Abstract
An UGA stop codon context which is inefficient because of the 3′-flanking context and the last two amino acids in the gene protein product has a negative effect on gene expression, as shown using a model protein A′ gene. This is particularly true at low mRNA levels, corresponding to a high intracellular ribosome/mRNA ratio. The negative effect is smaller if this ratio is decreased, or if the distance between the initiation and termination signals is increased. The results suggest that an inefficient termination codon can cause ribosomal pausing and queuing along the upstream mRNA region, thus blocking translation initiation of short genes. This cis control effect is dependent on the stop codon context, including the C-terminal amino acids in the gene product, the translation initiation signal strength, the ribosome/mRNA ratio and the size of the mRNA coding region. A large proportion of poorly expressed natural Escherichia coli genes are small, and the weak termination codon UGA is under-represented in small, highly expressed E.coli genes as compared with the efficient stop codon UAA.
Keywords: gene expression/protein A′/translation assay/translation initiation/translation termination
Introduction
The bacterial cell has developed a number of controls to regulate gene expression at the translational level. Translation initiation is governed by the three initiation factors IFs 1–3 together with fMet-tRNAfMet (Kozak, 1999), but initiation efficiency and thereby gene expression is also determined by the base sequences that flank the initiation codon in the mRNA. The well known Shine–Dalgarno (SD) sequence (Shine and Dalgarno, 1975) normally is located 5–8 bases upstream of the initiation codon (Chen et al., 1994). This sequence anchors the mRNA to the 30S sequence by complementary base pairing to the anti-SD, close to the 3′ end of the 16S rRNA. Besides the canonical start codon AUG, the near-cognate codons UUG, GUG and AUU can also function as start signals. The AUU codon is used in Escherichia coli only in the case of the gene infC coding for initiation factor IF3, where this non-canonical initiation codon is used in an auto-regulatory control circuit (Sacerdot et al., 1996).
The mRNA secondary structures that involve the regions that flank the initiation region and/or the initiation codon (de Smit and van Duin, 1994) can affect gene expression. Since the ribosome decodes the codons at various speeds, codon usage is also important and translation is faster for highly expressed, than for poorly expressed genes (Sorensen et al., 1989; Sorensen and Pedersen, 1991). Optimal codons and suboptimal codons, which are not necessarily rare codons, are evenly spaced in the different regions of genes. However, some rare codons are over-represented among the first 25 codons of E.coli genes (Chen and Inouye, 1990). In particular, the 5–6 codons that follow the initiation codon are important for gene expression levels. At least part of this effect originates from peptidyl-tRNA drop-off (Heurgue-Hamard et al., 2000) that reduces the effective gene expression level. The importance of the +2 codon for gene expression at the translational level has been studied extensively but is not fully understood (Looman et al., 1987; Stenström et al., 2001b).
In normal wild-type E.coli there are no tRNAs that decode the three stop codons UAA, UAG and UGA. Instead, the stop codon UAG is decoded by release factor 1 (RF1). UGA is recognized by RF2. UAA is recognized by both RF1 and RF2 (Capecchi, 1967; Caskey et al., 1969). The termination reaction at all three stop codons is stimulated by RF3, although this factor is not essential for bacterial viability (Milman et al., 1969; Grentzmann et al., 1994; Mikuni et al., 1994). The efficiency of a stop codon, especially UGA, in promoting termination is very sensitive to the contexts on both sides of the stop codon itself. Altogether, the termination signal consists of the 2–3 codons upstream of the stop codon and the codon that follows at the 3′ side, together constituting as many as 12–15 bases (Buckingham et al., 1990; Kopelowitz et al., 1992; Björnsson and Isaksson, 1993; Mottagui-Tabar et al., 1994; Poole et al., 1995; Björnsson et al., 1996). The effects of the codons upstream of the termination codon itself are indirect since the effect on the termination reaction originates from the P-site tRNA (Zhang et al., 1996) and the last two amino acid residues in the protein product, that are encoded by the –1 and –2 codons. These amino acid residues probably affect binding of the RFs (Zhang et al., 1996). As a result, the last two amino acid residues of the gene product affect the efficiency of decoding the stop codon that terminates its own gene. This is particularly true for the weak stop codon UGA (Mottagui-Tabar et al., 1994; Björnsson et al., 1996). Thus, certain C-terminal dipeptide residues, together with an A-base after a UGA stop codon, can produce a very inefficient termination, giving up to 30% readthrough of UGA by the UGG reader tRNATrp. On the contrary, other dipeptide residues together with a U base following the stop codon give essentially complete termination (Björnsson and Isaksson, 1993).
The bases in the codon downstream of the termination codon are directly involved in binding of the release factors. The frequency of the bases at the 3′ end of the stop codon is biased, being U>A>G>C for UGA, U>G>A>C for UAA and U>C=G>A for UAG (Bossi, 1983; Brown et al., 1990). Thus, U is over-represented and it also promotes the most efficient termination. In E.coli, UAA U and UAA G are major stop signals, whereas UGA U and UGA A are less common in highly expressed genes (Bossi, 1983; Brown et al., 1990; Tate and Brown, 1992).
It has been observed that stop codon contexts that give inefficient termination, as revealed by high readthrough, gave a lower output of protein product than did efficient contexts, thus suggesting some kind of cis control (Björnsson and Isaksson, 1993). The effect could be explained partly by the less stable full-length mRNA in the case of the low efficiency stop codon context. However, the effect of mRNA degradation was not large enough to explain fully the observed stop codon context effect on gene expression at the protein level (Björnsson and Isaksson, 1996).
To study further the combined influences of initiation and termination signals on gene expression, we have used a useful model gene for the investigation based on the protein A′ reporter gene derived from the protein A gene in Staphylococcus aureus (Björnsson et al., 1996, 1998).
Our results suggest that inefficient termination can cause ribosome queuing that has a negative influence on translational initiation some 400 bases upstream of the mRNA termination signal. The influence of the termination signal on gene expression is dependent on the initiation codon context, the length of the gene and the intracellular level of mRNA. Translation termination efficiency that can be influenced by the C-terminal amino acid residues in the gene product can thus act as a cis control device of gene expression at the translation initiation level.
Results
Technical evaluation of the 3A′ in vivo assay system
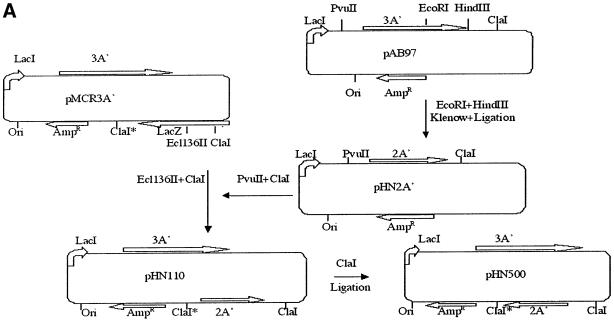
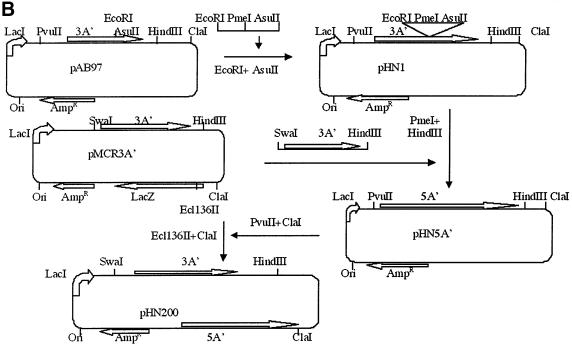
A′ gene-derived proteins are stable, non-functional and non-toxic for the E.coli host bacterium (Björnsson et al., 1998). The protein yield from an isopropyl-β-d-thiogalactopyranoside (IPTG)-induced 3A′ gene in the plasmid used here is ∼1% or more of the level of total cellular proteins (Björnsson et al., 1998). A series of protein A′ gene variants were constructed, in order to evaluate effects on A′ gene expression levels. Of these plasmids, pMCR3A′, carried 3A′ only. pHN110 carried the 3A′ test gene plus the 2A′ internal control gene (Figure 1A). pHN200 carried 3A′ as a test gene and 5A′ as an internal control gene (Figure 1B). The test gene 3A′ and control genes 2A′ or 5A′ are under separate control by the Ptrc promoter. This IPTG-inducible promoter can be repressed by the repressor encoded by lacI, which is also located in the plasmid. The 3A′ gene is preceded by a canonical, and thus highly efficient, SD sequence (SD+). The AUG initiation codon in the 3A′ gene in plasmids pMCR3A′, pHN110 and pHN200 is followed by a downstream region (DR) sequence (DR-A: AAA GCA AUU UUC GUA) that enables a high level of gene expression. Altogether, a high level of gene expression should be achieved for such 3A′ gene variants in the presence of IPTG.
Fig. 1. Plasmid cloning procedures for vector constructions. (A) Plasmid pHN110 carries a 3A′ gene as a test gene and a 2A′ gene as a standard gene. pHN500 has the 2A′ standard gene in the inverse orientation, as indicated. (B) Plasmid pHN200 has the 3A′ gene as test gene and a 5A′ gene as a standard gene. The directions of transcription of the test and internal control genes are indicated. AmpR, ampicillin resistance gene: Ori, plasmid origin of replication; lacZ, gene encoding β-galactosidase; lacI, repressor gene of lacZ.
In order to analyse the influence of the A′ gene induction levels on bacterial growth, we measured the growth rates of plasmid-carrying cells in the presence or absence of the inducer IPTG. Despite different loads on cellular metabolism by the plasmids pMCR3A′, pHN110 and pHN200, all the E.coli strains grew at similar rates, with a doubling time of ∼30 min in LB medium, in the presence of IPTG (not shown).
In the absence of IPTG, the A′ genes are expressed at very low levels, as revealed by SDS–PAGE (not shown). For cells induced with IPTG, significant expression is obtained. Cells with plasmids pMCR3A′, pHN110 or pHN200, growing in the presence of IPTG, were harvested in mid-log phase, and the A′ proteins were purified by one-step affinity chromatography on IgG–Sepharose and analysed by SDS–PAGE. Results presented in Figure 2 illustrate the separation obtained for the various A′ proteins that are expressed in the different plasmid-carrying strains. The molar expression ratios 3A′/2A′ and 3A′/5A′ are similar (0.48 and 0.49, respectively), showing that the nature of the internal control gene product, either 2A′ or 5A′, does not affect the expression level of the test gene 3A′. Since the initiation regions of the 2A′ and 5A′ genes are the same, being different from that of the 3A′ gene, this probably explains why the 2A′ and 5A′ genes can be interchanged as control genes.
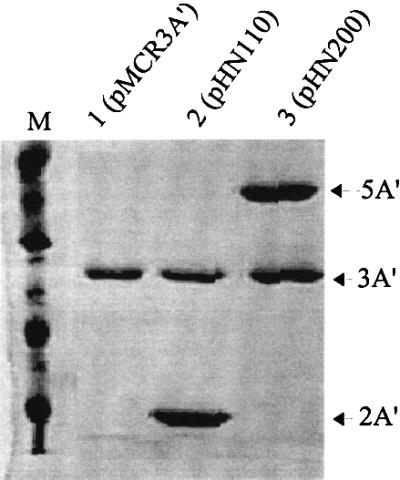
Fig. 2. SDS–PAGE analysis of A′ proteins encoded by plasmids pMCR3A′, pHN110 and pHN200. Proteins 5A′, 3A′ and 2A′ are indicated by arrows. The A′ proteins were isolated from similar amounts of IPTG-induced (1 mM) plasmid-containing cells, as described in Materials and methods. Protein bands were scanned for determination of expression. Molecular markers are indicated. Molar units are given to compensate for differences in molecular weights of the different A′ proteins. Lane 1, pMCR3A′ (3A′: 6.9 ± 0.4 U); lane 2, pHN110 (3A′: 6.9 ± 0.3 U; 2A′, 14.5 ± 0.5 U; relative expression ratio 3A′/2A′ = 0.48 ± 0.17); lane 3, pHN200 (5A′, 15.3 ± 1 U; 3A′, 7.5 ± 0.6 U; relative expression ratio 3A′/5A′ = 0.49 ± 0.19). M = molecular markers. Standard errors were determined by analysis of four samples of the same protein preparation.
The inverted direction of transcription from the test gene and the standard gene possibly could affect each other. To investigate this possibility, a plasmid was constructed where the direction of transcription of the internal standard gene 2A′ was inverted to be in the same direction as the test gene by using the two ClaI sites at both ends of the 2A′ gene. Relative 3A′ gene expression in pHN110 and pHN500 (Figure 1A) is very similar (0.47 and 0.49, respectively) irrespective of whether transcription of the test and control A′ genes is in the same or the inverted orientation.
Influence of different stop codon contexts on gene expression
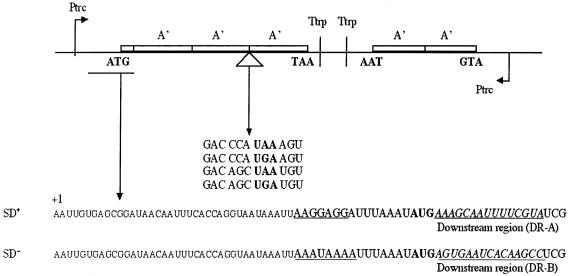
Earlier results had indicated that the stop codon context could influence gene expression (Björnsson and Isaksson, 1993). In order to evaluate this effect further, two gene variants were made that were preceded by a strong (SD+; 5′-AAGGAGGU-3′) or weak (SD–; 5′-AAAUAAAU-3′) SD sequence followed by an efficient DR-A or a less efficient DR-B penta-codon sequence, respectively, downstream of the initiation codon. These two gene variants should give high or low levels of translation initiation, respectively. Furthermore, in the spacer region between the second and third A′-encoding sequences, a weak stop signal (CCA UGA A) and several strong stop signals (CCA UAA A, AGC UGA U or AGC UAA U) (Björnsson and Isaksson, 1993) were introduced (Figure 3). The eight variants thus obtained were analysed for A′ protein production (Figure 4). Besides the internal standard 2A′ protein encoded by the control gene, one would expect to find a 3A′ protein that results from readthrough of the stop codon in the linker region in the test gene. In addition, a 2A′′ protein corresponding to two A′ units plus a few amino acids encoded by the linker sequence downstream of the start codon should be formed as the result of translation termination at the inserted stop codon. Since the three proteins are different in size, they can be separated from each other using SDS–PAGE and quantified by scanning of the stained protein bands in the gel (Figure 4).
Fig. 3. Translation assay system used in this study. Ptrc, promoter of transcription; Ttrp, transcription termination; A′, one protein A′ domain-encoding region; ATG and TAA are gene translational start and stop sites, respectively. mRNA sequences of start and stop regions are shown. The SD region is underlined. The AUG codon in the SD+ variant is followed by the DR-A penta-codon downsteam region. The AUG codon in the SD– variant is followed by the intermediately strong DR-B penta-codon sequence. The 2A′ gene mRNA starts with the sequence AATTGTGAGCGGATAA CAATTTCACACAGGAAACAGACCATGGAATTGCAACACGAT and the stop codon context is AAGTAAGTA. The +1 base of the mRNA transcript is indicated in the figure.
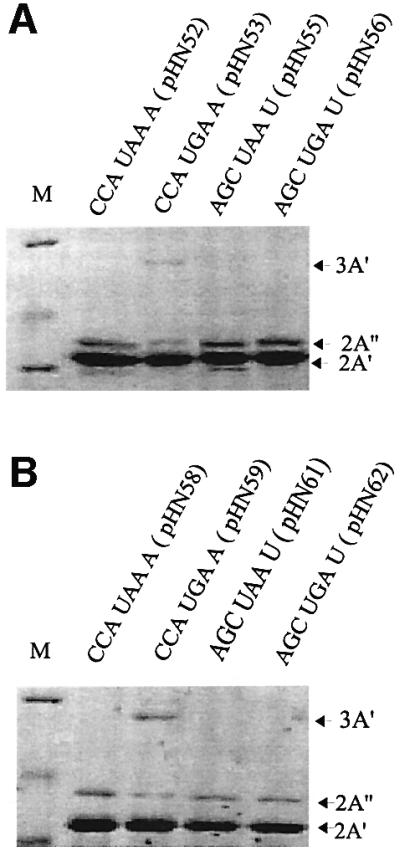
Fig. 4. Termination/readthrough of strong and weak stop codon contexts. Stop codon contexts of the A′ gene variants are indicated. The 3A′ protein band results from stop codon readthrough of the internal stop codon in the 3A′ gene. The 2A′′ band results from termination at this stop codon. The 2A′ protein is the gene product of the 2A′ internal standard gene. Analysis of gene variants with the indicated stop codon contexts and with SD+, DR-A (A) or SD–, DR-B (B) are shown.
Expression of the eight different gene variants (SD+ and SD– together with each of the four different stop codon contexts described above) was studied in connection with full induction by 1 mM IPTG. Protein A′ expression was quantified by measuring the product ratio (3A′ + 2A′′)/2A′, where 2A′′ represents the termination product, 3A′ the readthrough product and 2A′ the control product. For the stop codon contexts CCA UAA A (pHN52), AGC UAA U (pHN55) and AGC UGA U (pHN56), the stop signal was strong enough to prevent any measurable appearance of the 3A′ readthrough product on gels, and only the 2A′′ protein was expressed from the test gene (Table I). However, for the weak stop signal GAC CCA UGA A, both the 3A′ readthrough and the 2A′′ termination products were found, giving a transmission value (T; 3A′/2A′′) of 0.64 for the SD+ variant in pHN53 and 0.80 for the SD– variant in pHN59. Measurements of total gene expression indicated a slightly lower expression level [(3A′ + 2A′′)/2A′] for the variants with the inefficient stop codon context GAC CCA UGA A (Table I), than for those with the more efficient stop codon contexts. This was true no matter if a strong (SD+, DR-A) or weak (SD–, DR-B) start site was present in the 3A′ gene.
Table I. Protein expression with different translation and stop contexts.
| Plasmids | Constructions | Gene expression |
Transmission T (3A′/2A′′) | Stop codon effect UAA/UGA | ||
|---|---|---|---|---|---|---|
| 3A′ | 2A′′ | |||||
| pHN52 | SD+-AUG-2A′-CCAUAA A | – | 0.22 ± 0.03 | – | ||
| pHN53 | SD+-AUG-2A′-CCAUGA A | 0.07 ± 0.01 | 0.11 ± 0.01 | 0.64 ± 0.05 | pHN52/pHN53 | 1.2 |
| pHN55 | SD+-AUG-2A′-AGCUAA U | – | 0.25 ± 0.03 | – | ||
| pHN56 | SD+-AUG-2A′-AGCUGA U | – | 0.24 ± 0.02 | – | pHN55/pHN56 | 1.0 |
| pHN58 | SD–-AUG-2A′-CCAUAA A | – | 0.097 ± 0.01 | – | ||
| pHN59 | SD–-AUG-2A′-CCAUGA A | 0.035 ± 0.01 | 0.044 ± 0.01 | 0.80 ± 0.08 | pHN58/pHN59 | 1.2 |
| pHN61 | SD–-AUG-2A′-AGCUAA U | – | 0.105 ± 0.02 | – | ||
| pHN62 | SD–-AUG-2A′-AGCUGA U | 0.104 ± 0.02 | – | pHN61/pHN62 | 1.0 | |
Protein A′ expressed in M9 minimal medium in 1 mM IPTG. The 3A′ values are normalized to 2A′ obtained by correction for the molecular weight of the A′ proteins (Björnsson et al., 1998). All contexts have GAC as the –2 stop codon and DR-A penta-codon sequence downstream of the initiation codon of the SD+ group. The DR-B penta-codon sequence was used in the SD– group. SD+ is AAGGAGG and SD– is AAAUAAA (Figure 3). All data were obtained from at least four independent experiments.
As can also be seen in Table I, the expression levels associated with the SD+ sequences are about double those of their SD– counterparts, confirming the positive effect on translation initiation by a canonical SD sequence. Furthermore, since the level of readthrough of the weak termination context GAC CCA UGA A is rather similar in the presence or absence of a strong SD, this suggests that the level of initiation does not influence termination efficiency in these cases.
Influence of different induction levels on gene expression
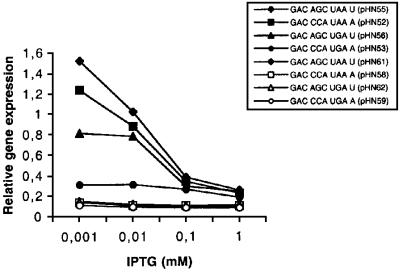
Since the induction by 1 mM IPTG used above should give a high level of mRNA, i.e. a low ratio between the number of free ribosomes and mRNA molecules in the cell, the effects of lowering the IPTG concentrations were analysed. This treatment should decrease the mRNA level, thus increasing the ribosomes/mRNA ratio. As can be seen in Figure 5, a low concentration of IPTG gave a higher total gene expression (3A′ + 2A′′) for several of the analysed gene variants relative to the internal 2A′ standard. Expression values for the three gene variants with SD+, DR-A together with an efficient stop signal (pHN52, pHN55 and pHN56) are increased, whereas the one with a weak stop signal (pHN53) is unaffected. As a result, for the GAC CCA UPuA A stop codon contexts, the level of normalized expression was almost four times higher for UAA than for its UGA counterpart. For the context GAC AGC UPuA U, it was almost double. Thus, up to a 5-fold difference in gene expression during low induction can be obtained, depending on the nature of the stop signal. The DR-B variants together with SD– (pHN58, pHN59, pHN61 and pHN62) have a weak start site and they are unaffected by a lowered IPTG induction (Figure 5). As a result, expression of these SD–, DR-B variants is only ∼10% compared with the strongest expression found for the SD+, DR-A variant in pHN55. Thus, if the mRNA carries a strong start site and a strong translational stop signal, an increased protein production per mRNA molecule is obtained if the ribosome/mRNA ratio in the cell is increased by lowered IPTG induction. The results suggest that expression is dependent on the stop signal some 140 codons further down the gene, the strength of the start site and the ribosome/mRNA ratio. Ribosomal queuing is a possible explanation for the observed results.
Fig. 5. Effect of stop codon context and gene induction by IPTG on relative gene expression. The analysed stop codon contexts are indicated. Expression is given as (3A′ + 2A′)/2A′, where 3A′ is the readthrough product, 2A′′ is the termination product and 2A′ is the product of the standard gene. The gene variants were preceded by SD+, DR-A (filled symbols) or SD–, DR-B (open symbols) as indicated in Table I.
Influence of UV irradiation or increased distance between start and stop sites on gene expression
The strain CSR603C is sensitive to UV irradiation, resulting in the destruction of DNA. In such a strain with a high copy number plasmid, any plasmid molecule that escapes the UV damage will continue to replicate, thus giving rise to a new population of the high copy number plasmid. Thus, chromosomal DNA is inactivated but plasmid-borne gene products are still expressed and enriched in the cell after the UV irradiation (Sancar et al., 1979). Since the gross mRNA concentration is decreased by such treatment, the effect should be an increased ratio of ribosomes over mRNA in the cell. The remaining functional mRNAs should be derived preferentially from plasmid genes being loaded efficiently by translating ribosomes. As a result, the chance of ribosome queuing should increase. However, for mRNAs with an initiation region that is covered by queued ribosomes due to slow termination, free ribosomes would nevertheless not give any increased initiation. This is true even if the free ribosomes are present in excess.
The results of an experiment where non-induced cells had been exposed to UV irradiation are shown in Table II. Even though the 3A′ and 2A′ bands are normally very weak, using a sample of cells that had not been induced by IPTG, both the 3A′ and 2A′ protein bands can be clearly recognized in the gels after UV irradiation in the absence of induction (not shown). This result confirms the enrichment and preferential translation of the plasmid-borne 3A′ and 2A′ genes after UV irradiation. From the data, it is clear that the stop codon context variant with the most inefficient stop signal GAC CCA UGA A (pHN53) indeed gives a much lower expression level than the other three more efficient stop signal variants.
Table II. Protein expression in the mutant strain CSR603C.
| Plasmids | Constructions | Gene expression | Stop codon effect | |
|---|---|---|---|---|
| |
|
(3A′ + 2A′′)/2A′ |
(UAA/UGA) |
|
| pHN52 | SD+-AUG-2A′-CCAUAA A | 2.7 ± 0.3 | ||
| pHN53 | SD+-AUG-2A′-CCAUGA A | 0.3 ± 0.02 | pHN52/pHN53 | 9 |
| pHN55 | SD+-AUG-2A′-AGCUAA U | 3.1 ± 0.4 | ||
| pHN56 | SD+-AUG-2A′-AGCUGA U | 2.5 ± 0.4 | pHN55/pHN56 | 1.2 |
CSR 603C is a mutant strain in which chromosomal DNA is inactivated and the plasmid test gene is overexpressed by UV irradiation. The initiation and termination codons are separated by a 2A′ gene sequence, as indicated. The SD preceding the AUG initiation codon is the SD+ variant, as shown in Figure 3. Cells were grown in M9 minimal medium without the inducer IPTG. The 3A′ values are normalized to 2A′ obtained by correction for the molecular weight of the A′ proteins (Björnsson et al., 1998). All contexts have GAC as the –2 codon (Figure 3). All data are based on at least four independent experiments.
It is quite possible that the UV irradiation causes some destruction not only of DNA but also of ribosomes. However, expression here is estimated as a relative value comparing it with the standard 2A′ gene. A decreased ribosome pool as the result of destruction by UV should also affect the expression of the 2A′ gene, i.e. the ratio 3A′/2A′ as a measure of relative expression is still relevant. A queuing effect should be apparent for low IPTG induction conditions giving a high ribosome/mRNA ratio. In this case and in the absence of UV treatment, relative expression values of 1.2 and 0.3 (pHN52 and pHN53, respectively, in Figure 5) are found. In the case of UV treatment, in the absence of induction, the expression values for these two constructs are 2.7 and 0.3 (Table II), respectively. Even if there is some destruction of ribosomes by the UV treatment, the increase in relative expression for pHN52 from 1.6 to 2.7 suggests that the treatment gives an increased ribosome/mRNA ratio, in line with the rationale behind the experiment. The significant difference in gene expression (9-fold) if gene variants ending with strong or weak termination signals are compared in connection with UV irradiation (Table II) supports a queuing model.
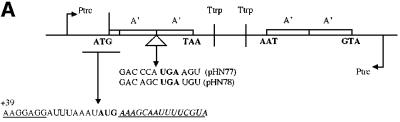
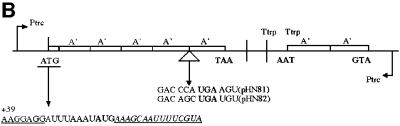
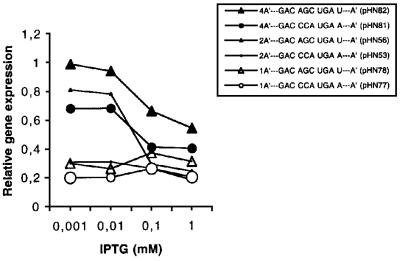
Even if a stop signal context favours ribosome queuing upstream of the stop codon, the effect should be less pronounced if the distance between the initiation and termination codons is increased. The weak stop context GAC CCA UGA A and the strong stop signal GAC AGC UGA U were therefore analysed in constructs where they were preceded by one (pHN77 and pHN78), two (pHN53, pHN56) or four (pHN81 and pHN82) A′-encoding units (Figures 5 and 6), giving 2A′, 3A′ and 5A′ proteins as the readthrough products, respectively. All gene variants were SD+ with the DR-A sequence, thus ensuring efficient initiation. As can be seen in Figure 7, during low induction (high ribosome/mRNA ratio), gene expression levels are 4A′>2A′>1A′ for the weak stop signal (GAC CCA UGA A), suggesting gene size-dependent queuing effects. For the stronger stop signal (GAC AGC UGA U), only the smallest gene variant (1A′) in pHN78 failed to give higher expression during low induction, thus indicating queuing in this case. In contrast, the 2A′ (pHN56) and 4A′ (pHN82) variants give increased expression (4A′>2A′) as a response to lowered induction. Taken together, the results presented in Figure 7 suggest that an increased distance between the stop and start signals decreases ribsomal queuing up to the initiation region. Such queuing is counteracted by an efficient stop signal, and the ribosome/mRNA ratio is also important.
Fig. 6. Gene variants with different distances between start and stop codons. (A) Short (1A′) (pHN77 and pHN78) and (B) long distances (4A′) (pHN81 and pHN82) are shown. Ptrc, promoter of transcription; Ttrp, transcription termination; the protein A′-encoding regions are shown. The initiation codon AUG (ATG) and the termination codon UGA (TGA) are indicated. mRNA sequences of start and stop codon contexts are shown. All gene variants are preceded by SD+ and the initiation codon is followed by the DR-A penta-codon downstream sequence. Base +39 of the mRNA transcript is indicated.
Fig. 7. Effect of stop codon context and gene induction by IPTG on relative gene expression. Plasmids are specified in Figure 6. Expression values include the readthrough product, if any. Data for pHN56 and pHN53 have been presented in Figure 5, but they have been inserted here, using small symbols, for the sake of comparison.
One A′ protein unit is comprised of 57–67 amino acids, corresponding to some 200 bases in the mRNA. The distance from the stop codon after the second A′-encoding unit up to the initiation codon is therefore almost 400 bases. If a ribosome covers ∼30–40 bases in the mRNA (Steitz and Jakes, 1975; Beyer et al., 1994), this means that queuing would block further initiation for polysomes containing ∼10 ribosomes, or more.
Influence of stop codon contexts on 3A′ gene expression is not the result of altered mRNA levels or secondary structures
It has been found that the half-life of a mRNA that uses an inefficient stop codon context is shorter by a factor of two than the half-life of a corresponding mRNA with an efficient stop codon context (Björnsson and Isaksson, 1996). It appeared possible that different pools of mRNA could explain the higher expression associated with some analysed efficient stop codon contexts, compared with the inefficient stop codon context CCA UGA A, during conditions of low induction by IPTG (Figure 5). Therefore, the steady-state levels of mRNA for the gene variants with the stop contexts GAC CCA UGA A, GAC AGC UAA U, GAC AGC UGA U and GAC CCA UAA A in weakly induced cells, as described above in Figure 5, were subjected to northern blot analysis. The 2A′ mRNA acted as an internal control. For these experiments, cells were cultivated together with 0.01 mM IPTG. As discussed above, this low level of induction, resulting in a high ribosome/mRNA ratio, gives significantly different gene expression values at the protein level when the four stop codon context variants were compared (Figure 5). However, according to the northern blot results shown in Figure 8, the relative level of 3A′ mRNA is similar for all four analysed stop codon context variants. Thus, the 4-fold difference in protein levels found for these stop codon context variants at low induction, as compared with the control gene 2A′ (Figure 5), cannot be explained by different mRNA steady-state levels. It is possible that the earlier results could reflect the fact that the A′ gene variants used for those experiments carried a short signal peptide-encoding sequence, as the beginning of the gene (Björnsson and Isaksson, 1993). This sequence is not included in the constructs used here.
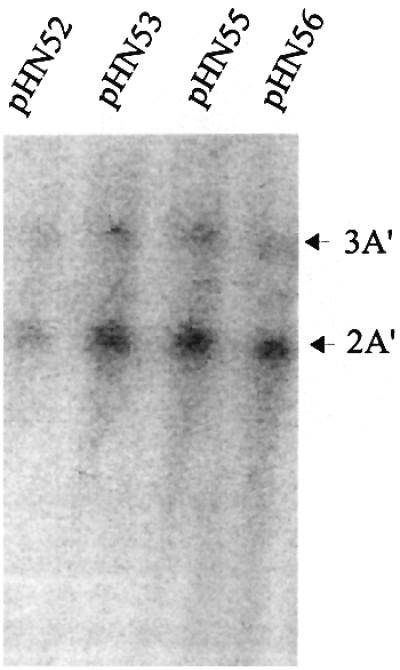
Fig. 8. mRNA steady-state levels of gene variants with different stop codon contexts. The 32P-labelled deoxyoligonucleotide probe is complementary to each A′-encoding domain in the 3A′ test gene and 2A′ control gene. The positions of the 3A′ and 2A′ mRNAs are indicated by arrows. The mRNA levels, as measured by the molar ratio (3A′/2A′) associated with the four stop codon context variants, were 0.37 (pHN52), 0.29 (pHN53), 0.31 (pHN55) and 0.26 (pHN56).
It was observed that the mRNA level ratio between the test gene 3A′ and control gene 2A′ is ∼0.3 for all four analysed gene variants (Figure 8), suggesting that all the different 3A′ mRNAs are at a lower steady-state level compared with the 2A′ mRNA. This ratio is comparable with the protein expression ratio found for maximum IPTG induction conditions (Table I; Figure 5).
Secondary structures involving the initiation region, including the SD sequence, can have profound effects on translation initiation (de Smit and van Duin, 1994). However, a search using the mFold (3.1) program (Zuker and Jacobson, 1998) did not reveal any correlation between possible secondary structures and gene expression levels (not shown).
It is conceivable that apparent gene expression values could be obscured by peptidyl-tRNA drop-off effects. For this reason, experiments analogous to those described in Figure 5 were carried for the gene variants in plasmids pHN52, pHN53, pHN55 and pHN56 in a pth temperature-sensitive (Ts) mutant strain. At 37°C, this strain is disturbed in growth because of impaired degradation of released peptidyl-tRNA (Heurgue-Hamard et al., 2000), and excessive release should give an increased inhibition of growth. However, no difference in growth inhibition was found for the pth(Ts) strain for the different plasmids at either 37 or 30°C, with or without IPTG induction (data not shown). The results therefore suggest that there is no difference in accumulation of peptidyl-tRNA as the result of drop-off, when the gene variants in these plasmids are compared.
Discussion
The test system
Here we have studied the effects on gene expression in vivo by stop codon signals of various strengths, using a new variant of the previously described protein A′ translation assay system as a model gene (Björnsson and Isaksson, 1993; Björnsson et al., 1998; Stenström et al., 2001b). It has several technical advantages for subcloning purposes, and expression of the test A′ gene variants can be controlled by IPTG. The test and 2A′ internal standard genes, both in the same plasmid, are built up of protein A′-encoding units, each unit containing 57–67 codons. Since the test and standard genes are both under separate control by the same promoter and terminator sequences, the expression at the transcriptional level should be similar. Therefore, any change in expression that results from manipulations of the test gene should originate at the post-transcriptional level.
Even by changing only the first codon in the DR (the +2 codon), relative gene expression can vary by 20-fold for gene variants that are SD–. These effects of the altered DRs were reported to be similar for both the β-galactosidase and A′ systems, suggesting that the 3A′ gene is a useful model system for expression studies (Stenström et al., 2001b).
Determinants of stop codon efficiency
The termination efficiency at the termination codons UAA, UAG or UGA is dependent on the codon context, as well as the codon itself. The 3′-flanking nucleotides interact directly with the termination factor (Brown et al., 1990; Tate and Mannering, 1996), and a pausing effect is obtained if the stop codon is UGA followed by an A base (Björnsson and Isaksson, 1996). The P-site tRNA and the last two amino acid residues in the gene protein product, encoded by the –2 and –1 codons, are also determinants (Mottagui-Tabar et al., 1994; Björnsson et al., 1996; Zhang et al., 1996). The stop codon context GAC CCA UGA A analysed here is weak because of the 3′-flanking A base and the last two amino acid residues in the gene protein product (Mottagui-Tabar et al., 1994; Björnsson et al., 1996). Together, this gives inefficient recognition by the termination factor RF2. This results in an extended ribosomal pausing and an increased readthrough by tRNATrp, which is a near-cognate reader of UGA, having UGG as its cognate codon (Curran and Yarus, 1988; Björnsson and Isaksson, 1996; Pavlov et al., 1998).
Abortive effects of ribosome pausing
Transcription termination factor Rho can give transcriptional polarity and a decrease in gene expression (Stanssens et al., 1986; Alifano et al., 1988; Deana et al., 1998) for the commonly used lacZ reporter gene. The semi-synthetic A′ gene used here is not subject to such transcriptional polarity (Björnsson et al., 1998). Decreased gene expression could also be the result of peptidyl-tRNA drop-off at an early codon, giving rise to abortion of translation (Dincbas et al., 1999; Heurgue-Hamard et al., 2000). However, pausing at the stop codon studied here, located after the second A′-encoding unit and thus being a late codon, does not trigger any peptidyl-tRNA drop-off as measured in a peptidyl-tRNA hydrolase conditionally deficient strain.
Ribosome pausing can give translational frameshifting (Weiss et al., 1988) and aminoacyl-tRNA-dependent readthrough of stop codons by a near-cognate tRNA (Curran and Yarus, 1988; Buckingham et al., 1990; Adamski et al., 1993). However, we did not find any significant ribosomal frameshifting in this study, since the frameshift products should have been revealed in the assay system if they appear at significant levels, regardless of whether they are the result of a +1 or a –1 frameshift (not shown).
Stop codon readthrough products normally arise at low levels, often below the level of detection for genes with an efficient stop signal. A readthrough product can, however, be detected in the case of the inefficient stop codon context GAC CCA UGA A analysed here by direct physical measurements of protein levels using gel electrophoresis. For this codon context, the ratio of readthrough/termination is as high as 0.6–0.8 if the weak stop codon context is preceded by one, two or four A′-encoding regions. This level of stop codon readthrough is independent of the strength of the start site. We have noticed that readthrough of this context that should give a 3A′ protein is non-detectable if the stop codon context is located only three or nine codons downstream of the initiation codon (not shown). One reason could be increased ribosome drop-off during expression of such a mini-gene (Dincbas et al., 1999; Heurgue-Hamard et al., 2000). It is also possible that the ribosome that pauses at such an early stop signal also blocks the translation initiation site, thus giving a low yield of a 3A′ readthrough protein despite a high readthrough of the stop codon.
Ribosome pausing and queuing
The initiation codon DR is very important for gene expression at the translational level, and some of these DRs, including DR-A used here, can compensate for the lack of a strong SD sequence (not shown). If the induction level is decreased by lowering the IPTG concentration, giving a higher ribosome/mRNA ratio, the relative expression of the SD+, DR-A gene variants is increased, except for the variant with a weak stop signal (pHN53). This result is in line with a queuing model implying that the mRNA start site in the latter case is covered by queuing ribosomes, as the result of slow termination.
The SD+ together with DR-A gives ∼2-fold higher expression compared with SD–, DR-B during high induction by IPTG. During low induction, this difference increases to 10-fold (Figure 5). The difference in expression can be even higher, since it has been reported (Stenström et al., 2001a) that the combination of SD+ with DR-A compared with SD– with DR-B in the β-galactosidase system can give 50-fold higher expression. This large difference was found in the absence of IPTG induction, i.e. at a low level of mRNA. Altogether, the results suggest that the effect of a start site on gene expression is very dependent on the intracellular ribosome/mRNA ratio.
The inefficient stop context GAC CCA UGA A analysed here causes inefficient termination and ribosomal pausing, which is long enough even to stabilize a degradation-related fragment of the mRNA (rpRNA; ribosome pausing RNA) in growing bacteria (Björnsson and Isaksson, 1996). A pausing ribosome can cause the following ribosomes to form a queue along the upstream mRNA (Wolin and Walter, 1988; Lesnik et al., 2000). As suggested here, this ribosomal queue could even block the upstream binding of other ribosomes at the initiation region. Since the pausing is affected by the last two amino acid residues in the gene protein product (Mottagui-Tabar et al., 1994; Björnsson et al., 1996), this means that queuing, translation initiation and gene expression can also be affected by the gene protein product itself.
If the termination step is rapid, in particular if the gene is expressed at a high level, allowing for only a limited number of ribosomes per mRNA molecule, less queuing should take place. Queuing effects should be more pronounced for short than for long genes, as indeed is observed since expression can be ranked as 4A′>2A′>1A′ (Figure 7). Ribosome queue formation should be favoured by efficient initiation signals such as SD+, together with a favourable DR, thus increasing the number of ribosomes that become recruited for translation of a particular mRNA molecule (Figure 5). The postulated queuing effect by a weak termination context should also be most favoured if the mRNA is at a low steady-state level, compared with the pool of free ribosomes. Thus, a ribosome queuing model could explain why expression is poor in the case of the weak stop codon context GAC CCA UGA A under conditions of low induction, whereas expression is higher for gene variants with other stronger terminator sequences under these conditions (Figure 5).
UV irradiation inactivates expression of a number of chromosomal genes in the E.coli strain CSR603 (Sancar et al., 1979), thus decreasing the gross mRNA level and increasing the pool of free ribosomes. If this strain carries a plasmid, any plasmid molecule that escapes the UV damage will continue to replicate, thus giving rise to a new plasmid population. Thus, chromosomal DNA is inactivated but mRNAs from plasmid-borne genes are still expressed and mRNA encoded by the 3A′ and 2A′ genes will be enriched in the cell after the UV irradiation. The 3A′ and 2A′ proteins cannot be found on gels using samples from cells that are not induced by IPTG. On the contrary, the proteins are visible using uninduced samples from UV-irradiated cells (Table II). This suggests an enrichment and preferential translation of the 3A′- and 2A′-encoding mRNAs as the result of UV irradiation, in line with expectation, since the genes are carried by the plasmid (Sancar et al., 1979).
Genes can be grouped with respect to their expression levels according to their codon adaptation index (CAI) (Sharp and Li, 1987). For natural genes, one of the consequences of the suggested queuing model should be a positive correlation between low levels of expression and small gene size, especially for genes that terminate with UGA, which is a weak stop codon in many contexts. Likewise, genes expressed at a high level should be large and/or terminate with a strong stop codon like UAA. As shown in Table III, for the poorly expressed genes (CAI <0.2), 75 out of a total of 145 genes (52%) are smaller than 150 codons, and for highly expressed genes (CAI >0.4) 147 out of a total of 754 (19%) are smaller than 150 codons. Thus, the proportion of small genes is higher in the poorly expressed than in the highly expressed group. An inspection of the distribution of UAA and UGA usage in genes smaller than 150 codons (Table III) suggests that the ratio UAA/UGA is 1.6 for the group with the lowest expression but is 5.7 for the group with the highest expression. Thus, UAA is over-represented in highly expressed, as compared with poorly expressed, genes. All these data are in support of a queuing model.
Table III. Use of termination codons in E.coli genes with different lengths and CAI values.
| Gene length (codons) | Codon adaptation index (CAI) |
Total | |||||
|---|---|---|---|---|---|---|---|
| <0.2 |
0.2–0.4 |
>0.4 |
|||||
| UAA | UGA | UAA | UGA | UAA | UGA | ||
| <150 | 46 (6) | 29 (4) | 338 (42) | 232 (29) | 125 (16) | 22 (3) | 792 (100) |
| 150–500 | 46 (2) | 17 (<1) | 1414 (54) | 687 (26) | 371 (14) | 82 (3) | 2617 (100) |
| >500 | 3 (<1) | 4 (<1) | 264 (46) | 131 (24) | 122 (22) | 32 (6) | 556 (100) |
All E.coli genes terminated by stop codons UAA and UGA were divided into groups with respect to gene length and gene expression level. The gene length was grouped as short genes (<150 codons), medium sized genes (150–500 codons) and long genes (>500 codons). Gene expression levels were assigned by their CAI (Sharp and Li, 1987) as low (CAI <0.2), intermediate (CAI 0.2–0.4) or high (CAI >0.4). Numbers of analysed genes are indicated, with percentage proportion in the respective gene size group in parentheses. The data were obtained from the E.coli genome bank of Wisconsin-Madison University (http://www.genome.wisc.edu/pub/analysis/).
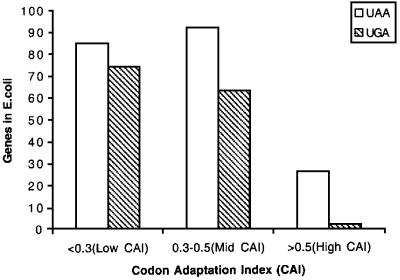
Figure 9 shows an analysis of very small E.coli genes (<100 codons) ending with a strong (UAA) or a weak (UGA) stop signal. As can be seen in the figure, UGA is almost as common as the strong UAA in poorly expressed genes. UGA becomes less common for the genes expressed at a medium level. For the highly expressed genes (CAI >0.5), UAA is ∼10 times as common as the two UGA examples actually found. These two UGA cases have flanking sequences (the –2, –1 and downstream codons) that support high termination efficiency at the otherwise weak termination codon UGA (Mottagui-Tabar et al., 1994; Björnsson et al., 1996). Thus, these data further suggest that short and highly expressed genes present in the E.coli genome avoid weak termination signals. Altogether, the analysis of natural E.coli genes suggests that ribosome queuing is a phenomenon that operates for normal genes and not only for the A′ model gene used here.
Fig. 9. Relationship between expression levels and use of termination codons UAA and UGA in short genes in E.coli. All 342 short genes smaller than 100 codons were analysed for the distribution of the termination codons UAA and UGA. The gene expression levels were grouped according to their codon adapation index, where a high value reflects a high level of expression. The genes were divided into three groups with low (<0.3), medium (0.3–0.5) or high (>0.5) CAI values (Sharp and Li, 1987). The data were obtained from the E.coli genome bank of Wisconsin University (http://www.genome.wisc.edu/pub/analysis/).
Conclusions
The results obtained here show that an inefficient termination signal can introduce a significant cis effect to lower gene expression at the translational level. Some important features for this control circuit are the strength of the SD sequence, the sequence of a few codons downstream of the initiation codon (DR) and the nature of the stop codon context involving at least 12 bases including the stop codon itself. This effect is particularly important in the case of UGA, since certain combinations of the two C-terminal amino acid residues in the protein product, encoded by the –2 and –1 codons, promote low termination efficiency. The length of the gene can also have a profound effect on gene expression since a higher proportion of poorly expressed natural genes are small, as compared with highly expressed genes. Small natural highly expressed genes are biased towards use of UAA instead of UGA. The results presented here illustrate how gene regulation at the translational level can be affected by the physiological conditions that affect the ribosome/mRNA ratio, together with the physical construction of a number of gene characteristics.
Materials and methods
Strains
The E.coli strain used for primary cloning is MC1061 [ara D139, ΔlacX74, galU, galK, rpsL, Δ(araABC-leu7697), hsdR, mcrB, thi] (Sambrook et al., 1989). Strain XAC [Δ(lacproB), argE, ara, gyrA, rpoB, thi] (Stenström et al., 2001b) is used for protein expression. The dam-strain HB101 [supE44, hsdS20, (rB– mB–), recA1, ara-14, proA2, lacY1, galK2, rspL20, xyl-5, mtl-1] (Sambrook et al., 1989) was used to obtain an unmodified ClaI site for later cleavage by this enzyme. Strain MG1655 with additional markers [zdh-925::Tn10, pth(Ts)] was used for peptidyl-tRNA drop-off experiments. This strain grows very slowly at 37°C because it is unable to hydrolyse released peptidyl-tRNA at high temperature, as the result of the pth(Ts) mutation. Because of this inability, growth is even more inhibited in connection with excessive peptidyl-tRNA drop-off. CSR603 (recA, uvrA6, phr-1) was used for UV irradiation giving chromosomal DNA inactivation and overexpression of plasmid gene products (Sancar et al., 1979).
Plasmid constructions
Plasmid DNA was isolated by using a Magic mini-prep kit (Promega). Treatment with restriction enzymes and T4 ligase was performed using established procedures. Transformation was made into competent E.coli cells (MC1061). Selection for ampicillin resistance was at 37°C on LB plates with 200 µg/ml ampicillin. Plasmid candidates as judged by analytical cleavage were DNA sequenced for confirmation and finally transformed into the E.coli strain XAC for protein A′ assay.
A modified 3A′ translation assay system was constructed based on plasmid pTrp99A (Amann et al., 1988). Plasmids pMCR3A′ (Figure 1A) and pAB97 (Björnsson et al., 1998) were used as sources of 3A′ and 2A′ genes, respectively. The plasmid pAB97 was cleaved with EcoRI and HindIII to remove the third of the A′-encoding sequences. After a Klenow treatment and re-ligation, a new plasmid (pHN2A′) with a 2A′ gene was created. The 2A′-encoding fragment was then taken from pHN2A′ by cleaving at the PvuII and ClaI sites. The 2A′ fragment was inserted next, to serve as an internal control gene, into pMCR-3A′ that had been cleaved at the Ecl136II and ClaI sites, thus giving plasmid pHN110. This vector carries lacI and A′ genes that are under control of Ptrc. A similar cloning principle was used starting with pHN1 that was extended with a 2A′ fragment to give a 5A′ gene. This gene was inserted, to serve as an internal control gene, thus giving plasmid pHN200, corresponding to pHN110 (Figure 1B). The final plasmids have one 3A′ gene together with one 2A′ (pHN110 derivatives) or 5A′ gene (pHN200 derivatives) as the internal control gene. In order to construct a plasmid where the direction of transcription of the internal standard gene 2A′ was inverted to be in the same direction as the test gene, the two ClaI sites at both ends of 2A′ were used. One of these sites can be modified by intracellular methylation. This plasmid was transformed into the methylation-deficient strain HB101, leaving the ClaI site unmodified and sensitive to ClaI cleavage. The plasmid was cleaved with ClaI and re-ligated to obtain a candidate where both the 3A′ and 2A′ genes were in the same transcriptional direction (pHN500) (Figure 1A).
Cell growth and protein A′ purification
Plasmid-harbouring XAC cells were cultured overnight at 37°C in M9 medium with 0.2% glucose, thiamine and all amino acids at recommended concentrations and with ampicillin (200 µg/ml). A 100-fold dilution was used as innoculum for growth in the same medium with or without IPTG, at the indicated concentrations, and growth was followed by spectrophotometer measurements. For induction, IPTG was added during 30 min in the mid-log phase of growth. Cell samples were cooled and harvested by centrifugation, followed by re-suspension in 1 ml of 10× TST buffer (0.5 M Tris pH 7.4, 2.5 M sodium chloride, 0.5% Tween-20) (Björnsson et al., 1998). Cells were lysed by incubation at 95°C for 10 min, and cell debris was eliminated by centrifugation. Protein A′ was purified from the supernatant fraction using IgG–Sepharose (Pharmacia) mini-columns and a vacuum mini-fold system (Promega). A′ proteins were eluted with 0.1 ml of 0.5 M HAc at pH 3.2. The eluant was dried in a vacuum Speed-Vac (Techtum). Protein samples were dissolved in sample loading buffer, after denaturation at 95°C for 2 min. Analyses of the A′ proteins were done by using SDS–PAGE, and protein bands were scanned and quantified as described previously (Björnsson et al., 1998).
For analysis of P-site peptidyl-tRNA drop-off experiments, a derivative of strain MG1655 with a temperature-sensitive peptidyl-tRNA hydrolase (see above) was transformed with the plasmids pHN52, pHN53, pHN55 and pHN56 and grown overnight at 30°C. Next day at mid-log phase of growth, the culture was divided into subcultures and growth was followed at 37 and 30°C, with or without IPTG at both temperatures.
Preparation and northern blot analysis of mRNA
Total cellular RNA from the E.coli strain XAC with plasmids was prepared by using the Totally RNA kit (Ambion Inc.). The 10 ml cultures were harvested in mid-log phase of growth and 0.01 mM IPTG was added 30 min before cell harvest. RNA samples were separated in 1% agarose gels, and transferred to a Hybond-N nylon membrane (Amersham Life Science). The transferred RNA was hybridized 16–20 h with the 32P-labelled deoxyoligonucleotide probe ABP01 (5′-CGTTGTTCTTCG TTTAAGTTAGG-3′) which is complementary to each of the A′-encoding RNA sequences. The radioactive signals on the films were analysed by scanning (FujiFilm FLA-3000).
Acknowledgments
Acknowledgements
This work has been supported by grants from the Swedish Natural Science Foundation (NFR), the Swedish Research Council for Engineering Sciences (TFR) and the Foundation for Strategic Research (SSF) to L.A.I.
References
- Adamski F.M., Donly,B.C. and Tate,W.P. (1993) Competition between frameshifting, termination and suppression at the frameshift site in the Escherichia coli release factor-2 mRNA. Nucleic Acids Res., 21, 5074–5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alifano P., Ciampi,M.S., Nappo,A.G., Bruni,C.B. and Carlomagno,M.S. (1988) In vivo analysis of the mechanisms responsible for strong transcriptional polarity in a ‘sense’ mutant within an intercistronic region. Cell, 55, 351–360. [DOI] [PubMed] [Google Scholar]
- Amann E., Ochs,B. and Abel,K.J. (1988) Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene, 69, 301–315. [DOI] [PubMed] [Google Scholar]
- Beyer D., Skripkin,E., Wadzack,J. and Nierhaus,K.H. (1994) How the ribosome moves along the mRNA during protein synthesis. J. Biol. Chem., 48, 30713–30717 [PubMed] [Google Scholar]
- Björnsson A. and Isaksson,L.A. (1993) UGA codon context which spans three codons. Reversal by ms2i6A37 in tRNA, mutation in rpsD(S4) or streptomycin. J. Mol. Biol., 232, 1017–1029. [DOI] [PubMed] [Google Scholar]
- Björnsson A. and Isaksson,L.A. (1996) Accumulation of a mRNA decay intermediate by ribosomal pausing at a stop codon. Nucleic Acids Res., 24, 1753–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björnsson A., Mottagui-Tabar,S. and Isaksson,L.A. (1996) Structure of the C-terminal end of the nascent peptide influences translation termination. EMBO J., 15, 1696–1704. [PMC free article] [PubMed] [Google Scholar]
- Björnsson A., Mottagui-Tabar,S. and Isaksson,L.A. (1998) The analysis of translational activity using a reporter gene constructed from repeats of an antibody-binding domain from protein A. Methods Mol. Biol., 77, 75–91. [DOI] [PubMed] [Google Scholar]
- Bossi L. (1983) Context effects: translation of UAG codon by suppressor tRNA is affected by the sequence following UAG in the message. J. Mol. Biol., 164, 73–87. [DOI] [PubMed] [Google Scholar]
- Brown C.M., Stockwell,P.A., Trotman,C.N. and Tate,W.P. (1990) Sequence analysis suggests that tetra-nucleotides signal the termination of protein synthesis in eukaryotes. Nucleic Acids Res., 18, 6339–6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham R.H., Sorensen,P., Pagel,F.T., Hijazi,K.A., Mims,B.H., Brechemier-Baey,D. and Murgola,E.J. (1990) Third position base changes in codons 5′ and 3′ adjacent UGA codons affect UGA suppression in vivo. Biochim. Biophys. Acta, 1050, 259–262. [DOI] [PubMed] [Google Scholar]
- Capecchi M.R. (1967) A rapid assay for polypeptide chain termination. Biochem. Biophys. Res. Commun., 28, 773–778. [DOI] [PubMed] [Google Scholar]
- Caskey T., Scolnick,E., Tompkins,R., Goldstein,J. and Milman,G. (1969) Peptide chain termination, codon, protein factor and ribosomal requirements. Cold Spring Harbor Symp. Quant. Biol., 34, 479–488. [DOI] [PubMed] [Google Scholar]
- Chen G.F. and Inouye,M. (1990) Suppression of the negative effect of minor arginine codons on gene expression; preferential usage of minor codons within the first 25 codons of the Escherichia coli genes. Nucleic Acids Res., 18, 1465–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Bjerknes,M., Kumar,R. and Jay,E. (1994) Determination of the optimal aligned spacing between the Shine–Dalgarno sequence and the translation initiation codon of Escherichia coli mRNAs. Nucleic Acids Res., 22, 4953–4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran J.F. and Yarus,M. (1988) Use of tRNA suppressors to probe regulation of Escherichia coli release factor 2. J. Mol. Biol., 203, 75–83. [DOI] [PubMed] [Google Scholar]
- de Smit M.H. and van Duin,J. (1994) Control of translation by mRNA secondary structure in Escherichia coli. A quantitative analysis of literature data. J. Mol. Biol., 244, 144–150. [DOI] [PubMed] [Google Scholar]
- Deana A., Ehrlich,R. and Reiss,C. (1998) Silent mutations in the Escherichia coli ompA leader peptide region strongly affect transcription and translation in vivo. Nucleic Acids Res., 26, 4778–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dincbas V., Heurgue-Hamard,V., Buckingham,R.H., Karimi,R. and Ehrenberg,M. (1999) Shutdown in protein synthesis due to the expression of mini-genes in bacteria. J. Mol. Biol., 291, 745–759. [DOI] [PubMed] [Google Scholar]
- Grentzmann G., Brechemier-Baey,D., Heurgue,V., Mora,L. and Buckingham,R.H. (1994) Localization and characterization of the gene encoding release factor RF3 in Escherichia coli. Proc. Natl Acad. Sci. USA, 91, 5848–5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurgue-Hamard V., Dincbas,V., Buckingham,R.H. and Ehrenberg,M. (2000) Origins of minigene-dependent growth inhibition in bacterial cells. EMBO J., 19, 2701–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopelowitz J., Hampe,C., Goldman,R., Reches,M. and Engelberg-Kulka,H. (1992) Influence of codon context on UGA suppression and readthrough. J. Mol. Biol., 225, 261–269. [DOI] [PubMed] [Google Scholar]
- Kozak M. (1999) Initiation of translation in prokaryotes and eukaryotes. Gene, 234, 187–208. [DOI] [PubMed] [Google Scholar]
- Lesnik T., Solomovici,J., Deana,A., Ehrlich,R. and Reiss,C. (2000) Ribosome traffic in E.coli and regulation of gene expression. J. Theor. Biol., 202, 175–185. [DOI] [PubMed] [Google Scholar]
- Looman A.C., Bodlaender,J., Comstock,L.J., Eaton,D., Jhurani,P., de Boer,H.A. and van Knippenberg,P.H. (1987) Influence of the codon following the AUG initiation codon on the expression of a modified lacZ gene in Escherichia coli. EMBO J., 6, 2489–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikuni O., Ito,K., Moffat,J., Matsumura,K., McCaughan,K., Nobukuni,T., Tate,W. and Nakamura,Y. (1994) Identification of the prfC gene, which encodes peptide-chain-release factor 3 of Escherichia coli. Proc. Natl Acad. Sci. USA, 91, 5798–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milman G., Goldstein,J., Scolnick,E. and Caskey,T. (1969) Peptide chain termination. 3. Stimulation of in vitro termination. Proc. Natl Acad. Sci. USA, 63, 183–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottagui-Tabar S., Björnsson,A. and Isaksson,L.A. (1994) The second to last amino acid in the nascent peptide as a codon context determinant. EMBO J., 13, 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov M.Y., Freistroffer,D.V., Dincbas,V., MacDougall,J., Buckingham,R.H. and Ehrenberg,M. (1998) A direct estimation of the context effect on the efficiency of termination. J. Mol. Biol., 284, 579–590. [DOI] [PubMed] [Google Scholar]
- Poole E.S., Brown,C.M. and Tate,W.P. (1995) The identity of the base following the stop codon determines the efficiency of in vivo translational termination in Escherichia coli. EMBO J., 14, 151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacerdot C., Chiaruttini,C., Engst,K., Graffe,M., Milet,M., Mathy,N., Dondon,J. and Springer,M. (1996) The role of the AUU initiation codon in the negative feedback regulation of the gene for translation initiation factor IF3 in Escherichia coli. Mol. Microbiol., 21, 331–346. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch ,E.F. and Maniatis,T., (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sancar A., Hack,A.M. and Rupp,W.D. (1979) Simple method for identification of plasmid-coded proteins. J. Bacteriol., 137, 692–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P.M. and Li,W.H. (1987) The codon adaptation index—a measure of directional synonymous codon usage bias and its potential applications. Nucleic Acids Res., 15, 1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J. and Dalgarno,L. (1975) Terminal-sequence analysis of bacterial ribosomal RNA. Correlation between the 3′-terminal-polypyrimidine sequence of 16-S RNA and translational specificity of the ribosome. Eur. J. Biochem., 57, 221–230. [DOI] [PubMed] [Google Scholar]
- Sorensen M.A. and Pedersen,S. (1991) Absolute in vivo translation rates of individual codons in Escherichia coli. The two glutamic acid codons GAA and GAG are translated with a threefold difference in rate. J. Mol. Biol., 222, 265–280. [DOI] [PubMed] [Google Scholar]
- Sorensen M.A., Kurland,C.G. and Pedersen,S. (1989) Codon usage determines translation rate in Escherichia coli. J. Mol. Biol., 207, 365–377. [DOI] [PubMed] [Google Scholar]
- Stanssens P., Remaut,E. and Fiers,W. (1986) Inefficient translation initiation causes premature transcription termination in the lacZ gene. Cell, 44, 711–718. [DOI] [PubMed] [Google Scholar]
- Steitz J.A. and Jakes,K. (1975) How ribosomes select initiator regions in mRNA: base pair formation between the 3′ terminus of 16S rRNA and the mRNA during initiation of protein synthesis in Escherichia coli. Proc. Natl Acad. Sci. USA, 72, 4734–4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenström C.M., Holmgren,E. and Isaksson,L.A. (2001a) Cooperative effects by the initiation codon and its flanking regions on translation initiation. Gene, 273, 259–265. [DOI] [PubMed] [Google Scholar]
- Stenström C.M., Jin,H., Major,L.L., Tate,W.P. and Isaksson,L.A. (2001b) Codon bias at the 3′-side of the initiation codon is correlated with translation initiation efficiency in Escherichia coli. Gene, 263, 273–284. [DOI] [PubMed] [Google Scholar]
- Tate W.P. and Brown,C.M. (1992) Translational termination: ‘stop’ for protein synthesis or ‘pause’ for regulation of gene expression. Biochemistry, 31, 2443–2450. [DOI] [PubMed] [Google Scholar]
- Tate W.P. and Mannering,S.A. (1996) Three, four or more: the translational stop signal at length. Mol. Microbiol., 21, 213–219. [DOI] [PubMed] [Google Scholar]
- Weiss R., Lindsley,D., Falahee,B. and Gallant,J. (1988) On the mechanism of ribosomal frameshifting at hungry codons. J. Mol. Biol., 203, 403–410. [DOI] [PubMed] [Google Scholar]
- Wolin S.L. and Walter,P. (1988) Ribosome pausing and stacking during translation of a eukaryotic mRNA. EMBO J., 7, 3559–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Ryden-Aulin,M. and Isaksson,L.A. (1996) Functional interaction between release factor one and P-site peptidyl-tRNA on the ribosome. J. Mol. Biol., 261, 98–107. [DOI] [PubMed] [Google Scholar]
- Zuker M. and Jacobson,A.B. (1998) Using reliability information to annotate RNA secondary structures. RNA, 4, 669–679. [DOI] [PMC free article] [PubMed] [Google Scholar]