Abstract
It is generally accepted that the damage recognition complex of nucleotide excision repair in Escherichia coli consists of two UvrA and one UvrB molecule, and that in the preincision complex UvrB binds to the damage as a monomer. Using scanning force microscopy, we show here that the damage recognition complex consists of two UvrA and two UvrB subunits, with the DNA wrapped around one of the UvrB monomers. Upon binding the damage and release of the UvrA subunits, UvrB remains a dimer in the preincision complex. After association with the UvrC protein, one of the UvrB monomers is released. We propose a model in which the presence of two UvrB subunits ensures damage recognition in both DNA strands. Upon binding of the UvrA2B2 complex to a putative damaged site, the DNA wraps around one of the UvrB monomers, which will subsequently probe one of the DNA strands for the presence of a lesion. When no damage is found, the DNA will wrap around the second UvrB subunit, which will check the other strand for aberrations.
Keywords: damage recognition/Escherichia coli/nucleotide excision repair/scanning force microscopy/UvrB
Introduction
The UvrABC nucleotide excision repair pathway of Escherichia coli is responsible for the removal of a wide variety of structurally unrelated lesions from the DNA. Following damage recognition, the damaged strand is cleaved, first 3′ then 5′ to the lesion. The oligonucleotide containing the damage is removed and the resulting gap is filled by DNA synthesis (Van Houten, 1990; Goosen et al., 1998). Damage recognition is a multi-step process in which a complex of UvrA and UvrB initially searches the DNA for abnormalities in the DNA helix. When the UvrB protein has verified the presence of damage, via a β-hairpin structure that is rich in hydrophobic residues (Moolenaar et al., 2001; Skorvaga et al., 2002), UvrA dissociates to leave a UvrB–DNA complex. During the damage recognition process the DNA is wrapped around the UvrB protein and this DNA wrap is dependent on ATP binding by UvrB (Verhoeven et al., 2001). DNA wrapping is expected to cause local melting of the DNA helix, thereby facilitating insertion of the β-hairpin of UvrB between the DNA strands. From mutational analysis of the β-hairpin it was proposed that UvrB scans the DNA for damage by trying to flip nucleotides out of the DNA helix, thereby probing for differences in base stacking (Moolenaar et al., 2001). When no damage is present, the non-damaged nucleotide will be held in place by its neighbours and a subsequent clashing of this nucleotide with the hydrophobic residues at the base of the hairpin will prevent stable binding of the UvrB protein. Only when a damage is detected can the UvrB–DNA preincision complex be formed. This complex is subsequently bound by the UvrC protein, which catalyses both incisions (Lin and Sancar, 1992; Verhoeven et al., 2000).
The UvrB protein plays a central role in the repair reaction as it is the final damage-recognizing factor (Moolenaar et al., 2000) and interacts with both UvrA and UvrC (Orren and Sancar, 1989; Moolenaar et al., 1995). The crystal structure of UvrB revealed the fold of the ATP binding region to be closely related to that of the helicases PcrA, NS3 and Rep (Machius et al., 1999; Nakagawa et al, 1999; Theis et al., 1999), suggesting that in UvrB, ATP binding and hydrolysis is coupled to domain movement. UvrB presumably interacts with UvrA via a region that shares homology with the transcription repair coupling factor (TRCF) (Selby and Sancar, 1993). The C-terminal part of the UvrB protein shows high homology with an internal region of UvrC. It has been shown that this domain is important for the interaction between the two proteins, since deletion of the last 43 C-terminal amino acids results in the UvrB* protein that is no longer capable of binding UvrC (Moolenaar et al., 1995). X-ray crystallography (Sohi et al., 2000) and nuclear magnetic resonance (NMR) spectroscopy (Alexandrovich et al., 1999) show that the C-terminal region of UvrB adopts a helix–loop– helix conformation capable of dimerization in a head-to-head fashion. It has been suggested that the interaction between UvrB and UvrC is similar to that observed between the two UvrB fragments in the crystal struc ture (Sohi et al., 2000). Using native gradient gels and dimethyl suberimidate (DMS) crosslinking, Hildebrand and Grossman (1999) have shown that UvrB can form dimers in solution and that dimerization is stabilized by the presence of the C-terminal region, since dimerization of the UvrB* protein lacking this domain is severely reduced. In contrast, from previous gel filtration and sedimentation studies, it was concluded that UvrB is a monomer in solution and that this monomer associates with a UvrA dimer to form the UvrA2B complex (Orren and Sancar, 1989).
To determine unambiguously the stoichiometry of the different protein–DNA complexes in the UvrABC nucleotide excision repair reaction, we have used scanning force microscopy (SFM). This technique uniquely allows determination of the multimer state of single protein complexes on DNA, and simultaneous assessment of the functional state of the same protein–DNA complex (in our case either wrapped or non-wrapped Uvr–DNA complexes). Volume measurements of the UvrAB complexes in search of damage suggest that it contains two UvrA as well as two UvrB molecules. Furthermore, we show that UvrB is a dimer in the UvrB–DNA preincision complex. We propose that the presence of two UvrB molecules plays an important role in the damage recognition process.
Results
UvrB binds the damage as a dimer in the UvrB–DNA preincision complex
We have studied the stoichiometry of the different protein–DNA complexes that are formed during the UvrABC nucleotide excision repair reaction by measuring their volumes from SFM images. To determine the size of the protein of interest based on its SFM measured volume, it is necessary to include another protein of known size in all depositions as a standard. The Ku70/80 heterodimer (155 kDa) bound to a 1500 bp DNA fragment was chosen as an internal standard, and the measured molecular weights of the various protein–DNA complexes were estimated using the average measured volume of the 155 kDa Ku70/80 complex within the same scan field as a reference.
Previously, we have shown that incubation of UvrA and UvrB with a 1020 bp substrate with a site-specific lesion at one-third of its length results in damage-specific UvrB– DNA complexes (Verhoeven et al., 2001). Contour length analysis showed that the DNA is still wrapped around the UvrB protein in these preincision complexes as long as UvrB has bound ATP (Verhoeven et al., 2001). The calculated size of a potential UvrB dimer (152 kDa) is almost exactly the size of the Ku70/80 heterodimer (155 kDa), therefore UvrB–DNA complexes with a volume similar to the Ku70/80 dimer can be determined reliably as UvrB dimers. Both complexes can easily be distinguished by DNA contour length analysis, since UvrB and the Ku70/80 complex are bound to a 1020 bp and a 1500 bp DNA fragment, respectively. No protein complexes other than those bound to the damaged site were observed on the 1020 bp damaged DNA fragment, indicating that no exchange of the Ku70/80 protein complexes occurs during the deposition procedure (see Materials and methods).
For the isolation of ATP-containing UvrB–DNA complexes, the DNA fragment was incubated with UvrA and UvrB, followed by washing with buffer containing high salt (to remove the UvrA protein) and ATP (to prevent dissociation of the cofactor from the UvrB protein). Contour length analysis of UvrB–DNA complexes from three independent isolations shows that in the majority (50/56) of the complexes, the DNA is wrapped (Table I, A). The average volume of the wrapped UvrB–DNA complexes was slightly greater than the average volume of the Ku70/80–DNA complexes, from which we estimate a molecular weight of 185 ± 26 kDa for the UvrB–DNA complexes (Figure 1A). This value is slightly above the expected size of two UvrB molecules (152 kDa), indicating that in the preincision complex, UvrB is bound to the damage as a dimer. The fact that the estimated size is bigger than the calculated value can be explained by the additional volume contributed by the DNA wrapped around UvrB.
Table I. Contour length and protein volumes of UvrB–DNA and UvrB*–DNA complexes.
| No. of molecules | Class I | Class II | |||||
|---|---|---|---|---|---|---|---|
| No. of molecules | Volume (kDa) | Wrap (nm) | No. of molecules | Volume (kDa) | Wrap (nm) | ||
| A. UvrB–DNA complexes | |||||||
| Washed with ATP | |||||||
| Wrapped | 50 | 0/50 (0%) | – | – | 50/50 (100%) | 185 ± 26 | 25 |
| Not wrapped | 6 | 3/6 (50%) | n.d.a | 0 | 3/6 (50%) | n.d.a | – |
| Washed without ATP | |||||||
| Wrapped | 0 | – | – | – | – | – | – |
| Not wrapped | 115 | 45/115 (39%) | 76.4 ± 11 | 0 | 70/115 (61%) | 151 ± 28 | 0 |
| Reintroduction of ATP | |||||||
| Wrapped | 64 | 24/64 (37%) | 87.4 ± 10 | 26 | 40/64 (63%) | 168 ± 14 | 22 |
| Not wrapped | 0 | – | – | – | – | – | – |
| B. UvrB*–DNA complexes | |||||||
| Washed with ATP | |||||||
| Wrapped | 27 | 0/27 (0%) | – | – | 27/27 (100%) | 135 ± 18 | 23 |
| Not wrapped | 56 | 50/56 (89%) | 61.7 ± 9 | 0 | 6/56 (11%) | n.d.a | 0 |
| Washed without ATP | |||||||
| Wrapped | 0 | – | – | – | – | – | – |
| Not wrapped | 36 | 33/36 (92%) | 61.7 ± 10 | 0 | 3/36 (8%) | n.d.a | 0 |
| Reintroduction of ATP | |||||||
| Wrapped | 20 | 17/20 (85%) | 77.1 ± 5 | 23 | 3/20 (15%) | 161 ± 9 | 24 |
| Not wrapped | 51 | 43/51 (84%) | 68.2 ± 8 | 0 | 8/51 (16%) | 143 ± 6 | 0 |
All measured molecules were first divided in two groups; one group containing wrapped complexes, the other containing unwrapped complexes. Of both these, the protein volume and the amount of DNA involved in the wrapping were determined.
aAmount of complexes too low to determine size accurately.
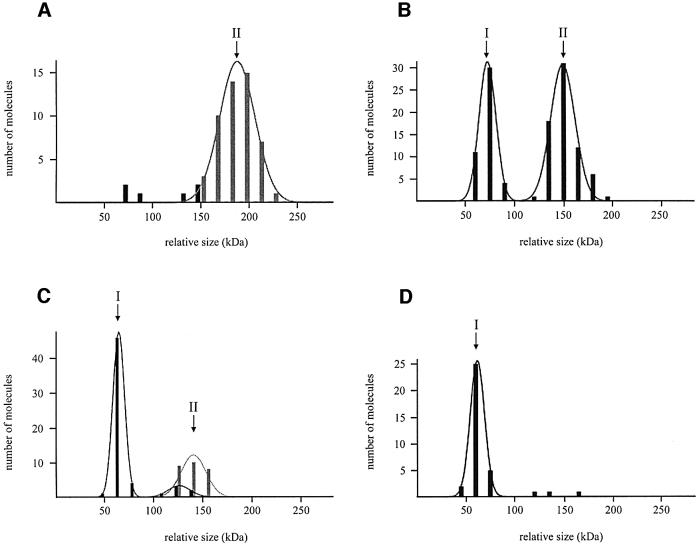
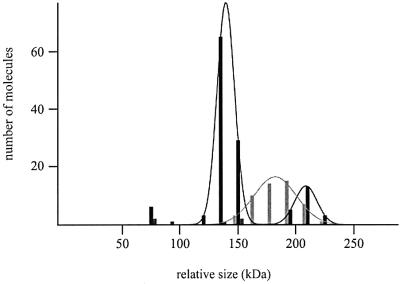
Fig. 1. Volume distributions of UvrB/UvrB*–DNA complexes. The lines represent the Gaussian fitting of the distribution and the values obtained are reported in Table I. In all cases, wrapped complexes are represented by grey bars and unwrapped complexes by black bars. The numbers above the distributions refer to the classes mentioned in Table I. (A) UvrB–DNA complexes isolated in the presence of ATP. (B) UvrB–DNA complexes isolated in the absence of ATP. (C) UvrB*–DNA complexes isolated in the presence of ATP. (D) UvrB*–DNA complexes isolated in the absence of ATP.
The residual small group of unwrapped complexes is divided into two classes (Figure 1A). The first class clusters around a volume close to half the Ku70/80 volume and the second class around a volume close to one Ku70/80 volume. However, the number of complexes in these two groups is too low (six in total) to determine a significant average volume. Since these unwrapped complexes are expected to have lost the ATP molecule, we decided to analyse these complexes further by determining the volumes of UvrB–DNA complexes that were isolated by washing in the absence of ATP. As expected, no wrapped complexes were observed after removal of the ATP (Table I, A), and the volume analysis now clearly showed two distinct classes (Figure 1B). Class I, comprising 39% of the complexes, has an average volume that is 0.49 times the Ku70/80 volume, and class II, comprising 61% of the complexes, has an average volume that is 0.97 times the Ku70/80 volume. The molecular weights of the UvrB–DNA complexes in class I and class II are therefore estimated at 76 ± 11 kDa and 151 ± 28 kDa, respectively (Table I, A). These values correspond very well to the expected sizes of a UvrB monomer and dimer, respectively. The smaller complexes can only logically be UvrB monomers and effectively serve as an additional internal size standard. Comparisons of the volume of the larger UvrB–DNA complexes to the UvrB monomer and Ku70/80 are both consistent with the conclusion that UvrB is present as a dimer in these larger complexes. The measured volume for the unwrapped dimer is smaller compared with the size of the dimers isolated in the presence of ATP, showing that the wrapped DNA does indeed contribute to the size of the UvrB–DNA complex. The three-dimensional representation of the SFM images clearly shows the two classes of molecules (Figure 2C), of which the largest complexes resemble the size of the Ku70/80 dimer. Taken together, the measurements of the UvrB–DNA complexes show that UvrB binds to the damage as a dimer. This dimer is stabilized by ATP binding and/or DNA wrapping, since more monomeric complexes are found after removal of the ATP.
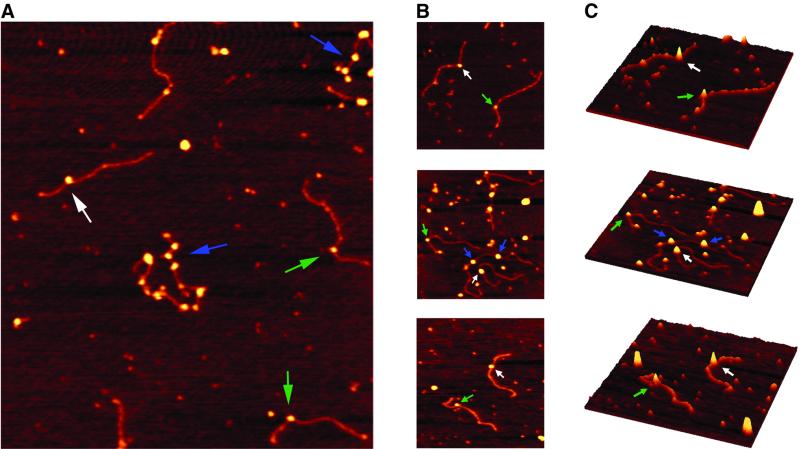
Fig. 2. Scanning force images of the Ku70/80–DNA and UvrB–DNA complexes isolated in the absence of ATP. The colour scale ranges from 0.0 to 3.0 nm (dark to bright). The UvrB monomers are indicated by green arrows. The white arrows indicate UvrB dimers and the Ku70/80 complexes are marked by blue arrows. (A) Characteristic scan field containing the UvrB monomer, UvrB dimer and the Ku70/80 marker complex. (B) Zoomed images of UvrB monomers and dimers. The Ku70/80 complexes are marked as well. (C) Three-dimensional pictures of images shown in (B).
As shown before (Verhoeven et al., 2001), addition of ATP to the UvrB–DNA complexes that were obtained by washing in buffer without ATP restores the DNA wrap (Table I, A). The monomer:dimer ratio is the same as before the introduction of ATP (Table I, A). The amount of DNA that becomes wrapped again is similar for both the monomer (26 nm) and the dimer (22 nm), which means that in the dimer the DNA is wrapped around one UvrB subunit only.
No unwrapped UvrB–DNA complexes were detected after reintroduction of ATP into the complexes that were initially isolated in the absence of ATP. This differs from the small number (6/56) of unwrapped complexes found after isolation of UvrB–DNA complexes in the presence of ATP. This might be explained by assuming that the ATP in the UvrB–DNA complexes can be hydrolysed to ADP. Addition of ADP to UvrB–DNA complexes without a cofactor indeed does not restore DNA wrapping (Table II). If some of the complexes isolated in the presence of ATP do contain ADP, wrapping can only be restored by exchanging ADP for ATP, which might be a slow process. In contrast, the complexes that were washed in the absence of ATP have lost all bound cofactors, which might make ATP binding easier upon addition of this cofactor. The fact that these newly bound ATP molecules are not hydrolysed again suggests that this hydrolysis can only occur in the presence of UvrA. In the complexes isolated in the presence of ATP, hydrolysis could have occurred before the washing step when UvrA was still present. For the UvrB–DNA complexes isolated in the absence of ATP, the washing step not only removes all cofactors but also UvrA. Hence, when ATP is added to these complexes it can no longer be hydrolysed.
Table II. Contour length of UvrB–DNA and UvrB*–DNA complexes.
| No. of molecules | DNA involved in wrap (nm) | |
|---|---|---|
| UvrB–DNA complexes | ||
| Addition of ATPγS | ||
| Wrapped | 61/84 (73%) | 25 |
| Not wrapped | 23/84 (27%) | 0 |
| Addition of ATP | ||
| Wrapped | 64/64 (100%) | 24 |
| Not wrapped | 0/64 (0%) | 0 |
| Addition of ADP | ||
| Wrapped | 0/52 (0%) | – |
| Not wrapped | 52/52 (100%) | 0 |
| UvrB*–DNA complexes | ||
| Addition of ATPγS | ||
| Wrapped | 34/47 (72%) | 24 |
| Not wrapped | 13/47 (28%) | 0 |
| Addition of ATP | ||
| Wrapped | 20/71 (28%) | 24 |
| Not wrapped | 51/71 (72%) | 0 |
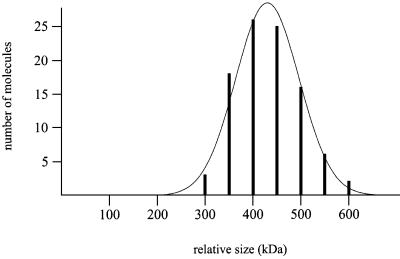
Since UvrB is a dimer in the UvrB–DNA preincision complex, we wished to determine whether UvrB contributes two subunits to the UvrAB complex as well. For this purpose, we incubated UvrA and UvrB with a 1020 bp undamaged DNA substrate. The UvrAB complexes bound to the undamaged DNA reflect complexes in search of a lesion (Verhoeven et al., 2001). It has been shown that in these complexes the DNA is wrapped around the UvrB protein (Verhoeven et al., 2001), resulting in a ‘loss’ of ∼23 nm of the contour length of the DNA fragment. Therefore, the UvrAB–DNA complexes could be distinguished from the UvrA–DNA complexes by measuring the contour length of the DNA. The average volume for the UvrAB complex was determined by measuring 96 wrapped complexes from three independent experiments (Figure 3). From these experiments, an average volume of 2.8 Ku70/80 equivalents was found for the UvrAB complex bound to DNA. The molecular weight estimated from this value (430 ± 64 kDa) exceeds the calculated size of a UvrA2B1–DNA complex (296 kDa), but also that of a UvrA2B2–DNA complex (372 kDa). As shown above for UvrB–DNA complexes, the DNA wrapped around the UvrB protein also contributes to the size of the complex, thereby increasing the measured volume over that expected based on proteins alone. Moreover, comparison of the volumes of wrapped and unwrapped UvrB–DNA complexes shows that the DNA can contribute ∼0.2 Ku70/80 volume equivalents (± 30 kDa). Taking this into account, the estimated size of the UvrAB–DNA complex is close to the calculated size of a UvrA2B2–DNA complex. In combination with the results, which show that UvrB binds as a dimer in the UvrB–DNA preincision complex, these data strongly suggest that UvrB contributes two subunits to the UvrAB complex in search of a damage. The amount of DNA involved in the wrapping is the same as observed in the UvrB–DNA preincision complex, where DNA is wrapped around one UvrB molecule. This strongly suggests that also in the UvrA2B2 complexes in search of a damage the DNA is wrapped around one of the two UvrB subunits. Taken together, our results indicate that, when searching for DNA damage, UvrA and UvrB bind DNA as a UvrA2B2 complex with the DNA wrapped around one of the UvrB subunits, and that the UvrB protein remains as a dimer in the UvrB–DNA preincision complex.
Fig. 3. Volume distribution of UvrAB–DNA complexes. The line represents the Gaussian fitting of the distribution.
The C-terminal region of UvrB is important for dimer stability on the DNA
Since it has been shown that the C-terminal region of UvrB is important for dimerization in solution (Hildebrand and Grossman, 1999), we analysed the preincision complexes formed by the UvrB* protein lacking this C-terminal domain. Contour length analysis of UvrB*–DNA preincision complexes isolated in the presence of ATP shows far fewer wrapped complexes (27/83) compared with wild-type UvrB (50/56) (Table I, B). Unwrapping in the UvrB*–DNA complexes again must result from the absence of bound ATP in these complexes, as was observed for the wild-type protein. This would imply that either ATP hydrolysis by the UvrB* protein is increased compared with that of wild-type UvrB, or that the affinity of UvrB* for ATP is reduced. To discriminate between these two possibilities, we added ATPγS to the UvrB*–DNA complexes and the wild-type UvrB–DNA complexes from which the cofactor was removed. Introduction of ATPγS restored DNA wrapping in the UvrB*–DNA complexes to the same extent as in the wild-type UvrB–DNA complexes (Table II), indicating that ATPγS binds with the same affinity to wild-type UvrB and UvrB*. It must therefore be increased ATP hydrolysis that causes a higher percentage of unwrapped complexes in the case of UvrB*. Introduction of ATPγS did not restore the wrapping to 100% of the complexes, in contrast to the reintroduction of ATP in the wild-type UvrB–DNA complex [compare Tables I (A) and II]. This is probably caused by the presence of ADP in the ATPγS solution, which results from degradation of ATPγS during storage. Addition of ATP to the UvrB*–DNA complexes from which the cofactor and UvrA were initially removed restored DNA wrapping in only 28% of the complexes, compared with 100% for the wild-type protein (Table II). This indicates that in contrast to wild-type UvrB, the UvrB*–DNA complex is capable of hydrolysing ATP in the absence of UvrA. This UvrA-independent ATPase activity explains the increased ATP hydrolysis observed when the UvrB*–DNA complexes were isolated in the presence of ATP, since during the washing procedure in this case, when UvrA is removed, ATP hydrolysis can still continue. Increased ATP hydrolysis of the UvrB* protein in the absence of UvrA and the presence of single-strand DNA (ssDNA) has been reported before (Caron and Grossman, 1988; Hildebrand and Grossman, 1998). In these studies it was suggested that the conformation of the ATPase site is altered upon removal of the C-terminal region of UvrB, resulting in a hyperactive ATPase site. It remains unclear whether this property of the UvrB* protein underlies its UvrA-independent ATPase activity when bound to a damaged site.
Another striking difference between UvrB* and wild-type UvrB is the reduced amount of dimers found with UvrB* after washing with ATP. Volume measurements show that all the wrapped complexes fall into one class, with an estimated size of 135 ± 18 kDa (Figure 1C; Table I, B). This is in good agreement with the calculated size of the UvrB* dimer (137 kDa). As mentioned above, however, in a large fraction of the UvrB*–DNA preincision complexes (56/83), the DNA is not wrapped around the protein (Table I, B). In this group of unwrapped complexes, only 11% seem to have the size of the dimer (Figure 1C). The majority fall into the group with an average size of 62 ± 9 kDa, corresponding to the size of the UvrB* monomer (68.5 kDa) (Figure 1C; Table I, B). Since the wrapped complexes represent the original situation immediately after the formation of the preincision complex, these results suggest that UvrB* binds the damage as a dimer like wild-type UvrB. During the washing procedure however, the dimer falls apart more rapidly compared with wild-type UvrB. Apparently, the C-terminal region of UvrB is not required for dimerization per se, but plays an important role in the stabilization of the dimer. This is confirmed by measuring the volumes of UvrB*–DNA complexes isolated in the absence of ATP. Whereas for wild-type UvrB the dimer:monomer ratio was 39%:61% (Table I, A), under the same conditions almost all UvrB*–DNA complexes (92%) contain only one UvrB* molecule (Table I, B; Figure 1D). This shows that under conditions where the dimer is less stable due to loss of ATP and/or DNA wrapping, the C-terminal domain of the protein becomes even more important. As expected, after reintroduction of ATP into these predominantly monomeric complexes, the amount of B* dimers remains very low (Table I, B).
Dissociation of one of the UvrB subunits upon binding of UvrC
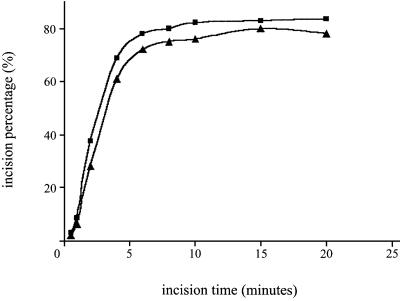
As shown above, the C-terminal region of UvrB is important for the stability of the dimer in the preincision complex, indicating that these domains of the two monomers interact. However, the same C-terminal region of UvrB is also important for the interaction with UvrC (Moolenaar et al., 1995, 1997). Since the C-terminal domain of UvrB involved in dimer stabilization would not be available for interaction with UvrC, we have tested whether the UvrB1–DNA and the UvrB2–DNA complexes are incised with the same efficiency. This was done by isolating wild-type UvrB–DNA preincision complexes in the absence of ATP (resulting in 61% dimers and 39% monomers) or in the presence of ATP (resulting in 95% dimers and 5% monomers), and determining the rate of incision by UvrC. No differences were detected in the incision kinetics of the two UvrB–DNA complex preparations when 5 nM of UvrC was used (Figure 4). Using lower (0.25 nM) or higher (50 nM) UvrC concentrations, we did not observe any differences either (data not shown). This shows, on the one hand, that the presence of two UvrB molecules is not essential for the incision reaction. On the other hand, dimerization of UvrB does not seem to disturb the action of UvrC, possibly because one of the UvrB molecules is released after binding of UvrC.
Fig. 4. Kinetics of the incision of UvrB–DNA complexes by UvrC. Complexes were isolated in the presence (rectangles) or absence (triangles) of ATP.
To test this latter possibility, we analysed the volumes of UvrBC–DNA complexes by adding UvrC to the preincision complexes. It has been shown before that both the 3′ and the 5′ incisions are induced by the binding of only one UvrC molecule (Moolenaar et al., 2000). This leaves two options for the stoichiometry of the UvrBC– DNA incision complex: a UvrB1C1–DNA complex (144 kDa) or a UvrB2C1–DNA complex (220 kDa). The UvrB–DNA complexes, which were isolated in the presence of ATP, were incubated with 100 nM UvrC(R42A). This UvrC is a 3′ catalytic site mutant and therefore incision, which might destabilize the complex, cannot occur. In contrast to the small amount of unwrapped molecules observed in the depositions of UvrB–DNA complexes isolated in the presence of ATP, no unwrapped complexes were detected upon addition of UvrC (Table III). This suggests that binding of UvrC to the preincision complex stabilizes the wrapped DNA conformation, perhaps by stimulating the exchange of ADP (resulting from ATP hydrolysis) for ATP from solution.
Table III. Contour length and protein volumes of UvrB–DNA and UvrBC–DNA complexes.
| No. of molecules | DNA involved in wrap (nm) | Class I volume (kDa) | Class II volume (kDa) | Class III volume (kDa) | |
|---|---|---|---|---|---|
| UvrB–DNA complexes | |||||
| UvrB–DNA | |||||
| Wrapped | 50 | 25 | – | 185 ± 26 (100%) | – |
| Not wrapped | 6 | 0 | n.d.a | n.d.a | – |
| UvrBC–DNA complexes | |||||
| UvrBC–DNA | |||||
| Wrapped | 124 | 23 | n.d.a (5%) | 141 ± 11 (78%) | 210 ± 10 (17%) |
| Not wrapped | 0 | 0 | – | – | – |
aAmount of complexes too low to determine size accurately.
The protein–DNA complexes formed after addition of UvrC can be sorted into three classes based on their volume (Figure 5; Table III). The first class, consisting of 5% of all complexes has an average size of ∼0.5 Ku70/80 equivalents, very likely being a UvrB monomer. The second and largest group, containing 78% of all complexes, shows an estimated molecular weight of 141 ± 11 kDa (Table III). From this value we cannot directly conclude whether this class consists of UvrB2– DNA or UvrB1C1–DNA complexes because of the small theoretical difference between their respective volumes (152 and 144 kDa, respectively). However, comparing the size distribution of these complexes with the size distribution of the UvrB–DNA complexes from which these complexes were formed shows that the addition of UvrC causes a significant shift towards a smaller volume (Figure 5). This strongly suggests that this class mainly consists of UvrB1C1–DNA complexes, although the presence of a low amount of UvrB2–DNA complexes cannot be ruled out.
Fig. 5. Volume distribution of UvrB–DNA complexes (grey bars) and protein–DNA complexes formed after the addition of UvrC(R42A) to these purified UvrB–DNA preincision complexes (black bars). The lines represent the Gaussian fitting of the distribution and the values obtained are reported in Table III.
The third class, which contains 17% of all complexes, has an average volume of 210 kDa (Table III), which roughly corresponds to the calculated size of a UvrB2C1 complex (220 kDa). The size distribution of this group is clearly shifted towards a larger volume when compared with the size distribution of the original UvrB–DNA complexes (Figure 5). Apparently, UvrC is capable of binding to a UvrB dimer. The UvrB2C1 fraction, however, is considerably smaller (17%) than the group containing the UvrB1C1–DNA complexes (78%), although the initial UvrB–DNA preparation mainly consisted of UvrB2–DNA complexes. This suggests that UvrC initially binds to the UvrB2–DNA complex, yielding an intermediate UvrB2C1–DNA complex, and that subsequently one of the UvrB monomers is released to form the UvrB1C1– DNA incision complex.
Discussion
For over 10 years now it has been generally believed that during nucleotide excision repair in E.coli, an asymmetric UvrA2B complex is responsible for damage recognition and that the UvrB–DNA preincision complex contains only one UvrB subunit. This was based upon studies by Orren and Sancar (1989), who concluded from gel filtration and sedimentation experiments that UvrA is a dimer and UvrB a monomer in solution, and that these proteins associate to form a UvrA2B complex. Moreover, by isolating UvrB–DNA complexes on ultraviolet (UV)-irradiated DNA they calculated that one UvrB molecule is bound per photoproduct. In their assays, however, they quantified the individual protein subunits by SDS–PAGE followed by Coomassie Brilliant Blue or silver staining. Such concentration estimates easily vary by a factor of two or more due to differences in staining efficiencies between the protein standard and the protein studied. In addition, the estimated number of TT-dimers in the UV-irradiated DNA introduced another variable into their assay, thereby lending even more uncertainty to their conclusion. In contrast to the conclusions drawn by Orren and Sancar, Hildebrand and Grossman (1999) more recently showed using hydrodynamic and crosslinking studies that UvrB is a dimer in solution and that it is even able to form higher order oligomers.
Here, we show unambiguously by SFM analysis of single molecules that UvrB binds to a damaged site as a dimer. The combined observations that UvrB binds to the damage as a dimer and forms a dimer in solution (Hildebrand and Grossman, 1999) makes it very likely that the UvrAB damage recognition complex contains two UvrB molecules as well. Indeed, our SFM analysis of the size of UvrAB complexes in search of damage is only consistent with the presence of a UvrA2B2 complex on DNA.
The UvrB protein is the ultimate damage-recognizing factor in the UvrABC nucleotide excision repair reaction. The current model for damage recognition by UvrB is that, following initial detection of a distortion in the helix by UvrA, UvrB inserts its β-hairpin between the two DNA strands (Theis et al., 1999; Skorvaga et al., 2002). Next, UvrB will probe the DNA for damages by trying to flip nucleotides out of the helix. Base damages cause impaired base stacking and therefore flipping will occur more readily at damaged nucleotides. When no damage is present the nucleotide will be held in place by its neighbours and a subsequent clashing of this nucleotide with tyrosine residues at the base of the hairpin will prevent stable binding of UvrB to the DNA. When a lesion is present the damaged nucleotide will be flipped out and the vacated space will be occupied by the tyrosine residues leading to a stable UvrB–DNA complex from which UvrA is released (Moolenaar et al., 2001). It has been proposed that in the UvrB–DNA complex the strand opposite the damage will become locked between the β-hairpin and domain IB of the protein (Theis et al., 1999). This model implies that only one UvrB molecule is directly involved in damage-specific binding.
We propose that the seemingly redundant second UvrB molecule plays a very important role in the early damage- recognition steps. In the model described above, probing of the DNA for the presence of a damage will be limited to one of the DNA strands. The orientation in which UvrB binds the DNA will determine in which of the two strands the nucleotide flipping occurs. The presence of two UvrB molecules in the UvrA2B2 complex allows an alternate scanning of both DNA strands. We have shown that in the UvrA2B2 complex in search of a damage the DNA is wrapped around one of the UvrB subunits. Most likely this subunit has inserted its β-hairpin into the DNA helix, where it probes one of the DNA strands for a lesion. When no damage is detected and no stable UvrB–DNA complex can be formed, the DNA might be able to wrap around the other UvrB subunit. Subsequently, the β-hairpin of this UvrB protein will be inserted into the helix to probe the other DNA strand. How can this alternate wrapping of the DNA around one of the UvrB subunits be regulated? We have shown that UvrB can only wrap the DNA when bound to ATP and not when bound to ADP or in the absence of a cofactor. This implies that the UvrB subunit of the UvrA2B2 complex that wraps the DNA is in the ATP-bound form. Most likely, the second UvrB subunit is in the ADP-bound form, thereby directing the wrap to the first UvrB molecule only. We propose that when the first UvrB molecule fails to detect a damage, the resulting clashing of the undamaged nucleotide with the tyrosines of the β-hairpin will induce hydrolysis of the ATP in this UvrB subunit. This ATP hydrolysis will result in unwrapping of the DNA. If, simultaneously, ATP hydrolysis by the first UvrB will stimulate ATP binding by the second UvrB, the DNA can now wrap around this other molecule, allowing scanning of the other strand. As soon as one UvrB molecule has found a damaged nucleotide it will stably bind the DNA and the other UvrB protein will have no direct interaction with the damaged site.
A similar asymmetric dimer with coordinated ATPase activities has been found for the Rep helicase. This helicase uses a subunit switching mechanism during DNA unwinding and translocation in which both subunits alternate binding to double-stranded (ds)- and ssDNA. The alternating DNA affinities are mediated by the ATPase activities of the two Rep monomers. ATP hydrolysis in one of the subunits stimulates dissociation of ssDNA from the other subunit, which then presumably binds to dsDNA (Bjornson et al., 1996; Hsieh et al., 1999). We propose a similar coordination between the ATPase activities and DNA binding properties for UvrB, in which ATP hydrolysis by one subunit stimulates ATP binding and subsequent DNA wrapping by the other UvrB subunit.
When one of the UvrB subunits has bound a damaged site, the UvrA subunits are released but the second UvrB molecule remains associated. Removal of ATP from the protein resulted in dissociation of this second UvrB in 39% of the complexes, indicating that ATP binding and/or DNA wrapping stabilize the dimerization of UvrB on the damaged site. This could be the result of a specific protein conformation induced by binding of ATP. Alternatively, the DNA wrapping itself might cause the stabilization. Wrapping of the DNA around one monomer might present the DNA to the other subunit for additional protein–DNA contacts.
We have also shown that with the UvrB* protein, which lacks the C-terminal region of UvrB, the second UvrB molecule dissociates more readily from the damage-bound complex than with wild-type UvrB. It was found by Hildebrand and Grossman, (1999) that dimerization of the UvrB* protein in solution is also reduced compared with wild-type UvrB. Apparently, the C-terminal domain of UvrB constitutes an important dimer-interaction domain. However, UvrB* dimers on a damaged site are still observed when the UvrB*–DNA complexes are isolated in the presence of ATP. This means that additional domains of UvrB must be involved in the dimer contacts.
Both the crystal (Sohi et al., 2000) and NMR structure (Alexandrovich et al., 1999) of the C-terminal part of UvrB showed that this domain adopts a helix–loop–helix fold. Two of these domains bind each other in a head-to-head fashion via interactions between residues in, and close to, the loop region. These contacts most likely represent the stabilizing interaction of the C-terminal domains in the UvrB dimer when bound to a damage as described above. The same C-terminal region, however, has also been shown to be important for interaction with a homologous region in UvrC, most likely via the same residues (Moolenaar et al., 1995, 1997). Therefore it seems very unlikely that the C-terminal domain of UvrB can provide stabilizing interactions for the UvrB dimer, and simultaneously serves as a binding site for UvrC. Our SFM data suggest that binding of UvrC to the UvrB2–DNA complex initially leads to an intermediate UvrB2C1–DNA complex from which one of the UvrB molecules is subsequently released, resulting in a UvrB1C1–DNA complex. There are two possible ways in which this UvrB2C1 complex may be formed. First, the UvrC protein might compete directly with the second UvrB subunit for binding to the C-terminal region of the damage-bound UvrB subunit, thereby disrupting the UvrB–UvrB contacts of this domain. If the C-terminal region of UvrB, however, would be the sole contact for UvrC, it is hard to envisage how UvrC can discriminate between the two UvrB subunits for binding. We therefore prefer the second possibility, where UvrC initially binds to another domain of UvrB. This domain might have the proper conformation for making contacts with UvrC in the damage-bound UvrB, but not in the second subunit. Subsequently, UvrC will make additional contacts with the C-terminal region of the same UvrB to which it is already bound. In such a two-step binding mode it is also possible that the first interaction between UvrC and UvrB already destabilizes the interaction between the C-terminal domains of the two UvrB subunits, thereby facilitating the binding of UvrC to this domain. As yet, a second UvrB domain for contact with UvrC has not yet been identified, but incision data using the truncated UvrB* protein strongly point towards the presence of such a domain. The 3′ incision of UvrB–DNA complexes formed by UvrB* is severely disturbed, but residual incision can be detected, indicating that UvrC can bind to UvrB–DNA complexes in the absence of the C-terminal domain (Moolenaar et al., 1995). Moreover UvrB*–DNA complexes that are pre-nicked at the 3′ incision site are normally incised at the 5′ site by UvrC (Moolenaar et al., 1995). This means that, at least for 5′ incision, but possibly for 3′ incision as well, UvrC binds to another domain of UvrB. The result of the UvrC binding, either in a one- or two-step fashion, is a disruption of the dimerization of the C-terminal domain of UvrB, finally leading to dissociation of the UvrB molecule that lacks the DNA contacts with the damaged site.
In conclusion, we propose the following chain of events during nucleotide excision repair in E.coli. A complex consisting of two UvrA and two UvrB subunits scans the two DNA strands for presence of a damage by alternate wrapping of the DNA around either UvrB subunit. Upon detection of damage, one of the UvrB subunits is firmly bound to the damaged site. The second UvrB remains associated with this complex, mainly via interaction between their respective C-terminal domains, and the UvrA subunits are released. UvrC then associates with the UvrB that is bound to the damage, the second UvrB is released and the UvrBC–DNA incision complex is formed.
Materials and methods
Proteins and DNA substrates
The UvrA, UvrB and UvrC proteins (Visse et al., 1992), the UvrB* protein (Moolenaar et al., 1995) and the UvrC(R42A) mutant (Verhoeven et al., 2000) were purified as described previously. The Ku70/80 complex was a generous gift from Mauro Modesti (Erasmus University).
The biotinylated 1020 bp SFM substrate with or without a site-specific cholesterol lesion at one-third of its length was constructed on streptavidin-coated magnetic beads (Dynal M-280) as described previously (Verhoeven et al., 2001).
The DNA fragment for Ku binding was produced by a PCR reaction on the UvrC gene, yielding a 1500 bp DNA substrate.
Scanning force microscopy
For the formation of UvrAB–DNA complexes with undamaged DNA, the non-damaged substrate (200 fmol) was cut from the beads with SmaI prior to incubation with 50 nM UvrA and 200 nM UvrB in 10 µl of Uvr-endo 1 buffer (15 mM Tris–HCl pH 7.5, 10 mM MgCl2, 40 mM KCl and 1 mM ATP).
To obtain purified preincision complexes, the immobilized SFM substrate containing a damage at one-third of the fragment (250 fmol) was incubated with 20 nM UvrA and 400 nM UvrB or UvrB* in 20 µl of Uvr-endo 2 buffer [50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 100 mM KCl, 0.1 µg bovine serum albumin (BSA)/µl and 1 mM ATP]. After 15 min at 37°C, the mixture was transferred to the magnetic particle concentrator and the formed complexes were purified in the presence or absence of ATP as described previously (Verhoeven et al., 2001). For this purification the complexes were first washed three times with 50 µl of wash buffer 1 (50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 0.5 M KCl, 0.1 µg/µl BSA) with or without ATP to remove UvrA, and then twice with 50 µl of wash buffer 2 (50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 0.1 M KCl, 0.1 µg/µl BSA) with or without ATP. After purification, the preincision complexes were incubated with SmaI in 10 µl of SmaI buffer (10 mM Tris-acetate pH 7.5, 10 mM Mg-acetate and 50 mM K-acetate) to cut the protein–DNA complexes from the beads. For reintroduction of the cofactor, a final concentration of 1 mM ATP, ATPγS or ADP was added to the purified complexes and the mixture was incubated for 5 min at room temperature. UvrBC–DNA complexes were formed by incubating the purified UvrB–DNA complexes that were cut from the beads with 100 nM UvrC(R42A) and 1 mM ATP for 10 min at 37°C. The Ku70/80 heterodimer (50 nM) was incubated with a 1500 bp DNA fragment (200 fmol) in 10 µl containing 50 mM HEPES–HCl pH 8.0, 10 mM MgCl2 and 50 mM KCl.
For the simultaneous deposition of the Uvr–DNA and Ku70/80–DNA complexes, 2 µl of each separate reaction mixture were combined at the dilution step. Dilution was performed in 16 µl of deposition buffer (5 mM HEPES–KOH pH 7.8, 5 mM MgCl2). The mixture was instantly deposited on freshly cleaved mica. After ∼30 s the mica was washed with 4 ml of glass distilled water and dried in a stream of air.
The complexes were imaged using a Nanoscope IIIa (Digital Instruments, Santa Barbara, CA) in the tapping mode. In general, the efficiency of complex formation varied between 65% and 90%. DNA length and the size of the protein complexes were measured using the Image SXM (v 1.62) software (an NIH Image version modified by Dr Steve Barrett, Surface Science Research Centre, University of Liverpool, UK). The volume of protein complexes was determined as described before (Wyman et al., 1997). In short, the complex was manually traced and the area and average height were determined, and subsequently a background volume of the same traced area at an adjacent position including DNA was subtracted. The volumes were calculated using the size of the Ku70/80 marker protein bound to a 1500 bp fragment within the same scan field only, to prevent large standard deviations due to variations in tip-induced aberrations between different scan fields. All data were collected from at least two independent depositions. Estimated molecular weights of the Uvr complexes were determined by multiplying the measured number of Ku70/80 equivalents for each complex with the size of the Ku70/80 protein (155 kDa).
Incision assay
To study the incision rate, the SFM substrate was terminally labelled at the 5′ side as described previously (Verhoeven et al., 2001). The fragment was incubated with 2.5 nM UvrA and 100 nM UvrB in 20 µl of Uvr-endo 2 buffer. After UvrB–DNA complex formation, the mixture was divided into two equal parts and the complexes were isolated either in the presence or absence of ATP. Subsequently, 5 nM UvrC was added to both parts and the incision reactions were stopped at different times by the addition of 2 µl of 0.5 M EDTA. The incision products were isolated by eluting the top strand with 10 µl of 0.1 M NaOH. After adding 3 µl of formamide, the samples were incubated at 95°C for 3 min and run on a 3.5% denaturing acrylamide gel. The amount of incision at each time point was quantified using a phosphoimager.
Acknowledgments
Acknowledgements
We kindly thank Mauro Modesti for his generous gift of the Ku70/80 protein. Dejan Ristic is acknowledged for determining the optimal reaction conditions for Ku70/80 binding. This work was supported by the J. A. Cohen Institute for Radiopathology and Radiation Protection (IRS) and an EC grant on ‘Quality of Life and Management of Living Resources’ (QLG1-CT-1999-00008).
References
- Alexandrovich A., Sanderson,M.R., Moolenaar,G.F., Goosen,N. and Lane,A.N. (1999) NMR assignments and secondary structure of the UvrC binding domain of UvrB. FEBS Lett., 451, 181–185. [DOI] [PubMed] [Google Scholar]
- Bjornson K.P., Wong,I. and Lohman,T.M. (1996) ATP hydrolysis stimulates binding and release of single-stranded DNA from alternating subunits of the dimeric E. coli Rep helicase. J. Mol. Biol., 263, 411–412. [DOI] [PubMed] [Google Scholar]
- Caron P.R. and Grossman,L. (1988) Involvement of a cryptic ATPase activity of UvrB and its proteolysis product, UvrB* in DNA repair. Nucleic Acids Res., 16, 10891–10902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goosen N., Moolenaar,G.F., Visse,R. and Van de Putte,P. (1998) Nucleic Acids and Molecular Biology: DNA Repair. Springer Verlag, Berlin.
- Hildebrand E.L. and Grossman,L. (1998) Introduction of a tryptophan reporter group into the ATP binding motif of the Escherichia coli UvrB protein for the study of nucleotide binding and conformational dynamics. J. Biol. Chem., 273, 7818–7827. [DOI] [PubMed] [Google Scholar]
- Hildebrand E.L. and Grossman,L. (1999) Oligomerisation of the UvrB nucleotide excision repair protein of Escherichia coli. J. Biol. Chem., 274, 27885–27890. [DOI] [PubMed] [Google Scholar]
- Hsieh J., Moore,K.J.M. and Lohman,T.M. (1999) A two-site kinetic mechanism for ATP binding and hydrolysis by E. coli Rep helicase dimer bound to a single-stranded oligodeoxynucleotide. J. Mol. Biol., 288, 255–274. [DOI] [PubMed] [Google Scholar]
- Lin J.-J. and Sancar,A. (1992) Active site of (A)BC excinuclease. I. Evidence for 5′ incision by UvrC through a catalytic site involving Asp399, Asp438, Asp466 and His538 residues. J. Biol. Chem., 267, 17688–17692. [PubMed] [Google Scholar]
- Machius M., Henry,L., Palnitkar,M. and Deisenhofer,J. (1999) Crystal structure of the DNA nucleotide excision repair enzyme UvrB from Thermus thermophilus. Proc. Natl Acad. Sci. USA, 96, 11717–11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moolenaar G.F., Franken,K.L., Dijkstra,D.M., Thomas-Oates,J.E., Visse,R., van de Putte,P. and Goosen,N. (1995) The C-terminal region of the UvrB protein of Escherichia coli contains an important determinant for UvrC binding to the preincision complex but not the catalytic site for 3′-incision. J. Biol. Chem., 270, 30508–30515. [DOI] [PubMed] [Google Scholar]
- Moolenaar G.F., Franken,K.L., van de Putte,P. and Goosen,N. (1997) Function of the homologous regions of the Escherichia coli DNA excision repair proteins UvrB and UvrC in stabilization of the UvrBC–DNA complex and in 3′-incision. Mutat. Res., 385, 195–203. [DOI] [PubMed] [Google Scholar]
- Moolenaar G.F., Herron,M.F., Monaco,V., van Der Marel,G.A., van Boom,J.H., Visse,R. and Goosen,N. (2000) The role of ATP binding and hydrolysis by UvrB during nucleotide excision repair. J. Biol. Chem., 275, 8044–8050. [DOI] [PubMed] [Google Scholar]
- Moolenaar G.F., Hoglund,L. and Goosen,N. (2001) Clue to damage recognition by UvrB: residues in the β-hairpin structure prevent binding to non-damaged DNA. EMBO J., 20, 6140–6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa N., Sugahara,M., Masui,R., Kato,R., Fukuyama,K. and Kuramitsu,S. (1999) Crystal structure of Thermus thermophilus HB8 UvrB protein, a key enzyme of nucleotide excision repair. J. Biochem., 126, 986–990. [DOI] [PubMed] [Google Scholar]
- Orren D.K. and Sancar,A. (1989) The (A)BC excinuclease of Escherichia coli has only the UvrB and UvrC subunits in the incision complex. Proc. Natl Acad. Sci. USA, 86, 5237–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby C.P. and Sancar,A. (1993) Molecular mechanism of transcription–repair coupling. Science, 260, 53–58. [DOI] [PubMed] [Google Scholar]
- Skorvaga M., Theis,K., Mandavilli,B.S., Kisker,C. and Van Houten,B. (2002) The β-hairpin of UvrB is essential for DNA binding, damage processing and UvrC mediated incisions. J. Biol. Chem., 277, 1553–1559. [DOI] [PubMed] [Google Scholar]
- Sohi M., Alexandrovich,A., Moolenaar,G.F., Visse,R., Goosen,N., Vernede,X., Fontecilla-Camps,J.C., Champness,J. and Sanderson,M.R. (2000) Crystal structure of Escherichia coli UvrB C-terminal domain and a model for UvrB–UvrC interaction. FEBS Lett., 465, 161–164. [DOI] [PubMed] [Google Scholar]
- Theis K., Chen,P.J., Skorvaga,M., Van Houten,B. and Kisker,C. (1999) Crystal structure of UvrB, a DNA helicase adapted for nucleotide excision repair. EMBO J., 18, 6899–6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houten B. (1990) Nucleotide excision repair in Escherichia coli. Microbiol. Rev., 54, 18–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeven E.E.A., van Kesteren,M., Moolenaar,G.F., Visse,R. and Goosen,N. (2000) Catalytic sites for 3′ and 5′ incision of Escherichia coli nucleotide excision repair are both located in UvrC. J. Biol. Chem., 275, 5120–5123. [DOI] [PubMed] [Google Scholar]
- Verhoeven E.E.A., Wyman,C., Moolenaar,G.F., Hoeijmakers,J.H. and Goosen,N. (2001) Architecture of nucleotide excision repair complexes: DNA is wrapped by UvrB before and after damage recognition. EMBO J., 20, 601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visse R., de Ruijter,M., Moolenaar,G.F. and van de Putte,P. (1992) Analysis of UvrABC endonuclease reaction intermediates on cisplatin-damaged DNA using mobility shift gel electrophoresis. J. Biol. Chem., 267, 6736–6742. [PubMed] [Google Scholar]
- Wyman C., Rombel,I., North,A.K., Bustamante,C. and Kustu,S. (1997) Unusual oligomerisation required for activity of NtrC, a bacterial enhancer-binding protein. Science, 275, 1658–1661. [DOI] [PubMed] [Google Scholar]