Abstract
Hepatitis C virus (HCV) is the major causative pathogen associated with liver cirrhosis and hepatocellular carcinoma. The virus has a positive-sense RNA genome encoding a single polyprotein with the virion components located in the N-terminal portion. During biosynthesis of the polyprotein, an internal signal sequence between the core protein and the envelope protein E1 targets the nascent polypeptide to the endoplasmic reticulum (ER) membrane for translocation of E1 into the ER. Following membrane insertion, the signal sequence is cleaved from E1 by signal peptidase. Here we provide evidence that after cleavage by signal peptidase, the signal peptide is further processed by the intramembrane-cleaving protease SPP that promotes the release of core protein from the ER membrane. Core protein is then free for subsequent trafficking to lipid droplets. This study represents an example of a potential role for intramembrane proteolysis in the maturation of a viral protein.
Keywords: aspartic protease/HCV/lipid metabolism/SPP/viral polyprotein processing
Introduction
Chronic infection with hepatitis C virus (HCV) is the leading cause of liver disease that can culminate in cirrhosis and hepatocellular carcinoma (Kuo et al., 1989). The genome of HCV consists of a positive-sense, single-stranded RNA molecule that has a single open reading frame encoding a polyprotein of ∼3000 amino acids (Grakoui et al., 1993). The polyprotein can be broadly separated into two regions; the N-terminal third encodes the structural components of the virion (core and two envelope glycoproteins E1 and E2; Figure 1A) with the remaining two-thirds comprising the non-structural proteins (NS2-NS5B; for reviews, see Clarke, 1997; Lindenbach and Rice, 2001). A small hydrophobic protein, p7, which lies between E2 and NS2, may also be a virion component. During protein translation, the mature viral products are generated from the polyprotein by a series of cleavage events. The structural components are produced by cellular proteases, while maturation of the non-structural proteins requires virus-encoded proteases.
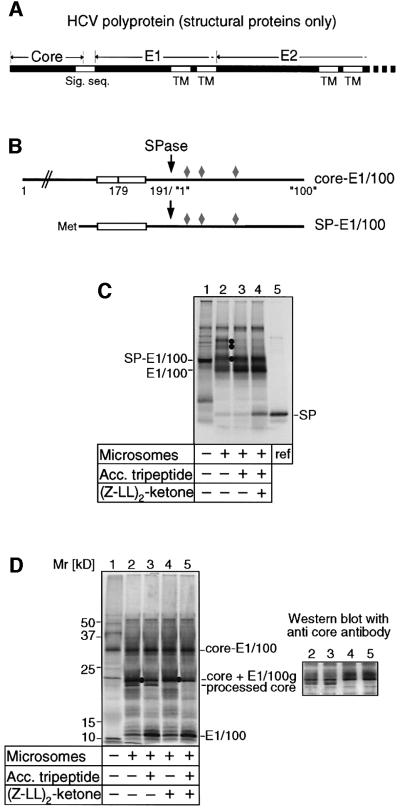
Fig. 1. Analysis of core protein processing in vitro. (A) Diagram of the N-terminal portion of the HCV polyprotein comprising the structural proteins, core, E1 and E2. Sig. seq. indicates the signal sequence at the core-E1 junction; TM, transmembrane regions of the envelope proteins. (B) Diagram of core-E1 constructs used for the in vitro assays. Open bars indicate the hydrophobic region of the signal sequence; arrows, the signal peptidase cleavage site; diamonds, the N-glycosylation sites in E1. (C) Synthesis and processing of SP-E1/100 by in vitro translation in the presence of rough microsomes (lanes 2–4), acceptor tripeptide to inhibit N-glycosylation (lanes 3 and 4), and SPP inhibitor (Z-LL)2- ketone (lane 4). Dots indicate 1x, 2x and 3x glycosylated E1/100; lane 5 shows the reference signal peptide (SP). (D) Synthesis and processing of core-E1/100 by in vitro translation in the presence of rough microsomes (lanes 2–5), acceptor tripepetide to inhibit N-glycosylation (lanes 3 and 5) and SPP inhibitor (lanes 4 and 5). Dots indicate the position of glycosylated E1/100 (E1/100g). The panel to the right shows a corresponding western blot for lanes 2–5 probed with a core-specific antibody.
Core protein, which is considered to constitute the virion capsid, is synthesized as the most N-terminal component of the polyprotein and is followed by the signal sequence of the E1 envelope glycoprotein (Figure 1A). The signal sequence targets the nascent polypeptide chain to the endoplasmic reticulum (ER) membrane and induces translocation of E1 into the ER lumen. There, cleavage by signal peptidase liberates the N-terminal end of E1, leaving core protein anchored in the ER membrane by the signal peptide. Maturation of core protein is considered to involve at least one additional cleavage event within or immediately N-terminal to the signal sequence by an unidentified microsomal protease (Santolini et al., 1994; Hussy et al., 1996; Moradpour et al., 1996; Liu et al., 1997; Yasui et al., 1998).
Signal sequences play a key role in the biosynthesis of secretory and membrane proteins. In eukaryotic cells, they target newly synthesized proteins to the ER membrane for entry into the secretory pathway (Walter and Johnson, 1994). Thereafter, signal sequences are usually cleaved from the pre-protein by signal peptidase (Evans et al., 1986). Recent findings indicated that some signal peptides are subsequently cleaved by signal peptide peptidase (SPP), a presenilin-type intramembrane-cleaving aspartic protease located in the ER membrane (Weihofen et al., 2002). N-terminal portions of processed signal peptides are released into the cytosol, and some are suspected to promote post-targeting functions in the cell (Martoglio and Dobberstein, 1998; Lemberg et al., 2001). The discovery that intramembrane proteolysis by SPP promotes the release of signal peptide fragments from ER membranes prompted us to investigate whether this mechanism is utilized for the processing of HCV core protein.
Results
The signal sequence at the HCV core-E1 junction is a substrate for subsequent cleavage by signal peptidase and SPP
Previous in vitro studies have indicated that the HCV core protein is processed in the presence of microsomal membranes, suggesting proteolysis by signal peptidase and/or other microsomal proteases (Santolini et al., 1994; Hussy et al., 1996; Moradpour et al., 1996; Liu et al., 1997; Yasui et al., 1998). This processing event was re-examined first in an in vitro translation/translocation system (Weihofen et al., 2000). Two substrates were employed (Figure 1B). One contained the C-terminal 30 residues of the core protein including the signal sequence of E1, and the N-terminal 100 amino acid residues of E1 (SP-E1/100). The second substrate consisted of the entire core protein, the signal sequence and the N-terminal 100 residues of E1 (core-E1/100). These polypeptides were synthesized in vitro in the presence of ER-derived rough microsomes and [35S]methionine (Martoglio et al., 1998). Microsomes were subsequently isolated and analysed for retention of radiolabelled translation products.
Translation of SP-E1/100 mRNA in the presence of microsomes yielded two products in addition to precursor protein (Figure 1C): first, an E1/100 species, with lower molecular weight than precursor protein, indicating cleavage of the signal peptide by signal peptidase at residue 191, and secondly, a pair of slower migrating proteins that did not appear in the presence of glycosylation inhibitor N-benzoyl-Asn-Leu-Thr-methyamide and thus represented glycosylated E1/100 peptides (Figure 1C, compare lanes 2 and 3). The presence of glycosylated products indicated translocation of E1/100 into the microsomes. The released signal peptide, which should have an apparent molecular weight of ∼4 kDa and co-migrate with in vitro translated reference signal peptide, was barely detectable, suggesting that it had been further processed and released from the membrane.
Previously, we have reported that signal peptides can be processed by SPP, a reaction that is blocked by (Z-LL)2-ketone (Weihofen et al., 2000). On addition of the inhibitor, the signal peptide generated from SP-E1/100 was readily detected, and co-migrated with the reference peptide indicating that cleavage by SPP had been blocked (Figure 1C, lane 4). These results demonstrate that the signal sequence at the junction of core-E1 functions as a typical ER targeting signal and is a substrate for both signal peptidase and SPP.
We next assayed the function of the signal sequence at the core-E1 junction in the context of the entire core protein. Translation of core-E1/100 in the presence of microsomes yielded several products including a non-translocated precursor (core-E1/100), and a 12 kDa product that corresponded to unglycosylated E1/100, generated through cleavage by signal peptidase at residue 191 (Figure 1D, lane 2). Two other major bands migrated with apparent molecular weights of 21 and 19 kDa. To identify these products, reactions were either performed in the presence of acceptor tripeptide to inhibit N-glycosylation, or (Z-LL)2-ketone to inhibit processing by SPP, or both. Addition of acceptor tripeptide reduced the intensity of the 21 kDa species while the amount of E1/100 increased (Figure 1D, lanes 3 and 5). This indicated that the product at 21 kDa contained glycosylated E1/100. Upon addition of SPP inhibitor, the 19 kDa product disappeared, suggesting that it might be processed core protein.
We next analysed where core protein migrated on the gel by western blot analysis with a core-specific antibody (Figure 1D, compare corresponding lanes in left and right panels). When no SPP inhibitor was added (Figure 1D, right panel, lanes 2 and 3), two core species were recognized that corresponded to the 21 and 19 kDa products described above. Their migration was unaffected by the addition of acceptor tripeptide, but production of the 19 kDa species was abolished in the presence of SPP inhibitor (Figure 1D right panel, compare lanes 2 and 3 with 4 and 5). From these results, we conclude that the 21 kDa species is a core product generated by cleavage by signal peptidase at residue 191 and thus contains the signal peptide at the C-terminus. The 19 kDa product results from cleavage at the signal peptide by SPP. Together with the findings presented above, these data indicate that the signal peptide at the core-E1 junction is first cleaved by signal peptidase and subsequently processed by SPP.
Mutations within the signal peptide block SPP cleavage in vitro and in tissue culture cells
Intramembrane proteolysis of signal peptides is abolished when helix-breaking and -bending residues in the trans membrane region are replaced by leucines or valines (Lemberg et al., 2001). Residues Ala180 and Ser183Cys184 were therefore replaced by Val and LeuVal, respectively (Figure 2A), and the resulting signal peptide mutant (spmt) was tested for protein targeting, translocation, cleavage by signal peptidase and processing by SPP. In vitro translation of spmtSP-E1/100 mRNA in the presence of microsomes and acceptor tripeptide revealed that identical levels of E1/100 were produced as compared with the substrate containing the wild-type (wt) signal sequence (Figure 2B, compare lanes 2 and 3 with 6 and 7). Hence, the substitutions in the signal sequence did not affect targeting, translocation and signal peptidase cleavage of the protein resulting from the spmt construct. However, the mutant signal peptide that had been cleaved from the precursor was not processed, and remained associated with the membrane irrespective of whether SPP inhibitor was added or not (Figure 2B and C). This showed that the mutations abolished processing of the core-E1 signal peptide by SPP in vitro.
Fig. 2. Mutations affecting processing of the core-E1 signal peptide. (A) Illustration of the mutations in the transmembrane region (underlined) of the signal sequence at the core-E1 junction. Exchanged residues in the mutant, spmt, are indicated. (B) In vitro translation and processing of SP-E1/100 (wt, lanes 1–3) and spmtSP-E1/100 (spmt, lanes 5–7) in the presence of rough microsomes and inhibitor of glycosylation (lanes 2, 3, 6 and 7), and SPP inhibitor (lanes 3 and 7). Lanes 4 and 8 show the respective reference signal peptide (SP). (C) Quanti fication of signal peptide processing using the IQMac v1.2 software (Molecular Dynamics). Grey bars indicate the relative amount of signal peptide obtained in lanes 2 and 6 (no SPP inhibitor) compared with the amount obtained in lanes 3 and 7, respectively, where SPP was inhibited (set to 0% processing). Dark bars indicate the efficiency of processing obtained with the core-E1/100 constructs [(D); lanes 3 wt and spmt]. Relative processing efficiencies were determined accordingly by comparing the amount of processed core protein in lane 3 with that obtained in the presence of SPP inhibitor (lane 4) of the respective panel. Each value was calculated from three independent experiments; the amounts of signal peptide were corrected for variations in translocated E1/100 (for SP-E1/100 constructs). (D) Comparison of core protein produced in cells expressing CE1E2 (wt and spmt) (lane 2) with core protein generated by in vitro translation of core-E1/100 (wt and spmt) in the presence of rough microsomes (lanes 3 and 4) and SPP inhibitor (lane 4). Lanes 1 and 5 show in vitro translated reference peptides corresponding to the N-terminal 179 and 191 residues of the HCV polyprotein, respectively. Dots indicate processed core protein. Note that the wt proteins (left panel) were resolved on Tris–glycine acrylamide gels, which did not resolve spmt proteins of 179 and 191 residues. The latter were analysed on Tris–bicine gels (right panel), where the 191 residue reference peptide showed greater mobility than the shorter 179 residue peptide. After electrophoresis, proteins were transferred to PVDF membranes and probed with a core-specific antibody.
We next tested the effect of the mutations in the signal sequence on the processing of core protein in tissue culture cells to verify that the above data were not restricted to the in vitro system. The signal peptide mutations were therefore transferred into the Semliki Forest virus (SFV) construct pSF/CE1E2, which encodes the core-E1-E2 polyprotein (CE1E2). Fragmentation of this portion of the HCV polyprotein relies solely on microsomal proteases (Lindenbach and Rice, 2001). Upon maturation, both E1 and E2 are retained at the membrane through transmembrane domains at their C-termini (Cocquerel et al., 1998, 1999; Flint and McKeating, 1999).
Extracts from cells electroporated with mRNA coding for CE1E2 and CspmtE1E2, respectively, were first examined by western blot analysis alongside corresponding products obtained from in vitro translation/translocation reactions with core-E1/100 mRNA (wt and spmt). When wt CE1E2 was expressed in cells, mature core protein was obtained and migrated with a reference peptide of 179 residues (Figure 2D, left panel, compare lanes 1 and 2). The in vitro experiments yielded two core protein species that migrated with reference peptides of 191 and 179 residues, respectively (Figure 2D, left panel, lane 3). The 191 residue product originates from cleavage by signal peptidase at Ala191. Production of the 179 residue species is indicative of further processing in the transmembrane region of the signal peptide. The latter product did not appear in the presence of (Z-LL)2-ketone, indicating inhibition of cleavage by SPP (Figure 2D, left panel, compare lanes 3 and 4). Co-migration of core products of 179 amino acids, generated both in tissue culture cells and in vitro, was indicative of cleavage of the HCV polyprotein by SPP in both systems.
When CspmtE1E2 was expressed in cells, the resulting core protein co-migrated with the 191 residue reference peptide (Figure 2D, right panel, compare lanes 2 and 5). This core protein species was also exclusively obtained in vitro, irrespective of whether SPP inhibitor was added or not, demonstrating that the mutations abolished signal peptide processing both in vitro and in tissue culture cells (Figure 2D, right panel, compare lanes 3 and 4). From all the above comparative data, we concluded that proteolysis occurs within the signal peptide at the core-E1 junction, and that the signal peptide is a substrate for cleavage by SPP in a cellular environment.
Disruption of signal peptide processing blocks trafficking of core protein to lipid droplets
A defining feature of core protein in mammalian cells after processing from the HCV polyprotein is its association with cytosolic lipid droplets (Barba et al., 1997). Wt and spmt versions of core protein were therefore expressed in baby hamster kidney (BHK) cells, and the products analysed by immunofluorescence. We first tested whether the glycoproteins were properly generated. Glycosylated E1 was equally produced by both wt and spmt constructs (Figure 3A, lanes 1 and 2), and, when treated with endoglycosidase H, the deglycosylated species had the same electrophoretic mobility on gels (Figure 3A, lanes 3 and 4). There was no evidence that uncleaved core-E1 species could be detected (data not shown). Also, immunofluorescence analysis did not detect any difference in the localization of E1 (data not shown) and E2 (Figure 3B) made from the two constructs, and is consistent with their association with the ER membrane. These results indicated once more that the mutations in the signal sequence between core protein and E1 did not affect its function in protein targeting and signal peptidase cleavage.
Fig. 3. Trafficking of core protein to lipid droplets requires intra membrane cleavage of the core-E1 signal peptide. CE1E2 polyproteins of the wt and spmt were expressed in BHK cells. (A) Cell extracts were probed for E1 by western blot analysis either directly (lanes 1 and 2) or upon treatment with endoglycosidase H (lanes 3 and 4). (B) Analysis of cells by immunofluorescence with an E2-specific antibody. (C) Immunofluorescence of core protein combined with Oil Red O staining of lipid droplets. (D) As for (C) except that a wider field is shown. (E) As for (C) except that the cells used were Huh7 cells.
By immunofluorescence, we next investigated the effects of signal sequence mutations on the intracellular localization of core protein. The wt protein was detected at the surface of cytosolic lipid droplets that had been stained with Oil Red O as reported previously (Hope and McLauchlan, 2000) (Figure 3C and D). In contrast, the spmt core protein was distributed in a reticular pattern throughout the cell and showed no co-localization with lipid droplets (Figure 3C and D). The same results with wt and spmt core were obtained when the HCV proteins were expressed in the human hepatoma cell line Huh7 (Figure 3E). The distribution of the mutant form of core protein was indistinguishable from those for E1 and E2, suggesting localization at the ER membrane (data not shown). These findings indicated that inhibiting cleavage in the transmembrane region of the signal peptide abolished trafficking of core protein to lipid droplets.
Mutations affecting signal peptide processing prevent release of core protein from the ER membrane
To examine whether preventing proteolysis within the signal peptide altered the membrane-binding properties of core protein, we tested the ability of alkali to release the protein from membranes (Fujiki et al., 1982). Crude membranes from cells electroporated with SFV RNAs encoding either CE1E2 or CspmtE1E2, were treated with sodium carbonate and the resultant soluble and insoluble fractions were analysed for both core protein and E2, which is anchored in the lipid bilayer by C-terminal transmembrane domains (Figure 1A) (Cocquerel et al., 1998; Duvet et al., 1998). Alkali extraction of membranes from cells electroporated with RNA from either construct did not release E2, as expected, and the protein was found almost exclusively in pelleted material (Figure 4A, left panel, compare lanes 3 and 4). Analysis of the same samples for core protein indicated that the wt protein was released by sodium carbonate (Figure 4A, left panel, lane 3). By contrast, the mutant protein was found in the pellet fraction, consistent with anchorage in the ER membrane via the non-processed signal peptide at its C-terminus (Figure 4A, right panel, lane 4).
Fig. 4. Mutations affecting intramembrane cleavage of the signal peptide at the core-E1 junction retain core protein at the ER membrane. (A) Wt and spmt CE1E2 polyproteins were expressed in BHK cells. Membranes were isolated from cells and extracted with sodium carbonate (lanes 3 and 4). The distribution of core protein and E2 were visualized by western blot analysis with a core- and an E2-specific antibody, respectively. (B) CE1E2 (lane 1), ΔCE1E2 (lanes 2 and 3) and ΔCspmtE1E2 (lanes 4 and 5) polyproteins were expressed in BHK cells in the absence or presence of proteasome inhibitor MG132 as indicated. Membranes were isolated and extracted with sodium carbonate, and the distributions of E2 (left panel) and core/ΔC protein (right panel) were visualized by western blot analysis with a core- and an E2-specific antibody, respectively. (C) Immunofluorescence of cells probed with a core- and an E2-specific antibody. Where indicated, proteasome inhibitor MG132 was present during expression of ΔCE1E2.
To further test whether the release of core protein from the ER membrane is affected when intramembrane proteolysis of the signal peptide cannot occur, we made use of a variant of core protein, ΔC, which lacks amino acid residues 125–144, and is degraded by the proteasome upon cleavage from E1 (Hope and McLauchlan, 2000). We hypothesized that blocking processing of the signal peptide may reduce degradation since core protein is expected to be retained at the ER membrane by its unprocessed signal peptide (Weihofen et al., 2000; Lemberg et al., 2001). Hence, a signal peptide mutant, ΔCspmtE1E2, was made that lacked residues 125–144 and had the same mutations in the signal sequence as spmt described above.
Comparing levels of protein made by cells either expressing CE1E2, ΔCE1E2 or ΔCspmtE1E2, did not reveal any differences in the amounts of E1 (data not shown) and E2 detected (Figure 4B, left panel). However, there was a considerable difference in the relative amounts of core protein. In cells expressing ΔCE1E2, core protein was barely detected yet it was readily recognized in cells expressing CE1E2 and ΔCspmtE1E2 (Figure 4B, right panel). To confirm that cells expressing ΔCE1E2 did nevertheless produce ΔC, cells were incubated in the presence of the proteasome inhibitor MG132. In agreement with our previous observations (Hope and McLauchlan, 2000), enhanced levels of ΔC were detected upon treatment with MG132 (Figure 4B, right panel). The difference in detectable amounts of ΔC is also evident from indirect immunofluorescence analysis (Figure 4C). In contrast, MG132 did not increase the amount of core protein produced from ΔCspmtE1E2 (Figure 4B, right panel).
Expression and intracellular localization of ΔC and ΔCspmt were also analysed by indirect immunofluorescence. ΔC, which was detected only in the presence of MG132, was distributed throughout the cell, including the nucleus (Figure 4C). ΔCspmt, in contrast, strictly co-localized with E2 at the ER membrane and was excluded from the nucleus. Furthermore, ΔCspmt was indistinguishably detected, irrespective of whether MG132 was added or not, suggesting that retention of the protein at the ER membrane did not expose it to degradation in the cytosol. Taken together, these results are consistent with cleavage of ΔCspmt from ΔCspmtE1E2 by signal peptidase, but not processing by SPP, leading to anchoring of non-processed ΔCspmt in the ER membrane, a finding that was confirmed by extraction with alkali (data not shown).
Discussion
HCV core protein is released from the viral polyprotein by the action of host proteases at the signal sequence between core protein and the envelope glycoprotein E1 (Hijikata et al., 1991; Ralston et al., 1993). In common with typical signal sequences, this region functions in protein targeting to the ER membrane (Walter and Johnson, 1994). It directs the nascent HCV polypeptide to translocation sites at the ER membrane where cleavage by signal peptidase in the ER lumen separates core protein and generates the N-terminal end of E1. Based on the size of core protein produced in tissue culture cells, it was proposed that the signal peptide was removed from the C-terminus by a second proteolytic event catalysed by an unidentified microsomal protease (Santolini et al., 1994; Hussy et al., 1996; Moradpour et al., 1996; Liu et al., 1997; Yasui et al., 1998). In the present study, we have provided evidence from both in vitro and cellular systems that this second protease is SPP, a recently identified presenilin-type aspartic protease that promotes intramembrane proteolysis of signal peptides at the ER membrane (Weihofen et al., 2002).
The signal sequence at the junction of core protein and E1 is processed in the centre of the transmembrane region as was reported previously for other signal sequences (Weihofen et al., 2000; Lemberg et al., 2001). The reaction was sensitive to (Z-LL)2-ketone, a compound that efficiently inhibits SPP in vitro (Weihofen et al., 2000). Moreover, signal sequence mutations known to abolish processing by SPP (Lemberg et al., 2001), also abolished processing of the core-E1 signal sequence both in vitro and in live cells. The amino acids that were mutated are conserved in strains from all genotypes examined thus far, including those that are infectious in chimpanzees (Bukh et al., 1994; Kolykhalov et al., 1997; Yanagi et al., 1997, 1999; Beard et al., 1999). Therefore, we conclude that intramembrane proteolysis within the core-E1 signal sequence is a general feature in the maturation of HCV core protein.
Core protein is thought to constitute the HCV capsid and it would thus be an integral component of the virion. At the present stage of research, HCV cannot be propagated in mammalian tissue culture cells and there is no system in such cells to demonstrate virus assembly. This includes human Huh7 cells, which harbour full-length genomes that can support constitutive HCV RNA replication but not virion production (Pietschmann et al., 2002). Hence, it is not possible to directly show that SPP cleavage to generate mature core protein is a prerequisite for virus production.
A characteristic feature of the intracellular distribution of core protein is its association with lipid droplets. This was not only demonstrated in tissue culture cells, as reported herein and elsewhere, but also in hepatocytes from an HCV-infected chimpanzee (Moradpour et al., 1996; Barba et al., 1997; Yasui et al., 1998; Hope and McLauchlan, 2000; Pietschmann et al., 2002). Such localization is found on expression of either the entire HCV polyprotein, truncated versions or the core-coding region alone. Therefore, it is independent of other HCV proteins. The consistency with which association with lipid droplets is observed in various cell types suggests that this localization may be important in the virus assembly pathway.
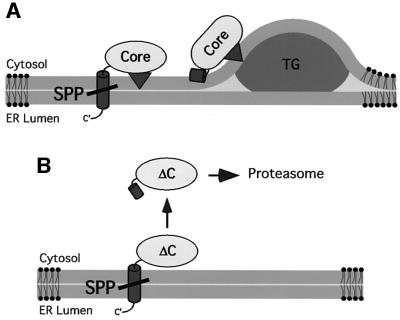
A hydrophobic domain, domain 2, not found in the capsid protein of related pesti- and flaviviruses, mediates targeting to lipid droplets (Hope and McLauchlan, 2000). We speculate that domain 2 integrates into the cytosolic leaflet of the ER membrane (Figure 5A) (Murphy and Vance, 1999; Brown, 2001; Ostermeyer et al., 2001). Upon intramembrane cleavage of the transmembrane signal peptide, the processed core protein may then be capable of trafficking along the lipid bilayer from the site of biosynthesis to zones at the ER, where lipid droplets are produced. Lipid droplet formation is proposed to occur at the ER, where an accumulation of neutral lipids in the lipid bilayer forces the leaflets of a bilayer apart leading to a bulge that may release budding droplets (Brown, 2001). Attachment of core protein could occur while droplets are generated. By contrast, the mutant core protein, which is not processed by SPP, may be unable to traffic to regions where lipid droplets are formed, or cannot transfer from the ER to lipid droplets as a consequence of the membrane-spanning signal peptide anchor at its C-terminus (Brown, 2001; Ostermeyer et al., 2001). Our data also indicate that deletion of domain 2 leads to degradation of the protein upon SPP cleavage (Figure 5B). Disabling intramembrane cleavage by SPP, leading to retention at the ER, prevented its degradation. Thus, the ability of domain 2 to mediate attachment of the protein to lipid droplets may act as a protective mechanism against degradation.
Fig. 5. Model for the release of HCV core protein from the ER membrane and its association with lipid droplets. (A) The central hydrophobic domain 2 (triangle) of HCV core protein anchors the protein in the phospholipid monolayer surrounding lipid droplets. During biosynthesis of the HCV polyprotein, core protein is cleaved from the nascent polypeptide by signal peptidase. Subsequent processing by SPP within the centre of the membrane-spanning portion of the signal peptide (barrel) liberates core protein from the membrane. Core protein, with domain 2 integrated into the cytosolic leaflet of the bilayer, is now free for entering the zone of lipid droplet formation. TG, triacylglycerol. (B) Mutant ΔC, which lacks domain 2, is targeted to the ER membrane and cleaved by signal peptidase as wt core protein. Processing by SPP liberates ΔC from the ER membrane leading to degradation by the proteasome.
The same genomic arrangement of the structural proteins in HCV is also found in other positive-sense RNA viruses such as members of the alpha-, rubi-, and flaviviruses (Schlesinger and Schlesinger, 2001). In each case, the capsid protein, which remains in the cytosol, is synthesized at the N-terminal end of a polyprotein and is followed by an envelope protein, which is translocated into the ER lumen. Targeting to the ER membrane and translocation into the ER lumen are mediated in all cases by a signal sequence that lies between the capsid and envelope proteins. Despite these organizational similarities and the common translocation function inherent in the signal sequences, members of the above viruses have evolved different strategies for processing at signal peptides to generate mature capsid proteins (Melancon and Garoff, 1987; Amberg and Rice, 1999; Law et al., 2001). From the available data, HCV core is the only example for which cleavage by SPP is required for maturation. This suggests that the processes to produce core proteins have followed distinct evolutionary pathways for viruses with common organizational and functional elements. Presumably, such evolution is driven by individual features of the life cycles of each of these viruses.
As reported herein, HCV is an example of a pathogen that exploits a host’s intramembrane cleaving protease for the processing of its proteins. Processing of core protein and trafficking to lipid droplets may be critical for the virus life cycle, but may also affect cellular lipid metabolism. Expression of core protein in animal model systems is associated with the development of liver pathologies such as steatosis and liver carcinoma that are features of HCV infection in humans (Dhillon and Dusheiko, 1995; Moriya et al., 1998; Perlemuter et al., 2002). Additional studies will be required to test whether inhibitors of SPP have the potential to affect the HCV life cycle and reduce any impact of HCV on associated disease.
Materials and methods
Plasmid constructs
For in vitro transcription and translation reactions, the EcoRI–HindIII fragment of pgHCV/CE1E2 (Hope and McLauchlan, 2000) was inserted into vector pSV-Sport1 (Life Technologies) under the control of the SP6 promoter to give pSV/CE1E2. The plasmid encoding signal sequence mutant, pSVspmt, was generated by overlap extension PCR using pSV/CE1E2 as the template. For expression in tissue culture cells, the BglII fragment of pgHCV/CE1E2 was inserted into the BamHI site of a Semliki Forest virus vector pSFV1 (Life Technologies) to give pSF/CE1E2. To generate the signal peptide mutant in the pSFV1 vector, pSF/spmt, the BstEII–NruI fragment of pSV/spmt was transferred firstly into pgHCV/CE1E2 and thereafter, the BglII fragment of the resulting plasmid was transferred into pSFV1. Likewise, the BstEII–NruI fragment of pSV/spmt was transferred into pgHCV/ΔCE1E2 (Hope and McLauchlan, 2000) and thereafter the BglII fragment of the resulting plasmid was transferred into pSFV1.
In vitro transcription, translation and signal peptide processing
To prepare mRNA coding for core-E1/100 precursors (wt and spmt), the respective coding region from the appropriate pSV-Sport1 plasmid was first amplified by PCR using Pfu DNA polymerase (Stratagene), SP6 primer and a reverse primer starting with 5′-NNNNNNNNNCTA to introduce a TAG stop codon at position ‘+101’ of the E1 sequence. PCR-amplified DNA fragments were transcribed in vitro with SP6 RNA polymerase at 42°C in the presence of 500 µM m7G(5′)ppp(5′)G CAP analog (Weihofen et al., 2000). mRNA coding for SP-E1/100 (wt and spmt) was prepared accordingly. For the latter PCR, the forward primer encoded the SP6 promotor, the Kozak initiation sequence and codons 161–167 of HCV open reading frame.
Translations of mRNAs were performed in 25 µl rabbit reticulocyte lysate (Promega) containing [35S]methionine (Amersham-Pharmacia) and, where indicated, two equivalents of nuclease-treated rough microsomes prepared from dog pancreas (Martoglio et al., 1998), 10 µM (Z-LL)2-ketone (Weihofen et al., 2000) and 30 µM N-benzoyl-Asn-Leu-Thr-methyamide (Martoglio et al., 1998). Samples were incubated for 30 min at 30°C. Membranes were next treated with 500 mM KOAc, recovered by centrifugation through a 500 mM sucrose cushion, and prepared for SDS–PAGE as described previously (Weihofen et al., 2000).
Expression in tissue culture cells, treatment with MG132 and deglycosylation
Prior to electroporation, the appropriate SpeI linearized pSFV-1 plasmids were transcribed in vitro with SP6 RNA polymerase (Hope and McLauchlan, 2000). Typical reactions were carried out in a volume of 20 µl and contained 40 mM Tris–HCl pH 7.5, 6 mM MgCl2, 2 mM spermidine, 10 mM NaCl, 1 mM dithiothreitol (DTT), 1 mM ATP, 1 mM CTP, 1 mM UTP, 0.5 mM GTP, 1 mM m7G(5′)ppp(5′)G CAP analog, 40 U RNasin, 40 U SP6 RNA polymerase and 1.5 µg linearized DNA. Reactions were performed at 37°C for 2 h.
BHK C13 cells were grown and maintained in Glasgow minimal Eagle’s medium supplemented with 10% new-born calf serum (NCS), 4% tryptose phosphate broth and 100 IU/ml penicillin/streptomycin (ETC10). Huh7 cells were propagated in Dulbecco’s modified Eagle’s Medium supplemented with 10% fetal calf serum, 2 mM l-glutamine, non-essential amino acids and 100 IU/ml penicillin/streptomycin. For electro poration, cells were washed and treated with trypsin for detachment from tissue culture containers. Detached cells were suspended in 20 ml of growth medium and centrifuged at 400 g for 5 min at room temperature. Cell pellets were suspended in 50 ml of PBSA [phosphate-buffered saline (PBS) solution that lacks Ca2+ and Mg2+ salts] and centrifuged as before. Pellets were suspended in PBSA at a final concentration of ∼2 × 107 cells/ml. Competent cells (0.8 ml) were mixed with in vitro transcribed RNA in an electroporation cuvette (0.4 cm gap) and pulsed once either at 1.2 kV, 25 µF (for BHK cells) or 0.36 kV, 950 µF (for Huh7 cells) (Hope and McLauchlan, 2000). Following electroporation, cells were diluted in growth medium, seeded onto either tissue culture dishes or coverslips in 24-well tissue culture plates and incubated at 37°C for 12 h. When treated with MG132 (Boston Biochem), cells were incubated for 5 h after electroporation at 37°C and supplemented with fresh medium containing the protease inhibitor at a final concentration of 2.5 µg/ml. Incubation was continued at 37°C for a further 12 h before the cells were either solubilized in sample buffer and prepared for SDS–PAGE and western blot analysis, or fixed for indirect immunofluorescence.
For deglycosylation, cells were dissolved in 0.5% SDS, 1% β-mercaptoethanol at 100°C for 10 min at a concentration of ∼6 × 106 cell equivalents/ml. Sodium citrate was added to 50 mM followed by 2000 U of endoglycosidase H (Roche Bioscience), and reactions were incubated at 37°C for 1 h (Martoglio et al., 1998). Sample buffer was added to dilute the extract to ∼4 × 106 cell equivalents/ml and samples were heated to 100°C as described above.
Immunofluorescence and staining of lipids
Cells grown on 13 mm coverslips were fixed in either methanol at –20°C for 15 min, or 4% paraformaldehyde and 0.1% Triton X-100 in PBS at 4°C for 30 min. Following washing with PBS and blocking with PBS/NCS (PBS containing 1% NCS), cells were incubated with primary antibody (1/200 for JM122 and ALP98; 1/1000 for R308) for 2 h at room temperature. Cells were washed with PBS/NCS and then incubated with conjugated secondary antibody (either anti-mouse or anti-rabbit IgG raised in goat) for 2 h at room temperature (Patel et al., 1999; Hope and McLauchlan, 2000). Following incubation with both antibodies and washing, lipid droplets were stained in paraformaldehyde-fixed cells by briefly rinsing coverslips in 60% propan-2-ol followed by incubation with 0.5 ml of 60% propan-2-ol containing 1% Oil Red O for 1.5–2 min at room temperature (Hope and McLauchlan, 2000). Coverslips were briefly rinsed with 60% propan-2-ol, washed with PBS and H2O and mounted on slides using Citifluor. Samples were analysed using a Zeiss LSM confocal microscope.
Alkali extraction of tissue culture cells
Tissue culture dishes (150 mm) of Huh7 cells, electroporated with RNA from the relevant pSF construct, were incubated at 37°C for 15 h. Cells were washed twice and scraped into PBS. After pelleting at 300 g for 5 min at 4°C, cells were swollen in 600 µl of buffer A (10 mM HEPES–KOH pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT) for 10 min at 4°C and pelleted as before. Cell pellets were resuspended in 600 µl of buffer A and homogenized by passage 10 times through a 0.45 gauge needle. Intact cells and nuclei that survived homogenization were pelleted by centrifugation at 1000 g for 10 min at 4°C. The resultant supernatant was centrifuged at 100 000 g for 15 min at 4°C. This generated crude membranes that were resuspended in 100 µl of freshly prepared 100 mM Na2CO3 (pH 11.3) and incubated on ice for 15 min. Soluble and insoluble material was separated by centrifugation through 200 µl of sucrose cushion (100 mM Na2CO3, 250 mM sucrose) at 130 000 g for 15 min at 4°C. For uniformity, all samples were delipidated and proteins precipitated by addition of acetone to 25% and trichloroacetic acid to 15%, respectively. Precipitated proteins were recovered by centrifugation at 13 000 g for 5 min, washed with acetone, dried and resuspended in sample buffer prior to electrophoresis on polyacrylamide gels.
Antibodies
Antibodies used to detect HCV (monoclonal antibody JM122 and rabbit antisera R308), E1 (rabbit antisera R528) and E2 proteins (monoclonal antibody ALP98) have been described previously (Hope and McLauchlan, 2000; Patel et al., 2001).
SDS–PAGE and western blotting
Proteins and peptides were analysed by SDS–PAGE using either Tris–glycine acrylamide gels (14% T, 2.7% C for Figure 1D; 13% T, 2.7% C for Figures 2D, 3A, 4A and B) (Laemmli, 1970) or Tris–bicine acrylamide gels (15% T, 5% C, 8 M urea for Figures 1C and 2B; 9% T, 5% C, for Figure 2D) (Wiltfang et al., 1997; Weihofen et al., 2000). Labelled proteins were visualized by a STORM phosphorImager (Molecular Dynamics; IQMac v1.2). For western blot analysis, proteins transferred onto PVDF membranes were blocked with 5% BSA in 20 mM Tris–HCl pH 7.6, 140 mM NaCl and 0.1% Tween-20 for 1 h at room temperature. Thereafter, membranes were incubated with the primary antibodies (1/500 for JM122 and ALP98; 1/1000 for R528 and R308). The appropriate secondary antibodies conjugated with horseradish peroxidase were used at a dilution of 1/10–50 000, and bound antibody was detected by enhanced chemiluminescence (Amersham-Pharmacia).
Acknowledgments
Acknowledgements
We would like to thank Bernhard Dobberstein, Ari Helenius and Andreas Weihofen for advice and critical comments on the manuscript. This work was supported by grants from ETHZ and the Swiss National Science Foundation to B.M., and a Boehringer Ingelheim fellowship to M.K.L.
References
- Amberg S.M. and Rice,C.M. (1999) Mutagenesis of the NS2B-NS3-mediated cleavage site in the flavivirus capsid protein demonstrates a requirement for coordinated processing. J. Virol., 73, 8083–8094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barba G. et al. (1997) Hepatitis C virus core protein shows a cytoplasmic localization and associates to cellular lipid storage droplets. Proc. Natl Acad. Sci. USA, 94, 1200–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard M.R. et al. (1999) An infectious molecular clone of a Japanese genotype 1b hepatitis C virus. Hepatology, 30, 316–324. [DOI] [PubMed] [Google Scholar]
- Brown D.A. (2001) Lipid droplets: proteins floating on a pool of fat. Curr. Biol., 11, R446–R449. [DOI] [PubMed] [Google Scholar]
- Bukh J., Purcell,R.H. and Miller,R.H. (1994) Sequence analysis of the core gene of 14 hepatitis C virus genotypes. Proc. Natl Acad. Sci. USA, 91, 8239–8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke B. (1997) Molecular virology of hepatitis C virus. J. Gen. Virol., 78, 2397–2410. [DOI] [PubMed] [Google Scholar]
- Cocquerel L., Meunier,J.C., Pillez,A., Wychowski,C. and Dubuisson,J. (1998) A retention signal necessary and sufficient for endoplasmic reticulum localization maps to the transmembrane domain of hepatitis C virus glycoprotein E2. J. Virol., 72, 2183–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquerel L., Duvet,S., Meunier,J.C., Pillez,A., Cacan,R., Wychowski, C., and Dubuisson,J. (1999) The transmembrane domain of hepatitis C virus glycoprotein E1 is a signal for static retention in the endoplasmic reticulum. J. Virol. 73, 2641–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon A.P. and Dusheiko,G.M. (1995) Pathology of hepatitis C virus infection. Histopathology, 26, 297–309. [DOI] [PubMed] [Google Scholar]
- Duvet S., Labiau,O., Mir,A.M., Kmiecik,D., Krag,S.S., Verbert,A. and Cacan,R. (1998) Cytosolic deglycosylation process of newly synthesized glycoproteins generates oligomannosides possessing one GlcNAc residue at the reducing end. Biochem. J., 335, 389–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E.A., Gilmore,R. and Blobel,G. (1986) Purification of microsomal signal peptidase as a complex. Proc. Natl Acad. Sci. USA, 83, 581–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint M., and McKeating,J.A. (1999) The C-terminal region of the hepatitis C virus E1 glycoprotein confers localization within the endoplasmic reticulum. J. Gen. Virol., 80, 1943–1947. [DOI] [PubMed] [Google Scholar]
- Fujiki Y., Hubbard,A.L., Fowler,S. and Lazarow,P.B. (1982) Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol., 93, 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grakoui A., Wychowski,C., Lin,C., Feinstone,S.M. and Rice,C.M. (1993) Expression and identification of hepatitis C virus polyprotein cleavage products. J. Virol., 67, 1385–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijikata M., Kato,N., Ootsuyama,Y., Nakagawa,M. and Shimotohno,K. (1991) Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc. Natl Acad. Sci. USA, 88, 5547–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope R.G. and McLauchlan,J. (2000) Sequence motifs required for lipid droplet association and protein stability are unique to the hepatitis C virus core protein. J. Gen. Virol., 81, 1913–1925. [DOI] [PubMed] [Google Scholar]
- Hussy P., Langen,H., Mous,J. and Jacobsen,H. (1996) Hepatitis C virus core protein: carboxy-terminal boundaries of two processed species suggest cleavage by a signal peptide peptidase. Virology, 224, 93–104. [DOI] [PubMed] [Google Scholar]
- Kolykhalov A.A., Agapov,E.V., Blight,K.J., Mihalik,K., Feinstone,S.M. and Rice,C.M. (1997) Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science, 277, 570–574. [DOI] [PubMed] [Google Scholar]
- Kuo G. et al. (1989) An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science, 244, 362–364. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Law L.M., Duncan,R., Esmaili,A., Nakhasi,H.L. and Hobman,T.C. (2001) Rubella virus E2 signal peptide is required for perinuclear localization of capsid protein and virus assembly. J. Virol., 75, 1978–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemberg M.K., Bland,F.A., Weihofen,A., Braud,V.M. and Martoglio,B. (2001) Intramembrane proteolysis of signal peptides: an essential step in the generation of HLA-E epitopes. J. Immunol., 167, 6441–6446. [DOI] [PubMed] [Google Scholar]
- Lindenbach B.D. and Rice,C.M. (2001) Flaviviridae: the viruses and their replication. In Knipe,D.M. and Howley,P.M. (eds), Fields Virology, 4th ed., Vol. 1. Lippincott Williams and Wilkins, Philadelphia, PA, pp. 991–1041.
- Liu Q., Tackney,C., Bhat,R.A., Prince,A.M. and Zhang,P. (1997) Regulated processing of hepatitis C virus core protein is linked to subcellular localization. J. Virol., 71, 657–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martoglio B. and Dobberstein,B. (1998) Signal sequences: more than just greasy peptides. Trends Cell Biol., 8, 410–415. [DOI] [PubMed] [Google Scholar]
- Martoglio B., Hauser,S. and Dobberstein,B. (1998) Cotranslational translocation of proteins into microsomes derived from the rough endoplasmic reticulum of mammalian cells. In Celis,J.E. (ed.), Cell Biology: A Laboratory Handbook, Vol. 2. Academic Press, San Diego, CA, pp. 265–274.
- Melancon P. and Garoff,H. (1987) Processing of the Semliki Forest virus structural polyprotein: role of the capsid protease. J. Virol., 61, 1301–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradpour D., Englert,C., Wakita,T. and Wands,J.R. (1996) Characterization of cell lines allowing tightly regulated expression of hepatitis C virus core protein. Virology, 222, 51–63. [DOI] [PubMed] [Google Scholar]
- Moriya K. et al. (1998) The core protein of hepatitis C virus in duces hepatocellular carcinoma in transgenic mice. Nat. Med., 4, 1065–1067. [DOI] [PubMed] [Google Scholar]
- Murphy D.J. and Vance,J. (1999) Mechanisms of lipid-body formation. Trends Biochem. Sci., 24, 109–115. [DOI] [PubMed] [Google Scholar]
- Ostermeyer A.G., Paci,J.M., Zeng,Y., Lublin,D.M., Munro,S. and Brown,D.A. (2001) Accumulation of caveolin in the endoplasmic reticulum redirects the protein to lipid storage droplets. J. Cell Biol., 152, 1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel J., Patel,A.H. and McLauchlan,J. (1999) Covalent interactions are not required to permit or stabilize the non-covalent association of hepatitis C virus glycoproteins E1 and E2. J. Gen. Virol., 80, 1681–1690. [DOI] [PubMed] [Google Scholar]
- Patel J., Patel,A.H. and McLauchlan,J. (2001) The transmembrane domain of the hepatitis C virus E2 glycoprotein is required for correct folding of the E1 glycoprotein and native complex formation. Virology, 279, 58–68. [DOI] [PubMed] [Google Scholar]
- Perlemuter G. et al. (2002) Hepatitis C virus core protein inhibits microsomal triglyceride transfer protein activity and very low density lipoprotein secretion: a model of viral-related steatosis. FASEB J., 16, 185–194. [DOI] [PubMed] [Google Scholar]
- Pietschmann T., Lohmann,V., Kaul,A., Krieger,N., Rinck,G., Rutter,G., Strand,D. and Bartenschlager,R. (2002) Persistent and transient replication of full-length hepatitis C virus genomes in cell culture. J. Virol., 76, 4008–4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston R. et al. (1993) Characterization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia viruses. J. Virol., 67, 6753–6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santolini E., Migliaccio,G. and La Monica,N. (1994) Biosynthesis and biochemical properties of the hepatitis C virus core protein. J. Virol., 68, 3631–3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger S. and Schlesinger,M.J. (2001) Togaviridae: the viruses and their replication. In Knipe,D.M. and Howley,P.M. (eds), Fields Virology, 4th ed., Vol. 1. Lippincott Williams and Wilkins, Philadelphia, PA, pp. 895–990.
- Walter P. and Johnson,A.E. (1994) Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu. Rev. Cell. Biol., 10, 87–119. [DOI] [PubMed] [Google Scholar]
- Weihofen A., Lemberg,M.K., Ploegh,H.L., Bogyo,M. and Martoglio,B. (2000) Release of signal peptide fragments into the cytosol requires cleavage in the transmembrane region by a protease activity that is specifically blocked by a novel cysteine protease inhibitor. J. Biol. Chem., 275, 30951–30956. [DOI] [PubMed] [Google Scholar]
- Weihofen A., Binn,K., Lemberg,M.K., Ashman,K. and Martoglio,B. (2002) Identification of signal peptide peptidase, a presenilin-type aspartic protease. Science, 296, 2215–2218. [DOI] [PubMed] [Google Scholar]
- Wiltfang J. et al. (1997) Improved electrophoretic separation and immunoblotting of β-amyloid (Aβ) peptides 1–40, 1–42, and 1–43. Electrophoresis, 18, 527–532. [DOI] [PubMed] [Google Scholar]
- Yanagi M., Purcell,R.H., Emerson,S.U. and Bukh,J. (1997) Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc. Natl Acad. Sci. USA, 94, 8738–8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagi M., Purcell,R.H., Emerson,S.U. and Bukh,J. (1999) Hepatitis C virus: an infectious molecular clone of a second major genotype (2a) and lack of viability of intertypic 1a and 2a chimeras. Virology, 262, 250–263. [DOI] [PubMed] [Google Scholar]
- Yasui K., Wakita,T., Tsukiyama-Kohara,K., Funahashi,S.I., Ichikawa, M., Kajita,T., Moradpour,D., Wands,J.R. and Kohara,M. (1998) The native form and maturation process of hepatitis C virus core protein. J. Virol., 72, 6048–6055. [DOI] [PMC free article] [PubMed] [Google Scholar]