Abstract
The basic function of the Src homology 3 (SH3) domain is considered to be binding to proline-rich sequences containing a PxxP motif. Recently, many SH3 domains, including those from Grb2 and Pex13p, were reported to bind sequences lacking a PxxP motif. We report here that the 22 residue peptide lacking a PxxP motif, derived from p47phox, binds to the C-terminal SH3 domain from p67phox. We applied the NMR cross-saturation method to locate the interaction sites for the non-PxxP peptides on their cognate SH3 domains from p67phox, Grb2 and Pex13p. The binding site of the Grb2 SH3 partially overlapped the conventional PxxP-binding site, whereas those of p67phox and Pex13p SH3s are located in different surface regions. The non-PxxP peptide from p47phox binds to the p67phox SH3 more tightly when it extends to the N-terminus to include a typical PxxP motif, which enabled the structure determination of the complex, to reveal that the non-PxxP peptide segment interacted with the p67phox SH3 in a compact helix–turn–helix structure (PDB entry 1K4U).
Keywords: Grb2/NADPH oxidase/p67phox/Pex13p/SH3 domain
Introduction
The Src homology 3 (SH3) domain is found frequently in proteins involved in signal transduction and cytoskeletal networks, and mediates protein–protein interactions (Kay et al., 2000; Mayer, 2001). SH3 domains bind to proline-rich sequences containing a conserved PxxP motif, where P is proline and x is any amino acid (Kay et al., 2000; Mayer, 2001). The PxxP sequences occupy the canonical PxxP-binding site of SH3 domains in a type II polyproline helix (PPII) conformation with two helical orientations, class I and class II (Feng et al., 1994; Lim et al., 1994). Feng et al. (1995) reported that a few residues flanking the PxxP motif substantially increased the binding affinities of both class I and class II proline-rich peptides to the Src SH3 domain, by adding a new interaction between these residues and the Src SH3 specificity pocket. The specificity pocket of an SH3 domain is defined as a shallow concave surface between the RT loop and the n-Src loop. Ghose et al. (2001) also reported that, in addition to the interaction via a PxxP motif, the interaction of the specificity pocket of the Csk SH3 with two hydrophobic residues flanking the PxxP motif in the PEP protein provides the high affinity and high specificity of the Csk–PEP interaction (Gregorieff et al., 1998; Ghose et al., 2001). A similar mode of interaction is seen in the complex of the Fyn R96I SH3 domain and HIV-1 Nef, though the tip of the RT loop of the Fyn SH3 domain rather than the specificity pocket is involved (Lee et al., 1996). It has not been reported that these flanking sequences alone can bind to the Src, Csk or Fyn SH3 domains, independent of the PxxP motifs.
Recently, sequences lacking a typical PxxP motif have been reported to bind to a variety of SH3 domains. Sequences with the consensus PxxDY, derived from the e3b1/Abi-1 and RN-tre proteins, interact with the Eps8 SH3 domain (Mongiovi et al., 1999). Sequences with the consensus Px[VI][DN]RxxKP, from the UBPY, Gab1, AMSH and SLP-76 proteins, bind to the SH3 domains in the Hbp, STAM and Grb2 proteins (Kato et al., 2000; Lewitzky et al., 2001). These two consensus patterns contain at least one proline residue, but some lacking a proline residue have also been reported. Sequences with the consensus RKxxYxxY, from SKAP55, bind to the SH3 domains in the Fyb, Fyn and Lck proteins (Kang et al., 2000), and sequences with the WxxxFxxLE pattern from Pex5p interact with the Pex13p SH3 domain (Barnett et al., 2000; Urquhart et al., 2000). Some of these SH3 domains are known to interact with peptides containing typical PxxP motifs, e.g. the Grb2, Fyn, Lck and Pex13p SH3s. Thus, the binding sites of the unconventional motifs on SH3 domains are of great interest. The PxxDY and RKxxYxxY motifs have been suggested to interact at sites overlapping the canonical PxxP-binding sites of the Eps8 and Fyn SH3 domains, respectively (Mongiovi et al., 1999; Kang et al., 2000). In contrast, the WxxxFxxLE sequence is thought to interact at a site on the Pex13p SH3 domain that is distinct from the classical PxxP-binding site (Barnett et al., 2000).
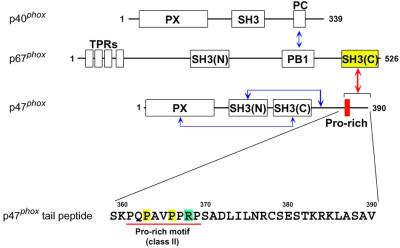
The phagocyte NADPH oxidase plays a crucial role in host defense against microbial infection by producing superoxide. As shown in Figure 1, the three soluble cytosolic components of NADPH oxidase contain five SH3 domains, some of which have been elucidated to regulate the assembly and activity of the oxidase (Sumimoto et al., 1994; Finan et al., 1994; Shiose and Sumimoto, 2000; Hiroaki et al., 2001). The C-terminal SH3 domain [SH3(C)] of the cytosolic activator, p67phox, interacts with the PxxP sequence in the C-terminal tail region of another soluble factor, p47phox (Finan et al., 1994). This interaction is involved in the oxidase activation (de Mendez et al., 1994; Hata et al., 1997). We have shown that the SH3–PxxP interaction is significantly enhanced by the extension of ∼20 residues C-terminal to the PxxP motif in two scenarios: the PxxP sequence in the full-length p47phox protein and that in a 30 residue peptide (this study; K.Mizuki, R.Takeya, F.Kuribayashi, I.Nobuhisa, D.Kohda, H.Nunoi, K.Takeshige and H.Sumimoto, submitted). More complete characterization of this unique interaction, including in vivo experiments, will be reported elsewhere (K.Mizuki, R.Takeya, F.Kuribayashi, I.Nobuhisa, D.Kohda, H.Nunoi, K.Takeshige and H.Sumimoto, submitted). We have also shown that this ∼20 residue extension lacking a PxxP motif alone can bind to the p67phox SH3(C) with a µM range affinity, thereby adding the p67phox SH3(C) to the list of SH3 domains that bind to non-PxxP sequences.
Fig. 1. Domain structure of the cytosolic activators of human NADPH oxidase and its intermolecular and intramolecular interactions. Schematic drawing of the domains and motifs contained in the p47phox, p67phox and p40phox proteins. The p67phox SH3(C) constitutively interacts with the C-terminal tail region of p47phox (red arrow). The other interactions in the resting state (blue arrows) are described (Shiose and Sumimoto, 2000; Hiroaki et al., 2001; Ito et al., 2001). PX, phox homology domain; PB1, phox and bem homology 1; PC, phox and cdc24 motif; TPR, tetratrico peptide repeat. The amino acid sequence of the p47phox tail region, which contains a class II SH3-binding consensus, PxxPxR, is shown.
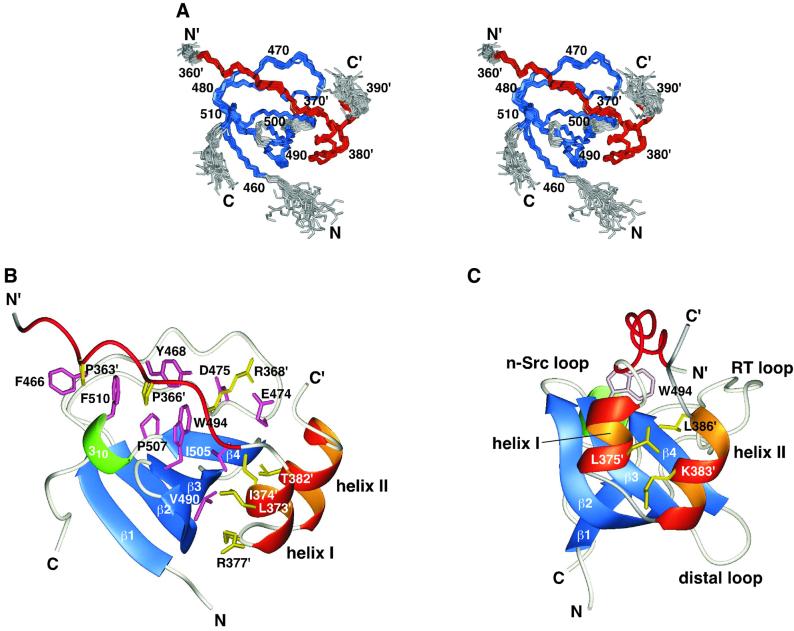
We selected SH3 domains from three proteins, p67phox, Grb2 and Pex13p, as examples of non-PxxP-binding SH3s, and investigated the interactions, using a recently developed NMR method, cross-saturation (Takahashi et al., 2000). The interaction sites are all different from each other: a region overlapping the canonical PxxP-binding site, the specificity pocket, and another region that has not yet been identified. In the case of the p67phox SH3(C), we also determined its three-dimensional structure in complex with the p47phox C-terminal tail peptide, which consists of the PxxP sequence and the non-PxxP sequence. The complex structure determined indicates that the non-PxxP portion of the tail peptide adopts a compactly folded helix–turn–helix (HTH) structure in the specificity pocket of the p67phox SH3(C).
Results and discussion
Enhancement of the PxxP motif binding by the C-terminally flanking region of p47phox
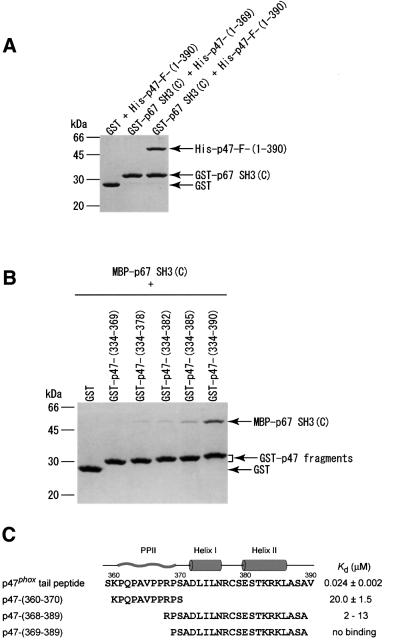
The three proteins p67phox, p47phox and p40phox form a ternary complex in the cytosol of neutrophils (Figure 1). The interaction between p67phox and p47phox can be studied by a pull-down assay using purified proteins in vitro. The p67phox SH3(C) fused to glutathione S-transferase (GST) was pulled-down with glutathione beads. The full-length p47phox protein (with an N-terminal His tag) was co-precipitated with GST–p67phox SH3(C) (Figure 2A). The truncation of the C-terminal 21 residues of p47phox resulted in a failure to detect co-precipitation even though the PxxP motif still remained, showing a considerable reduction in the interaction. Next, we prepared a series of C-terminally truncated p47phox fragments with a GST tag, and pulled them down with glutathione beads. The p67phox SH3(C) [fused with maltose binding protein (MBP)] was co-precipitated with the p47phox fragments (Figure 2B). The five residue deletion from the C-terminus weakened the intermolecular interaction substantially, and deletion of additional three or seven residues reduced the interaction further. No interaction was detected in the case of the 21 residue deletion of the p47phox fragment, which is consistent with the pull-down assay using the full-length p47phox lacking the C-terminal 21 residues (Figure 2A). These results indicate that almost the entire sequence flanking the PxxP motif in p47phox (residues 370–390) is necessary for the stable interaction of p47phox with the p67phox SH3(C).
Fig. 2. Effects of the deletions of the C-terminal tail region of p47phox on the interactions with the p67phox SH3(C). The proteins were pulled-down with glutathione beads, and analyzed by SDS–PAGE followed by staining with Coomassie Brilliant Blue. (A) The full-length and the C-terminally truncated form (residues 1–369) of p47phox (with a His tag) were tested for the interaction with the p67phox SH3(C) fused to GST. (B) A series of C-terminally truncated fragments of p47phox fused to GST were tested for the interaction with an MBP-tagged p67phox SH3(C). (C) Amino acid sequence of the p47phox tail peptide, on which the secondary structures in the complex with the p67phox SH3(C) are shown; PPII helix, the first α-helix and the second α-helix. The dissociation constants (Kd) of the p47phox tail peptide and p47-(360–370) for the p67phox SH3(C) were determined by tryptophan fluorescence perturbation. The binding of p47-(368–389) and p47-(369–389) was monitored with the shifts of the [1H,15N]HSQC cross-peaks of the p67phox SH3(C). The analysis of the NMR signals provided a rough estimate of the dissociation constants. The p47phox tail peptide was produced as a GST fusion protein and was cleaved with thrombin, and thus an extra glycine is attached to the N-terminus. The other peptides were prepared chemically, and contain an acetyl group at the N-terminus and an amide group at the C-terminus.
We determined the dissociation constants by fluorescence perturbation of the SH3 tryptophan residue (Figure 2C). The 32 residue p47phox tail peptide, residues 359–390, binds to the p67phox SH3(C) with a Kd of 24 nM. To our knowledge, this is the strongest interaction among naturally occurring peptide ligands. The 11 residue peptide corresponding to the PxxP motif, residues 360–370, interacts with the p67phox SH3(C) with a Kd of 20 µM, which is a typical value for SH3 domain interactions with proline-rich peptides.
New non-PxxP type ligand for the C-terminal SH3 domain of p67phox
As described above, the sequence flanking the PxxP motif of p47phox enhances the binding affinity for p67phox SH3(C) by ∼1000-fold. We next examined whether the flanking sequences alone, without the aid of the PxxP motif, could interact with the p67phox SH3(C), by means of NMR titration experiments as these did not cause fluorescence perturbation. A peptide, p47phox-(368–389), binds to the p67phox SH3(C) with a Kd of ∼10 µM (Figure 2C). Thus, the non-PxxP peptide, p47phox-(368–389), serves as a novel ligand targeting the C-terminal SH3 domain of p67phox. Interestingly, the binding of the peptide to the p67phox SH3(C) was lost on removal of the N-terminal Arg368 residue.
Binding sites for non-PxxP type ligands
Several consensus sequences without a typical PxxP pattern have been reported to interact with the SH3 domains from a variety of proteins (Mongiove et al., 1999; Barnett et al., 2000; Kang et al., 2000; Kato et al., 2000; Urquhart et al., 2000; Lewitzky et al., 2001). However, little is known about the structural basis of the interactions between the SH3 and non-PxxP motifs. We studied the binding sites for non-PxxP motifs in cognate SH3 domains using NMR spectroscopy. We selected the SH3 domains from Grb2 and Pex13p in addition to the p67phox SH3(C). The C-terminal SH3 domain of the Grb2 adaptor protein can bind to a non-PxxP peptide from SLP-76 (Lewitzky et al., 2001), in addition to a typical PxxP peptide from the SOS protein (Kohda et al., 1994). The Pex13p SH3 interacts with a non-PxxP peptide from Pex5p as well as a PxxP peptide from Pex14p (Barnett et al., 2000; Urquhart et al., 2000). It was reported for the Saccharomyces cerevisiae Pex proteins that Pex5p (peptide) and Pex14p interact simultaneously with the Pex13p SH3, suggesting two different binding sites for these two ligands (Barnett et al., 2000).
To identify the binding site of the non-PxxP peptides, we first tried to detect direct intermolecular nuclear Overhauser effects (NOEs) by a conventional NOESY experiment, but it was unsuccessful due to the weak interactions. The chemical shift perturbation method is widely used in such weakly interacting systems, but it is not as quantitative as NOESY experiments. Here, we used a recently developed NMR method, cross-saturation (Takahashi et al., 2000), which can identify the interfaces reliably based on the saturation transfer from a non-labeled to a labeled protein (Wüthrich, 2000).
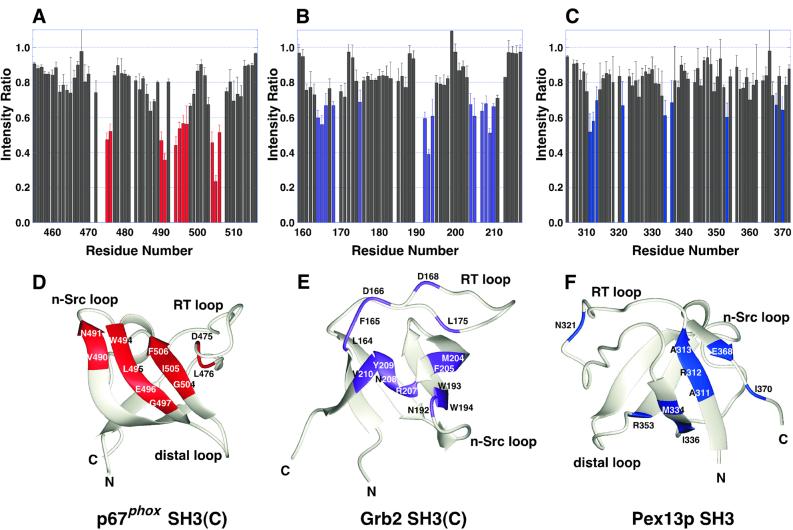
The cross-saturation decreases the intensity of the 1H–15N cross-peaks corresponding to the residues at the molecular interface. Figure 3A shows the 11 residues of the p67phox SH3(C) most affected by cross-saturation from the non-PxxP peptide, p47phox-(368–389). These residues cover the concave surface between the RT loop and the n-Src loop of the p67phox SH3(C), which corresponds to the specificity pocket of the SH3 domain (Figure 3D). We then examined the interaction of the peptide, APSIDRSTKPP, from SLP-76 (the consensus residues are underlined) with the C-terminal SH3 domain of Grb2 [Grb2 SH3(C)], and that of the peptide, PWTDQFEKLEKEVSEN, from Pex5p with the Pex13p SH3 domain. Figure 3B and C shows the residues in which the intensity of the 1H–15N cross-peak was affected by the cross-saturation. The effects of the cross-saturation were not only within the canonical PxxP-binding site but also spread to the specificity pocket of the Grb2 SH3(C), suggesting that the SLP-76 peptide lies across the boundary between the PxxP motif-binding site and the specificity pocket of the Grb2 SH3(C) (Figure 3E). As for the Pex system, the affected residues of the Pex13p SH3 formed a patch on the β-sheets containing the N- and C-termini (Figure 3F). This region partially overlaps the binding region suggested by the suppressor mutagenesis study of Barnett et al. (2000). In summary, the three different SH3 domains studied here, p67phox SH3(C), Grb2 SH3(C) and Pex13p SH3, accommodate the cognate non-PxxP peptides with different regions on the molecular surfaces (Figure 4). This result indicates that the sequences with unconventional motifs occupy a different site on each SH3 domain. In this context, it is interesting that a novel interaction mode between the two SH3 domains from the Grb2 and Vav proteins has been reported (Nishida et al., 2001).
Fig. 3. Cross-saturation experiments for the identification of the non-PxxP peptide-binding sites on three SH3 domains. (A) Intensity changes of the 1H–15N cross-peaks of the p67phox SH3(C) by the cross-saturation from the truncated p47phox tail peptide, p47-(368–389). The residues with intensity ratios <0.6 are selected and colored red. (B) Intensity changes of the 1H–15N cross-peaks of human Grb2 SH3(C) by the cross-saturation from the peptide, Ac-APSIDRSTKPP-NH2, of human SLP-76 (residues 232–242). The residues with intensity ratios <0.7 are selected and colored purple. (C) Intensity changes of the 1H–15N cross-peaks of the S.cerevisiae Pex13p SH3 by the cross-saturation from the peptide, Ac-PWTDQFEKLEKEVSEN-NH2, of the S.cerevisiae Pex5p (residues 203–218). The residues with intensity ratios <0.7 are selected and colored blue. (D) p67phox SH3(C) residues that contact the truncated p47phox tail peptide are Asp475, Leu476, Val490, Asn491, Trp494, Leu495, Glu496, Gly497, Gly504, Ile505 and Phe506. (E) Grb2 SH3(C) residues that contact the SLP-76 peptide are Leu164, Phe165, Asp166, Asp168, Leu175, Asn192, Trp193, Trp194, Met204, Phe205, Arg207, Asn208, Tyr209 and Val210. The Kd of the peptide for the Grb2 SH3(C) was estimated to be 10–20 µM by the curve fitting of the 1H–15N cross-peak shifts in the titration. The coordinates of the Grb2 SH3(C) were obtained from the PDB (entry 1GFC). (F) Pex13p SH3 residues that contact the Pex5p peptide are Ala311, Arg312, Ala313, Asn321, Met334, Ile336, Arg353, Glu368 and Ile370. The structure of the Pex13p SH3 was modeled by the SWISS MODEL server (Guex et al., 1999), on the basis of the coordinates of the mouse P38 crk SH3 (PDB entry 1B07) and the mouse C-crk SH3 (1CKA). The Kd of the peptide for the Pex13p SH3 was estimated to be ∼1 mM by the curve fitting of the 1H–15N cross-peak shifts in the titration.
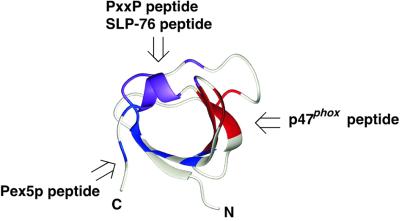
Fig. 4. Summary of the binding sites on three SH3 domains for peptides lacking a PxxP motif. The three SH3 domains from p67phox, Grb2 and Pexp13p accommodate the cognate non-PxxP peptides with different regions on the molecular surfaces. The SLP-76 peptide-binding site on the Grb2 SH3(C) domain partially overlaps the conventional PxxP motif-binding site.
Complex structure of the p67phox SH3(C) and the p47phox tail peptide
The binding of the non-PxxP peptide, p47phox-(368–389), to the p67phox SH3(C) is not tight enough for structure determination. The N-terminal extension of the p47phox tail peptide by ∼10 residues to include the PxxP motif enhances the binding affinity by ∼1000-fold (Figure 2C). The tight binding enabled us to determine the three-dimensional structure of the complex of the p67phox SH3(C) and the p47phox C-terminal tail peptide [residues 359′–390′: hereafter, a prime (′) denotes the numbering of the p47phox peptide] by the conventional NOE-based NMR method (Figure 5A and Table I). The p67phox SH3(C) maintains a typical SH3 fold in the complex. The p47phox tail peptide in the bound state contains three secondary structures (Figure 5B). The eight N-terminal residues corresponding to the PxxP motif, Gln362′–Pro369′, adopt a type II polyproline helix conformation, and occupy the canonical PxxP-binding site of the p67phox SH3(C) in a class II orientation (Figure 5B). In the remaining C-terminal segment, Ser370′–Val390′, of the p47phox tail peptide, two α-helices, Asp372′–Asn376′ (helix I) and Glu380′–Leu386′ (helix II), are linked by a turn, Arg377′–Ser379′. The two α-helices are packed together in an antiparallel manner to form a compact HTH structure (Figure 5C). The side chain of Leu375′ in helix I, and those of Lys383′ and Leu386′ in helix II, fill the space between the two α-helices. Note that NMR measurements suggested no stable folded conformation of the p47phox tail peptide in the absence of the p67phox SH3(C). No structural evidence has been obtained for the Grb2 SH3(C) or Pex13p SH3, but a helical conformation was suggested for the Pex5p peptide when bound to the Pex13p SH3 (Barnett et al., 2000).
Fig. 5. Structure of the complex of the p67phox SH3(C) and the p47phox tail peptide. (A) Overlay of the 22 NMR structures. The residues used for superimposing the different structures are colored blue (SH3) and red (the tail peptide), and the other residues are in gray. Residue numbers of the p47phox tail peptide are labeled with a prime (′). (B) Ribbon representation of the lowest energy structure. The β-strands of the p67phox SH3(C) are colored blue: β1, residues 460–463; β2, 483–491; β3, 494–498; β4, 503–506; and the 310-helix is colored green: residues 508–510. The PxxP motif and the two α-helices of the p47phox tail peptide are drawn in red and orange, respectively. Side chains located within the binding interface are shown in pink (SH3) and in yellow (the tail peptide). (C) The same structure viewed from a different angle. The side chains of Leu375′, Lys383′ and Leu386′, colored yellow, fill the space between the two α-helices. The positions of the three SH3 loops are shown.
Table I. Statistics for the 22 final structures of the p67phox SH3(C)–p47phox tail peptide complex.
| R.m.s. deviations from experimental distance constraints (Å) (611)a | 0.029 ± 0.001 | |
| No. of distance constraint violations >0.4 Å | 0 (max. 0.33) | |
| R.m.s. deviations from experimental dihedral constraints (°) (82)a | 0.335 ± 0.041 | |
| No. of dihedral constraint violations >2.5° | 0 (max. 2.4) | |
| R.m.s. deviations from idealized geometry | ||
| Bonds (Å) | 0.0026 ± 0.0001 | |
| Angles (°) | 0.514 ± 0.011 | |
| Impropers (°) | 0.376 ± 0.015 | |
| Ramachandran plot analysisb | ||
| Most favored regions | 69.1% | |
| Additionally allowed regions | 26.5% | |
| Generously allowed regions | 2.6% | |
| Disallowed regions | 1.7% | |
| R.m.s. deviations (Å) from the mean structurec | Backbone | All heavy atoms |
| P67phoxSH3(C) + p47phox tail peptide | 0.30 ± 0.08 | 0.73 ± 0.09 |
| p67phox SH3(C) | 0.29 ± 0.10 | 0.69 ± 0.13 |
| p47phox tail peptide | 0.23 ± 0.08 | 0.73 ± 0.09 |
aNumbers of constraints are indicated in parentheses.
bResidues analyzed with PROCHECK-NMR (Laskowski et al., 1996) were the same as those used for superposition. The residue in the disallowed regions is mainly Ala371′ of the p47phox tail peptide, which is located at the boundary between the PPII helix and helix I.
cResidues of the p67phox SH3(C) and the p47phox tail peptide used for superposition are residues 460–491, 494–498 and 503–511, and residues 361′–386′, respectively. The residues of the N- and C-terminal segments, and the unconstrained loops (the n-Src and distal loops) were excluded from the superposition.
The C-terminal five, eight and 12 residue deletions of the p47phox fragment remove the last residue, the C-terminal half and the whole of the second α-helix in the HTH structure, respectively, which substantially destabilized the bound state of the p47phox tail peptide as shown in Figure 2B. A residual interaction was still detected, indicating that the first α-helix alone can increase the affinity of the PxxP motif to some extent.
The non-PxxP portion of the 32 residue p47phox tail peptide in the complex occupies the specificity pocket of the p67phox SH3(C), which exactly corresponds to the binding surface suggested by the cross-saturation experiment using p47phox-(368′–389′) (compare Figures 3D and 5C). Thus, we consider that the 22 residue non-PxxP peptide, p47phox-(368′–389′), binds to the specificity pocket of the p67phox SH3(C) in a very similar HTH conformation.
Mutational analysis of the p47phox tail peptide
The complex structure indicated several peptide residues involved in the interaction with the p67phox SH3(C). As expected, the P363′A and P366′A mutations in the PxxP core motif resulted in 30- and 100-fold decreases in the affinity, respectively (Table II). The R368′A mutation impaired the binding substantially (625-fold). This arginine residue is a part of the class II type consensus, PxxPxR, and determines the orientation of the PPII helix through the interaction with the negatively charged residues of the SH3 RT loop (Feng et al., 1994; Lim et al., 1994). As mentioned above, Arg368′ is essential for the binding of the non-PxxP peptide, p47phox-(368′–389′) (Figure 2C). Therefore, Arg368′ is shared by both the PxxP and non-PxxP sequences in the p47phox tail peptide.
Table II. Effects of mutations in the p47phox tail peptide on binding to the p67phox SH3(C).
| Peptidesa | Kd (µM)b |
|---|---|
| Wild-type | 0.024 ± 0.002 |
| P363′A | 0.73 ± 0.07 |
| P366′A | 2.4 ± 0.2 |
| R368′A | 15. ± 2. |
| L373′A | 0.13 ± 0.01 |
| I374′A | 3.0 ± 0.5 |
| R377′A | 0.10 ± 0.02 |
| T382′A | 1.1 ± 0.5 |
| T382′V | 0.12 ± 0.02 |
aThe sequence of the wild-type peptide is shown in Figure 1. All the peptides were produced as GST fusion proteins and were cleaved with thrombin, and thus an extra glycine is attached to the N-terminus.
bKd values were determined by the perturbation of the SH3 tryptophan fluorescence, and reported values are the averages ± standard devi ations of four independent experiments.
As for the HTH structure of the p47phox tail peptide, Leu373′, Ile374′, Arg377′ and Thr382′ are found on the molecular interface (Figure 5B). The substitutions of Ile374′ in helix I and Thr382′ in helix II with alanine residues reduced the binding affinities by 125- and 46-fold, respectively, demonstrating that these residues are indispensable. In contrast, the substitution of Thr382′ with a valine residue did not change the binding greatly (5-fold), which suggests that the hydrophobic character of the threonine residue is important for the interaction. The contributions of Leu373′ in helix I and Arg377′ in the turn are not important for the binding, because their substitutions with alanine residues caused only small changes (∼5-fold) in the dissociation constants. In summary, Pro363′, Pro366′, Ile374′ and Thr382′ of the peptide contribute to hydrophobic interactions, whereas Arg368′ is essential for the interaction by localizing the peptide in the correct position via ionic interactions.
Comparison with the complex structure of the Csk SH3 and a PEP peptide
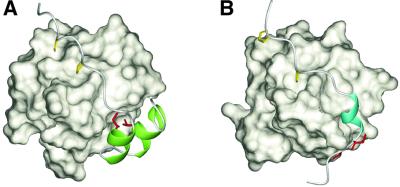
The SH3 domain of Csk interacts with a 25 residue peptide containing a class II consensus, PxxPxR, derived from the PEP protein with a Kd of 0.8 µM (Ghose et al., 2001). The high affinity compared with a Kd >10 µM for most SH3–peptide interactions was ascribed to two hydrophobic residues, isoleucine and valine, C-terminal to the PxxPxR motif (Gregorieff et al., 1998). The NMR structure of the complex of the Csk SH3 and the PEP peptide was determined recently (Ghose et al., 2001). There are three common characteristics between the complex of the Csk SH3–PEP peptide and that of the p67phox SH3(C)–p47phox tail peptide (Figure 6): (i) multivalent contacts with a PxxP motif and a region C-terminal to the PxxP motif increase the binding affinity; (ii) both of the non-PxxP regions occupy the specificity pockets of the cognate SH3 domains via hydrophobic interactions; and (iii) the non-PxxP sequence regions contain secondary structures. However, the structural roles of the secondary structures are quite different. The 310-helix structure in the PEP peptide forms a sharp turn in the polypeptide chain to position the two essential hydrophobic residues onto the binding site of the Csk SH3, whereas the two α-helices in the p47phox tail peptide are directly involved in the interactions with the p67phox SH3(C). The formation of a folded HTH structure by the two α-helices in the specificity pocket enables the non-PxxP segment alone to bind to the p67phox SH3(C).
Fig. 6. Comparison of the recognition of the non-PxxP segments outside the conventional PxxP motifs. (A) Ribbon representation of the p47phox tail peptide in complex with the p67phox SH3(C). The residues directly contacting the helix–turn–helix structure (Ile374′ and Thr382′) are colored red, and the two α-helices are in green. The PxxP core proline residues (Pro363′ and Pro366′) are shown in yellow. (B) Ribbon representation of the PEP-3BP1 peptide in complex with the Csk SH3 (PDB entry 1JEG). The hydrophobic residues that are essential for the tight binding (Ile625 and Val626 in full-length PEP) are colored red, and the 310-helix is in cyan. The PxxP core proline residues (Pro614 and Pro617) are shown in yellow.
Conclusions
We have shown that the 22 residue sequence (residues 368′–389′) derived from the C-terminal region of p47phox serves as a novel non-PxxP type SH3 ligand (Figure 1C). We have also shown that three different non-PxxP type peptide ligands from p47phox, SLP-76 and Pex5p bind to different surface regions on their cognate SH3 domains (Figure 4).
A great deal of attention has been paid to the PxxP motifs in SH3-mediated protein–protein interactions. In some situations, however, the interaction reliant on the PxxP motif may be insufficient for SH3 domains to achieve specific interactions (Ladbury and Arold, 2000). An SH3 domain can interact with two proteins simultaneously using a conventional PxxP motif-binding site and another binding site for a non-PxxP sequence, leading to the assembly of a multiprotein complex, as seen in S.cerevisiae Pex13p–Pex14p–Pex5p. Otherwise, an SH3 domain can serve as a signaling switch by a competition mechanism, for example the Grb2 SH3(C) as the binding site of the non-PxxP motif overlaps the PxxP motif-binding site (Figure 4). A PxxP motif and a non-PxxP motif in the same polypeptide chain can generate a protein ligand with high affinity and specificity via the multivalent binding mechanism, as seen in p67phox–p47phox and Csk–PEP. The data presented here provide the starting point for understanding the structural basis of SH3-mediated protein–protein interactions beyond the simple PxxP motif-based interactions.
Materials and methods
Sample preparation
The human p67phox SH3(C) (residues 455–516), the C-terminal peptide of human p47phox (residues 360–390), the human Grb2 SH3(C) (residues 159–217) and the S.cerevisiae Pex13p SH3 domain (residues 304–372) were overexpressed as GST fusion proteins in Escherichia coli BL21(DE3), using the vector pGEX-2T (Amersham Pharmacia Biotech). The p67phox SH3 mutant with double C499S/C514S substitutions was used throughout this study to avoid unfavorable aggregation through intermolecular S–S bond formation. The cells were cultured in M9 minimal media containing 15NH4Cl (1 g/l) and/or [U-13C]glucose (2.5 g/l) to prepare uniformly isotopically labeled proteins. For the SH3 domains uniformly labeled with 2H and 15N, the cells were grown in M9 minimal medium in 98% 2H2O containing 15NH4Cl and [2H]glucose (98 atom %). The GST fusion proteins were purified on a glutathione–Sepharose 4B column (Amersham Pharmacia Biotech), digested with thrombin and separated using a reversed-phase C18 column. An extra Gly–Ser sequence was attached to the N-terminus due to the thrombin cleavage. The p47phox tail peptide consists of residues 360–390, but residue 359 is serine, and thus we refer to the peptide as residues 359–390 in the text. Truncated peptides of the p47phox tail peptide were synthesized with an Advanced ChemTech 357 flexible multipeptide synthesizer, and were purified using a reversed-phase C18 column.
In vitro pull-down binding assays
For pull-down binding assays, GST and MBP fusion proteins, and His6-tagged proteins were expressed in E.coli strain BL21, and purified by glutathione–Sepharose 4B (Amersham Pharmacia Biotech), amylose resin (New England Biolabs) and His-bind resin (Novagen), respectively. The appropriate mixtures of the GST fusion (5 µg), MBP fusion (10 µg) and His6-tagged (10 µg) proteins were incubated in 400 µl of phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4 and 1.5 mM KH2PO4 pH 7.4) containing 5 mM dithiothreitol (DTT) and 0.5% (v/v) Triton X-100. Glutathione–Sepharose 4B beads were added to the solution, incubated for 1.5 h at 4°C, and then washed five times with PBS containing 5 mM DTT. The beads were collected by centrifugation, resuspended in the SDS sample buffer and boiled for 3 min. Proteins were separated on a 15% SDS–polyacrylamide gel, and stained with Coomassie Brilliant Blue.
Fluorescence measurement
Equilibrium fluorescence measurements were made using a Hitachi F-4500 fluorescence spectrophotometer at 25°C. The excitation wavelength was 278 nm (10 nm slit) and the emission wavelength was 345 nm (5 nm slit). Aliquots of the concentrated wild-type and mutated p47phox tail peptide solutions were added to a 2.0 ml solution of the p67phox SH3(C) (0.21 µM) in 50 mM MOPSO pH 7.1 containing 100 mM KCl. After the addition of each aliquot, the mixture was incubated in a stirred cuvette for 3 min, and the fluorescence intensity was measured. Dissociation constants were calculated by non-linear least square fitting (Nguyen et al., 1998). Peptide concentrations were determined by amino acid analysis.
NMR spectroscopy
NMR spectra were recorded at 25°C on a Bruker DMX600 or DMX750 spectrometer. A series of three-dimensional double and triple resonance experiments were recorded for spectral assignments (Clore and Gronenborn, 1994). Data were processed and analyzed using the nmrPipe/nmrDraw program (Delaglio et al., 1995). For the signal assignment of the p67phox SH3(C), NMR samples contained 0.5–0.7 mM [13C,15N]p67phox SH3(C) in complex with the non-labeled p47phox tail peptide (10% molar excess), dissolved in 1H2O/2H2O (9:1 v/v) or 2H2O containing 5 mM MOPSO pH 7.1, 30 mM d10-DTT and 100 mM KCl. For the assignment of the p47phox tail peptide, the sample contained 0.5 mM of [13C,15N]p47phox tail peptide and the non-labeled p67phox SH3(C) in a 10% molar excess. Stereospecific assignments of the methyl groups of the leucine and valine residues were obtained from the [1H,13C]HSQC spectrum recorded on 13C-biosynthetically directed labeled samples (Neri et al., 1989).
Cross-saturation experiments were carried out at 10°C according to Takahashi et al. (2000). The sample contained 0.5 mM [2H,15N]p67phox SH3(C) with the non-labeled peptide, residues 368–389, of p47phox (1:3 mol/mol) dissolved in 1H2O/2H2O (1:9 v/v) containing 50 mM phosphate pH 7.1, 30 mM d10-DTT and 150 mM KCl. The WURST-2 decoupling scheme (Kupce and Freeman, 1995) was used for the saturation of aliphatic protons with a sweep width of 1800 Hz and a 180° pulse duration of 10 ms. The saturation frequencies were set to 0.9 and –5.3 p.p.m. (for reference). The Grb2 SH3(C)–SLP-76 peptide sample contained 0.4 mM [2H,15N]Grb2 SH3(C) with the non-labeled SLP-76 peptide (1:2.2 mol/mol) dissolved in 1H2O/2H2O (1:9 v/v) containing 50 mM phosphate pH 7.1 and 150 mM KCl. For the Pex13p SH3–Pex5p peptide sample, 0.3 mM [2H,15N]Pex13p SH3 with the Pex5p peptide (1:4.2 mol/mol) was dissolved in 1H2O/2H2O (1:9 v/v) containing 50 mM phosphate pH 6.4 and 150 mM KCl. A recycle delay of 3.0 s (1.8 s for the saturation time) was employed for the p67phox SH3(C)–p47phox-(368′–389′) peptide and the Pex13p SH3–Pex5p peptide, and a 5.0 s delay (3.8 s for the saturation time) was used for the Grb2 SH3(C)–SLP-76 peptide. Three independent experiments were carried out, and the averages and the standard deviations of the cross-peak intensity ratios were calculated.
Structure calculation
Intramolecular NOEs were obtained from three-dimensional 15N-edited NOESY and 13C-edited NOESY experiments with a mixing time of 150 ms. Intermolecular NOEs between [13C,15N]-p67phox SH3(C) and the non-labeled p47phox tail peptide were collected from three-dimensional 15N-filtered-15N-edited and 13C-filtered-13C-edited NOESY experiments with a mixing time of 150 ms (Ogura et al., 1996). The 3JHN–Hα coupling constants were measured in a three-dimensional HNHA experiment with a correction factor of 1.1 (Kuboniwa et al., 1994). The 3JCγ–N and 3JCγ–C′ coupling constants of the aromatic residues, and the 3JCγ–N of the aliphatic residues were measured in two-dimensional spin-echo difference experiments (Bax et al., 1994; Hu et al., 1997).
The NOE intensities were converted to upper-limit distance constraints using the relationship of (NOE intensity) ∝ (distance)–5, taking into account the spin diffusion effects (Suri and Levy, 1993). No hydrogen bond constraints were used. The 3JHN–Hα coupling constants were used to constrain the dihedral angle φ in the range of –140° to –100° (>10 Hz), –160° to –80° (>8 Hz) and –90° to –40° (<5.5 Hz). The χ1 angle constraints were obtained from the 3JCγ–N and 3JCγ–C′ coupling constants and a three-dimensional HNHB experiment (Archer et al., 1991).
One hundred structures of the p67phox SH3(C)–p47phox tail peptide complex were calculated, using the program CNS version 1.0 with the dynamical simulated annealing protocol implemented in the program (Brünger et al., 1998). The 22 final structures have no NOE distance violations >0.4 Å or torsion angle violations >2.5°. All figures were generated using MOLMOL (Koradi et al., 1996). Atomic coordinates have been deposited in the Protein Data Bank with accession code 1K4U.
Acknowledgments
Acknowledgements
We are very grateful to Dr F.Inagaki (Hokkaido University) and Dr J.Schlessinger (New York University Medical Center) for providing the expression plasmid of human Grb2 SH3(C).
References
- Archer S.J., Ikura,M., Torchia,D.A. and Bax,A. (1991) An alternative 3D NMR technique for correlating backbone 15N with side chain Hβ resonances in larger proteins. J. Magn. Reson., 95, 636–641. [Google Scholar]
- Barnett P., Bottger,G., Klein,A.T.J., Tabak,H.F. and Distel,B. (2000) The peroxisomal membrane protein Pex13p shows a novel mode of SH3 interaction. EMBO J., 19, 6382–6391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bax A., Vuister,G.W., Grzesiek,S., Delaglio,F., Wang,A.C., Tschudin,R. and Zhu,G. (1994) Measurement of homo- and heteronuclear J couplings from quantitative J correlation. Methods Enzymol., 239, 79–105. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Clore G.M. and Gronenborn,A.M. (1994) Multidimensional hetero nuclear nuclear magnetic resonance of proteins. Methods Enzymol., 239, 349–363. [DOI] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek,S., Vuister,G.W., Zhu,G., Pfeifer,J. and Bax,A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX PIPES. J. Biomol. NMR, 6, 277–293. [DOI] [PubMed] [Google Scholar]
- de Mendez I., Garrett,M.C., Adams,A.G. and Leto,T.L. (1994) Role of p67-phox SH3 domains in assembly of the NADPH oxidase system. J. Biol. Chem., 269, 16326–16332. [PubMed] [Google Scholar]
- Feng S., Chen,J.K., Yu,H., Simon,J.A. and Schreiber,S.L. (1994) Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3–ligand interactions. Science, 266, 1241–1247. [DOI] [PubMed] [Google Scholar]
- Feng S., Kasahara,C., Rickles,R.J. and Schreiber,S.L. (1995) Specific interactions outside the proline-rich core of two classes of Src homology 3 ligands. Proc. Natl Acad. Sci. USA, 92, 12408–12415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finan P., Shimizu,Y., Gout,I., Hsuan,J., Truong,O., Butcher,C., Bennett,P., Waterfield,M.D. and Kellie,S. (1994) An SH3 domain and proline-rich sequence mediate an interaction between two components of the phagocyte NADPH oxidase complex. J. Biol. Chem., 269, 13752–13755. [PubMed] [Google Scholar]
- Ghose R., Shekhtman,A., Goger,M.J., Ji,H. and Cowburn,D. (2001) A novel, specific interaction involving the Csk SH3 domain and its natural ligand. Nat. Struct. Biol., 8, 998–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorieff A., Cloutier,J.-F. and Veillette,A. (1998) Sequence requirements for association of protein-tyrosine phosphatase PEP with the Src homology 3 domain of inhibitory tyrosine protein kinase p50csk. J. Biol. Chem., 273, 13217–13222. [DOI] [PubMed] [Google Scholar]
- Guex N., Diemand,A. and Peitsch,M.C. (1999) Protein modelling for all. Trends Biochem. Sci., 24, 364–367. [DOI] [PubMed] [Google Scholar]
- Hata K., Takeshige,K. and Sumimoto,H. (1997) Roles for proline-rich regions of p47phox and p67phox in the phagocyte NADPH oxidase activation in vitro. Biochem. Biophys. Res. Commun., 241, 226–231. [DOI] [PubMed] [Google Scholar]
- Hiroaki H., Ago,T., Ito,T., Sumimoto,H. and Kohda,D. (2001) Solution structure of the PX domain, a target of the SH3 domain. Nat. Struct. Biol., 8, 526–530. [DOI] [PubMed] [Google Scholar]
- Hu J.S., Grzesiek,S. and Bax,A. (1997) Two-dimensional NMR methods for determining χ1 angles of aromatic residues in proteins from three-bond JC′Cγ and JNCγ couplings. J. Am. Chem. Soc., 119, 1803–1804. [Google Scholar]
- Ito T., Matsui,Y., Ago,T., Ota,K. and Sumimoto,H. (2001) Novel modular domain PB1 recognizes PC motif to mediate functional protein–protein interactions. EMBO J., 20, 3938–3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H., Freund,C., Duke-Cohan,J.S., Musacchio,A., Wagner,G. and Rudd,C.E. (2000) SH3 domain recognition of a proline-independent tyrosine-based RKxxYxY motif in immune cell adapter SKAP55. EMBO J., 19, 2889–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Miyazawa,K. and Kitamura,N. (2000) A deubiquitinating enzyme UBPY interacts with the Src homology 3 domain of Hrs-binding protein via a novel binding motif PX(V/I)(D/N)RXXKP. J. Biol. Chem., 275, 37481–37487. [DOI] [PubMed] [Google Scholar]
- Kay B.K., Williamson,M.P. and Sudol,M. (2000) The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J., 14, 231–241. [PubMed] [Google Scholar]
- Kohda D., Terasawa,H., Ichikawa,S., Ogura,K., Hatanaka,H., Mandiyan,V., Ullrich,A., Schlessinger,J. and Inagaki,F. (1994) Solution structure and ligand-binding site of the carboxy-terminal SH3 domain of GRB2. Structure, 2, 1029–1040. [DOI] [PubMed] [Google Scholar]
- Koradi R., Billeter,M. and Wüthrich,K. (1996) MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph., 14, 52–55. [DOI] [PubMed] [Google Scholar]
- Kuboniwa H., Grzesiek,S., Delaglio,F. and Bax,A. (1994) Measurement of HN–Hα J couplings in calcium-free calmodulin using new 2D and 3D water-flip-back methods. J. Biomol. NMR, 4, 871–878. [DOI] [PubMed] [Google Scholar]
- Kupce E. and Freeman,R. (1995) Stretched adiabatic pulses for broadband spin inversion. J. Magn. Reson. A, 117, 246–256. [Google Scholar]
- Ladbury J.E. and Arold,S. (2000) Searching for specificity in SH domains. Chem. Biol., 7, R3–R8. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., Rullmannn,J.A., MacArthur,M.W., Kaptein,R. and Thornton,J.M. (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR, 8, 477–486. [DOI] [PubMed] [Google Scholar]
- Lee C.-H., Saksela,K., Mirza,U.A., Chait,B.T. and Kuriyan,J. (1996) Crystal structure of the conserved core of HIV-1 Nef complexed with a Src family SH3 domain. Cell, 85, 931–942. [DOI] [PubMed] [Google Scholar]
- Lewitzky M., Kardinal,C., Gehring,N.H., Schmidt,E.K., Konkol,B., Eulitz,M., Birchmeier,W., Schaeper,U. and Feller,S.M. (2001) The C-terminal SH3 domain of the adapter protein Grb2 binds with high affinity to sequences in Gab1 and SLP-76 which lack the SH3-typical P-x-x-P core motif. Oncogene, 20, 1052–1062. [DOI] [PubMed] [Google Scholar]
- Lim W.A., Richards,F.M. and Fox,R.O. (1994) Structural determinants of peptide-binding orientation and of sequence specificity in SH3 domains. Nature, 372, 375–379. [DOI] [PubMed] [Google Scholar]
- Mayer B.J. (2001) SH3 domains: complexity in moderation. J. Cell Sci., 114, 1253–1263. [DOI] [PubMed] [Google Scholar]
- Mongiovi A.M., Romano,P.R., Panni,S., Mendoza,M., Wong,W.T., Musacchio,A., Cesareni,G. and Fiore,P.P.D. (1999) A novel peptide–SH3 interaction. EMBO J., 18, 5300–5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri D., Szyperski,T., Otting,G., Senn,H. and Wüthrich,K. (1989) Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of the 434 repressor by biosynthetically directed fractional 13C labeling. Biochemistry, 28, 7510–7516. [DOI] [PubMed] [Google Scholar]
- Nguyen J.T., Turck,C.W., Cohen,F.E., Zuckermann,R.N. and Lim,W.A. (1998) Exploiting the basis of proline recognition by SH3 and WW domains: design of N-substituted inhibitors. Science, 282, 2088–2092. [DOI] [PubMed] [Google Scholar]
- Nishida M., Nagata,K., Hachimori,Y., Horiuchi,H., Ogura,K., Mandiyan,V., Schlessinger,J. and Inagaki,F. (2001) Novel recognition mode between Vav and Grb2 SH3 domains. EMBO J., 20, 2995–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura K., Terasawa,H. and Inagaki,F. (1996) An improved double-tuned and isotope-filtered pulse scheme based on a pulsed field gradient and a wide-band inversion shaped pulse. J. Biomol. NMR, 8, 492–498. [DOI] [PubMed] [Google Scholar]
- Shiose A. and Sumimoto,H. (2000) Arachidonic acid and phosphorylation synergistically induce a conformational change of p47phox to activate the phagocyte NADPH oxidase. J. Biol. Chem., 275, 13793–13801. [DOI] [PubMed] [Google Scholar]
- Sumimoto H., Kage,Y., Nunoi,H., Sasaki,H., Nose,T., Fukumaki,Y., Ohno,M., Minakami,S. and Takeshige,K. (1994) Role of Src homology 3 domains in assembly and activation of the phagocyte NADPH oxidase. Proc. Natl Acad. Sci. USA, 91, 5345–5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suri A.K. and Levy,R.M. (1993) Estimation of interatomic distances in proteins from NOE spectra at longer mixing times using an empirical two-spin equation. J. Magn. Reson. B, 101, 320–324. [Google Scholar]
- Takahashi H., Nakanishi,T., Kami,K., Arata,Y. and Shimada,I. (2000) A novel NMR method for determining the interfaces of large protein–protein complexes. Nat. Struct. Biol., 7, 220–223. [DOI] [PubMed] [Google Scholar]
- Urquhart A.J., Kennedy,D., Gould,S.J. and Crane,D.I. (2000) Interaction of Pex5p, the type 1 peroxisome targeting signal receptor, with the peroxisomal membrane proteins Pex14p and Pex13p. J. Biol. Chem., 275, 4127–4136. [DOI] [PubMed] [Google Scholar]
- Wüthrich K. (2000) Protein recognition by NMR. Nat. Struct. Biol., 7, 188–189. [DOI] [PubMed] [Google Scholar]