysis of oxidized Pseudomonas aeruginosa azurin at pH 5.5 and pH 9.0. A pH-induced conformational transition involves a peptide bond flip. J Mol Biol. 1991 Oct 5;221(3):765–772. doi: 10.1016/0022-2836(91)80173-r. [DOI] [PubMed] [Google Scholar]
- Richardson D. C., Richardson J. S. The kinemage: a tool for scientific communication. Protein Sci. 1992 Jan;1(1):3–9. doi: 10.1002/pro.5560010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoelson S. E., Lu Z. X., Parlautan L., Lynch C. S., Weiss M. A. Mutations at the dimer, hexamer, and receptor-binding surfaces of insulin independently affect insulin-insulin and insulin-receptor interactions. Biochemistry. 1992 Feb 18;31(6):1757–1767. doi: 10.1021/bi00121a025. [DOI] [PubMed] [Google Scholar]
- Smith J. L., Corfield P. W., Hendrickson W. A., Low B. W. Refinement at 1.4 A resolution of a model of erabutoxin b: treatment of ordered solvent and discrete disorder. Acta Crystallogr A. 1988 May 1;44(Pt 3):357–368. doi: 10.1107/s0108767388000303. [DOI] [PubMed] [Google Scholar]
- Smith J. L., Hendrickson W. A., Honzatko R. B., Sheriff S. Structural heterogeneity in protein crystals. Biochemistry. 1986 Sep 9;25(18):5018–5027. doi: 10.1021/bi00366a008. [DOI] [PubMed] [Google Scholar]
- Svensson L. A., Sjölin L., Gilliland G. L., Finzel B. C., Wlodawer A. Multiple conformations of amino acid residues in ribonuclease A. Proteins. 1986 Dec;1(4):370–375. doi: 10.1002/prot.340010410. [DOI] [PubMed] [Google Scholar]




