Abstract
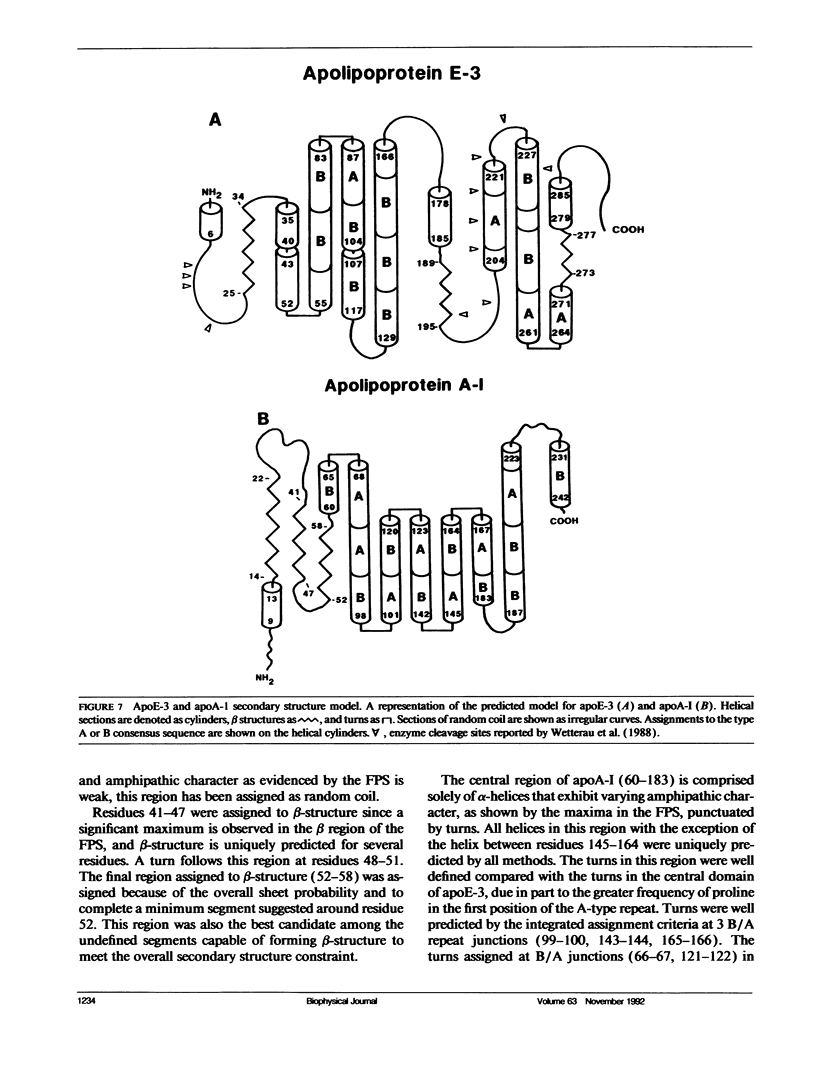
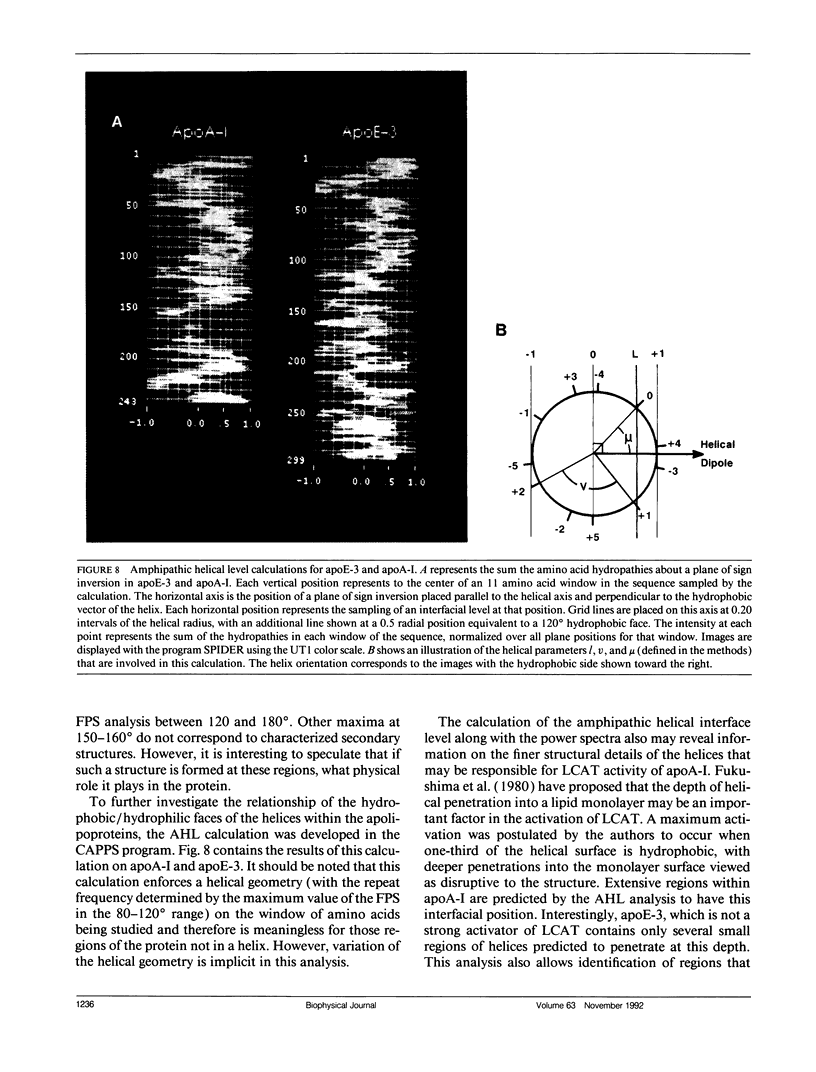
The primary and secondary structure of human plasma apolipoprotein A-I and apolipoprotein E-3 have been analyzed to further our understanding of the secondary and tertiary conformation of these proteins and the structure and function of plasma lipoprotein particles. The methods used to analyze the primary sequence of these proteins used computer programs: (a) to identify repeated patterns within these proteins on the basis of conservative substitutions and similarities within the physicochemical properties of each residue; (b) for local averaging, hydrophobic moment, and Fourier analysis of the physicochemical properties; and (c) for secondary structure prediction of each protein carried out using homology, statistical, and information theory based methods. Circular dichroism was used to study purified lipid-protein complexes of each protein and quantitate the secondary structure in a lipid environment. The data from these analyses were integrated into a single secondary structure prediction to derive a model of each protein. The sequence homology within apolipoproteins A-I, E-3, and A-IV is used to derive a consensus sequence for two 11 amino acid repeating sequences in this family of proteins.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anantharamaiah G. M., Venkatachalapathi Y. V., Brouillette C. G., Segrest J. P. Use of synthetic peptide analogues to localize lecithin:cholesterol acyltransferase activating domain in apolipoprotein A-I. Arteriosclerosis. 1990 Jan-Feb;10(1):95–105. doi: 10.1161/01.atv.10.1.95. [DOI] [PubMed] [Google Scholar]
- Andrews A. L., Atkinson D., Barratt M. D., Finer E. G., Hauser H., Henry R., Leslie R. B., Owens N. L., Phillips M. C., Robertson R. N. Interaction of apoprotein from porcine high-density lipoprotein with dimyristoly lecithin. 2. Nature of lipid-protein interaction. Eur J Biochem. 1976 May 1;64(2):549–563. doi: 10.1111/j.1432-1033.1976.tb10335.x. [DOI] [PubMed] [Google Scholar]
- Argos P., Hanei M., Wilson J. M., Kelley W. N. A possible nucleotide-binding domain in the tertiary fold of phosphoribosyltransferases. J Biol Chem. 1983 May 25;258(10):6450–6457. [PubMed] [Google Scholar]
- Atkinson D., Small D. M. Recombinant lipoproteins: implications for structure and assembly of native lipoproteins. Annu Rev Biophys Biophys Chem. 1986;15:403–456. doi: 10.1146/annurev.bb.15.060186.002155. [DOI] [PubMed] [Google Scholar]
- Biou V., Gibrat J. F., Levin J. M., Robson B., Garnier J. Secondary structure prediction: combination of three different methods. Protein Eng. 1988 Sep;2(3):185–191. doi: 10.1093/protein/2.3.185. [DOI] [PubMed] [Google Scholar]
- Boguski M. S., Freeman M., Elshourbagy N. A., Taylor J. M., Gordon J. I. On computer-assisted analysis of biological sequences: proline punctuation, consensus sequences, and apolipoprotein repeats. J Lipid Res. 1986 Oct;27(10):1011–1034. [PubMed] [Google Scholar]
- Bordo D., Argos P. Evolution of protein cores. Constraints in point mutations as observed in globin tertiary structures. J Mol Biol. 1990 Feb 20;211(4):975–988. doi: 10.1016/0022-2836(90)90087-3. [DOI] [PubMed] [Google Scholar]
- Bordo D., Argos P. Suggestions for "safe" residue substitutions in site-directed mutagenesis. J Mol Biol. 1991 Feb 20;217(4):721–729. doi: 10.1016/0022-2836(91)90528-e. [DOI] [PubMed] [Google Scholar]
- Brasseur R., De Meutter J., Vanloo B., Goormaghtigh E., Ruysschaert J. M., Rosseneu M. Mode of assembly of amphipathic helical segments in model high-density lipoproteins. Biochim Biophys Acta. 1990 Apr 17;1043(3):245–252. doi: 10.1016/0005-2760(90)90023-q. [DOI] [PubMed] [Google Scholar]
- Breiter D. R., Kanost M. R., Benning M. M., Wesenberg G., Law J. H., Wells M. A., Rayment I., Holden H. M. Molecular structure of an apolipoprotein determined at 2.5-A resolution. Biochemistry. 1991 Jan 22;30(3):603–608. doi: 10.1021/bi00217a002. [DOI] [PubMed] [Google Scholar]
- Brewer H. B., Jr, Fairwell T., LaRue A., Ronan R., Houser A., Bronzert T. J. The amino acid sequence of human APOA-I, an apolipoprotein isolated from high density lipoproteins. Biochem Biophys Res Commun. 1978 Feb 14;80(3):623–630. doi: 10.1016/0006-291x(78)91614-5. [DOI] [PubMed] [Google Scholar]
- Brouillette C. G., Jones J. L., Ng T. C., Kercret H., Chung B. H., Segrest J. P. Structural studies of apolipoprotein A-I/phosphatidylcholine recombinants by high-field proton NMR, nondenaturing gradient gel electrophoresis, and electron microscopy. Biochemistry. 1984 Jan 17;23(2):359–367. doi: 10.1021/bi00297a027. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Cladaras C., Hadzopoulou-Cladaras M., Nolte R. T., Atkinson D., Zannis V. I. The complete sequence and structural analysis of human apolipoprotein B-100: relationship between apoB-100 and apoB-48 forms. EMBO J. 1986 Dec 20;5(13):3495–3507. doi: 10.1002/j.1460-2075.1986.tb04675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornette J. L., Cease K. B., Margalit H., Spouge J. L., Berzofsky J. A., DeLisi C. Hydrophobicity scales and computational techniques for detecting amphipathic structures in proteins. J Mol Biol. 1987 Jun 5;195(3):659–685. doi: 10.1016/0022-2836(87)90189-6. [DOI] [PubMed] [Google Scholar]
- Dayhoff M. O., Barker W. C., Hunt L. T. Establishing homologies in protein sequences. Methods Enzymol. 1983;91:524–545. doi: 10.1016/s0076-6879(83)91049-2. [DOI] [PubMed] [Google Scholar]
- De Loof H., Rosseneu M., Brasseur R., Ruysschaert J. M. Functional differentiation of amphiphilic helices of the apolipoproteins by hydrophobic moment analysis. Biochim Biophys Acta. 1987 Jan 5;911(1):45–52. doi: 10.1016/0167-4838(87)90268-8. [DOI] [PubMed] [Google Scholar]
- De Loof H., Rosseneu M., Brasseur R., Ruysschaert J. M. Use of hydrophobicity profiles to predict receptor binding domains on apolipoprotein E and the low density lipoprotein apolipoprotein B-E receptor. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2295–2299. doi: 10.1073/pnas.83.8.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D., Schwarz E., Komaromy M., Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol. 1984 Oct 15;179(1):125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- Eisenberg D. Three-dimensional structure of membrane and surface proteins. Annu Rev Biochem. 1984;53:595–623. doi: 10.1146/annurev.bi.53.070184.003115. [DOI] [PubMed] [Google Scholar]
- Eisenberg D., Weiss R. M., Terwilliger T. C. The helical hydrophobic moment: a measure of the amphiphilicity of a helix. Nature. 1982 Sep 23;299(5881):371–374. doi: 10.1038/299371a0. [DOI] [PubMed] [Google Scholar]
- Fielding C. J., Shore V. G., Fielding P. E. A protein cofactor of lecithin:cholesterol acyltransferase. Biochem Biophys Res Commun. 1972 Feb 25;46(4):1493–1498. doi: 10.1016/0006-291x(72)90776-0. [DOI] [PubMed] [Google Scholar]
- Finer-Moore J., Stroud R. M. Amphipathic analysis and possible formation of the ion channel in an acetylcholine receptor. Proc Natl Acad Sci U S A. 1984 Jan;81(1):155–159. doi: 10.1073/pnas.81.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima D., Yokoyama S., Kroon D. J., Kézdy F. J., Kaiser E. T. Chain length-function correlation of amphiphilic peptides. Synthesis and surface properties of a tetratetracontapeptide segment of apolipoprotein A-I. J Biol Chem. 1980 Nov 25;255(22):10651–10657. [PubMed] [Google Scholar]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978 Mar 25;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Gascuel O., Golmard J. L. A simple method for predicting the secondary structure of globular proteins: implications and accuracy. Comput Appl Biosci. 1988 Aug;4(3):357–365. doi: 10.1093/bioinformatics/4.3.357. [DOI] [PubMed] [Google Scholar]
- Gibrat J. F., Garnier J., Robson B. Further developments of protein secondary structure prediction using information theory. New parameters and consideration of residue pairs. J Mol Biol. 1987 Dec 5;198(3):425–443. doi: 10.1016/0022-2836(87)90292-0. [DOI] [PubMed] [Google Scholar]
- Greenfield N., Fasman G. D. Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry. 1969 Oct;8(10):4108–4116. doi: 10.1021/bi00838a031. [DOI] [PubMed] [Google Scholar]
- Hopp T. P., Woods K. R. Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas A., Kézdy K. E., Wald J. H. Defined apolipoprotein A-I conformations in reconstituted high density lipoprotein discs. J Biol Chem. 1989 Mar 25;264(9):4818–4824. [PubMed] [Google Scholar]
- Jones D. D. Amino acid properties and side-chain orientation in proteins: a cross correlation appraoch. J Theor Biol. 1975 Mar;50(1):167–183. doi: 10.1016/0022-5193(75)90031-4. [DOI] [PubMed] [Google Scholar]
- Karathanasis S. K., Yunis I., Zannis V. I. Structure, evolution, and tissue-specific synthesis of human apolipoprotein AIV. Biochemistry. 1986 Jul 1;25(13):3962–3970. doi: 10.1021/bi00361a034. [DOI] [PubMed] [Google Scholar]
- Karathanasis S. K., Zannis V. I., Breslow J. L. Isolation and characterization of the human apolipoprotein A-I gene. Proc Natl Acad Sci U S A. 1983 Oct;80(20):6147–6151. doi: 10.1073/pnas.80.20.6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Levin J. M., Robson B., Garnier J. An algorithm for secondary structure determination in proteins based on sequence similarity. FEBS Lett. 1986 Sep 15;205(2):303–308. doi: 10.1016/0014-5793(86)80917-6. [DOI] [PubMed] [Google Scholar]
- Levitt M. Conformational preferences of amino acids in globular proteins. Biochemistry. 1978 Oct 3;17(20):4277–4285. doi: 10.1021/bi00613a026. [DOI] [PubMed] [Google Scholar]
- Li W. H., Tanimura M., Luo C. C., Datta S., Chan L. The apolipoprotein multigene family: biosynthesis, structure, structure-function relationships, and evolution. J Lipid Res. 1988 Mar;29(3):245–271. [PubMed] [Google Scholar]
- Luo C. C., Li W. H., Moore M. N., Chan L. Structure and evolution of the apolipoprotein multigene family. J Mol Biol. 1986 Feb 5;187(3):325–340. doi: 10.1016/0022-2836(86)90436-5. [DOI] [PubMed] [Google Scholar]
- Lux S. E., Hirz R., Shrager R. I., Gotto A. M. The influence of lipid on the conformation of human plasma high density apolipoproteins. J Biol Chem. 1972 Apr 25;247(8):2598–2606. [PubMed] [Google Scholar]
- Mahley R. W., Hui D. Y., Innerarity T. L., Weisgraber K. H. Two independent lipoprotein receptors on hepatic membranes of dog, swine, and man. Apo-B,E and apo-E receptors. J Clin Invest. 1981 Nov;68(5):1197–1206. doi: 10.1172/JCI110365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahley R. W., Innerarity T. L., Rall S. C., Jr, Weisgraber K. H. Plasma lipoproteins: apolipoprotein structure and function. J Lipid Res. 1984 Dec 1;25(12):1277–1294. [PubMed] [Google Scholar]
- Mao D., Wallace B. A. Differential light scattering and absorption flattening optical effects are minimal in the circular dichroism spectra of small unilamellar vesicles. Biochemistry. 1984 Jun 5;23(12):2667–2673. doi: 10.1021/bi00307a020. [DOI] [PubMed] [Google Scholar]
- Marcel Y. L., Provost P. R., Koa H., Raffai E., Dac N. V., Fruchart J. C., Rassart E. The epitopes of apolipoprotein A-I define distinct structural domains including a mobile middle region. J Biol Chem. 1991 Feb 25;266(6):3644–3653. [PubMed] [Google Scholar]
- Markwell M. A., Haas S. M., Bieber L. L., Tolbert N. E. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem. 1978 Jun 15;87(1):206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- Matthews B. W. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim Biophys Acta. 1975 Oct 20;405(2):442–451. doi: 10.1016/0005-2795(75)90109-9. [DOI] [PubMed] [Google Scholar]
- Nishikawa K., Ooi T. Amino acid sequence homology applied to the prediction of protein secondary structures, and joint prediction with existing methods. Biochim Biophys Acta. 1986 May 12;871(1):45–54. doi: 10.1016/0167-4838(86)90131-7. [DOI] [PubMed] [Google Scholar]
- Rall S. C., Jr, Weisgraber K. H., Mahley R. W. Human apolipoprotein E. The complete amino acid sequence. J Biol Chem. 1982 Apr 25;257(8):4171–4178. [PubMed] [Google Scholar]
- Roth R. I., Jackson R. L., Pownall H. J., Gotto A. M., Jr Interaction of plasma "arginine-rich" apolipoprotein with dimyristoylphosphatidylcholine. Biochemistry. 1977 Nov 15;16(23):5030–5036. doi: 10.1021/bi00642a014. [DOI] [PubMed] [Google Scholar]
- Scanu A., Toth J., Edelstein C., Koga S., Stiller E. Fractionation of human serum high density lipoprotein in urea solutions. Evidence for polypeptide heterogeneity. Biochemistry. 1969 Aug;8(8):3309–3316. doi: 10.1021/bi00836a027. [DOI] [PubMed] [Google Scholar]
- Schulz G. E., Barry C. D., Friedman J., Chou P. Y., Fasman G. D., Finkelstein A. V., Lim V. I., Pititsyn O. B., Kabat E. A., Wu T. T. Comparison of predicted and experimentally determined secondary structure of adenyl kinase. Nature. 1974 Jul 12;250(462):140–142. doi: 10.1038/250140a0. [DOI] [PubMed] [Google Scholar]
- Segrest J. P. Amphipathic helixes and plasma lipoproteins: thermodynamic and geometric considerations. Chem Phys Lipids. 1977 Jan;18(1):7–22. doi: 10.1016/0009-3084(77)90023-8. [DOI] [PubMed] [Google Scholar]
- Segrest J. P., De Loof H., Dohlman J. G., Brouillette C. G., Anantharamaiah G. M. Amphipathic helix motif: classes and properties. Proteins. 1990;8(2):103–117. doi: 10.1002/prot.340080202. [DOI] [PubMed] [Google Scholar]
- Segrest J. P., Feldmann R. J. Amphipathic helixes and plasma lipoproteins: a computer study. Biopolymers. 1977 Sep;16(9):2053–2065. doi: 10.1002/bip.1977.360160916. [DOI] [PubMed] [Google Scholar]
- Segrest J. P., Jackson R. L., Morrisett J. D., Gotto A. M., Jr A molecular theory of lipid-protein interactions in the plasma lipoproteins. FEBS Lett. 1974 Jan 15;38(3):247–258. doi: 10.1016/0014-5793(74)80064-5. [DOI] [PubMed] [Google Scholar]
- Sparrow J. T., Gotto A. M., Jr Apolipoprotein/lipid interactions: studies with synthetic polypeptides. CRC Crit Rev Biochem. 1982;13(1):87–107. doi: 10.3109/10409238209108710. [DOI] [PubMed] [Google Scholar]
- Steinmetz A., Utermann G. Activation of lecithin: cholesterol acyltransferase by human apolipoprotein A-IV. J Biol Chem. 1985 Feb 25;260(4):2258–2264. [PubMed] [Google Scholar]
- Teeter M. M., Whitlow M. Test of circular dichroism (CD) methods for crambin and CD-assisted secondary structure prediction of its homologous toxins. Proteins. 1988;4(4):262–273. doi: 10.1002/prot.340040405. [DOI] [PubMed] [Google Scholar]
- Walsh M. T., Atkinson D. Physical properties of apoprotein B in mixed micelles with sodium deoxycholate and in a vesicle with dimyristoyl phosphatidylcholine. J Lipid Res. 1986 Mar;27(3):316–325. [PubMed] [Google Scholar]
- Weisgraber K. H., Innerarity T. L., Harder K. J., Mahley R. W., Milne R. W., Marcel Y. L., Sparrow J. T. The receptor-binding domain of human apolipoprotein E. Monoclonal antibody inhibition of binding. J Biol Chem. 1983 Oct 25;258(20):12348–12354. [PubMed] [Google Scholar]
- Wetterau J. R., Aggerbeck L. P., Rall S. C., Jr, Weisgraber K. H. Human apolipoprotein E3 in aqueous solution. I. Evidence for two structural domains. J Biol Chem. 1988 May 5;263(13):6240–6248. [PubMed] [Google Scholar]
- Wetterau J. R., Jonas A. Effect of dipalmitoylphosphatidylcholine vesicle curvature on the reaction with human apolipoprotein A-I. J Biol Chem. 1982 Sep 25;257(18):10961–10966. [PubMed] [Google Scholar]
- Wilson C., Wardell M. R., Weisgraber K. H., Mahley R. W., Agard D. A. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science. 1991 Jun 28;252(5014):1817–1822. doi: 10.1126/science.2063194. [DOI] [PubMed] [Google Scholar]