Abstract
The role played by insertion sequence IS911 proteins, OrfA and OrfAB, in the choice of a target for insertion was studied. IS911 transposition occurs in several steps: synapsis of the two transposon ends (IRR and IRL); formation of a figure-of-eight intermediate where both ends are joined by a single-strand bridge; resolution into a circular form carrying an IRR–IRL junction; and insertion into a DNA target. In vivo, with OrfAB alone, an IS911-based transposon integrated with high probability next to an IS911 end located on the target plasmid. OrfA greatly reduced the proportion of these events. This was confirmed in vitro using a transposon with a preformed IRR–IRL junction to examine the final insertion step. Addition of OrfA resulted in a large increase in insertion frequency and greatly increased the proportion of non-targeted insertions. The intermolecular reaction leading to targeted insertion may resemble the intramolecular reaction involving figure-of-eight molecules, which leads to the formation of circles. OrfA could, therefore, be considered as a molecular switch modulating the site-specific recombination activity of OrfAB and facilitating dispersion of the insertion sequence (IS) to ‘non-homologous’ target sites.
Keywords: insertion sequence/insertion specificity/intermolecular transposition/synaptic complex/transposition switch
Introduction
The choice of insertion sites for transposable elements has important implications both for the transposable element itself and for the host. Insertion into a specific target DNA sequence can provide a secure refuge for the transposon. For example, by using a specific sequence in a highly conserved gene in such a way that insertion does not disrupt the gene, the transposon would be assured of an insertion site in many bacterial species and would not affect the fitness of the host. On the other hand, in excluding insertions into other DNA sequences, this strategy precludes the capacity of the element to generate mutations or to activate or sequester genes, and thus to generate genetic diversity.
On a genomic scale, most transposons appear to insert ‘randomly’. However, when analysed at the nucleotide level, many show some degree of target preference. This differs significantly, however, from element to element. Recognition can be sequence specific, with sequence lengths varying from a few to many nucleotides. Other elements exhibit regional preferences, e.g. GC- or AT-rich DNA segments, probably reflecting more global parameters such as local DNA structure. Other factors that have also been implicated are the degree of supercoiling, DNA replication, transcription, direction of conjugative transfer and protein-mediated targeting to, or exclusion from, transcriptional control regions (for reviews, see Craig, 1997; Chandler and Mahillon, 2002).
Clearly, these various insertion preferences must reflect constraints in the architecture of the synaptic complexes (nucleoprotein structures that are a prerequisite for insertion) formed between the transposon ends, the target DNA and the enzyme that catalyses insertion (the transposase).
An additional type of insertion ‘specificity’ has also been noted intermittently in the literature: insertion close to sequences resembling the transposon end (Casadaban et al., 1981; Polard et al., 1994; Olasz et al., 1997). Indeed, in the initial isolate of the insertion sequence (IS), IS911, insertion had occurred next to a sequence resembling the IS end located in the cI gene of bacteriophage λ (Prere et al., 1990). This neighbouring sequence carried 16 nucleotides identical to IRL, including the conserved terminal trinucleotide 5′-TCA-3′. The sequence was located 3 bp from IRR, forming a sequence resembling a transposon circle junction (see below), and further analysis demonstrated that the flanking λ DNA acted as a surrogate end in transposition (Polard et al., 1992, 1994).
Targeted insertions of this type have not been studied systematically in detail and the mechanisms leading to their formation, their frequency and biological relevance are not known. We have addressed some of these questions here by exploiting the multistep transposition pathway of IS911.
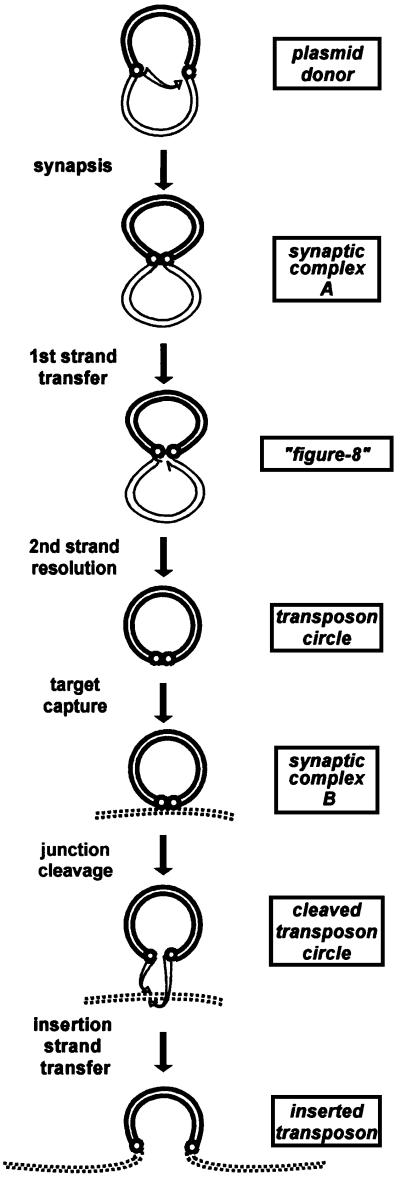
Transposition of many IS elements, including IS911 and other members of the large IS3 family, involves the formation of a transposon circle intermediate (for review, see Rousseau et al., 2002; Figure 1). During circle formation, the transposase recognizes both IS ends [IRL (left) and IRR (right)], assembles them into a first synaptic complex (synaptic complex A), and catalyses cleavage at the 3′ end of one inverted repeat (the donor IR) to liberate a 3′OH end, which is then used in a nucleophilic attack of the opposing end (the target IR). This asymmetric strand transfer event generally occurs at a position three bases from the tip of the second IR. This creates a single- strand bridge between the two ends, which, on a circular plasmid molecule, generates a figure-of-eight structure. Resolution of the second strand, presumably using host cell functions, generates a closed circular transposon molecule in which the two abutted IRs are separated by 3 bp. Insertion must then involve the formation of a second type of synaptic complex (synaptic complex B) in which the covalently joined IRL and IRR are paired with a target DNA.
Fig. 1. Proposed IS911 transposition pathway. Transposon DNA (bold lines), donor backbone sequences (fine lines), target DNA (dotted lines), IRL and IRR (small circles). The different steps of IS911 transposition are shown: synapsis of the ends in a plasmid donor to generate synaptic complex A; strand cleavage at one end (the donor end) and transfer to the other (the target end) to form a figure-of-eight structure in a reaction that requires only the transposase OrfAB; second strand resolution requiring host functions but no IS911 proteins to generate the transposon circle; synapsis with a target DNA molecule (synaptic complex B); cleavage of the IRR–IRL junction; and transfer into the target, which requires the transposase OrfAB. Final insertion requires OrfAB and is greatly stimulated by OrfA.
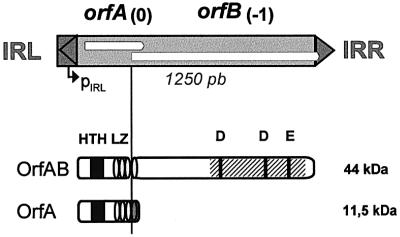
The two types of synapse necessitate two related IS911-encoded proteins, OrfA and OrfAB (Figure 2). OrfAB is a fusion protein produced by programmed translational frameshifting between two partially overlapping open reading frames (ORFs), orfA and orfB. OrfAB carries a classic DDE catalytic pocket in its C-terminal region (see Rousseau et al., 2002). Residues involved in binding the 36 bp terminal inverted repeats (IRs) and in ensuring protein multimerization are located in its N-terminal region (Haren et al., 1998, 2000; Normand et al., 2001). OrfAB is sufficient for generating figure-of-eight molecules both in vivo and in vitro (Polard and Chandler, 1995; Polard et al., 1996). OrfA, the product of the upstream reading frame orfA, shares its first 86 amino acids with OrfAB but differs in its terminal 14 amino acids. It can form homo- and heteromultimers with OrfAB (Haren et al., 1998, 2000). It strongly stimulates IS circle insertion in the presence of OrfAB (Ton-Hoang et al., 1998), and therefore appears to be implicated in synaptic complex B and in modulating insertion activity.
Fig. 2. Genetic organization and structure–function map of OrfAB and OrfA. Top: cartoon of IS911. Dark grey box, left terminal inverted repeat (IRL); triangle, right terminal inverted repeat (IRR); white boxes, reading frames orfA and orfB; 0 and –1, relative reading phases; pIRL, endogenous promoter; vertical line, point of translational frameshifting. Bottom: organization of IS911 proteins together with their molecular mass: black square, α-helix–turn–α-helix motif (HTH); ellipses, individual heptads of leucine zipper (LZ); cross-hatched box, catalytic domain; black vertical lines, DDE signature. Programmed translational frameshifting that fuses orfA and orfB to generate the transposase OrfAB occurs within the fourth heptad. The LZ of OrfA and OrfAB therefore differ in their fourth heptad.
We show here that insertion of IS911 circles in vivo, in the presence of OrfAB alone, occurs with high frequencies next to a resident IR into a target DNA molecule carrying inactivated IS911 ends. Co-production of OrfA with OrfAB in the same cells reduced the proportion of targeted insertions of this type. Similar results were obtained in vitro using a circular transposon carrying a preformed IRR–IRL junction. In these in vitro studies, OrfA was found to greatly stimulate insertion using the active junction, as observed previously (Ton-Hoang et al., 1998). This stimulation was found to be principally restricted to non-IR-targeted insertions. The results are discussed in terms of a model based on the known capacity of OrfAB to generate complexes including two transposon ends. Targeted insertion both in vivo and in vitro can be viewed as involving the pairing of two ends located on different DNA molecules: one of the ends of the IR–IR junction, together with an end on the target plasmid. This pairing may either resemble the synaptic complex involved in the formation of figure-of-eight molecules (synaptic complex A) or simply provide an anchor for localizing the incoming IR–IR junction (synaptic complex B). The capacity to switch target site specificity in this way provides a subtle mechanism for extending the range of recombination products. In the presence of both OrfAB and OrfA, the IS would simply tend to undergo dispersion.
Results
Preferential insertion of IS911 next to terminal IR sequences in vivo
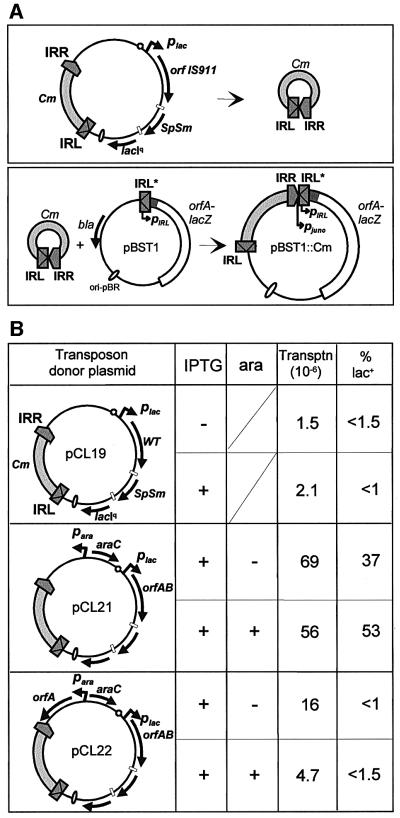
To assess whether IS911 generally prefers to insert next to sequences with homology to its ends, we initially used a plasmid-based in vivo system. Here, the transposon donor plasmid carried inducible IS911 genes and an artificial IS911-based transposon. The transposon must first be converted into a non-replicative circular form (Figure 3A, top panel) before insertion into the target molecule. The target plasmid carried the left end of IS911 (IRL) with the resident pIRL promoter and an orfA–lacZ fusion. Previous studies had shown that the resident promoter, pIRL, partially located in IRL was too weak to confer a lac+ phenotype. However, formation of an IRR–IRL junction creates a strong promoter, pjunc, composed of a –35 sequence located in IRR and a –10 sequence located in IRL and correctly positioned to drive expression of IS911 proteins (Ton-Hoang et al., 1998; Duval-Valentin et al., 2001). Insertions targeted to a resident IRL might therefore generate an IRR–IRL junction that could reconstitute pjunc and might be observed using a suitably located reporter gene (Figure 3A, bottom panel).
Fig. 3. Targeted insertion next to a single IR in vivo: influence of the IS911 proteins. (A) Experimental scheme. Top panel: circle formation from the donor plasmid. The general structure of the donor plasmids is indicated. They are derivatives of p15A and contain an artificial transposon composed of 52 bp of the left and 52 bp of the right IS911 end (dark grey boxes) flanking a gene for resistance to chloramphenicol (Cm; light grey box). The individual plasmids are described in more detail in (B). The cartoon shows the formation of transposon circle intermediates, which are subsequently inserted into the target plasmid (bottom panel). Bottom panel: structure of the target plasmid and the product of targeted insertion. Plasmid pBST1 carries a mutated IRL sequence (grey box) with the indigenous promoter pIRL and a gene fusion between the 5′ part of orfA and lacZ from the eighth codon (orfA–lacZ) (cross hatched box), a pBR322 origin of replication (unfilled oval) and the β-lactamase (bla) gene. The IR-targeted product that would result from the insertion of the CmR transposon 3 bp from the resident IRL* reconstitutes the strong pjunc promoter and activates the orfA–lacZ gene. The position and orientation of the two promoters are indicated by an arrow. This type of targeted IRL* insertion is observed, after transformation, on MacConkey lactose plates. (B) Structure of the donor plasmids, transposition frequency and percentage of lac+ colonies. The left column presents the different plasmids used to supply IS911 proteins and the artificial IS911 transposon. pCL19 carries the wild-type configuration of IS911 orfs, which drives expression of both OrfA and OrfAB proteins under the control of the lacUV5 promoter (plac). pCL21 and pCL22 carry the orfAB gene alone, where the A6G frameshift signal was exchanged for CA5G (Polard et al., 1992) to artificially fuse the orfA and orfB reading frames under the control of placUV5. It also carries the E.coli paraBAD and the araC gene, which encodes the arabinose-inducible repressor. In addition, pCL22 carries the orfA gene, which drives expression of OrfA alone (orfA), under the control of paraBAD. The direction of gene transcription is indicated by curved arrows, and the position and orientation of the promoter are indicated by an arrow. The small open circles correspond to transcription terminators of the E.coli rrnB operon, and the small white boxes represent phage T4 transcription and translation termination signals flanking the streptomycin/spectinomycin resistance cassette (SpSm). lacIq, the lacIq allele. The two right-hand columns indicate the transposition frequency (Transptn; ×106) and the percentage of lac+ (% lac+) colonies obtained on MacConkey lactose 1% Cm in the absence (–) or presence (+) of 1 mM IPTG, and in the absence (–) or presence (+) of 1% arabinose. The transposition frequency is expressed as the ratio of CmR transformants compared with ApR transformants and the percentage of lac+ is expressed as the number of CmR lac+ transformants as a percentage of total transformants (CmR).
The target plasmid, pBST1 (Figure 3A), was based on the pBR322 replicon and specified resistance to ampicillin (ApR). The single IRL carried by pBST1 included a mutation of the terminal CA dinucleotide to TC (IRL*) to eliminate its activity as a donor (Polard et al., 1996). All three transposon/transposase donor plasmids (pCL19, pCL21 and pCL22; Figure 3B) were p15A based. They included a spectinomycin/streptomycin resistance gene (SpSmR) and an artificial transposon composed of 52 bp of the left and 52 bp of the right IS911 ends flanking a chloramphenicol resistance gene (CmR). One, pCL19, carried the wild-type configuration of IS911 genes under the control of the plac promoter, which was itself controlled by a lacIq gene that was also located on the plasmids. The others, pCL21 and pCL22, carried a plac-driven orfAB gene in which orfA and orfB had been artificially fused to produce OrfAB constitutively (without frameshifting) (Polard et al., 1991). Plasmid pCL22 included, in addition, an orfA gene driven by the para promoter together with the araC gene under the control of its own promoter, while pCL21 carried only araC and para.
Escherichia coli JS238 carrying the target and one of the three donor plasmids was grown as described in Materials and methods, with induction of the plac-driven orfAB gene and, where appropriate, the para-driven orfA gene. Plasmid DNA was extracted, treated with appropriate restriction enzymes (to digest the donor plasmid) and used to transform the lac– host JS238. Selection was for CmR (for the transposon) on MacConkey lactose indicator plates, and the resulting colonies were screened for sensitivity to SpSm (SpSmS) to ensure loss of the donor plasmid backbone. The number of ApR colonies was also measured to determine the transformation efficiency of the target plasmid. Transposition frequencies were expressed as the number of CmR-SpSmS colonies compared with ApR colonies. The results are shown in Figure 3B. Plasmid pCL21, carrying only orfAB, exhibited a transposition frequency of between 56 and 69 × 10–6. Remarkably, between 37 and 53% of transposition events gave rise to CmR-SpSmS colonies with a lac+ phenotype. Similar results were obtained with a second donor plasmid devoid of para and araC (data not shown). The nucleotide sequence of the transposon-target junctions of six lac+ clones showed that insertion had occurred at 3 bp from the resident IRL*. Moreover, the 3 bp of sequence separating the two IRs in the newly formed IR–IR junction were those flanking the target IR, IRL*, as expected (see Rousseau et al., 2002).
A somewhat lower transposition frequency (16 × 10–6) was observed using pCL22, which includes the orfA gene. Induction of OrfA synthesis, under the control of the para promoter, reduced this frequency further to 4.7 × 10–6. Moreover, no colonies with a lac+ phenotype were detected (<1.5%).
Plasmid pCL19, in which plac drives a wild-type configuration of IS911 orfs (therefore requiring translational frameshifting to express OrfAB), also failed to generate detectable lac+ clones (<1.5%) and exhibited similarly reduced transposition frequencies to those of pCL22 both with and without plac induction.
These results therefore demonstrate that OrfAB alone can efficiently direct transposition events to a resident IS911 end in the target plasmid. Furthermore, the simultaneous presence of OrfA both reduces the frequency of targeted insertion and diminishes the overall frequency of transposition.
Preferential insertion of IS911 next to terminal IR sequences in vitro
To investigate the mechanism underlying this target site choice further, we used an in vitro assay for transposon circle insertion as described previously (Ton-Hoang et al., 1998). For these purposes we used a preformed IRL–IRR circle junction. This permits direct analysis of insertion and eliminates any effects of the IS911 proteins on the initial IRL–IRR synapsis, figure-of-eight and circle formation required for transposition in the in vivo system used above. Transposon circles, 3468 bp in length, composed of 55 bp of the left and 52 bp of the right IS911 ends flanking a chloramphenicol resistance gene (CmR), were produced in vivo from plasmid pAPT140.1 and purified as described in Materials and methods.
The results obtained in vivo addressed insertions directed to IRL only. In order to assess directed insertions at both IRL and IRR ends, pAPT182, the parent of pBST1 (used above in vivo), was employed as a target. This plasmid carries both IS911 ends in their natural relative orientation (Figure 4A) and an orfA–lacZ translational fusion. It includes the terminal 52 bp of the right end of IS911, with IRR in an inverted orientation with respect to IRL, inserted at the 3′ end of the gene fusion. pAPT182 was derived from pAPT166 (Polard and Chandler, 1995) by mutation of the terminal dinucleotide of both IRL (IRL*) and IRR (IRR*). IRR* carries the same mutation (CA to TC) as IRL*.
Fig. 4. Targeted insertion into a two-ended IS911 derivative in vitro. (A) Structure of the substrate and target, and a targeted insertion product. The cartoons show the transposon circle employed as substrate and pAPT182 employed as target plasmid together with an IRL*- targeted insertion product. The symbols are identical to those presented in Figure 3. The insertion reaction included OrfAB at 0.42 µM. (B) Junction structure of insertion products. The direction of gene (Cm and orfA–lacZ) transcription is indicated by arrows. The number of clones for each type of junction structure obtained is indicated. (C) Variation in junction spacing. Although the majority of insertions occur at a distance of 3 bp from the resident target IR, less frequent spacings of 2 and 4 bp are observed. The cartoons show how these two types of IR integration may be generated. The vertical arrows show the position of nucleophilic attack on the target plasmid. The black circles correspond to the 3 bp duplicated in the target plasmid.
In vitro insertion reactions were carried out between purified transposon circles and pAPT182 in the presence of OrfAB alone, purified as described previously (Ton-Hoang et al., 1998). The products were then transformed into the lac– indicator strain JS238 and CmR transformants were selected on MacConkey lactose indicator plates. Approximately 11% (124/1130) of CmR transformants were also lac+. Determination of the sequence neighbouring IRL* from several of the resulting plasmids confirmed that insertion of the CmR transposon circle had occurred such that the IRR was correctly placed with respect to IRL* to generate an active pjunc.
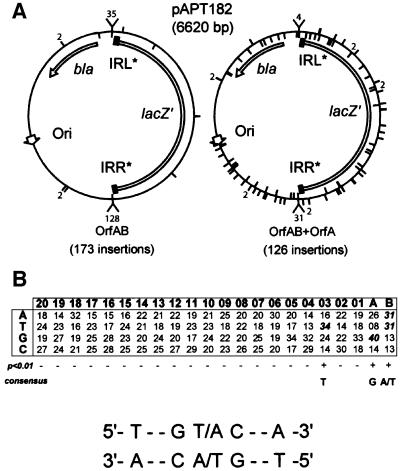
However, insertion resulting in pjunc formation (IRR– IRL*) represents only one of four such possible IR-targeted configurations. The other three, IRL–IRL*, IRR–IRR* and IRL–IRR*, would not be expected to generate an active pjunc. The sequence of 173 insertions, representing all CmR clones obtained in two independent experiments, was therefore determined. Of these, 163 (Figure 4B) occurred at one or another IR in the target plasmid. The remaining 10 insertions occurred at various alternative positions.
Of the 163 targeted insertions, 128 were adjacent to IRR* and 35 to IRL*. Thirty-nine of the 128 IRR*-targeted insertions carried an abutting IRR of the inserted transposon and 89 carried an abutting IRL. Eighteen of the 35 IRL*-targeted insertions carried an abutting IRR and 17 an abutting IRL. While most targeted insertions appeared to occur at a distance of 3 bp from the resident IR, several gave rise to 4- and 2-bp spacer lengths. The sequence of both the IR–IR* junction and the IR–plasmid junction of a sample of these insertions was determined. In all cases, including those with non-canonical spacer lengths, the target duplication was found to be 3 bp, as expected (see Rousseau et al., 2002). This implies that in the case of the 2 bp spacing, the initial strand transfer event occurred within the target IR between coordinates 1 and 2, and in the case of the 4 bp spacing, it occurred outside the IR between coordinates –1 and –2 (Figure 4C). In four cases, the mutated terminal dinucleotide of the targeted IR was found to have reverted to the wild-type sequence, presumably resulting from rare, host-mediated repair/recombination events. The nature of these events is at present under investigation.
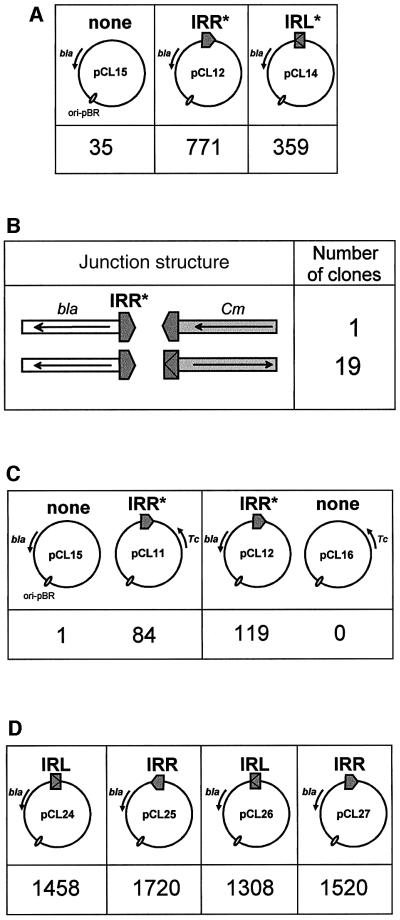
To study directed insertion into IR-carrying targets in more detail, a set of pBR322-based plasmids carrying single left or right ends was made (Figure 5A). Plasmids pCL12 and pCL14 carry IRR* and IRL*, respectively, cloned into pCL15 at the same position but inverted with respect to each other. In vitro insertion reactions were carried out using purified IS911 CmR transposon circles and purified OrfAB together with the target plasmid. The reaction products were used to transform JS238 with selection for CmR.
Fig. 5. Targeted insertion into a single IR derivative in vitro. (A) Efficiency of transposition using a target plasmid carrying mutated copies of IRR and IRL. The figure shows a physical map of the target plasmid. These were constructed from a pBR322-based plasmid and all included a selectable ampicillin resistance gene (bla). The right and left terminal inverted repeats, IRL and IRR, are represented by a grey box and a grey pointed box, respectively. Plasmids pCL12 and pCL14 carry IRR* and IRL*, in which the 5′-CA terminal dinucleotide has been mutated to 5′-TC, inverted with respect to each other. Both IRs were cloned at the same position in the plasmid. The parent plasmid pCL15 does not carry an IR. Insertion reactions were carried out with an OrfAB concentration of 0.32 µM and the products were introduced into JS238 by electroporation. The number of CmR colonies obtained in parallel is shown below for each target plasmid. These values are from a single experiment, although similar values were obtained in successive experiments. They are a measure of the relative efficiency of insertion of the CmR transposon circle. (B) Junction structure of targeted insertion products. The figure shows the configuration of IRs at the newly formed junction with pCL12 obtained from sequencing. bla and Cm indicate the plasmid- and transposon circle-associated antibiotic resistance genes, and the arrows show their direction of transcription. (C) Competition experiments using two target plasmids. pCL11 and pCL16 were constructed from a pBR322-based plasmid that included the selectable tetracycline resistance gene (Tc). pCL12 [see (A)] and pCL11 carry IRR* cloned in the same position and orientation. pCL15 [see (A)] and pCL16 are IR less. Insertion reactions were carried out with an OrfAB concentration of 0.32 µM and the products were introduced into DH5α by transformation. The values below the plasmids represent the absolute number of CmR colonies from one experiment that were also ApR or TcR. They represent the relative target efficiency of the two plasmids present in the reaction. (D) Efficiency of transposition using a target plasmid carrying wild-type copies of IRR and IRL. pCL24 and pCL26 each carry a wild-type IRL copy inverted with respect to each other, and pCL25 and pCL27 each carry an IRR inverted with respect to each other. The IRs were cloned at the same position in the plasmid as those in pCL12 and pCL14. Insertion reactions were carried out under the same conditions and in parallel with pCL12 and pCL14 [see (A)].
The results, presented in Figure 5A, indicated that the presence of a single end significantly increased the number of CmR transformants, confirming that a resident IR attracts transposon insertion. Similar results were obtained with another strain, DH5α (data not shown). Sequence analysis revealed that nearly 100% of insertions occurred next to the resident IR in the target plasmid. For pCL12, sequence analysis of 21 independent CmR clones revealed that 20 insertions were targeted to the resident IRR*, with 19 carrying an abutting IRL (Figure 5B). The remaining insertion had occurred within IRR*. Similar results were obtained with pCL14. Thus, as found in the previous experiments in vivo, a large fraction of insertions were targeted to the resident IR. Moreover, a majority were observed in a configuration where the opposite transposon IR was found abutting the resident end. These results clearly demonstrate that both IRL and IRR are strongly preferred IS911 integration sites.
This result was confirmed in competition experiments using an equimolar mixture of an IRR*-carrying target and a second target plasmid devoid of IS911 ends (Figure 5C). Plasmids carrying the tetracycline resistance gene (TcR) instead of ApR were constructed (pCL11, IRR*; pCL16, no end) and used as shown, in conjunction with pCL15 and pCL12. Since the standard strain used here, JS238, was TcR, the reaction products were used to transform the TcS strain DH5α with selection for CmR, and the resulting transformants were screened for ApR or TcR to determine which plasmid had received the insertion. The results are presented in Figure 5C. In both pairwise combinations, insertions into the IR-less plasmids were seen to occur at extremely low levels, as judged from these antibiotic resistance patterns. Similar results were obtained when the IRR*-carrying plasmids were substituted by identical plasmids carrying IRL* (data not shown). Thus, in competition with an end-less target, a single IS911 end attracted the majority of IS911 insertion events.
In the experiments described above (Figure 5A), the IRs carried by the target plasmid each contained a mutation of the terminal CA dinucleotide to inactivate their capacity as donors. Additional experiments were performed using a set of plasmids carrying a single wild-type IRL or IRR (Figure 5D). Several conclusions can be drawn from these results. The presence of a wild-type IR on the target plasmid resulted in a slight increase in insertion efficiency compared with a mutant target end (compare pCL12 with pCL27, and pCL14 with pCL26). This suggests that the activity of the wild-type target end contributes to the overall transposition reaction. Furthermore, the orientation of the IR had no notable effect on its efficiency as a target (compare pCL24 with pCL26, and pCL25 with pCL27). Moreover, although IRR* appeared to be used somewhat more frequently than IRL* (compare pCL12 with pCL14), no significant bias could be observed with the equivalent wild-type ends.
Together, the in vivo and in vitro results demonstrate that insertion of IS911 mediated by OrfAB alone is strongly directed to IS911 ends.
Effect of OrfA on targeting IS911 insertion in vitro
While recombination between IS911 ends to generate the figure-of-eight molecule is proficient in the presence of OrfAB alone, insertion of transposon circles is greatly stimulated by the addition of OrfA, both in vivo and in vitro (Ton-Hoang et al., 1998, 1999). Results obtained in vivo using pBST1 (Figure 3) indicated that the simultaneous production of OrfA with OrfAB greatly reduced the proportion of insertions that create pjunc. It was, therefore, of interest to determine the effect of different concentrations of OrfA on IS911 insertion specificity.
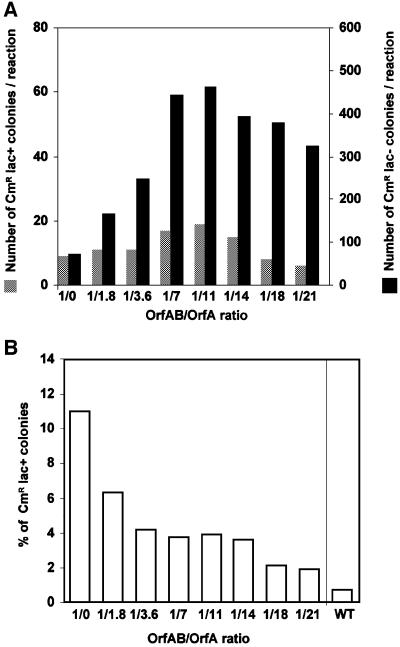
To address this, insertion reactions were carried out in vitro using the lacZ reporter plasmid pAPT182 as described above (Figure 4A). In this case, a constant quantity of OrfAB was supplemented with increasing levels of purified OrfA, which span the approximate molar ratio of 1/7 (OrfAB/OrfA), expected to be produced by natural frameshifting in vivo (Polard et al., 1991). The results presented in Figure 6A show that the level of both CmR lac+ and CmR lac– colonies was stimulated by OrfA. However, CmR lac– colonies increased ∼6-fold, while CmR lac+ colonies increased only by a factor of 2 at an optimal OrfAB/OrfA ratio of 1/11. It should be noted that a further increase in the amount of OrfA resulted in a small decrease in the number of colonies. This effect had already been observed in an analysis of the in vitro reaction products by gel electrophoresis (Ton-Hoang et al., 1998). The overall result (Figure 6B) was that the percentage of CmR lac+ colonies decreased from 11 to 1.9% as the levels of OrfA were increased. Moreover, although an enriched extract obtained from overexpression of the wild-type configuration of IS911 orfs (which contain OrfA and OrfAB produced by natural frameshifting) proved to be highly efficient in insertion, only 0.7% of CmR colonies were also lac+ (Figure 6B, WT).
Fig. 6. Targeted insertion into a two-ended IS911 derivative in vitro as a function of OrfAB and OrfA. (A) Efficiency of targeted and non- targeted insertion as a function of the OrfAB/OrfA ratio. The efficiency of targeted and non-targeted insertion was measured as the number of CmR lac+ and CmR lac– colonies, respectively, following transformation of JS238 with the reaction products. The concentration of OrfAB was kept constant (0.42 µM) and the concentration of OrfA was varied. The concentration of OrfA used was 0.0, 0.7, 1.4, 2.8, 4.2, 5.6, 7 and 8.4 µM, corresponding to the molar ratios of OrfAB/OrfA 1/0, 1/1.8, 1/3.6, 1/7, 1/11, 1/14, 1/18 and 1/21. (B) Percentage of IRL*-targeted insertion as a function of the OrfAB/OrfA ratio. The percentage of IRL*-targeted insertion is expressed as the number of CmR lac+ transformants as a percentage of total (CmR) transformants. A similar experiment performed with proteins obtained from an overexpressed wild-type (WT) configuration of IS911 orfs (0.6 µg) was also included.
The above experiment addresses the number of targeted insertions resulting in the formation of pjunc. To obtain a more accurate estimate of the overall proportion of targeted insertions, the sequence of the flanking DNA in a number of randomly selected insertions was determined. At an OrfAB/OrfA ratio of 1/1.8, the proportion of targeted insertions was reduced from 94% (163/173) with OrfAB alone (Figure 4B) to 39% (19/49), while at a ratio of 1/18 it was reduced even further to 21% (16/77). As observed in the absence of OrfA, there was clearly a strong bias towards insertions neighbouring IRR* (16/19 and 15/16, respectively). In the majority of targeted insertions, the two IRs were separated by the 3 bp flanking the target IR. However, several of these targeted insertions proved to carry non-canonical lengths of spacer between (2, 4 and 5 bp), as observed above (Figure 4C). All targeted insertions were consistent with a 3 bp flanking target duplication.
Sequence preferences of non-IR-targeted insertions
Localization of 126 insertions, examined at the DNA sequence level, showed that while 35 were targeted to an IS end, 91 were non-targeted and broadly distributed around the entire plasmid (Figure 7A, right), except in the region essential for replication, which includes RNA I and RNA II together with the origin of replication (see Wagner and Simons, 1994). This is to be compared with the distribution of 173 insertions obtained in the presence of OrfAB alone (Figure 7A, left). The insertions obtained in the presence of both OrfAB and OrfA appeared random, both in distribution and in orientation.
Fig. 7. Non-IR-targeted insertions. (A) Distribution of insertion sites in pAPT182. Genetic elements are indicated in the inner circles and include the following: gene encoding ampicillin resistance (bla), the ends IRR* and IRL*, the pBR322 origin of replication (Ori) and the reporter gene lacZ (see description of pAPT182, Figure 4A). The lines in the inner circle correspond to one orientation of insertion and those in the outer circle correspond to the opposite orientation. The numbers indicate the total number of insertion at a given site. The ‘funnels’ indicate IR-targeted insertions and numbers present the number of insertions found at a specific site. The left section shows the results obtained with OrfAB alone (0.42 µM), while the right section shows those obtained in the presence of both OrfAB (0.42 µM) and OrfA (0.7 and 7 µM). (B) Alignment of target sites used by IS911. Forty-four target sites and the flanking DNA sequences were aligned as 88 half-sites. The analysis was performed on a 20 bp window flanking the duplicated insertion site. A table was generated by compiling the sum of each nucleotide at each position in a 20 × 4 table. The significance of the nucleotide distribution at a given site was determined by the χ2 Pearson test [χ2 = Σ(o – c)2/c], where o and c are the observed and calculated values, respectively. It took into consideration the overall nucleotide composition of the target plasmid pAPT182 (22.59% A, 23% T, 27.28% G and 27.11% C) as calculated values. The positions at which a nucleotide bias occurs with >99% (P < 0.01) confidence levels are indicated by a ‘+’.
The sequences of both junctions of 61 of these examples were determined using primers that anneal within the ends of the Cm transposon circle. Of these, 44 exhibited a duplication of 3 bp of flanking target DNA while 17 had generated a 4 bp duplication. The presence of the flanking target duplication confirms that these insertions resulted from authentic transposition events. To determine whether there was a preferred target sequence with 2-fold symmetry, the 88 target half-sites of 44 insertions with a 3 bp target duplication were aligned as described for IS10 (Bender and Kleckner, 1992) and analysed as described by Hu and Derbyshire (1998). The results are presented in Figure 7B. Column B corresponds to the central base of the 3 bp target duplication, and column A to the first and third base. Columns 1–20 correspond to the base positions flanking the target duplication. The 22 × 4 table was generated by compiling the sum of each nucleotide at each position. The significance of the nucleotide distribution at a given site was determined by a χ2 test, which took into consideration the overall nucleotide composition of the target plasmid pAPT182 (22.59% A, 23% T, 27.28% G and 27.11% C). The positions at which a nucleotide bias occurs with >99% (P < 0.01) confidence levels are indicated by a ‘+’. This analysis failed to show significant conservation in the flanking regions, but revealed a consensus (T··G T/A C··A) for the flanking 3 bp direct repeat target sequence generated by the insertion.
Discussion
OrfAB and OrfA influence target choice in vivo
The results of in vivo experiments (Figure 3) using a donor plasmid carrying an artificial IS911-based transposon and supplying OrfAB alone showed a high level of IR-specific insertion. About 50% of transposition events were directed to a resident IS911 end in the target plasmid in such a way as to create the pjunc promoter, able to drive a β-galacto sidase reporter gene, giving rise to a lac+ phenotype. The DNA sequence of several of these clones confirmed that IRR and IRL* were separated, as expected, by 3 bp. The simultaneous presence of OrfA somewhat reduced the overall transposition frequency, but strongly reduced or eliminated (<1.5%) the formation of pjunc.
This assay depended on a complete IS911 transposition cycle, involving synapsis of IRL and IRR in the donor plasmid, figure-of-eight formation and resolution into transposon circles, and, finally, insertion of the transposon.
Although OrfA has been shown to stimulate the final insertion step in IS911 transposition both in vivo and in vitro (Ton-Hoang et al., 1998, 1999), it has a small inhibitory effect on the overall transposition pathway in vivo (Duval-Valentin et al., 2001). The protein may, therefore, influence earlier steps in IS911 transposition, e.g. by reducing the efficiency of synapsis of IRR and IRL in the figure-of-eight formation. The various possible ways in which OrfA may intervene in IS911 transposition are under investigation.
Targeted insertion in an in vitro integration assay
Target choice was investigated in a more controlled way, using an in vitro integration system that employed a preformed IRR–IRL junction as the transposon donor. With OrfAB alone, and a target plasmid carrying an artificial transposon with both IRL* and IRR* (inactivated for donor recombination activity by mutation of the terminal CA dinucleotide) in their natural orientation, 94% (163/173) of the insertions occurred at one end or the other (Figures 4B and 7A, left panel). Moreover, using target plasmids carrying either a single IRR* or IRL*, nearly all insertions were specifically directed to an end (Figure 5B). These insertions exhibited a strong preference for an orientation generating an IRL–IRR* or IRR–IRL* junction. However, IRL–IRL* and IRR–IRR* junctions were observed and proved to be functional in transposition (data not shown). In reactions containing a mixture of one of these plasmids (pCL11 or pCL12) and a second IR-less plasmid (pCL15 or pCL16), insertion into the IR-less plasmid was excluded (<1%; Figure 5C). Target plasmids carrying a functional IR, without the terminal mutated dinucleotide, exhibited a somewhat higher target efficiency than those carrying IRR* or IRL*. This observation suggests that the wild-type target IR contributes to the insertion reaction either by participating as a donor in the process or by permitting the formation of a more efficient synaptic complex. Interestingly, the results suggested that IRR* was a somewhat more efficient target than IRL*. This was observed when the IRs were located either on the same (pAPT182) or on different plasmids (pCL12 and pCL14). However, the effect was not observed when the target plasmid carried wild-type IRs. In view of the observed preference for forming IRR–IRL junctions in vitro (Figures 4B and 5B), this result might be explained if IRL was a more effective donor than IRR. When both target IRs are mutant, attack of IRR* by IRL in the IR–IR junction would be more efficient than attack of IRL* by IRR. However, when the target plasmid carries wild-type ends, these are also capable of contributing as donors. A reciprocal contribution by wild-type ends in the target plasmid would tend to favour the attack of IRR in the IR–IR junction by the ‘target’ IRL and thus mask the difference in activities revealed by use of a mutant partner.
OrfA modulates target choice in vitro
As had been observed previously (Ton-Hoang et al., 1998, 1999), the simultaneous presence of OrfA and OrfAB strongly stimulated insertion using a preformed IRL–IRR junction. Addition of OrfA had been shown to result in a large increase in species in which coincident or coordinated strand transfer of both transposon ends had occurred. The stimulation of insertion observed here was primarily due to an increase in non-targeted insertions compared with targeted insertions (Figure 6A and B). Although most of these insertions were observed to generate 3 bp flanking duplications in the target DNA, a relatively high proportion (39%) gave rise to flanking duplications of 4 bp. This is a similar proportion to those found in nature: of 12 insertions in the Shigella genome listed in the IS database (www-IS.biotoul.fr), 10 proved to have a duplication of 3 bp, while the remaining two exhibited a duplication of 4 bp. Although excluded from the essential replication region of the target plasmid, the non-IR-targeted insertions obtained here appeared to be otherwise distributed at random (Figure 7A, right panel). Indeed, the flanking sequences of insertions with 3 bp target duplications showed no obvious similarities. However, a clear preference was observed for the three duplicated base pairs (G T/A C; Figure 7B). Similar analysis of insertions that had generated 4 bp duplications failed to show a significant consensus target duplication (data not shown).
These results therefore indicate that OrfA is capable of extending the choice of target sequences that OrfAB is able to use.
Target choice: a model
These results can be interpreted by a model that invokes the two types of IS911 synaptic complex, A and B (Figure 1). In this model, targeted insertions would arise from the formation of an intermolecular synaptic complex between the circular transposon intermediate carrying the IRL–IRR junction and the IR carried by the target plasmid (Figure 8). This complex would resemble a type A complex in which the donor and target molecules are held together by OrfAB. Cleavage and strand transfer would generate the equivalent of the intramolecular figure-of-eight intermediate by the creation of an intermolecular single-strand bridge. To complete insertion, the second donor end might undergo OrfAB-mediated cleavage and strand transfer, as would be expected to occur during circle insertion into a non-homologous target or, alternatively, this branched molecule could undergo resolution via host recombination and replication functions. This model is attractive in its simplicity as it uses only the presently known properties of the IS911 transposition cycle.
Fig. 8. A proposed mechanism for IR-targeted insertion. Transposon circle DNA is shown (bold lines), as is target DNA (dotted lines). The small white circles represent IRL and IRR, and the small grey circles represent IRL or IRR carrying the terminal 5′-CA to 5′-TC mutation. Insertion next to the resident mutated IR in the target plasmid is proposed to initiate by the formation of an intermolecular OrfAB-mediated synaptic complex that resembles the intramolecular synaptic complex A (Figure 1). A single-strand cleavage of one end of the transposon circle and site-specific transfer to the target plasmid would create an intermolecular single-strand bridge. Simple cleavage at the second transposon end and intermolecular strand transfer would result in simple insertion of the transposon circle.
The transposase OrfAB has been shown to pair two IS911 ends (Haren et al., 1998). In vivo and in vitro, it is the only IS911 protein required to generate figure-of-eight molecules (Polard and Chandler, 1995; Polard et al., 1996). Moreover, studies with resected IS911 ends showed that to retain the capacity to participate in a type A synaptic complex and to act as a target for a wild-type partner end in a figure-of-eight formation, it is sufficient to retain simply the sequences necessary for OrfAB binding (Normand et al., 2001). Furthermore, OrfAB is not only capable of acting on two IRs carried by the same IS, but efficiently catalyses exchange between two appropriately oriented IRs carried by different copies of the IS located on the same DNA molecule (Turlan et al., 2000). These observations are supported by in vitro studies showing that OrfAB can pair two DNA fragments, which include wild-type ends (Haren et al., 2000) or resected ends carrying the OrfAB binding site (Normand et al., 2001). Finally, OrfAB can generate a paired complex between a DNA fragment carrying an IRL–IRR junction and one carrying a single IR (R.Alazard, unpublished).
The notion that strand transfer occurs between IRs on the donor and target molecules is supported by the observation that substitution of a mutant for a wild-type end on the target plasmid results in a significant increase in insertion. This suggests that the wild-type ‘target’ IR can contribute to the recombination reaction.
The effect of simultaneous production of OrfA would then be to favour formation of synaptic complex B (Figure 1) and concerted integration of both ends contained in the IRL–IRR junction into a non-homologous target sequence, as has been shown previously (Ton-Hoang et al., 1998). OrfA would therefore channel transposition from complexes of type A to generate type B synapses. This role could therefore be regarded as a molecular switch determining insertion specificity.
However, this view may be oversimplified. Although OrfA greatly stimulates non-IR-targeted insertions in vitro (Figure 6), it also has a slight stimulatory effect on targeted insertions. It is possible that the A-type synaptic complex we propose to be involved in intermolecular strand transfer between the target IR and an IR from the IR–IR transposon junction is modified in the presence of OrfA and that, in these circumstances, the second transposon end is inserted directly by transpositional strand cleavage and transfer, just as in the case of the non-targeted insertions catalysed by the type B synaptic complexes. We are currently testing this model.
The control of target choice adopted by IS911, and presumably by other members of the IS3 family, uses two proteins to modulate insertion specificity and differs considerably from that of other bacterial transposable elements studied to date. Although transposon Tn7 also employs two different proteins, these are employed separately to direct insertion either to a specific chromosomal site, attTn7, or elsewhere in a non-sequence-specific manner (Peters and Craig, 2001). Moreover, other elements have developed mechanisms that, rather than attracting insertions, actively exclude them from inserting close to resident copies. This has been demonstrated for the transposable bacteriophage Mu, for the transposon Tn7 and for members of the Tn3 family (Craig, 1997), in spite of earlier reports that Tn3 undergoes preferential insertion into regions with similarity to its ends (Casadaban et al., 1981). Control of IS911 target choice is also quite distinct from other IS elements, such as IS10, whose transposition involves a single protein and which appears to be attracted to target sites that are naturally ‘kinked’ (Pribil and Haniford, 2000).
Targeted insertion and IS911 biology
Insertion next to a surrogate end to form an IR–IR junction is expected to occur only rarely under natural conditions since it is unlikely that OrfAB would be produced in a natural situation without accompanying synthesis of OrfA. However, little is known about the relationship between synthesis of IS911 proteins in a natural situation and the influence of changing environments on the dynamics of transposition. Once formed in the target, the IR–IR junction would be highly active in transposition when both OrfAB and OrfA are available. Not only would it facilitate insertion of the entire target molecule into a second target, but intramolecular ‘insertion’ would generate deletions, as has been demonstrated in the case of a cloned IR–IR junction in vivo and in vitro (Ton-Hoang et al., 1997, 1998). This type of activity of the IS might, therefore, result in the formation of new transposable DNA segments and would be effective in generating a variety of genomic rearrangements. On the other hand, intervention of OrfA would favour the spread of the entire IS element to non-related sites.
IS911 is not alone in possessing an activity that directs insertion next to a DNA sequence that resembles part of its terminal IRs. Similar observations were made in early studies with Tn3 (Casadaban et al., 1981), and more recently with IS30 (Olasz et al., 1997). In vitro studies with IS10 have also uncovered intermolecular synaptic complexes, called paired end complexes, of the type proposed here to be involved in targeted IS911 insertions (Chalmers and Kleckner, 1996). However, in contrast to IS911, these complexes are unlikely to play a role in vivo because transposition of IS10 involves cleavage of both DNA strands at the ends of the IS and separation from flanking DNA (Kennedy et al., 1998). Moreover, the IS10 transposase does not have the capacity for covalent end joining (formation of an IR–IR junction) essential for this type of targeted insertion. This would preclude productive transposition events using an intermolecular IS10 paired end complex.
IS911 and other members of the IS3 family have adopted a variety of novel mechanisms to control their transposition activity. These include the formation of circular transposition intermediates in which, in certain cases, the formation of the IR–IR circle junction assembles a strong promoter to amplify the synthesis of element-encoded proteins, and programmed translational frameshifting, providing related proteins with complementary functions. The ability of IS911 to choose between two alternative types of target sequence represents yet another example of the many levels of fine-tuning adopted by this element.
Materials and methods
Bacterial strains and media
Cell extracts were prepared from Mi898 (Polard et al., 1996). The bacterial strains used in all other assays were JS238 (Polard et al., 1992) and DH5α. Cultures were grown in Terrific Broth (Sambrook et al., 1989) supplemented, where necessary, with ampicillin (Ap; 100 µg/ml) and methicillin (Met; 1000 µg/ml), kanamycin (Km; 25 µg/ml), chloramphenicol (Cm; 30 µg/ml), tetracycline (Tc; 12.5 µg/ml), spectinomycin (Sp; 30 µg/ml) and streptomycin (Sm; 20 µg/ml). Selection was on L plates supplemented with appropriate antibiotics. Standard MacConkey indicator plates (Miller, 1972) were supplemented with 1% lactose and appropriate antibiotics.
Plasmids
Plasmids used for in vivo assays. Target plasmid pBST1 carries the same orfA–lacZ fusion as pAPT182 (below), but is deleted for IRR* (Ton-Hoang et al., 1997). Details of the construction of plasmids pCL19, pCL21 and pCL22 (p15A-based), derived from pAPT111 (pCL21, pCL22) or pAPT112 (pCL19), can be obtained upon request. The relevant features of these plasmids were verified by DNA sequencing (Amersham thermosequenase cycle sequencing protocol).
Plasmids used for in vitro assays. pAPT140.1 (Polard et al., 1992) was used to generate transposon circles in vivo. Plasmid pAPT182, used as a target, was derived from pAPT166 (Polard and Chandler, 1995) by directed mutagenesis of the terminal two base pairs of each IR of the IS911-derived transposon.
Details of the construction of pBR322-derived target plasmids pCL11, pCL12, pCL14, pCL15, pCL16, pCL24, pCL25, pCL26 and pCL27 can be obtained upon request. For pCL11 and pCL16, the resident bla gene was removed by EcoRI–PstI digestion, and either replaced by two complementary oligonucleotides with EcoRI and PstI termini, which constitute the mutated IRR*, to generate pCL11, or recircularized after blunting to generate pCL16. These plasmids are therefore TcR and ApS.
For pCL12, pCL14, pCL24, pCL25, pCL26 and pCL27, the resident TcR gene was removed by EcoRI–EagI digestion and replaced by two complementary oligonucleotides with EcoRI and EagI termini to reconstitute: IRR*(pCL12); IRL*(pCL14); IRL(pCL24); IRR(pCL25); IRL(pCL26); IRR(pCL27) (see Figure 5 for IR orientation). For pCL15, the digested plasmid was treated with the DNA blunting kit (Amersham) and recircularized. These plasmids are therefore TcS and ApR.
Plasmids used for IS911 protein production. Plasmids pAPT158, pAPT155 and pAPT156 (pBR322 based) were used to prepare OrfAB, the wild-type configuration of proteins or OrfA, respectively, as described previously (Ton-Hoang et al., 1998).
DNA procedures
Standard techniques were used for DNA manipulation and cloning (Sambrook et al., 1989). Restriction and DNA-modifying enzymes were purchased from New England Biolabs. DNA was isolated from agarose gels using the QIAquick gel extraction kit (Qiagen). Plasmid DNA was extracted using the miniprep or maxiprep kits (Qiagen). PCR products were purified using the QIAquick PCR purification kit (Qiagen). Oligonucleotides were radiolabelled for use in sequencing as described previously (Ton-Hoang et al., 1998).
Cell-free insertion system
Transposon circles were produced in vivo from pAPT140.1, gel purified and used in a standard reaction with purified IS911 proteins (Ton-Hoang et al., 1998).
In vivo transposition assay
JS238 carrying different combinations of the transposon donor and target plasmid was grown at 42°C with 1% glucose, in the presence of Ap and Cm. At an OD600 of 0.6, the culture was centrifuged, resuspended in Luria–Bertani medium supplemented with Cm and divided into two parts. In the case of pCL19 expressing the wild-type complement of proteins, one part was uninduced while the other was induced with 1 mM IPTG. For pCL21 (OrfAB) and pCL22 (OrfAB and OrfA), IPTG was added alone or together with 1% arabinose. Induction was for 2 h at 30°C.
Plasmid DNA was prepared, digested with ApaI and PmeI, which cleave only the donor plasmid, and the products were used to transform JS238. CmR transformants were selected on MacConkey lactose indicator plates and the resulting colonies were screened for sensitivity to SpSm (SpSmS) and for Ap resistance. Transposition frequency was determined as the ratio of CmR-SpSmS colonies to ApR colonies. The percentage of IRL*-targeted insertion corresponds to the percentage of red (lac+) colonies.
Acknowledgments
Acknowledgements
We thank G.Duval-Valentin, Z.Nagy and N.Larabi for discussions, G.Duval-Valentin, D.Lane and A.Carpoussis for critically reading the manuscript, and B.Cointin for technical assistance. C.L. was supported by a grant from the Ministère de l’Education Nationale. This work benefited from grants from the CNRS (UMR5100), the Programme Physique et Chimie du vivant (CNRS), the Programme microbiologie (MENRST), l’ARC, le Groupement de Recherche (GDR 2157), Évolution des Éléments Transposables: du Génome aux Populations and the Institut Fédératif de Recherche (IFR 109).
References
- Bender J. and Kleckner,N. (1992) Tn10 insertion specificity is strongly dependent upon sequences immediately adjacent to the target-site consensus sequence. Proc. Natl Acad. Sci. USA, 89, 7996–8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M.J., Chou,J., Lemaux,P., Tu,C.P. and Cohen,S.N. (1981) Tn3: transposition and control. Cold Spring Harb. Symp. Quant. Biol., 45, 269–273. [DOI] [PubMed] [Google Scholar]
- Chalmers R.M. and Kleckner,N. (1996) IS10/Tn10 transposition efficiently accommodates diverse transposon end configurations. EMBO J., 15, 5112–5122. [PMC free article] [PubMed] [Google Scholar]
- Chandler M. and Mahillon,J. (2002) Insertion sequences revisited. In Craig,N.L., Craigie,R., Gellert,M. and Lambowitz,A. (eds), Mobile DNA II. American Society of Microbiology, Washington, DC, pp. 305–366.
- Craig N.L. (1997) Target site selection in transposition. Annu. Rev. Biochem., 66, 437–474. [DOI] [PubMed] [Google Scholar]
- Duval-Valentin G., Normand,C., Khemici,V., Marty,B. and Chandler,M. (2001) Transient promoter formation: a new feedback mechanism for regulation of IS911 transposition. EMBO J., 20, 5802–5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haren L., Polard,P., Ton-Hoang,B. and Chandler,M. (1998) Multiple oligomerisation domains in the IS911 transposase: a leucine zipper motif is essential for activity. J. Mol. Biol., 283, 29–41. [DOI] [PubMed] [Google Scholar]
- Haren L., Normand,C., Polard,P., Alazard,R. and Chandler,M. (2000) IS911 transposition is regulated by protein–protein interactions via a leucine zipper motif. J. Mol. Biol., 296, 757–768. [DOI] [PubMed] [Google Scholar]
- Hu W.Y. and Derbyshire,K.M. (1998) Target choice and orientation preference of the insertion sequence IS903. J. Bacteriol., 180, 3039–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy A.K., Guhathakurta,A., Kleckner,N. and Haniford,D.B. (1998) Tn10 transposition via a DNA hairpin intermediate. Cell, 95, 125–134. [DOI] [PubMed] [Google Scholar]
- Miller J.H. (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Normand C., Duval-Valentin,G., Haren,L. and Chandler,M. (2001) The terminal inverted repeats of IS911: requirements for synaptic complex assembly and activity. J. Mol. Biol., 308, 853–871. [DOI] [PubMed] [Google Scholar]
- Olasz F., Farkas,T., Kiss,J., Arini,A. and Arber,W. (1997) Terminal inverted repeats of insertion sequence IS30 serve as targets for transposition. J. Bacteriol., 179, 7551–7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J.E. and Craig,N.L. (2001) Tn7: smarter than we thought. Nat. Rev. Mol. Cell Biol., 2, 806–814. [DOI] [PubMed] [Google Scholar]
- Polard P. and Chandler,M. (1995) An in vivo transposase-catalyzed single-stranded DNA circularization reaction. Genes Dev., 9, 2846–2858. [DOI] [PubMed] [Google Scholar]
- Polard P., Prere,M.F., Chandler,M. and Fayet,O. (1991) Programmed translational frameshifting and initiation at an AUU codon in gene expression of bacterial insertion sequence IS911. J. Mol. Biol., 222, 465–477. [DOI] [PubMed] [Google Scholar]
- Polard P., Prere,M.F., Fayet,O. and Chandler,M. (1992) Transposase-induced excision and circularization of the bacterial insertion sequence IS911. EMBO J., 11, 5079–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polard P., Seroude,L., Fayet,O., Prere,M.F. and Chandler,M. (1994) One-ended insertion of IS911. J. Bacteriol., 176, 1192–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polard P., Ton-Hoang,B., Haren,L., Betermier,M., Walczak,R. and Chandler,M. (1996) IS911-mediated transpositional recombination in vitro. J. Mol. Biol., 264, 68–81. [DOI] [PubMed] [Google Scholar]
- Prere M.F., Chandler,M. and Fayet,O. (1990) Transposition in Shigella dysenteriae: isolation and analysis of IS911, a new member of the IS3 group of insertion sequences. J. Bacteriol., 172, 4090–4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pribil P.A. and Haniford,D. (2000) Substrate recognition and induced DNA deformation by transposase at the target-capture stage of Tn10 transposition. J. Mol. Biol., 303, 145–159. [DOI] [PubMed] [Google Scholar]
- Rousseau P., Normand,C., Loot,C., Turlan,C., Alazard,R., Duval-Valentin,G. and Chandler,M. (2002) Transposition of IS911. In Craig,N.L., Craigie,R., Gellert,M. and Lambowitz,A. (eds), Mobile DNA II. American Society of Microbiology, Washington, DC, pp. 267–383.
- Sambrook J., Fritsh,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Ton-Hoang B., Betermier,M., Polard,P. and Chandler,M. (1997) Assembly of a strong promoter following IS911 circularization and the role of circles in transposition. EMBO J., 16, 3357–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton-Hoang B., Polard,P. and Chandler,M. (1998) Efficient transposition of IS911 circles in vitro. EMBO J., 17, 1169–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton-Hoang B., Polard,P., Haren,L., Turlan,C. and Chandler,M. (1999) IS911 transposon circles give rise to linear forms that can undergo integration in vitro. Mol. Microbiol., 32, 617–627. [DOI] [PubMed] [Google Scholar]
- Turlan C., Ton-Hoang,B. and Chandler,M. (2000) The role of tandem IS dimers in IS911 transposition. Mol. Microbiol., 35, 1312–1325. [DOI] [PubMed] [Google Scholar]
- Wagner E.G. and Simons,R.W. (1994) Antisense RNA control in bacteria, phages, and plasmids. Annu. Rev. Microbiol., 48, 713–742. [DOI] [PubMed] [Google Scholar]