Abstract
Membrane-type 1 matrix metalloproteinase (MT1- MMP) localizes at the front of migrating cells and degrades the extracellular matrix barrier during cancer invasion. However, it is poorly understood how the polarized distribution of MT1-MMP at the migration front is regulated. Here, we demonstrate that MT1-MMP forms a complex with CD44H via the hemopexin-like (PEX) domain. A mutant MT1-MMP lacking the PEX domain failed to bind CD44H and did not localize at the lamellipodia. The cytoplasmic tail of CD44H, which comprises interfaces that associate with the actin cytoskeleton, was important for its localization at lamellipodia. Overexpression of a CD44H mutant lacking the cytoplasmic tail also prevented MT1-MMP from localizing at the lamellipodia. Modulation of F-actin with cytochalasin D revealed that both CD44H and MT1-MMP co-localize closely with the actin cytoskeleton, dependent on the cytoplasmic tail of CD44H. Thus, CD44H appears to act as a linker that connects MT1-MMP to the actin cytoskeleton and to play a role in directing MT1-MMP to the migration front. The PEX domain of MT1-MMP was indispensable in promoting cell migration and CD44H shedding.
Keywords: actin/CD44/invasion/lamellipodium/MT1-MMP
Introduction
Cell migration requires a continuous reorganization of the actin-based cytoskeleton within the cell and concerted action of adhesion molecules and proteases outside. Cell adhesion molecules play a particular role in gaining a foothold at the migration front and releasing it at the rear (Lauffenburger and Horwitz, 1996; Mitchison and Cramer, 1996). At the same time, cells need to remove the extracellular matrix (ECM) to open the migration pathway. Matrix metalloproteinases (MMPs), also referred to as matrixins, are responsible for the degradation of ECM components, and have therefore been implicated in this process (Stetler-Stevenson et al., 1993; Murphy and Gavrilovic, 1999; Nagase and Woessner, 1999; McCawley and Matrisian, 2000). To eliminate the ECM barrier efficiently, MMP activity is required to work at the cell migration front, although the specific mechanism to regulate this polarized action is poorly understood. While the majority of MMPs are in a soluble form, six are membrane-type proteins (MT-MMPs) having devices that enable integration into the plasma membrane (Egeblad and Werb, 2002), such as transmembrane domains (Cao et al., 1995) or glycosyl-phosphatidyl inositol (GPI) moieties (Itoh et al., 1999; Kojima et al., 2000). By having a potent pericellular proteolytic activity, MT-MMPs have strong invasion-promoting activity compared with soluble MMPs (Hotary et al., 2000). In particular, MT1-MMP is frequently expressed in invasive cancer cells (Sato et al., 1994; Seiki, 1999) and in endothelial cells during angiogenesis (Hiraoka et al., 1998; Galvez et al., 2001). The substrates of MT1-MMP include type I collagen, laminin and fibronectin (Egeblad and Werb, 2002). The enzyme also activates other proMMPs, including proMMP-2 and proMMP-13 (Sato et al., 1994; Knauper et al., 1996). Thus, MT1-MMP triggers MMP activation cascades on the cell surface.
Upon stimulation of cells to migrate, MT1-MMP relocates to the lamellipodium (Sato et al., 1997; Galvez et al., 2001; Itoh et al., 2001) and such a polarized localization seems particularly useful for the cells to invade tissue. The increased concentration of MT1-MMP at the lamellipodia also promotes the formation of homodimers via the hemopexin-like (PEX) domain. This dimer formation in turn facilitates proMMP-2 activation (Itoh et al., 2001), since this process requires at least two adjacent MT1-MMP molecules (Kinoshita et al., 1998); one MT1-MMP molecule acts as a receptor for the proMMP-2–TIMP-2 complex while the other acts as an activator. In addition to its role as a protease in ECM degradation, MT1-MMP is also a critical part of the mechanism of cellular locomotion. For example, cleavage of laminin 5 at the γ2 chain (Koshikawa et al., 2000) or CD44H (Kajita et al., 2001) by MT1-MMP is linked to its migration-stimulating activity. Thus, it is particularly important to understand the mechanism by which the localiza tion of MT1-MMP is regulated during cell migration.
Cell adhesion molecules should cooperate closely with ECM-degrading proteases during cell migration and recent findings support this idea. For example, MMP-1 associates with α2β1 integrin during epithelial cell movement on type I collagen in the wound-healing process (Dumin et al., 2001). MMP-2 binds αvβ3 integrin and promotes endothelial cell migration (Brooks et al., 1996). We have also observed that MT1-MMP shed CD44H from the cell surface and stimulated cell migration in conjunction with the CD44 shedding (Kajita et al., 2001). It also binds MMP-9 and promotes tumor invasion (Yu and Stamenkovic, 1999). CD44 is primarily a hyaluronan (hyaluronic acid, HA) receptor and binds collagen I, fibrin and chondroitin sulfate proteoglycans (Naot et al., 1997). CD44 has multiple isoforms containing inserts derived from alternatively spliced exons at the variable region (Naot et al., 1997). Within the cytoplasm, CD44 interacts with ezrin/radixin/moesin (ERM) (Yonemura et al., 1998) and/or ankyrin (Bourguignon et al., 1992), both of which bind F-actin (Kalomiris and Bourguignon, 1988; Tsukita et al., 1994). CD44 is expressed in many types of migratory and metastatic tumor cells (Gunthert et al., 1991; Naot et al., 1997; Sneath and Mangham, 1998) and promotes the migratory potential of these cells (Thomas et al., 1992, 1993; Henke et al., 1996; Ladeda et al., 1998).
CD44 is spontaneously released from the cell surface by proteolytic processing (Goebeler et al., 1996; Okamoto et al., 1999; Kajita et al., 2001). Interestingly, only CD44 shedding by metalloproteinases seems to play a role in cell migration because MMP inhibitors suppress the cell migration and shedding while serine proteinase inhibitors do not (Okamoto et al., 1999; Kajita et al., 2001; Shi et al., 2001). Multiple metalloproteinases appear to have CD44 shedding activity (Okamoto et al., 1999; Kajita et al., 2001), but only MT1-MMP has been clearly characterized as a shedding enzyme with migration-promoting activity (Kajita et al., 2001). Both MT1-MMP and CD44H localize at the migration front and we speculate that the events occurring here are critical for cell migration. Thus, it is of particular interest how CD44H and MT1-MMP co-localize at the lamellipodia when the cells migrate.
This study aims to elucidate how both MT1-MMP and CD44H are regulated to co-localize at the migration front. We found that MT1-MMP and CD44H form a complex mediated by the interaction between the PEX domain and the stem region of CD44H. Deletion of the PEX domain abolished the ability of the mutant MT1-MMP to localize at lamellipodia. Conversely, overexpression of the cytoplasmic-deletion mutant of CD44H, which shows no association with the actin cytoskeleton, prevented the localization of MT1-MMP to the lamellipodia. Thus, CD44H appears to link MT1-MMP to the actin cytoskeleton and regulate its localization. This interaction through the PEX domain is also critical for the shedding of CD44H and the cell migration-promoting activity of MT1-MMP. Our findings shed light on the close interrelationship between CD44 and MT1-MMP, both of which are implicated in cell migration and invasion.
Results
MT1-MMP forms a complex with CD44H
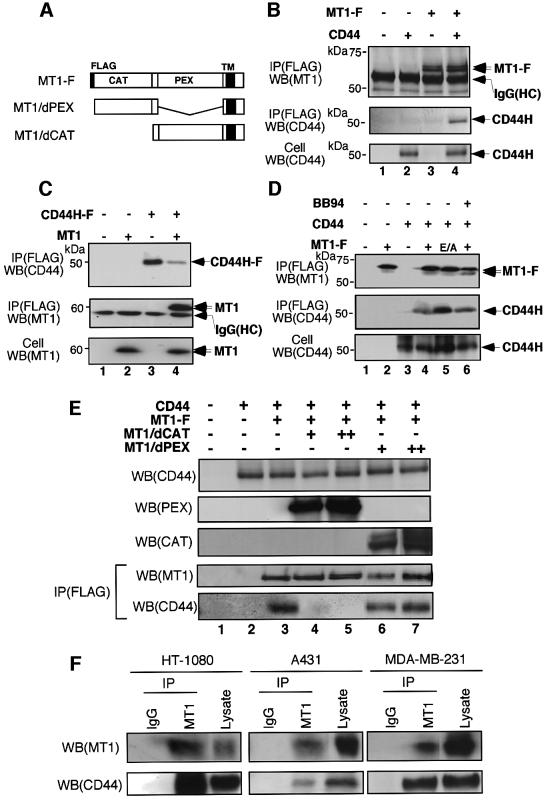
MT1-MMP co-localizes with CD44H at the cell migration front and releases CD44H via processing in the stem region (Kajita et al., 2001). Since the physical distance between the two molecules must be sufficiently close for such a cleavage reaction to occur, we examined the possibility that both MT1-MMP and CD44H are in the same complex. FLAG-tagged MT1-MMP (MT1-F) and CD44H were expressed in COS-1 cells to investigate whether the immunoprecipitation of either molecule led to co-precipitation of the other. As shown in Figure 1B, CD44H was precipitated by anti-FLAG antibody when it was co-expressed with MT1-F (lane 4) but not by itself (lane 2). Next, we tested whether MT1-MMP can be accompanied by precipitation of FLAG-tagged CD44H (CD44H-F). As shown in Figure 1C, MT1-MMP was precipitated by anti-FLAG antibody when it was co-expressed with CD44H-F (lane 4), but not by itself (lane 2). These results indicate that MT1-MMP and CD44H form a complex, either directly or indirectly.
Fig. 1. MT1-MMP and CD44H form a complex. (A) Schematic illustration of expressed MT1-MMP proteins. FLAG, FLAG epitope; CAT, catalytic domain; PEX, hemopexin-like domain; TM, transmembrane domain. (B–E) Complex formation between MT1-MMP and CD44H was examined by immunoprecipitating one component, followed by an analysis of the existence of the other in the precipitate by western blotting. (B) COS-1 cells were transiently transfected with expression plasmids for MT1-F and CD44H, as indicated. Immunoprecipitation was performed using anti-FLAG M2 mouse monoclonal antibody. Precipitates were then analyzed by western blotting using mouse monoclonal antibody (222-1D8) for MT1-MMP or mouse monoclonal antibody (2C5) for CD44. (C) Cells were transfected with plasmids for MT1-MMP and CD44H-F as indicated, and analyzed similarly. CD44H-F was immunoprecipitated with anti-FLAG M2 antibody. (D) Similar experiments to (B) were performed, using the (E/A) mutant of MT1-MMP or in the presence of BB94 (10 µM). (E) The role of the PEX and catalytic domains in complex formation was examined. To compete with MT1-F in the formation of a complex with CD44H, either MT1/dPEX or MT1/dCAT was co-expressed at increasingly higher doses (+, 8 µg; ++, 16 µg of plasmid per 100 mm dish). (F) Similar experiments to (B) were performed using HT1080, A431 and MDA-MB-231 cells. Immunoprecipitation was carried out using either rabbit IgG or rabbit polyclonal antibodies against the catalytic domain of MT1-MMP. These immunoprecipitates and cell lysate were then analyzed by western blotting using the antibodies indicated.
CD44H expressed in COS-1 cells appeared as a major 50 kDa band with an additional weak smear scattering between 60 and 95 kDa (Figure 1D, bottom column). This scattered molecular weight presumably represents variation in the glycosylation state of the protein. Upon co-precipitation with MT1-F, the 50 kDa band corresponding to the major product was observed (Figure 1B, lane 4 and D, lane 4). Since MT1-MMP can cleave CD44H, we examined whether the CD44H associating with MT1-MMP remains as an intact product or is processed. To do this, we used a mutant MT1-MMP that is deficient in proteolytic activity due to a Glu to Ala substitution at the catalytic site (E/A) (Atkinson et al., 1995). The CD44H that was co-immunoprecipitated with MT1-F(E/A) was the same size as that which co-immunoprecipitated with MT1-F (Figure 1D, lanes 4 and 5). The same result was obtained using the MMP inhibitor, BB94 (lane 6). Thus, we concluded that the CD44H in the complex is in an uncleaved form.
To identify the domain responsible for complex formation, a MT1-MMP mutant deleted of either the catalytic or PEX domain (MT1/dCAT and MT1/dPEX, respectively, see Figure 1A) was co-expressed with MT1-F and CD44H, and tested for its ability to compete with the formation of a complex between MT1-F and CD44H. As shown in Figure 1E, CD44H co-precipitated with MT1-F was dramatically reduced when increasing amounts of MT1/dCAT were co-expressed (lanes 4 and 5), but this was not the case for MT1/dPEX expression (lanes 6 and 7). Thus, the PEX domain of MT1-MMP is important for the formation of a complex with CD44H.
To confirm the complex formation of endogenous molecules, we used three human cell lines that express both MT1-MMP and CD44 (fibrosarcoma HT1080, breast carcinoma MDA-MB-231 and epidermoid carcinoma A431). Endogenous CD44 was detected in the MT1-MMP precipitates from cell lysate of the three cell lines (Figure 1F). However, MT1-MMP in the CD44 precipitates was difficult to detect presumably because CD44 is an abundant molecule and only a minor portion of CD44 forms a complex with MT1-MMP (data not shown).
The PEX domain of MT1-MMP binds directly to the stem region of CD44H
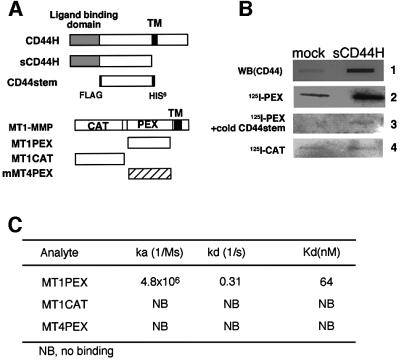
To further examine interactions between CD44H and MT1-MMP, a soluble fragment of CD44H (sCD44H) lacking the transmembrane and the cytoplasmic domains was constructed and expressed in COS-1 cells. The culture medium of the cells expressing sCD44H or that of mock-transfected cells was collected and immobilized on a membrane filter. The amount of sCD44 immobilized on the filter was detected using anti-CD44 antibody (Figure 2B, column 1). The small amount of CD44 observed in the control slot represents the endogenous shedded CD44 fragment derived from the mock-transfected cells. The filter was then blotted with either 125I-labeled PEX or CAT fragments. [125I]PEX bound the filter and the level was proportional to the amount of sCD44 immobilized there (column 2). On the other hand, 125I-labeled CAT did not bind the filter (column 3). To confirm the binding specificity of the PEX fragment for sCD44H, a competition experiment was performed using a purified CD44 fragment (CD44stem) that comprises the region between the ligand-binding and membrane-spanning domains (Figure 2A). Addition of the CD44stem fragment dramatically reduced the binding of [125I]PEX to the filter (Figure 2A, column 3), indicating that the PEX fragment binds directly to CD44stem.
Fig. 2. MT1-MMP binds directly to CD44H. (A) Schematic illustration of recombinant proteins used in the experiments. The proteins were expressed in E.coli and purified as described in Materials and methods. (B) Binding of recombinant MT1-MMP fragments to the ectodomain of CD44H (sCD44H) was examined by the ligand blot method. sCD44H was expressed in COS-1 cells and conditioned medium was collected and immobilized on a PVDF membrane filter. Anti-CD44 antibody (2C5), [125I]PEX or [125I]CAT was blotted onto the filter and bound ligands were visualized, either using secondary antibody conjugated to alkaline phosphatase or by radioautography. As a competitor, a 100-fold molar amount of purified CD44stem against [125I]PEX was used. (C) Kinetic analyses of the interactions between CD44stem and MT1-MMP. Interactions between CD44stem and either MT1PEX, MT1CAT, or MT4PEX were measured by SPR, as described in Materials and methods. CD44stem was immobilized on a sensor chip and interactions with the indicated analyte were examined at three different concentrations (2.5, 4.0 and 6.0 µM). Kinetic constants were calculated using the BIA evaluation program. NB, no binding was detected.
To verify the direct interaction between the two regions further, we analyzed the interactions between purified PEX and CD44stem fragments using surface plasmon resonance (SPR). CD44stem was immobilized on the sensor chip and MT-MMP fragments were used as analytes. Binding curves were obtained by flowing each analyte over the sensor chip at a flow rate of 5 µl/min with three different concentrations (2.5, 4 and 6 µM). The binding constant was obtained by analysis of the initial dissociation phase to obtain the Kd, which was then used for a global analysis of the association region of the curves, where a 1:1 interaction model was operative. MT1PEX but not MT1CAT or MT4PEX generated signals to bind CD44stem and the dissociation constant (Kd) between MT1PEX and CD44stem was calculated as 64 nM (Figure 2C). These results clearly indicate that MT1-MMP binds directly to the polypeptide core of CD44H at its stem region through the PEX domain.
Essential role of CD44H in directing MT1-MMP to lamellipodia
The actin cytoskeleton plays a pivotal role in controlling cell locomotion and affects the localization of cell surface molecules during cell migration. CD44 links the actin cytoskeleton within cells and localizes at the lamellipodia upon stimulation to induce cell migration. It is therefore possible that CD44 modulates the localization of MT1-MMP on the cell surface by forming a complex.
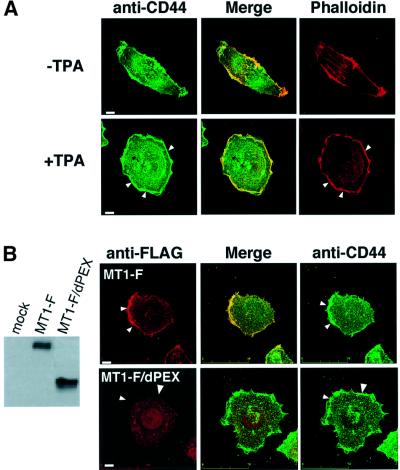
Human fibrosarcoma HT1080 is an invasive tumor cell line expressing both endogenous CD44H and MT1-MMP. Immunostaining of cell surface CD44 revealed that the pattern of distribution overlaps in part with that for F-actin, particularly at the cell periphery but not among the stress fibers (Figure 3A, –TPA). Upon stimulation of cells with 12-O-tetradecanoylphorbol-13-acetate (TPA), a ruffled membrane structure with dense cortical actin (lamellipodia) was detected at the edge and localization of CD44H to this area was also observed (+TPA). When MT1-F was expressed, it also localized to the ruffled area produced by TPA (Figure 3B, MT1-F) and this is consistent with the previous result reported for endogenous MT1-MMP (Itoh et al., 2001). If the localization of MT1-MMP at lamellipodia is regulated through binding to CD44H, it should be dependent on the PEX domain. As expected, MT1-F/dPEX did not localize at the lamellipodia (Figure 3B, MT1-F/dPEX). We have confirmed that deletion of other domains (catalytic, transmembrane and cytoplasmic domains) did not affect the localization (data not shown). Thus, we conclude that the PEX domain of MT1-MMP is critical in directing the protein to lamellipodia.
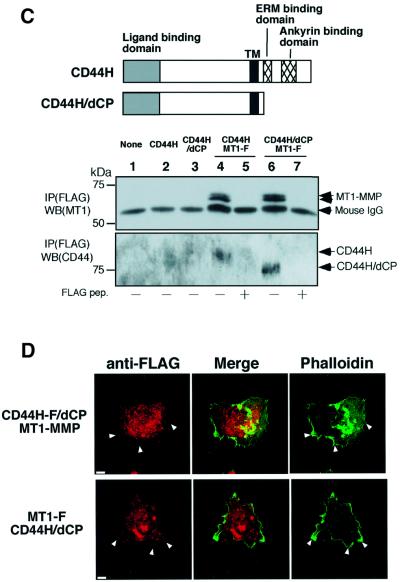
Fig. 3. Localization of CD44H and MT1-MMP to the lamellipodium. (A) HT-1080 cells were cultured on glass coverslips for 12 h. Cells were reacted with rabbit anti-CD44 antibody for 30 min. After a wash with PBS, cells were treated with TPA for 30 min. Following fixation, bound anti-CD44 antibody was visualized with Alexa488-conjugated anti-rabbit IgG. Actin cytoskeleton was identified with Alexa594-conjugated phalloidin. (B) HT-1080 cells expressing either MT1-F or MT1-F/dPEX were treated with anti-FLAG antibody for 30 min, followed by TPA. After fixation, antibodies reacting with MT1-F were visualized with Alexa488-conjugated anti-mouse IgG. The expression level of MT1-F and MT1-F/dPEX was confirmed by western blotting. (C) A mutant CD44H (CD44H/dCP) that lacks the cytoplasmic domain (as illustrated) was constructed. CHO-K1 cells were transiently transfected with the plasmids indicated and subjected to immunoprecipitation using anti-FLAG M2 antibody in the presence or absence of FLAG peptide (200 μg/ml). Precipitates were analyzed by western blotting using anti-CD44 antibody (2C5). (D) MT1-MMP + CD44H-F/dCP or MT1F + CD44H/dCP were expressed in HT-1080 cells. CD44H-F/dCP and MT1-F on the cell surface were visualized using anti-FLAG M2 antibody and Cy3- conjugated anti-mouse IgG. Cells were treated with TPA similarly to (A) before fixation. F-actin was stained with Alexa-594-conjugated phalloidin. Scale bar, 10 µm.
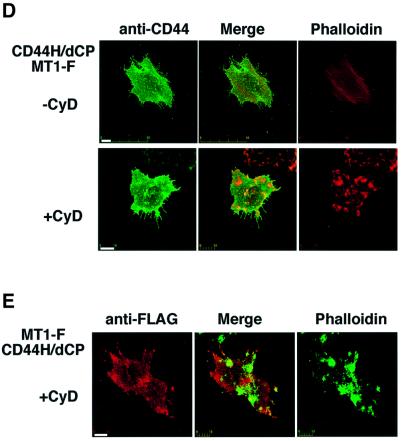
To examine the importance of CD44H in the localization of MT1-MMP to the lamellipodia, a mutant CD44H lacking the cytoplasmic domain, which is necessary for actin binding, was constructed (Figure 3C, CD44H/dCP). First, we confirmed the ability of the mutant to form a complex with MT1-F. MT1-F was co-expressed with either wild-type CD44H or CD44H/dCP, and immunoprecipitated using the anti-FLAG antibody. As shown in Figure 3C, CD44H was co-precipitated with MT1-F again (lane 3) and excess amounts of FLAG peptide inhibited the precipitation of both MT1-F and CD44H (lane 4). CD44H/dCP was also detected in the MT1-F precipitate (lane 6) and blocked by FLAG peptide (lane 7). Thus, CD44H/dCP retains the ability to form a complex with MT1-MMP just like wild-type CD44H.
Localization of FLAG-tagged CD44H/dCP (CD44H-F/dCP) in HT1080 cells was no longer observed at the ruffled area of TPA-treated cells (Figure 3D, CD44H-F/dCP + MT1-MMP). Thus, the cytoplasmic domain of CD44H is essential for its localization to the lamellipodia. Cells expressing CD44H/dCP formed a somewhat irregular ruffled edge compared with the parental cells, although dense cortical actin was still observed at the edge. When the localization of MT1-F co-expressed with CD44H/dCP was examined (Figure 3D, MT1-F + CD44H/dCP), MT1-F was no longer observed at the lamellipodia. At the same time, dissociation of the distribution patterns of MT1-F and CD44H/dCP from that of actin became clear. These results strongly suggest that CD44H plays a critical role in directing MT1-MMP to the lamellipodia.
CD44H mediates the linkage between MT1-MMP and the actin cytoskeleton
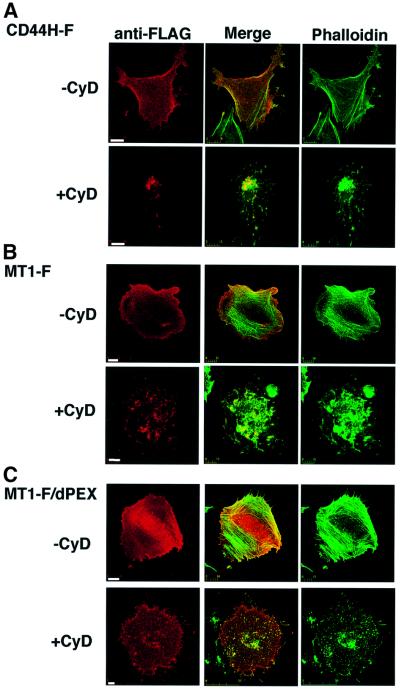
The localization of CD44H is regulated through its cytoplasmic tail, presumably by its association with the actin cytoskeleton. Accordingly, CD44H appears to regulate the localization of MT1-MMP that forms a complex with CD44H. To further verify the relationship between CD44H, MT1-MMP and the actin cytoskeleton, we modified the actin cytoskeleton using cytochalasin D (CyD), which disrupts polymerized F-actin (Morris and Tannenbaum, 1980).
CHO-K1 cells were used for the assay because they were adherent even after CyD treatment. Treatment of the cells with CyD disrupted the actin structure and caused aggregates to form within each cell (Figure 4). When CD44H-F was expressed in the cells, it co-localized in part with the actin structure before the CyD treatment (Figure 4A, –CyD). Upon treatment of the cells with CyD, CD44H-F moved to the site of actin aggregates (+CyD). The distribution of MT1-F was similar to that of CD44H-F before and after CyD treatment (Figure 4B). Interestingly, MT1-F/dPEX, which cannot form a complex with CD44H, failed to co-localize with actin cytoskeleton, both before and after CyD treatment (Figure 4C), while MT1-F mutants lacking domains other than the PEX behaved like wild-type MT1-F (data not shown).
Fig. 4. Actin cytoskeleton regulates localization of CD44H and MT1-MMP. CHO-K1 cells were transiently transfected with expression plasmids for CD44H-F (A), MT1-F (B), MT1-F/dPEX (C) and CD44H/dCP + MT1-F (D and E). Transfected cells were seeded on fibronectin-coated glass coverslips for 1 h and treated with anti-FLAG antibody (A–C and E) or anti-CD44 antibody (D) for 30 min at 37°C. After excess antibody had been washed off with PBS, the cells were treated with CyD (1 µg/ml) for 30 min at 37°C. They were then fixed, and the localization of the bound antibody and actin cytoskeleton was visualized with Cy3-conjugated anti-mouse IgG (red) and Alexa488-conjugated phalloidin (A–C and E) or Alexa488-conjugated anti-rabbit IgG and Alexa594-conjugated phalloidin (D), respectively. Scale bar, 10 µm.
To confirm that the cytoplasmic tail of CD44H mediates the co-localization of MT1-MMP and CD44H with F-actin, CD44H/dCP and MT1-F were co-expressed in the cells and examined in a similar fashion. As shown in Figure 4D and E, neither CD44H/dCP nor MT1-F co-localized with F-actin before or after CyD treatment. From these results, it is apparent that the localization of CD44H is regulated by the actin cytoskeleton through its cytoplasmic tail. Eventually, the actin cytoskeleton regulates the localization of MT1-MMP through CD44H.
MT1-MMP can be moved via CD44H on the cell surface
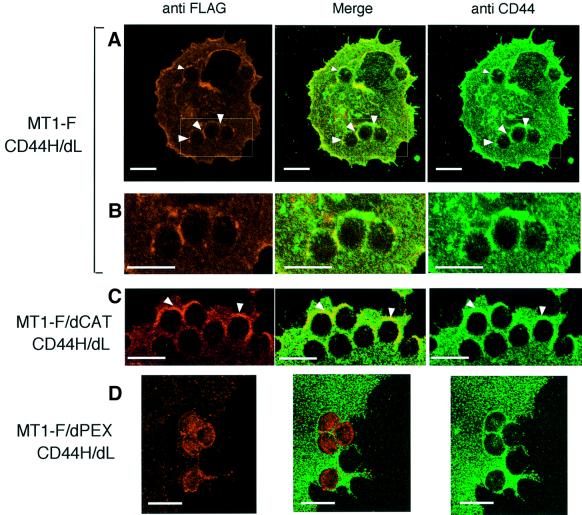
The above experiments suggest that CD44H functions as a molecular linker that connects MT1-MMP to the actin cytoskeleton, and eventually determines the localization of the metalloproteinase on the cell surface. To examine whether the complex is stable enough to move MT1-MMP via CD44H on the cell surface, CD44H was forced to move using antibody-conjugated microbeads. To avoid the effects of ligands in the culture, we constructed a CD44H mutant in which the ligand-binding domain was substituted with a Myc-tag sequence (CD44H/dL). Anti-Myc antibody-conjugated microbeads were used to bind the mutant protein around the beads. CD44H/dL was expressed in COS-1 cells, together with either MT1-F or MT1-F/dPEX. The ability of CD44H/dL to form a complex with MT1-F was confirmed by immunoprecipitation assay and the localization was similar to that of wild-type CD44H before and after CyD treatment (data not shown). The antibody-conjugated beads bound tightly to cells expressing CD44H/dL (Figure 5A), but not to mock-transfected cells (data not shown). Immunostaining of the cells with anti-CD44 antibody (Figure 5) revealed clustering of CD44H/dL surrounding the beads. As a control, IgG-conjugated beads were overlaid on the cells, which did not bind tightly to the cells or induce clustering of CD44 around the beads (data not shown). A similar localization of MT1-F to CD44/dL was observed around the antibody-conjugated beads (Figure 5A and B). Thus, the complex containing MT1-MMP and CD44H is stable enough to determine the localization of MT1-MMP by regulating CD44H expression on the cell surface. A similar co-localization with CD44HdL was observed for MT1-F/dCAT (Figure 5C) but not MT1-F/dPEX, which cannot bind to CD44H (Figure 5D). In Figure 5D, the amount of MT1-F/dPEX looks somewhat greater under the beads, but it is clear that the localization pattern of CD44HdL is different from that of MT1-F/dPEX (Figure 5D, Merge).
Fig. 5. MT1-MMP movement by pulling CD44H on the cell surface. A Myc-tagged mutant CD44H lacking the ligand-binding domain (CD44H/dL) was expressed together with either MT1-F (A and B), MT1F/dCAT (C) or MT1-F/dPEX (D) in COS-1 cells. After 48 h transfection, cells on glass coverslips were treated with anti-Myc-conjugated microbeads for 30 min. CD44 was then visualized with anti-CD44 rabbit polyclonal antibody and Alexa488-conjugated anti-rabbit IgG antibody. MT1-F, MT1-F/dCAT and MT1-F/dPEX were visualized with anti-FLAG M2 antibody and Alexa568-conjugated anti-mouse IgG antibody. The boxed areas in the MT1-F column (A) are magnified in the lower column (B). Only magnified pictures are presented for MT1-F/dCAT (C) and MT1-F/dPEX (D). Scale bar, 10 µm.
Role of the complex in CD44H processing and promotion of cell migration
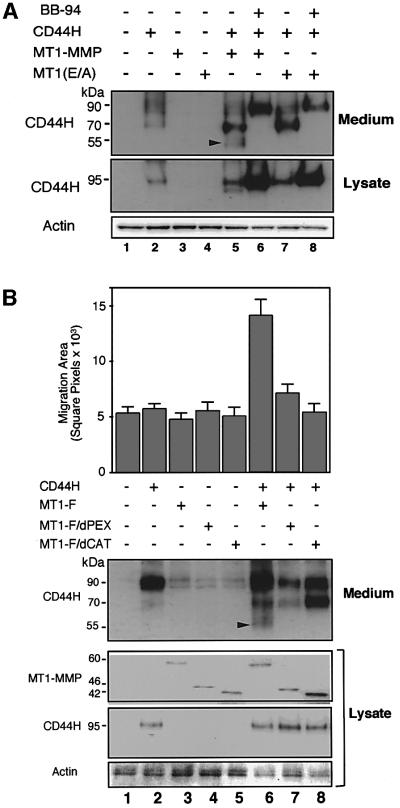
Since the PEX domain of MT1-MMP has the ability to form a complex with CD44H, we examined the role of this domain in CD44H processing and cell migration-promoting activity. A human osteosarcoma cell line (MG63) expressing low levels of endogenous CD44 and MT1-MMP was employed in this study. Exogenously expressed CD44H generated a 95 kDa band repre senting the highly glycosylated form, and the cells shed CD44 fragments of 90 and 70 kDa spontaneously (Figure 6A). Enforced expression of MT1-MMP with CD44H generated an additional 50 kDa fragment in the medium (lane 5). Expression of an inactive MT1-MMP (E/A) mutant failed to shed the 50 kDa fragment (lane 7), indicating that the shedding of this fragment depends on the catalytic activity of MT1-MMP. The MMP inhibitor, BB94, inhibited the shedding of both 70 and 50 kDa fragments (lane 6).
Fig. 6. Role of the PEX domain in CD44H processing and cell migration. CD44H was co-expressed with various MT1-MMP constructs in MG-63 cells and CD44H shedding into the medium and cell migration were analyzed. After transfection, cells were cultured for 24 h in the presence of 10% serum and then used for each assay. For the analysis of shedding (A and B), the cells were cultured for an additional 24 h in serum-free medium. The conditioned media were subjected to western blot analysis after TCA precipitation. For the phagokinetic assay (B), the cells were seeded onto colloidal gold-coated coverslips and allowed to migrate for 16 h in the serum plus culture condition. (A) CD44H shedding by MT1-MMP was examined. CD44H, MT1-MMP and MT1-MMP(E/A) were expressed as indicated. BB94 (10 µM) was also used to inhibit MT1-MMP activity. The CD44H fragment shed by MT1-MMP is indicated by an arrowhead. (B) Effects of MT1-MMP mutants on CD44H shedding and cell migration. A phagokinetic track assay on colloidal gold-coated coveslips was performed using the transfected cells. The cells were detected using green fluorescent protein (GFP) co-expressed with the effector proteins. The migration area is indicated by square pixels. An average of 30 cells ± SEM is shown in the upper panel. The lower panel displays proteins detected by western blotting and CD44H shed in the medium. The arrowhead signifies the CD44H fragment released by MT1-MMP.
In addition to the shedding of the 50 kDa CD44H fragment, the expression of MT1-MMP- and CD44H-stimulated cell migration was analyzed by a phagokinetic track assay (Figure 6B, lane 6). Although the expression of CD44H alone generated both 90 and 70 kDa fragments in the medium, this shedding was not sufficient to stimulate cell migration (lane 2). As reported previously (Itoh et al., 2001), MT1/dPEX is expressed as a catalytically active enzyme on the cell surface, but it alone was not enough to shed the 50 kDa CD44H fragment (lane 7) and to stimulate cell migration. MT1-MMP or CD44H alone could not promote cell migration (lanes 2 and 3). Thus, the PEX domain, which is necessary for formation of a complex with CD44H, is also critical for the processing of CD44H and stimulation of cell migration.
Discussion
MT1-MMP forms a complex with CD44H on the cell surface
Our results demonstrate that MT1-MMP and CD44H form a complex and co-precipitate in an immunoprecipitation assay. This interaction is not due to the overexpression of the proteins, because complex formed from endogenous CD44 and MT1-MMP was detected in three different cell lines (Figure 1F). The PEX domain of MT1-MMP binds directly to the stem region of CD44H and the binding does not require the glycosylation of this region. The core polypeptide of CD44H is 40 kDa, but its molecular weight varies between 50 and 95 kDa, depending on the glycosylation state of the protein. Glycosylation patterns of CD44H vary from cell to cell. In COS-1 cells, the major form of CD44H was a 50 kDa fragment that co-precipitated with MT1-MMP. However, in CHO-K1 cells, a 95 kDa band was detected in MT1-MMP precipitates. The difference in molecular weight is probably due to the glycosylation state of the ligand-binding domain, since mutations at possible glycosylation sites of the stem region do not substantially affect the molecular weight (Bartolazzi et al., 1996).
Role of complex formation in cell migration and invasion
When cells are stimulated to migrate, the first step involves the formation of microspikes and lamellipodia in the direction of migration (Lauffenburger and Horwitz, 1996; Mitchison and Cramer, 1996). The formation of these structures is regulated by the Rho family of GTPases, in particular, Cdc42 and Rac1 (Ridley et al., 1992). Overexpression of constitutively active Rac1 (Rac1V12) induces membrane ruffling, while the expression of constitutively active Rho A (RhoAV14) stimulates the formation of actin stress fibers and inhibits the formation of lamellipodia (Ridley et al., 1992). Okamoto et al. (1999) demonstrated that the treatment of U251MG cells with TPA leads to the redistribution of cell surface CD44 in the ruffled area. This redistribution process was inhibited when RhoAV14 was overexpressed. Thus, the localization of CD44 at the lamellipodia is regulated by the Rho family GTPases, presumably through reorganization of the actin cytoskeleton. The cytoplasmic tail of CD44 contains binding sites for ERM proteins (Tsukita et al., 1994) and ankyrin (Bourguignon et al., 1999), both of which mediate the association with F-actin. In our experiments, the co-localization of CD44H with the actin cytoskeleton was dependent on the cytoplasmic tail of the protein, which was also indispensable in directing CD44H to the lamellipodia in TPA-treated cells. Thus, by forming a complex with MT1-MMP, CD44H links MT1-MMP to the actin cytoskeleton and places it downstream of the signaling by the Rho family GTPases. In support of this, the localization of MT1-MMP to the lamellipodia was dependent on the PEX domain of MT1-MMP and the cytoplasmic domain of CD44H. The regulation of MT1-MMP by CD44 and the actin cytoskeleton is presumably an important part of cellular invasiveness. Disruption of the MT1-MMP/CD44/actin axis by overexpression of either MT1/dCAT (Itoh et al., 2001) or CD44H/dCP (data not shown) in HT1080 cells resulted in the inhibition of matrigel invasion. Although these results support the idea mentioned above, it is also difficult to exclude the possibility that MT1/dCAT or CD44H/dCP affected other machinery required for invasion because of their multifunctional properties. In relation to this, an interesting paper appeared recently in which the importance of CD44 in tumor invasion and metastasis was demonstrated using CD44 knockout mice (Weber et al., 2002). Tumors developed in the CD44 null mice when they also had a mutation in either the APC gene or the p53 gene. The absence of CD44 did not affect tumor incidence, but virtually abolished metastasis (Weber et al., 2002). Part of the effect of a CD44 deficiency on metastasis might be explained by a defect in the regulation of MT1-MMP by CD44. In addition, MT1-MMP is not the only metalloproteinase that binds CD44. Recently, MMP-9 and MMP-7 were both reported to bind to CD44 on the cell surface (Yu and Stamenkovic, 1999; Yu et al., 2002). Thus, CD44 seems to be a platform on which to assemble multiple MMPs on the cell surface, especially at the migration front. Additionally, MT1-MMP is reported to activate proMMP-9 indirectly through the activation of MMP-2 and MMP-13 (Cowell et al., 1998). Therefore, it is also possible that CD44 facilitates activation of proMMP-9.
CD44 is expressed in a wide variety of cell types and therefore, the above-mentioned mechanism possibly works in cells that additionally express MT1-MMP. Although it is difficult to find cell lines that do not express CD44, an osteosarcoma cell line, MG63, expresses this molecule at very low levels. In this particular cell line, exogenously expressed MT1-MMP did not co-localize with actin as long as CD44H was not expressed (data not shown). However, these experiments do not exclude the possibility that other cell adhesion molecules have a similar function to CD44H and regulate the localization of MT1-MMP. It is also possible that additional cell surface molecules participate in the formation and stabilization of the complex in cooperation with CD44H. The binding affinity between PEX and CD44stem expressed as the dissociation constant is 64 nM, which is not particularly high. However, the complex is stable enough to move MT1-MMP by pulling CD44 as indicated in Figure 5.
Complex formation, CD44 processing and stimulation of cell migration
In a previous study, we found that the catalytic fragment of MT1-MMP can cleave CD44H, at least in vitro (Kajita et al., 2001). However, MT1/dPEX expressed on the cell surface failed to shed CD44H, as shown in Figure 6. Thus, the PEX domain, which is needed to form a complex with CD44H, is clearly indispensable to the shedding event. On the other hand, processed CD44H fragments were not detected in the CD44H–MT1-MMP complex. One possible explanation for this apparent discrepancy is that MT1-MMP initially binds CD44H at the PEX domain, cleaves the CD44H and releases the cleaved fragment rapidly. If this is the case, the cleavage reaction in the complex may not be very efficient as significant amounts of intact CD44H are still present in the complex. This slow cleavage reaction may make it possible to maintain MT1-MMP at the sites where CD44H is localized. Another scenario is that heterogeneous glycosylation of CD44H at the stem region may lead to two types of molecule, one that binds MT1-MMP but is either not cleavable or cleaved very slowly, and one that binds MT1-MMP and is very sensitive to cleavage.
In this study, we showed that the PEX domain of MT1-MMP is important for the shedding of CD44H, stimulation of cell migration and localization to the migration front. However, many proteinases, including serine proteinases and metalloproteinases, can also shed CD44 (Okamoto et al., 1999; Alpaugh et al., 2000; Kajita et al., 2001). Although it is evident from a previous study (Kajita et al., 2001) that the shedding by MT1-MMP promoted cell migration, the roles of the shedding of CD44 by other proteinases are not fully understood yet. The role of the shedding in cellular function may differ depending on the responsive proteinases and cell types as in the following examples. Although serine proteinase inhibitors do not suppress cellular locomotion (Okamoto et al., 1999; Kajita et al., 2001), a chymotrypsin-like enzyme shed CD44 in myoepithelial cells and may modulate the adhesion and migration properties of the cells (Alpaugh et al., 2000). In U251MG cells, the main enzyme for CD44 shedding is a membrane-bound MMP that is sensitive to TIMP-1 but not TIMP-2 and appears to be responsible for the cell migration as well (Okamoto et al., 1999). Although these proteinases have not yet been identified, they may also associate with CD44 and localize at the cell migration front. MG63 cells, one of the cell lines used in this study, expresses proteinases that shed exogenously expressed CD44H into the medium as 90 and 70 kDa fragments (Figure 6). Shedding of the 70 kDa fragment was sensitive to BB94, but it was not sufficient to promote cell migration upon CD44H expression. Expression of MT1-MMP together with CD44H generated an additional 50 kDa fragment, and stimulated cell migration. However, it is currently unclear why the shedding of CD44 by the endogenous metalloproteinases in MG63 does not affect cell migration while MT1-MMP does. The identification of each proteinase responsible for shedding may provide an answer to this question. Shedding enzymes other than MT1-MMP may localize at particular compartments on the cell surface that are different from the migration front and regulate different CD44 functions there.
For cell migration and invasion, cell surface molecules must be regulated precisely in cooperation with multiple partners in order to function at the right site and time. The net amount of proteolytic activity by ECM-degrading enzymes is not enough to support efficient cell migration and invasion in tissue. In this study, we shed light on the mechanisms regulating the localization of MT1-MMP, one of the key enzymes responsible for cellular invasiveness. Since MT1-MMP is an important component of the invasion machinery of tumor cells, details of its regulatory mechanism will facilitate the development of advanced therapeutics for malignant cancer.
Materials and methods
Cell culture and transfection
Chinese hamster ovary cells, CHO-K1, were obtained from the Japanese Collection of Research Bioresources (Tokyo, Japan). Human fibrosarcoma (HT-1080), human osteosarcoma (MG-63), human epidermoid carcinoma (A431), human breast carcinoma (MDA-MB-231) and green monkey kidney (COS-1) cells were obtained from the American Type Culture Collection (Manassas, VA). CHO-K1 were maintained in Ham’s F12 medium (Sigma, St Louis, MO) supplemented with 10% fetal bovine serum (FBS) and non-essential amino acids (Life Technologies, MD) and other cell lines in Dulbecco’s modified Eagle’s medium (Sigma) with 10% FBS. For transfection, cells were seeded in six-well plates (at 1.0 × 105 cells/well) and transfected with expression plasmid using FuGENE6 (Roche Molecular Biochemicals, Basel, Switzerland), according to the manufacturer’s instructions.
Antibodies
Rabbit polyclonal antibody against human CD44 was purchased from Santa Cruz (Santa Cruz, CA); mouse anti-FLAG M2 antibody and alkaline phosphatase-conjugated antibodies (goat anti-mouse IgG and goat anti-rabbit IgG) from Sigma; mouse anti-c-Myc antibody from Roche Molecular Biochemicals; goat Cy3-conjugated anti-mouse IgG from Jackson ImmunoResearch Laboratories (USA); goat Alexa488-conjugated anti-rabbit IgG from Molecular Probes (USA); and mouse anti-MT1-MMPPEX monoclonal antibody (222-1D8) and anti-MT1-MMPCAT monoclonal antibody (113-5B7) from Daiichi Fine Chemical Co. (Takaoka, Japan). Anti-c-Myc antibody-conjugated beads were prepared using a carbodiimide kit for carboxylated microparticles (Polysciences, Inc.), according to the manufacturer’s instructions.
Construction of expression plasmids
All the mutants were constructed using a polymerase chain reaction (PCR)-based method. The cDNA sequences used as templates were as follows: human MT1-MMP (DDBJ/EMBL/GenBank accession No. D26512), MT4-MMP (accession No. AB021225) and CD44H (accession No. U40373). A mammalian expression vector, pSG5 (Stratagene, La Jolla, CA), was used to express the gene products. MT1-F with a FLAG-tag at the N-terminus of the activated form of the enzyme was constructed as reported previously (Itoh et al., 1999). Mutants were constructed as described previously (Uekita et al., 2001) and had the following structures: MT1-F/GPI mutant comprises the ectodomain of MT1-F (Met1–Cys508) and the GPI signal derived from MT4-MMP (Gly526–Leu607); MT1-F/dCAT is a catalytic domain-deleted mutant (Tyr112–Pro312) of MT1-F; MT1-F/dPEX lacks a PEX domain (Ile318–Gly535); PEX–GPI is a chimera of MT1-F/dCAT and the GPI-anchoring signal of MT4-MMP. CD44H/dL lacks a ligand-binding domain (Gln21–Cys129) and contains a Myc-tag sequence downstream of the signal peptide (Ala20). CD44 H/dCP contains Met1–Ser291 without a cytoplasmic tail. All cDNA constructs were confirmed by DNA sequencing.
Immunostaining
Cells were cultured on a glass coverslip and reacted with antibodies [rabbit anti-CD44 polyclonal antibody (1:500) or mouse anti-FLAG M2 monoclonal antibody (3 µg/ml)] in culture medium for 30 min at 37°C. After three washes with phophate-buffered saline (PBS), cells were fixed with 4% paraformaldehyde in PBS at room temperature for 10 min. Cy3-conjugated anti-mouse IgG and Alexa488-conjugated anti-rabbit IgG were employed as secondary antibodies. To disrupt the F-actin structure, cells were treated with 10 µg/ml of CyD (Sigma) for 30 min, then immunostained. F-actin was visualized with Alexa594-conjugated phalloidin (Molecular Probes) or Alexa488-conjugated phalloidin (Molecular Probes). Signals were subsequently analyzed using a Bio-Rad MRC-1024 confocal laser microscope.
Immunoprecipitation and western blot analysis
Cells (8 × 105 cells) were seeded in 100 mm dishes and transfected with expression plasmid (8 µg) using FuGENE6 After 48 h, the cells were solubilized in RIPA buffer (1% Triton X-100, 50 mM Tris–HCl pH 7.6, 150 mM NaCl, 1% deoxycholic acid and 0.1% SDS) in the presence of a protease inhibitor cocktail (Roche). Cell lysates were clarified by centrifugation at 16 000 g for 15 min, and the supernatant was incubated with anti-FLAG M2 antibody-conjugated agarose beads (Sigma) for 2 h. The beads were then washed three times with RIPA buffer, and the immune complex was solubilized in SDS sample buffer and subjected to western blot analysis.
Cell lysate or medium precipitated with trichloroacetic acid (TCA) was separated by SDS–PAGE, and proteins were transferred to a PVDF membrane. Following blockage with 10% fat-free dried milk in Tris-buffered saline, the membrane was probed with primary antibodies specific to each protein. The membrane was further probed with alkaline phosphatase-conjugated goat anti-mouse IgG to visualize bands.
Expression of recombinant proteins in Escherichia coli
The cDNA fragments encoding the catalytic domain (Ser24–Gly284) of MT1-MMP and PEX domain (Cys319–Gly535) of MT1-MMP or that of mouse MT4-MMP (Cys336–Cys527) were expressed using the bacterial expression plasmid, pET3a (Stratagene), in E.coli BL21(DE3) pLysS. Protein expression was induced by the addition of 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to the culture medium. Expressed proteins were isolated and folded according to the procedure of Huang et al. (1996). CD44 stem (Thr130–Glu268) tagged with FLAG at the N-terminus and His6 at the C-terminus was expressed in E.coli and purified as described previously (Kajita et al., 2001).
Kinetic analyses of protein interactions by SPR
Interactions between CD44stem and either MT1PEX, MT1CAT or MT4PEX were analyzed using BIAcore 1000 (BIAcore, Sweden). CD44stem was immobilized to the CM5 sensor chip using an amino coupling kit (BIAcore). Binding reactions were performed in 10 mM HEPES pH 7.4, 150 mM NaCl, 10 mM CaCl2 and 0.05% Tween-20. Surface plasmon resonance was measured at a flow-rate of 5 µl/min at 25°C. Parameters were calculated from the data at three different concentrations of analyte (2.5, 4 and 6 µM), using the BIA evaluation program (BIAcore).
Phagokinetic motility assay
Cell migration was evaluated using a phagokinetic track motility assay, as described previously (Albrecht-Buehler, 1977; Kajita et al., 2001; Uekita et al., 2001). Colloidal gold-coated coverslips were placed in a 12-well plate and transfected cells (3 × 103 per well) were seeded. After a 12 h incubation, phagokinetic tracks were observed under dark-field illumination in a confocal laser microscope (Bio-Rad). The area of each track was measured with NIH Image software.
Acknowledgments
Acknowledgements
We thank Drs Hong Zhu and Erik Thompson for critical reading of the manuscript and Ms Noriko Ito for technical assistance. This work was supported by the Special Coordination Fund for promoting Science and a grant-in-aid for Cancer Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- Albrecht-Buehler G. (1977) The phagokinetic tracks of 3T3 cells. Cell, 11, 395–404. [DOI] [PubMed] [Google Scholar]
- Alpaugh M.L., Lee,M.C., Nguyen,M., Deato,M., Dishakjian,L. and Barsky,S.H. (2000) Myoepithelial-specific CD44 shedding contributes to the anti-invasive and antiangiogenic phenotype of myoepithelial cells. Exp. Cell Res., 261, 150–158. [DOI] [PubMed] [Google Scholar]
- Atkinson S.J., Crabbe,T., Cowell,S., Ward,R.V., Butler,M.J., Sato,H., Seiki,M., Reynolds,J.J. and Murphy,G. (1995) Intermolecular autolytic cleavage can contribute to the activation of progelatinase A by cell membranes. J. Biol. Chem., 270, 30479–30485. [DOI] [PubMed] [Google Scholar]
- Bartolazzi A., Nocks,A., Aruffo,A., Spring,F. and Stamenkovic,I. (1996) Glycosylation of CD44 is implicated in CD44-mediated cell adhesion to hyaluronan. J. Cell Biol., 132, 1199–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourguignon L.Y., Lokeshwar,V.B., He,J., Chen,X. and Bourguignon,G.J. (1992) A CD44-like endothelial cell transmembrane glycoprotein (GP116) interacts with extracellular matrix and ankyrin. Mol. Cell. Biol., 12, 4464–4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourguignon L.Y., Zhu,H., Shao,L., Zhu,D. and Chen,Y.W. (1999) Rho-kinase (ROK) promotes CD44v(3,8-10)–ankyrin interaction and tumor cell migration in metastatic breast cancer cells. Cell Motil. Cytoskeleton, 43, 269–287. [DOI] [PubMed] [Google Scholar]
- Brooks P.C., Stromblad,S., Sanders,L.C., von Schalscha,T.L., Aimes,R.T., Stetler-Stevenson,W.G., Quigley,J.P. and Cheresh,D.A. (1996) Localization of matrix metalloproteinase MMP-2 to the surface of invasive cells by interaction with integrin αvβ3. Cell, 85, 683–693. [DOI] [PubMed] [Google Scholar]
- Cao J., Sato,H., Takino,T. and Seiki,M. (1995) The C-terminal region of membrane type matrix metalloproteinase is a functional transmembrane domain required for pro-gelatinase A activation. J. Biol. Chem., 270, 801–805. [DOI] [PubMed] [Google Scholar]
- Cowell S., Knauper,V., Stewart,M.L., D’Ortho,M.P., Stanton,H., Hembry,R.M., Lopez-Otin,C., Reynolds,J.J. and Murphy,G. (1998) Induction of matrix metalloproteinase activation cascades based on membrane-type 1 matrix metalloproteinase: associated activation of gelatinase A, gelatinase B and collagenase 3. Biochem. J., 331, 453–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumin J.A., Dickeson,S.K., Stricker,T.P., Bhattacharyya-Pakrasi,M., Roby,J.D., Santoro,S.A. and Parks,W.C. (2001) Pro-collagenase-1 (matrix metalloproteinase-1) binds the α2β1 integrin upon release from keratinocytes migrating on type I collagen. J. Biol. Chem., 276, 29368–29374. [DOI] [PubMed] [Google Scholar]
- Egeblad M. and Werb,Z. (2002) New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer, 2, 161–174. [DOI] [PubMed] [Google Scholar]
- Galvez B.G., Matias-Roman,S., Albar,J.P., Sanchez-Madrid,F. and Arroyo,A.G. (2001) Membrane type 1-matrix metalloproteinase is activated during migration of human endothelial cells and modulates endothelial motility and matrix remodeling. J. Biol. Chem., 276, 37491–37500. [DOI] [PubMed] [Google Scholar]
- Goebeler M., Kaufmann,D., Brocker,E.B. and Klein,C.E. (1996) Migration of highly aggressive melanoma cells on hyaluronic acid is associated with functional changes, increased turnover and shedding of CD44 receptors. J. Cell Sci., 109, 1957–1964. [DOI] [PubMed] [Google Scholar]
- Gunthert U. et al. (1991) A new variant of glycoprotein CD44 confers metastatic potential to rat carcinoma cells. Cell, 65, 13–24. [DOI] [PubMed] [Google Scholar]
- Henke C.A., Roongta,U., Mickelson,D.J., Knutson,J.R. and McCarthy,J.B. (1996) CD44-related chondroitin sulfate proteoglycan, a cell surface receptor implicated with tumor cell invasion, mediates endothelial cell migration on fibrinogen and invasion into a fibrin matrix. J. Clin. Invest., 97, 2541–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka N., Allen,E., Apel,I.J., Gyetko,M.R. and Weiss,S.J. (1998) Matrix metalloproteinases regulate neovascularization by acting as pericellular fibrinolysins. Cell, 95, 365–377. [DOI] [PubMed] [Google Scholar]
- Hotary K., Allen,E., Punturieri,A., Yana,I. and Weiss,S.J. (2000) Regulation of cell invasion and morphogenesis in a three-dimensional type I collagen matrix by membrane-type matrix metalloproteinases 1, 2 and 3. J. Cell Biol., 149, 1309–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Suzuki,K., Nagase,H., Arumugam,S., Van Doren,S.R. and Brew,K. (1996) Folding and characterization of the amino-terminal domain of human tissue inhibitor of metalloproteinases-1 (TIMP-1) expressed at high yield in E. coli. FEBS Lett., 384, 155–161. [DOI] [PubMed] [Google Scholar]
- Itoh Y., Kajita,M., Kinoh,H., Mori,H., Okada,A. and Seiki,M. (1999) Membrane type 4 matrix metalloproteinase (MT4-MMP, MMP-17) is a glycosylphosphatidylinositol-anchored proteinase. J. Biol. Chem., 274, 34260–34266. [DOI] [PubMed] [Google Scholar]
- Itoh Y., Takamura,A., Ito,N., Maru,Y., Sato,H., Suenaga,N., Aoki,T. and Seiki,M. (2001) Homophilic complex formation of MT1-MMP facilitates proMMP-2 activation on the cell surface and promotes tumor cell invasion. EMBO J., 20, 4782–4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajita M., Itoh,Y., Chiba,T., Mori,H., Okada,A., Kinoh,H. and Seiki,M. (2001) Membrane-type 1 matrix metalloproteinase cleaves CD44 and promotes cell migration. J. Cell Biol., 153, 893–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalomiris E.L. and Bourguignon,L.Y. (1988) Mouse T lymphoma cells contain a transmembrane glycoprotein (GP85) that binds ankyrin. J. Cell Biol., 106, 319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T., Sato,H., Okada,A., Ohuchi,E., Imai,K., Okada,Y. and Seiki,M. (1998) TIMP-2 promotes activation of progelatinase A by membrane-type 1 matrix metalloproteinase immobilized on agarose beads. J. Biol. Chem., 273, 16098–16103. [DOI] [PubMed] [Google Scholar]
- Knauper V., Will,H., Lopez-Otin,C., Smith,B., Atkinson,S.J., Stanton,H., Hembry,R.M. and Murphy,G. (1996) Cellular mechanisms for human procollagenase-3 (MMP-13) activation. Evidence that MT1-MMP (MMP-14) and gelatinase A (MMP-2) are able to generate active enzyme. J. Biol. Chem., 271, 17124–17131. [DOI] [PubMed] [Google Scholar]
- Kojima S., Itoh,Y., Matsumoto,S., Masuho,Y. and Seiki,M. (2000) Membrane-type 6 matrix metalloproteinase (MT6-MMP, MMP-25) is the second glycosyl-phosphatidyl inositol (GPI)-anchored MMP. FEBS Lett., 480, 142–146. [DOI] [PubMed] [Google Scholar]
- Koshikawa N., Giannelli,G., Cirulli,V., Miyazaki,K. and Quaranta,V. (2000) Role of cell surface metalloprotease MT1-MMP in epithelial cell migration over laminin-5. J. Cell Biol., 148, 615–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladeda V., Aguirre Ghiso,J.A. and Bal de Kier Joffe,E. (1998) Function and expression of CD44 during spreading, migration and invasion of murine carcinoma cells. Exp. Cell Res., 242, 515–527. [DOI] [PubMed] [Google Scholar]
- Lauffenburger D.A. and Horwitz,A.F. (1996) Cell migration: a physically integrated molecular process. Cell, 84, 359–369. [DOI] [PubMed] [Google Scholar]
- McCawley L.J. and Matrisian,L.M. (2000) Matrix metalloproteinases: multifunctional contributors to tumor progression. Mol. Med. Today, 6, 149–156. [DOI] [PubMed] [Google Scholar]
- Mitchison T.J. and Cramer,L.P. (1996) Actin-based cell motility and cell locomotion. Cell, 84, 371–379. [DOI] [PubMed] [Google Scholar]
- Morris A. and Tannenbaum,J. (1980) Cytochalasin D does not produce net depolymerization of actin filaments in HEp-2 cells. Nature, 287, 637–639. [DOI] [PubMed] [Google Scholar]
- Murphy G. and Gavrilovic,J. (1999) Proteolysis and cell migration: creating a path? Curr. Opin. Cell Biol., 11, 614–621. [DOI] [PubMed] [Google Scholar]
- Nagase H. and Woessner,J.F.,Jr (1999) Matrix metalloproteinases. J. Biol. Chem., 274, 21491–21494. [DOI] [PubMed] [Google Scholar]
- Naot D., Sionov,R.V. and Ish-Shalom,D. (1997) CD44: structure, function and association with the malignant process. Adv. Cancer Res., 71, 241–319. [DOI] [PubMed] [Google Scholar]
- Okamoto I., Kawano,Y., Tsuiki,H., Sasaki,J., Nakao,M., Matsumoto,M., Suga,M., Ando,M., Nakajima,M. and Saya,H. (1999) CD44 cleavage induced by a membrane-associated metalloprotease plays a critical role in tumor cell migration. Oncogene, 18, 1435–1446. [DOI] [PubMed] [Google Scholar]
- Ridley A.J., Paterson,H.F., Johnston,C.L., Diekmann,D. and Hall,A. (1992) The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell, 70, 401–410. [DOI] [PubMed] [Google Scholar]
- Sato H., Takino,T., Okada,Y., Cao,J., Shinagawa,A., Yamamoto,E. and Seiki,M. (1994) A matrix metalloproteinase expressed on the surface of invasive tumour cells. Nature, 370, 61–65. [DOI] [PubMed] [Google Scholar]
- Sato T., del Carmen Ovejero,M., Hou,P., Heegaard,A.M., Kumegawa,M., Foged,N.T. and Delaisse,J.M. (1997) Identification of the membrane-type matrix metalloproteinase MT1-MMP in osteoclasts. J. Cell Sci., 110, 589–596. [DOI] [PubMed] [Google Scholar]
- Seiki M. (1999) Membrane-type matrix metalloproteinases. APMIS, 107, 137–143. [DOI] [PubMed] [Google Scholar]
- Shi M., Dennis,K., Peschon,J.J., Chandrasekaran,R. and Mikecz,K. (2001) Antibody-induced shedding of CD44 from adherent cells is linked to the assembly of the cytoskeleton. J. Immunol., 167, 123–131. [DOI] [PubMed] [Google Scholar]
- Sneath R.J. and Mangham,D.C. (1998) The normal structure and function of CD44 and its role in neoplasia. Mol. Pathol., 51, 191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetler-Stevenson W.G., Aznavoorian,S. and Liotta,L.A. (1993) Tumor cell interactions with the extracellular matrix during invasion and metastasis. Annu. Rev. Cell Biol., 9, 541–573. [DOI] [PubMed] [Google Scholar]
- Thomas L., Byers,H.R., Vink,J. and Stamenkovic,I. (1992) CD44H regulates tumor cell migration on hyaluronate-coated substrate. J. Cell Biol., 118, 971–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas L., Etoh,T., Stamenkovic,I., Mihm,M.C.,Jr and Byers,H.R. (1993) Migration of human melanoma cells on hyaluronate is related to CD44 expression. J. Invest. Dermatol., 100, 115–120. [DOI] [PubMed] [Google Scholar]
- Tsukita S., Oishi,K., Sato,N., Sagara,J. and Kawai,A. (1994) ERM family members as molecular linkers between the cell surface glycoprotein CD44 and actin-based cytoskeletons. J. Cell Biol., 126, 391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uekita T., Itoh,Y., Yana,I., Ohno,H. and Seiki,M. (2001) Cytoplasmic tail-dependent internalization of membrane-type 1 matrix metalloproteinase is important for its invasion-promoting activity. J. Cell Biol., 155, 1345–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber G.F., Bronson,R.T., Ilagan,J., Cantor,H., Schmits,R. and Mak,T.W. (2002) Absence of the CD44 gene prevents sarcoma metastasis. Cancer Res., 62, 2281–2286. [PubMed] [Google Scholar]
- Yonemura S., Hirao,M., Doi,Y., Takahashi,N., Kondo,T. and Tsukita,S. (1998) Ezrin/radixin/moesin (ERM) proteins bind to a positively charged amino acid cluster in the juxta-membrane cytoplasmic domain of CD44, CD43 and ICAM-2. J. Cell Biol., 140, 885–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q. and Stamenkovic,I. (1999) Localization of matrix metalloproteinase 9 to the cell surface provides a mechanism for CD44-mediated tumor invasion. Genes Dev., 13, 35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W.H., Woessner,J.F.,Jr, McNeish,J.D. and Stamenkovic,I. (2002) CD44 anchors the assembly of matrilysin/MMP-7 with heparin-binding epidermal growth factor precursor and ErbB4 and regulates female reproductive organ remodeling. Genes Dev., 16, 307–323. [DOI] [PMC free article] [PubMed] [Google Scholar]