Abstract
Flowering in Arabidopsis is controlled by endogenous and environmental signals relayed by distinct genetic pathways. The MADS-box flowering-time gene SOC1 is regulated by several pathways and is proposed to co-ordinate responses to environmental signals. SOC1 is directly activated by CONSTANS (CO) in long photoperiods and is repressed by FLC, a component of the vernalization (low-temperature) pathway. We show that in transgenic plants overexpressing CO and FLC, these proteins regulate flowering time antagonistically and FLC blocks transcriptional activation of SOC1 by CO. A series of SOC1::GUS reporter genes identified a 351 bp promoter sequence that mediates activation by CO and repression by FLC. A CArG box (MADS-domain protein binding element) within this sequence was recognized specifically by FLC in vitro and mediated repression by FLC in vivo, suggesting that FLC binds directly to the SOC1 promoter. We propose that CO is recruited to a separate promoter element by a DNA-binding factor and that activation by CO is impaired when FLC is bound to an adjacent CArG motif.
Keywords: Arabidopis/CONSTANS/FLC/flowering/SOC1
Introduction
In plants, the transition from vegetative growth to flowering occurs in response to both environmental stimuli and endogenous signals. Genetic analyses of the control of flowering in Arabidopsis thaliana identified four major floral promotion pathways (reviewed in Simpson et al., 1999; Reeves and Coupland, 2000; Araki, 2001). The photoperiod and vernalization pathways mediate the response to environmental signals, whereas the autonomous and gibberellin (GA) pathways appear to act independently of these signals (Koornneef et al., 1991).
The photoperiod pathway mediates the promotion of flowering by daylength. Arabidopsis is a facultative long-day plant, flowering more rapidly under long-day (LD) conditions of 16 h of light than in short days (SDs) of 10 h light. CONSTANS (CO) and FT were placed in this pathway because mutations in these genes delay flowering in LDs, but not in SDs, and thereby modulate the response to photoperiod (reviewed in Reeves and Coupland, 2000; Araki, 2001). CO encodes a putative transcription factor (Putterill et al., 1995; Robson et al., 2001), whilst FT encodes a protein with similarity to RKIP proteins (Kardailsky et al., 1999; Kobayashi et al., 1999; Pnueli et al., 2001).
The vernalization pathway promotes flowering in response to extended exposures to low temperature. This pathway acts redundantly with the autonomous pathway. Both of these pathways promote flowering by preventing accumulation of the FLOWERING LOCUS C (FLC) mRNA (Michaels and Amasino, 1999, 2001; Sheldon et al., 1999, 2000). FLC acts synergistically with FRI to repress flowering in late-flowering accessions (Koornneef et al., 1994; Sanda and Amasino, 1996; Johanson et al., 2000; Sheldon et al., 2000), and encodes a MADS-domain transcription factor that represses flowering when overexpressed in transgenic plants (Michaels and Amasino, 1999; Sheldon et al., 1999). Mutation of FLC accelerates flowering in LDs and SDs, and is epistatic to mutations in the autonomous pathway and to dominant alleles of FRI (Michaels and Amasino, 2001). The abundance of FLC mRNA and protein is elevated by mutations in the autonomous pathway and is reduced by vernalization, suggesting that modulation of FLC expression is central to the control of flowering time (Michaels and Amasino, 1999; 2001; Sheldon et al., 2000), but it is not essential for a vernalization response (Michaels and Amasino, 2001).
The floral promotion pathways ultimately converge to regulate the expression and function of the floral meristem identity genes that control flower development (Blázquez and Weigel, 2000; Borner et al., 2000; Lee et al., 2000; Samach et al., 2000; Rouse et al., 2002). For example, the floral meristem identity gene LEAFY (LFY) is regulated both by CO, a component of the photoperiod pathway, and GA (Blázquez and Weigel, 2000). These act through different motifs within the LFY promoter, although CO probably does not directly activate LFY, and the transcription factor that regulates LFY in response to GA is not yet known (Blázquez and Weigel, 2000; Samach et al., 2000).
The flowering-time genes FT and SOC1 (or AGL20) are also common targets of distinct pathways and are proposed to function upstream of the floral meristem identity genes. SOC1 and FT were shown to be direct targets of CO by using plants that overexpressed a translational fusion of CO to the ligand-binding domain of the glucocorticoid receptor (35S::CO:GR) (Samach et al., 2000). In agreement with this, FT expression is reduced in co mutants (Kardailsky et al., 1999; Kobayashi et al., 1999; Samach et al., 2000; Suárez-López et al., 2001), whilst SOC1 expression responds to photoperiod and is slightly reduced in co mutants (Borner et al., 2000; Lee et al., 2000; Samach et al., 2000). In addition, ft or soc1 mutations partially suppress the early flowering of 35S::CO plants (Onouchi et al., 2000). Thus, FT and SOC1 act downstream of CO in the photoperiod pathway and CO promotes their expression. However, FT and SOC1 also act downstream of the floral inhibitor FLC, which is a component of the autonomous/vernalization pathway and does not affect CO expression. For example, SOC1 mRNA abundance is reduced in genotypes with high levels of FLC and is increased in flc mutants (Borner et al., 2000; Lee et al., 2000; Samach et al., 2000; Michaels and Amasino, 2001). These observations are consistent with the proposal that FLC represses SOC1.
Thus, the antagonistic effect of transcription factors CO and FLC on the expression of downstream genes FT and SOC1 may represent a direct convergence of signalling pathways and provide a means of co-ordinating the control of flowering by daylength and temperature. Here we further characterize how CO and FLC interact to generate antagonistic effects on SOC1 expression.
Results
Phenotypes associated with overexpression of FLC are suppressed by overexpression of CONSTANS
To examine the antagonistic effect of CO and FLC on flowering, the phenotypes of plants overexpressing both genes from the strong CaMV 35S promoter were examined. A 35S::CO 35S::FLC line was generated by crossing plants carrying 35S::CO (Onouchi et al., 2000) with those containing 35S::FLC (Michaels and Amasino, 1999). The flowering times of wild-type (Ler), 35S::CO, 35S::CO 35S::FLC, and 35S::FLC plants were scored in LDs. 35S::CO plants flowered earlier than wild type, whilst 35S::FLC plants flowered much later (Table I; Figure 1A). The 35S::CO 35S::FLC plants flowered much earlier than 35S::FLC and at a time between that of 35S::CO and wild-type plants (Table I). CO and FLC, therefore, have antagonistic effects on flowering time and overexpression of CO can largely overcome the delay in flowering caused by overexpression of FLC.
Table I. Effect of overexpression of CO and FLC on flowering time in LDs.
| Genotype | No. of rosette leaves | No. of cauline leaves | Total No. of leaves |
|---|---|---|---|
| Ler | 5.5 ± 0.5 | 3.1 ± 0.3 | 8.6 ± 0.5 |
| 35S::CO | 3.0 ± 0.0 | 1.9 ± 0.5 | 4.9 ± 0.5 |
| 35S::CO 35S::FLC | 4.4 ± 0.5 | 2.5 ± 0.5 | 6.9 ± 0.6 |
| 35S::FLC | 33.3 ± 3.4 | 6.9 ± 0.7 | 40.2 ± 3.9 |
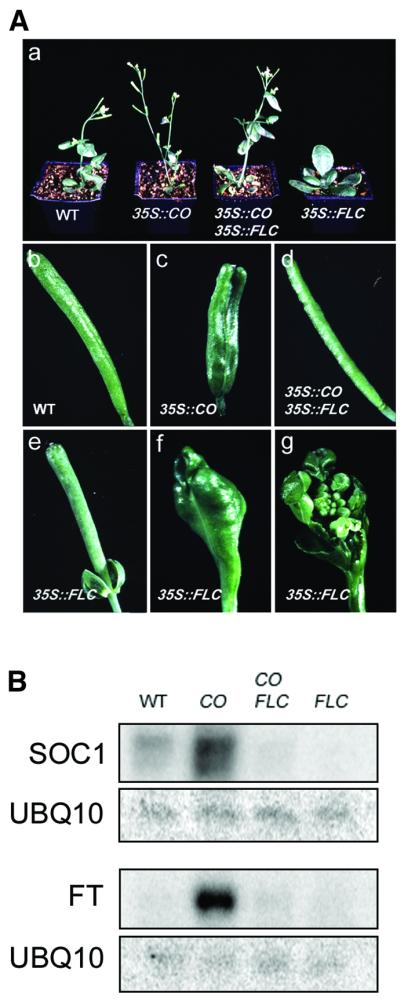
Fig. 1. Antagonistic effect of 35S::CO and 35S::FLC on flowering time, floral morphology, and expression of SOC1 and FT. (A) Phenotype of wild-type (WT), 35S::CO, 35S::CO 35S::FLC, and 35S::FLC plants. (a) Thirty-two-day-old plants grown in LDs; (b– f) morphology of siliques; (g) dissection of silique in (f) to show an ectopic inflorescence. (B) Northern analysis of SOC1 and FT mRNA in WT, 35S::CO, 35S::CO 35S::FLC, and 35S::FLC plants. One filter made with RNA harvested 8 h after dawn was hybridized with probes for SOC1 and UBQ10 (upper rows). A second filter made with RNA harvested 16 h after dawn was sequentially hybridized with probes for FT and UBQ10 (lower rows).
Overexpression of FLC also caused defects in floral morphogenesis. A proportion of flowers produced anthers with little or no pollen (data not shown), the petioles were often retained at the base of mature siliques (Figure 1A) and floral reversion caused development of an inflorescence inside some siliques (Figure 1A). These defects were absent in 35S::CO 35S::FLC lines (Figure 1A), although the mRNA expressed from 35S::FLC was still present in these plants. Similarly, defects associated with overexpression of CO, such as the presence of extra carpels and the short club-like appearance of siliques, were absent in 35S::CO 35S::FLC lines (see Onouchi et al., 2000; Figure 1), although 35S::CO mRNA was present.
This genetic analysis supports the notion that CO and FLC interact antagonistically to regulate flowering time, and indicates that this antagonism persists throughout development when these genes are overexpressed.
Antagonistic effect of CONSTANS and FLC on expression of target genes
Previously, SOC1 and FT were shown to be immediate targets of CO, and their mRNA levels correlate with the level of CO expression (Samach et al., 2000). SOC1 mRNA levels also correlate with the level of FLC expression (Lee et al., 2000; Michaels and Amasino, 2001). We compared the level of SOC1 and FT mRNAs in 35S::CO, 35S::CO 35S::FLC, 35S::FLC and wild-type plants to determine whether they correlate with flowering time. RNA was extracted from 10-day-old seedlings and subjected to northern analysis. As expected, SOC1 and FT mRNAs accumulated to a higher level in 35S::CO plants than in wild type and were not detected in 35S::FLC plants (Figure 1B). In 35S::CO 35S::FLC plants, the levels of SOC1 and FT mRNAs were dramatically reduced compared with those in 35S::CO plants. Thus, 8.3-fold less SOC1 mRNA and 11.3-fold less FT mRNA was detected in 35S::CO 35S::FLC plants compared with 35S::CO plants (Figure 1B). Nevertheless, 35S::CO 35S::FLC plants flowered only slightly later than 35S::CO plants (Figure 1A; Table I). Furthermore, SOC1 mRNA in 35S::CO 35S::FLC plants was 3-fold less abundant in comparison to wild-type plants, although 35S::CO 35S::FLC plants flowered earlier than wild type. Despite the lack of correlation between SOC1 mRNA levels and flowering time, our analysis confirmed that CO and FLC have antagonistic effects on SOC1 and FT expression.
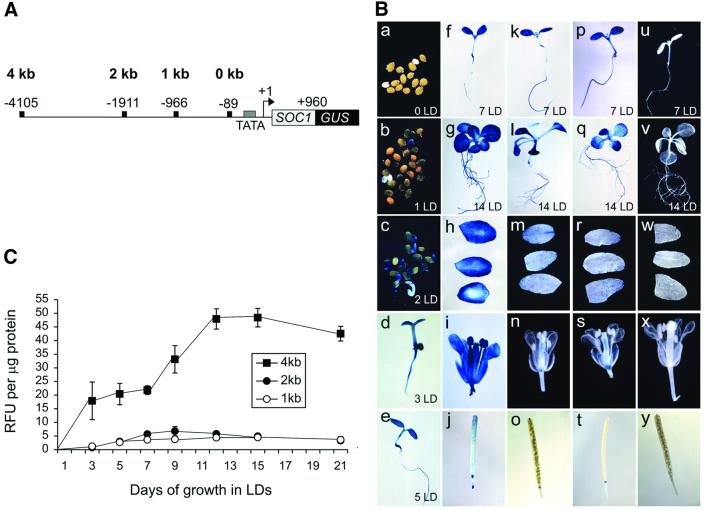
Expression of SOC1 in cauline leaves and flowers and accumulation of transcript with age require promoter sequences between nucleotides –4105 and –1911
Previously, SOC1 mRNA was shown to accumulate early in development and to be present in most tissues of mature plants (Borner et al., 2000; Lee et al., 2000; Samach et al., 2000). To identify SOC1 promoter sequences, expression of a SOC1::GUS reporter gene containing ∼4 kb of sequence upstream of the SOC1 transcriptional start site was monitored (Materials and methods). This was assumed to contain the full-length promoter since a genomic DNA fragment containing 1.4 kb of upstream sequence complemented the soc1 mutation (Samach et al., 2000). Twenty primary transformants carrying SOC1::GUS were analysed by β-glucuronidase (GUS) staining. Staining was first visible in the germinating seed after 1 day of growth in LDs, and was subsequently detected in the roots, apex and cotyledons of seedlings. GUS expression was observed in mature plant tissues such as rosette leaves, cauline leaves, inflorescences and flowers, but not in mature siliques or in seeds (Figure 2B). The pattern of expression of the 4 kb SOC1::GUS reporter gene was, therefore, similar to that of the endogenous SOC1 gene as monitored by RT–PCR (Borner et al., 2000; Lee et al., 2000).
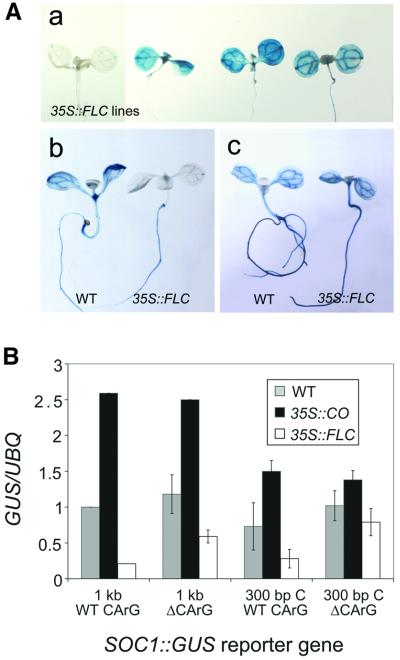
Fig. 2. Analysis of expression of SOC1::GUS reporter genes in wild-type plants. (A) Diagram of SOC1::GUS reporter genes with full-length (4 kb) or truncated promoters (2, 1 and 0 kb). The 5′ endpoints of constructs are numbered relative to the transcription start site (+1). (B) Expression of SOC1::GUS reporter genes in seedlings, cauline leaves, flowers and siliques as monitored by GUS staining. Days of growth are in the lower right of panels. Four kilobase SOC1::GUS reporter gene expression in (a–g) seedlings, (h) cauline leaves, (i) flowers and (j) siliques; 2 kb SOC1::GUS reporter gene expression in (k and l) seedlings, (m) cauline leaves, (n) flowers and (o) siliques; 1 kb SOC1::GUS expression in (p and q) seedlings, (r) cauline leaves, (s) flowers and (t) siliques; 0 kb SOC1::GUS expression in (u and v) seedlings, (w) cauline leaves, (x) flowers and (y) siliques. (C) Time course of 4, 2 and 1 kb SOC1::GUS reporter gene expression in wild-type seedlings as determined by GUS activity assays (see Materials and methods). RFU, relative fluorescence units.
To identify a minimal promoter sequence that would still mediate activation by CO and repression by FLC, the effects of sequential 5′ deletions of the 4 kb promoter on expression of SOC1::GUS were monitored in wild-type plants. SOC1::GUS reporter genes with deletion endpoints at nucleotide (nt) –1911 (2 kb SOC1::GUS), nt –966 (1 kb SOC1::GUS) and nt –89 (0 kb SOC1::GUS) were introduced into wild-type plants (Figure 2A; Materials and methods). Twenty primary transformants were obtained for each construct and analysed for activity by GUS staining. The patterns of expression for the 1 and 2 kb SOC1::GUS reporter genes were similar (Figure 2B). Staining was detected in seedlings and rosette leaves, but was reduced or absent in cauline leaves and was not detected in inflorescences or flowers. GUS staining was not detected for plants transformed with a 0 kb SOC1::GUS reporter gene (Figure 2B). Therefore, the 5′ boundary of promoter sequences required for SOC1 expression in cauline leaves and in flowers was located upstream of nt –1911, but sequences between nt –966 and –89 were sufficient for SOC1 expression in seedlings.
Previously, SOC1 mRNA was detected at a low level early in development and slowly accumulated over a 12 day period (Lee et al., 2000). To determine whether the 4 kb SOC1::GUS reporter gene was similarly expressed in developing seedlings and to monitor activity of the truncated 2 and 1 kb promoters, seedlings with 4, 2 or 1 kb SOC1::GUS reporter genes were monitored for GUS activity over 21 days (Figure 2C). Expression of 4 kb SOC1::GUS was detected early in development (day 3) and gradually increased until about day 12. For the 1 and 2 kb SOC1::GUS reporter genes, activity was also detected early in development (day 3), but at ∼10-fold lower levels than for 4 kb SOC1::GUS. Expression reached a maximum at about day 5 and remained constant until day 21. Therefore, truncation of the SOC1 promoter resulted in an overall decrease in the level of reporter gene expression and abolished the age-dependent increase in expression that is observed for the 4 kb promoter. The 5′ boundary of sequences required for maximal accumulation of SOC1 mRNA in seedlings must be located upstream of nt –1911.
The 1 kb SOC1::GUS reporter is activated by CO and repressed by FLC
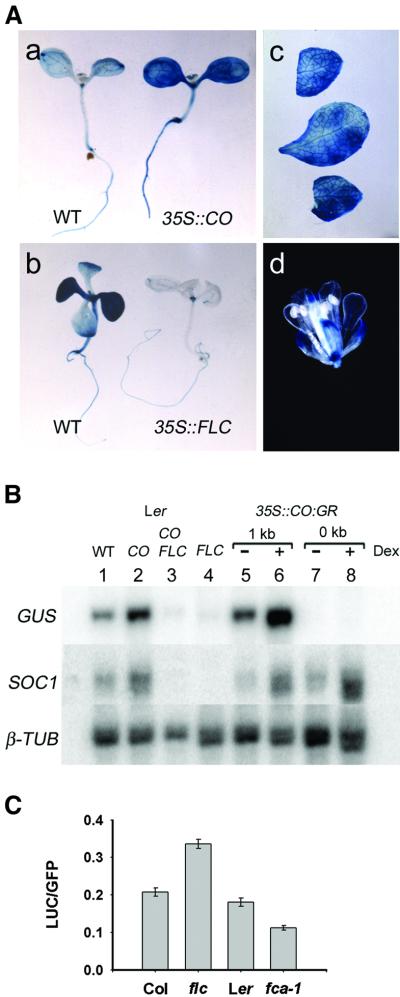
To test whether the 1 kb SOC1::GUS reporter retained the ability to be activated by CO and/or repressed by FLC, 1 kb SOC1::GUS was introduced into 35S::CO, 35S::CO 35S::FLC, and 35S::FLC plants by crossing. Expression of 1 kb SOC1::GUS in these lines was then monitored by GUS staining and northern blotting (Figure 3).
Fig. 3. One kilobase SOC1::GUS reporter gene expression is activated in 35S::CO lines and repressed by FLC. (A) Analysis of 1 kb SOC1::GUS expression in wild-type (WT), 35S::CO and 35S::FLC plants by GUS staining: (a) 10-day-old seedlings, 1 kb SOC1::GUS WT (left) and 1 kb SOC1::GUS 35S::CO (right); (b) 14-day-old seedlings, 1 kb SOC1::GUS WT (left) and 1 kb SOC1::GUS 35S::FLC (right); 1 kb SOC1::GUS 35S::CO in (c) cauline leaves and (d) flower. (B) Northern analysis of 1 and 0 kb SOC1::GUS expression. RNA was purified from 1 kb SOC1::GUS or 0 kb SOC1::GUS seedlings: wild type (WT), 35S::CO (CO), 35S::CO 35S::FLC (CO FLC), 35S::FLC (FLC) and 35S::CO:GR. Dexamethasone (Dex) treatment, (+) or (–). The filter was sequentially hybridized with probes for GUS, SOC1 and β-TUB (loading control). (C) Transient expression assays of 1 kb SOC1::LUC. Leaves of Columbia (Col), flc-1 (in Col background), Landsberg erecta (Ler) and fca-1 (in Ler) plants were bombarded with beads coated with DNA of plasmids carrying 1 kb SOC1::LUC and 35S::GFP (Materials and methods). The ratio of luciferase to GFP expression is shown for each genotype (Materials and methods). In each case, the column represents the mean value, with the standard error.
GUS activity was higher in all tissues of 35S::CO seedlings in comparison to wild type (Figure 3A). An increase in GUS activity was also detected in cauline leaves and in flowers relative to that of wild-type plants (Figures 2B and 3A). In contrast, GUS activity was dramatically reduced in 35S::FLC seedlings in comparison with wild-type seedlings. Repression was strongest in the leaves; a residual amount of expression was retained in the roots, at the apex and in the veins of the leaves (Figure 3A). GUS expression in 35S::CO 35S::FLC seedlings was similar to that in 35S::FLC seedlings (data not shown).
The accumulation of GUS mRNA in whole seedlings was also measured by northern blotting (Figure 3B). The 1 kb SOC1::GUS transcript was upregulated 3.1-fold in 35S::CO seedlings and repressed 5.1-fold in 35S::FLC seedlings in comparison with wild type (Figure 3B). In addition, a transient assay was developed to determine whether the 1 kb fragment of the SOC1 promoter responded to FLC in wild-type plants and in fca mutants, which contain elevated levels of FLC (Michaels and Amasino, 1999, 2001; Sheldon et al., 2000). A 1 kb SOC1::LUCIFERASE fusion was introduced into wild-type plants, flc loss-of-function mutants and fca mutants by microprojectile bombardment. Levels of luciferase expression were compared with those of co-bombarded 35S::GFP (Materials and methods). Luciferase expression was ∼1.75-fold higher in flc loss-of-function mutants than in wild-type plants, and ∼0.6-fold wild-type levels in fca mutants (Figure 3C). The differences between the mutants and wild-type plants were confirmed as significantly different (P < 0.001) using the Mann–Whitney rank sum test. The 1 kb SOC1 promoter fragment therefore also confers responses to FLC at levels of expression found in wild-type and fca mutant plants.
In 35S::CO 35S::FLC seedlings carrying 1 kb SOC1::GUS, GUS mRNA was 4.1-fold less abundant than in wild type, and was most similar to that in 35S::FLC (Figure 4B). Analysis of SOC1 mRNA demonstrated that the endogenous gene was upregulated in 35S::CO plants and repressed in 35S::FLC plants to a similar extent to 1 kb SOC1::GUS (Figure 3B).
Fig. 4. Identification of a 234 bp region of the SOC1 promoter that mediates activation by CO and repression by FLC. (A) Summary of expression of SOC1::GUS reporter genes in wild-type (WT), 35S::CO and 35S::FLC lines. Relative activities were determined by GUS staining. Top line, 1 kb SOC1::GUS reporter gene; second to fifth lines, 300 bp SOC1::GUS reporter genes containing overlapping fragments A, B, C or D from the SOC1 promoter. Fragments of 300 bp were each inserted upstream of the minimal 0 kb reporter gene at a unique BamHI site (see Materials and methods). Asterisk denotes that fragment D contains two copies of the minimal promoter region between –89 and +5. Bottom line, minimal 0 kb SOC1::GUS reporter gene that contains the TATA box and transcription start site (+1) for SOC1. (B) Promoter fragment C mediates activation in 35S::CO plants and repression in 35S::FLC plants. Expression of 1 kb SOC1::GUS (1 kb) and 300 bp SOC1::GUS (A, B, C, D) reporter genes was monitored in WT, 35S::CO and 35S::FLC seedlings by northern blotting using probes for GUS and UBQ10 (loading control). These data were quantified and are presented in histogram format. The amount of GUS/UBQ transcript in WT plants containing the 1 kb SOC1::GUS reporter gene was given an arbitrary value of 1. The relative level of transcript for each reporter gene construct in each background is presented as an average.
We also tested whether upregulation of 1 kb SOC1::GUS expression in 35S::CO:GR lines was likely to be mediated directly by CO (Samach et al., 2000). Expression of 1 kb SOC1::GUS was monitored in 35S::CO:GR plants by northern blotting (Figure 3B). A 3.5-fold increase in GUS mRNA abundance was detected after 4 h of dexamethasone (Dex) treatment in comparison to untreated control plants (Figure 3B), whereas no GUS mRNA was detected for the minimal 0 kb SOC1::GUS reporter gene in 35S::CO:GR plants (Figure 3B). The endogenous SOC1 gene was also analysed, demonstrating that treatment with Dex was sufficient for upregulation of endogenous SOC1 mRNA.
These experiments indicated that sequences between nt –966 and –89 in the SOC1 promoter mediate activation by CO and repression by FLC, and that 1 kb SOC1::GUS is regulated by CO and FLC in a similar manner to the endogenous gene.
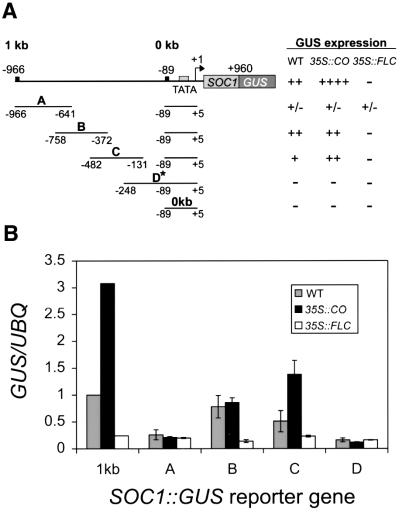
Overlapping 300 bp fragments in the 1 kb promoter of SOC1 mediate activation by CO and repression by FLC
To further define the sequences that mediate activation by CO and repression by FLC, the 1 kb promoter was subdivided into four overlapping fragments of ∼300 bp each. These fragments were cloned upstream of the minimal 0 kb SOC1::GUS reporter gene to generate the 300 bp A, B, C and D SOC1::GUS reporter genes (Figure 4A; Materials and methods). Each of these constructs was used to transform wild-type plants. Twenty primary transformants for each construct were obtained. Reporter genes present in these lines were introduced into 35S::CO and 35S::FLC lines by crossing.
The expression pattern of each construct was first analysed qualitatively by staining seedlings for GUS activity in the T2 generation (Figure 4A). In wild-type plants, constructs containing fragments B and C supported expression of the minimal 0 kb SOC1::GUS gene. In general, constructs containing fragment B supported higher levels of GUS activity than those containing fragment C. However, the GUS activity mediated by fragment C was strongly increased in 35S::CO lines, whereas that mediated by fragment B was not. The GUS expression caused both by fragment B and C was significantly repressed by 35S::FLC, indicating that the overlapping region between these two sequences was likely to contain sequences that mediate repression by FLC. Little or no GUS activity for constructs containing fragments A and D was detected in wild-type, 35S::CO or 35S::FLC lines.
The accumulation of GUS transcript in homozygous T3 lines was also monitored in whole seedlings by northern blotting. Figure 4B shows that for constructs containing fragment C, SOC1::GUS mRNA abundance increased by an average of 2.7-fold in 35S::CO seedlings. This increase is similar to that observed for 1 kb SOC1::GUS mRNA, which was increased 3.1-fold in 35S::CO seedlings in comparison to wild type. SOC1::GUS mRNA expressed from fragments A, B or D was not significantly increased in abundance in 35S::CO seedlings compared with wild type. Figure 4B also shows that constructs containing fragment B or C were repressed by an average of 5.6- or 2.1-fold, respectively, in 35S::FLC seedlings compared with wild type. The level of repression of fragment B is similar to that observed for the 1 kb SOC1::GUS reporter gene in 35S::FLC seedlings (Figure 4). The mRNA of SOC1::GUS constructs containing fragments A and D was not significantly reduced in abundance in 35S::FLC seedlings in comparison to wild type.
This analysis showed that overlapping fragments B and C support activity of a minimal 0 kb SOC1::GUS reporter gene in wild-type plants, and that this activity is repressed by 35S::FLC. Therefore, sequences between nt –482 and –372 of the SOC1 promoter mediate repression by FLC. Furthermore, only fragment C could support efficient activation of the minimal 0 kb SOC1::GUS reporter in 35S::CO lines, suggesting that the sequences required for this response are located between nt –372 and –248.
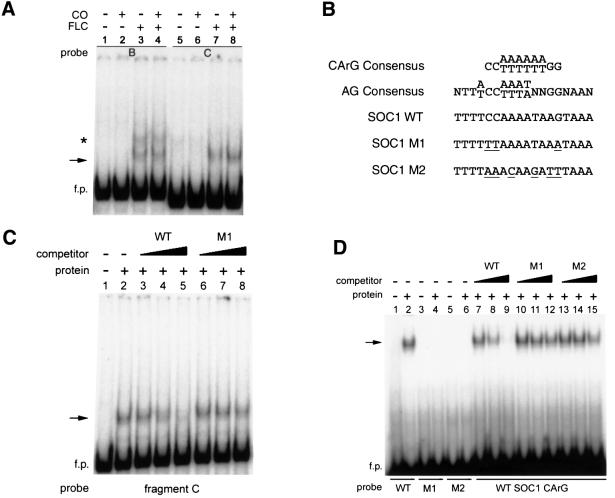
Specific in vitro binding of FLC to DNA containing the CArG box at nt –400 of the SOC1 promoter
To test whether CO and/or FLC bind directly to the SOC1 promoter, gel retardation experiments were performed using fragments B and C as probe. A protein–DNA complex was detected by gel retardation after incubation of recombinant FLC protein with labelled fragments B and C (Figure 5A). Formation of a similar complex was observed when FLC and CO proteins were present together in the reaction mixture, and no complex was observed when recombinant CO protein alone was incubated with the probes (Figure 5A).
Fig. 5. Specific binding of FLC protein to a CArG box at nt –400 of the SOC1 promoter. (A) FLC protein forms a gel retardation complex (arrow) with 300 bp fragments B and C. FLC and/or CO protein were incubated with promoter fragments B or C as probes. The protein contained in each reaction is indicated above the panel. f.p., free probe. The asterisk indicates an apparent non-specific protein–DNA complex that was not consistently observed and was competed with non-specific competitor. (B) Comparison of wild-type (WT) and mutant MADS-box protein binding sites (CArG boxes): includes CArG box at nt –400 of the SOC1 promoter (CCAAAATAAG), as well as mutant versions of SOC1 CArG box. (C) Specific binding of FLC protein to fragment C probe. The presence of FLC protein is indicated above the panels and competitor DNAs are described in the text. Lane 1, no protein and no competitor DNA; lanes 3–5, SOC1 WT fragment as competitor DNA; lanes 6–8, SOC1 M1 fragment as competitor DNA. Non-labelled DNA in molar excess was used as competitor in lanes 3 and 6 (10-fold), lanes 4 and 7 (100-fold), and lanes 5 and 8 (1000-fold). (D) Specific binding of FLC protein to a 30 bp fragment containing the CArG sequence at nt –400. FLC protein was incubated with 30 bp fragment probes as indicated below the panel. The presence of FLC protein is indicated above the panels and competitor DNAs are described in the text. Lanes 1, 3, 5, no protein and no competitor DNA; lanes 2, 4, 6, FLC and no competitor DNA; lanes 7–9, SOC1 WT fragment as competitor DNA; lanes 10–12, SOC1 M1 fragment as competitor DNA; lanes 13–15, SOC1 M2 fragment as competitor DNA. Non-labelled DNA in molar excess was used as competitor in lanes 7, 10 and 13 (10-fold), lanes 8, 11 and 14 (100-fold), and lanes 9, 12 and 15 (1000-fold).
Inspection of the overlapping region between fragments B and C revealed a sequence motif centred at nt –400 that resembled a CArG box, the site to which MADS-domain proteins bind (Figure 5B; reviewed in Shore and Sharrocks, 1995). This CArG sequence in the SOC1 promoter is most similar to the consensus binding site for AGAMOUS (AG) (Figure 5B; Huang et al., 1993; Shiraishi et al., 1993).
The specificity of the interaction of FLC protein with fragment C was tested (Figure 5C). For this, a 30 bp fragment (SOC1 WT) that spanned the CArG box at nt –400 was used as specific competitor (Figure 5B, third line). As non-specific competitor, a similar fragment with three base-pair changes in the CArG site was used (SOC1 M1; Figure 5B). Formation of a complex was observed when FLC protein was incubated with radioactively labelled fragment C (Figure 5C, lane 2). Formation of this complex was inhibited by the presence of excess non-labelled WT fragment (Figure 5C, lanes 3–5), but not by fragment M1. Therefore, this protein–DNA complex reflects a specific interaction of FLC with the CArG box within fragment C.
We also tested whether FLC protein could bind to 30 bp fragments containing either WT or mutant CArG sites: WT and M1 fragments are described above; fragment M2 contains six base-pair changes in the CArG site (Figure 5B). Formation of a complex was observed when FLC protein was incubated with WT 30 bp fragment, but not with the M1 or M2 fragments (Figure 5D). Formation of this complex was inhibited by the presence of excess non-labelled WT fragment, but not by M1 or M2 fragments (Figure 5D, compare lanes 7–9 with 10–15). These data indicate that the interaction of FLC protein with the CArG box at nt –400 in the SOC1 promoter is specific.
The CArG box at nt –400 is required for repression in vivo
To test whether the CArG box at nt –400 is important for FLC-mediated repression of SOC1 in vivo, site-directed mutagenesis was used to create 1 kb and 300 bp C SOC1 promoter fragments containing the changes present in M2 that prevented FLC binding (Figure 5B; Materials and methods). SOC1::GUS reporter genes with these mutant promoters, denoted 1 kb ΔCArG SOC1::GUS and 300 bp C ΔCArG SOC1::GUS, were introduced into wild-type plants. Reporter genes present in these lines were introduced into 35S::CO and 35S::FLC lines by crossing. The activity of the 1 kb ΔCArG and 300 bp C ΔCArG SOC1::GUS reporter genes was monitored by GUS staining and northern blotting (Figure 6).
Fig. 6. The CArG box at nt –400 of the SOC1 promoter mediates repression of SOC1::GUS expression in 35S::FLC lines. (A) Mutation of the CArG box at nt –400 leads to derepression of SOC1::GUS expression in 35S::FLC lines. Ten-day-old seedlings were stained for GUS activity. (a) One kilobase SOC1::GUS expression in a 35S::FLC control seedling (left); three independent lines showing 1 kb ΔCArG SOC1::GUS expression in 35S::FLC seedlings (right); (b) 300 bp C SOC1::GUS expression in a wild-type (WT) seedling and in a 35S::FLC seedling; (c) mutated 300 bp C ΔCArG SOC1::GUS expression in a WT seedling and in a 35S::FLC seedling. (B) Analysis of the effect of the CArG box mutation on SOC1::GUS activation in 35S::CO lines and repression in 35S::FLC lines. Expression of WT and mutated ΔCArG reporter genes was monitored in WT, 35S::CO and 35S::FLC seedlings by northern blotting using probes for GUS and UBQ10 (loading control). The data were quantified and are presented in histogram format.
In wild-type plants, both the 1 kb ΔCArG and 300 bp C ΔCArG mutant promoters supported strong expression of SOC1::GUS, which was efficiently upregulated in the corresponding 35S::CO line. In contrast, both the 1 kb ΔCArG and 300 bp C ΔCArG promoters failed to mediate efficient repression of SOC1::GUS in 35S::FLC lines (Figure 6A). Loss of repression was most evident in leaves, which did not stain for GUS activity in 35S::FLC plants carrying constructs containing wild-type promoters (Figure 6A). The accumulation of GUS transcript was also tested in whole seedlings by northern blotting. For constructs containing 300 bp ΔCArG fragment C, SOC1::GUS transcript was on average 1.4-fold more abundant in 35S::CO seedlings in comparison to wild type (Figure 6B). This ratio of activation appears lower than that observed for the WT 300 bp C SOC1::GUS reporter gene in 35S::CO seedlings in comparison to wild type because there is a higher level of GUS activity in WT ΔCArG 300 bp lines. GUS mRNA produced from constructs containing 300 bp ΔCArG fragment C was on average 1.3-fold lower in abundance in 35S::FLC seedlings compared with wild-type seedlings (Figure 6B). Repression of this construct by FLC is, therefore, much lower than that observed for the WT 300 bp fragment C SOC1::GUS reporter gene, in which GUS mRNA abundance was on average 2.1-fold less in 35S::FLC seedlings (see also Figure 4). Similarly, in plants carrying 1 kb ΔCArG SOC1::GUS, the level of GUS mRNA was reduced 2.2-fold compared with 5.1-fold in plants carrying 1 kb SOC1::GUS and 35S::FLC (Figure 6B).
In summary, this analysis indicates that mutation of the CArG box reduced the repression of SOC1 expression by 35S::FLC, whereas the level of activation of reporter gene expression in 35S::CO lines was not significantly affected.
Discussion
FT and SOC1 were recently shown to be common targets of multiple flowering-time pathways. In particular, they are positively regulated by the photoperiod pathway through the B-box transcription factor CO and negatively regulated by the MADS-domain transcription factor FLC, a component of the autonomous/vernalization pathway (reviewed in Araki, 2001). The antagonistic effect of these transcription factors on the expression of downstream genes FT and SOC1 may provide a means of co-ordinating the control of flowering time by daylength and temperature.
We explored the specific relationship between CO and FLC using plants overexpressing each of these transcription factors. This demonstrated the antagonistic activity of these transcription factors on flowering time and expression of SOC1 and FT, and identified a 351 bp region of the SOC1 promoter that mediates activation by CO and repression by FLC. A CArG box within this sequence was bound specifically by FLC in vitro and mediated repression by FLC in vivo, but was not required for activation by CO.
Flowering time is not completely correlated with SOC1 and FT mRNA level
Although the expression of SOC1 and FT was antagonistically regulated by CO and FLC, the flowering time of plants specifically overexpressing these transcription factors did not correlate with SOC1 and FT mRNA levels. For example, 35S::CO 35S::FLC plants flowered 1.6 leaves earlier than wild-type plants despite containing 3-fold less SOC1 mRNA and only 1.4-fold more FT mRNA in comparison to wild-type plants (Table I; Figure 1B). Similarly, 35S::CO plants flowered just two leaves earlier than 35S::CO 35S::FLC plants, despite having 11.3-fold more FT mRNA and 8.3-fold more SOC1 mRNA than 35S::CO 35S::FLC plants (Table I; Figure 1B). This suggests that the determination of flowering time by CO and FLC is not mediated solely by SOC1 and FT. Alternatively, a small increase in FT mRNA abundance may be responsible for the early flowering of 35S::CO 35S::FLC compared with wild type, and the effect of FT may reach saturation so that the higher levels present in 35S::CO compared with 35S::CO 35S::FLC do not promote greatly earlier flowering.
Ubiquitous expression of SOC1 requires a complex promoter with multiple elements
Analysis of SOC1 mRNA previously demonstrated that the gene has a broad pattern of expression and is regulated by multiple flowering-time pathways, suggesting that the regulatory sequences may be complex. Our analysis of 4, 2 and 1 kb SOC1::GUS expression patterns shows that the correct regulation of SOC1 requires up to 4 kb of promoter sequence. Specifically, promoter sequences between nt –1955 and –4105 are needed for the expression of SOC1 in cauline leaves and in flowers, for an age-dependent increase of expression in developing seedlings, and for full levels of expression in seedlings. A similar pattern of age-dependent regulation was reported for both LFY and FT (Blázquez et al., 1997; Kardailsky et al., 1999; Kobayashi et al., 1999).
A CArG box mediates repression by FLC in vivo
A CArG box motif at nt –400 in the SOC1 promoter is bound specifically by FLC in vitro and mediates repression by FLC in vivo. SOC1 may, therefore, be an immediate target of FLC, although it is also possible that expression of another MADS-domain protein is activated by FLC and this binds to the CArG box in the SOC1 promoter. Consistent with this notion, these two genes share a ubiquitous pattern of expression prior to the transition to flowering (Michaels and Amasino, 1999; Borner et al., 2000; Lee et al., 2000). However, mutation of the CArG box that is recognized in vitro by FLC in the promoter of SOC1 did not lead to full derepression of SOC1::GUS expression in vivo. No other CArG box motifs were apparent in the 300 bp fragment C. This may indicate that FLC retains some ability to interact with the mutated CArG site in vivo that was not detected in vitro. Alternatively, FLC may also repress SOC1 expression indirectly.
The gel shift data suggest that FLC can interact in vitro with the CArG box motif in the SOC1 promoter as a homodimer. FLC may also form complexes with other MADS-domain partners to interact with this motif as a heterodimer in vivo, as has been described for other MADS-domain proteins (reviewed in Riechmann and Meyerowitz, 1997). Phylogenetic analysis places FLC in a subfamily with two other MADS-box genes: FLM/MAF1/AGL27 and AGL31 (Alvarez-Buylla et al., 2000; Ratcliffe et al., 2001). FLM was recently demonstrated to function as a floral repressor, to have a pattern of expression similar to FLC and to be down-regulated by vernalization in some ecotypes (Ratcliffe et al., 2001; Scortecci et al., 2001). Another candidate for heterodimerization with FLC is the MADS-box protein encoded by SVP, which is also a negative regulator of the floral transition and has a pattern of expression that is similar to that of FLC (Hartmann et al., 2000). The effect of overexpression of these genes on FT and SOC1 expression is untested.
A separate motif is required for activation by CO
Analysis of the overlapping 300 bp fragments A, B, C and D from the 1 kb SOC1 promoter indicated that only fragment C (nt –482 to –131) could efficiently mediate upregulation of SOC1::GUS expression in 35S::CO lines. This suggests that the motif that mediates activation by CO lies downstream of the CArG box and is located between the 3′ endpoint of fragment B at nt –372 and the 5′ endpoint of fragment D nt –248. Consistent with the idea that a separate motif is required for activation by CO, mutation of the CArG box at nt –400 did not impair the ability of fragment C to mediate activation in 35S::CO lines.
CO probably does not bind directly to DNA to mediate activation of SOC1. We did not detect CO binding to the SOC1 promoter using yeast one-hybrid selection or gel retardation assays with either purified recombinant CO protein and/or nuclear extracts prepared from 35S::CO or 35S::CO:GR plants (Figure 5; S.R.Hepworth and G.Coupland, unpublished results). Also, sequence analysis shows that the zinc fingers of CO present at the N-terminus of the protein are most similar to those of B-box proteins (Robson et al., 2001). In animal proteins, the B-box is usually accompanied by a RING finger and closely followed (5–8 amino acids) by a predicted α-helical coiled coil. This RBCC motif is believed to mediate protein–protein interactions (Borden, 1998; Hassler and Richmond, 2001). The C-terminus region of CO, which contains a CCT domain, also appears to mediate protein– protein interactions with other transcription factors. For example, the CCT domains of CO and TOC1 will interact with the DNA-binding protein ABI3 (Kurup et al., 2000), suggesting that ABI3 or related proteins could recruit CO or its homologues to DNA.
Model for the regulation of SOC1 by CO and FLC
We propose that the antagonistic regulation of SOC1 by CO and FLC requires a CArG box at nt –400 that mediates repression by FLC, and a separate motif located between nt –372 and –248 that mediates activation by CO (Figures 4A and 7). Based on previous experiments utilizing 35S::CO:GR (Samach et al., 2000), we favour a model in which CO is recruited directly to the SOC1 promoter by a DNA-binding factor to activate the expression of SOC1 in LD photoperiods, and FLC binds directly to DNA to mediate signals from the autonomous and vernalization pathways. FLC may mediate repression through recruitment of a global repression complex to the promoter, although it remains possible that another MADS-domain protein mediates between FLC and the repression of SOC1 expression. The finding that CO and FLC act through separate motifs implies that other flowering-time genes may be regulated by only CO or FLC, but not both. Furthermore, deletion of the FLC binding site from the SOC1 promoter did not abolish repression of SOC1 by FLC, suggesting that in addition to the direct mechanism of repression described here, FLC can also repress SOC1 expression indirectly. Determining how widespread is the antagonism between CO and FLC, and whether direct repression of CO targets by FLC is a general mechanism, will require analysing the response of other CO target genes to FLC.
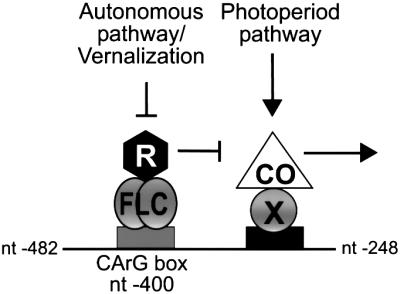
Fig. 7. Model for the antagonistic regulation of SOC1 expression by CO and FLC. The horizontal line represents a 234 bp region of the SOC1 promoter with the indicated nucleotide endpoints. Arrows indicate promotion and T-bars indicate repression of SOC1 expression. We propose that FLC binds directly to a CArG box motif at nt –400 of the SOC1 promoter and may recruit a global repression complex (R) to DNA to repress transcription. CO is proposed to be recruited by a sequence-specific DNA-binding factor (X) to an adjacent motif located between nt –372 and –248 in the SOC1 promoter to mediate activation. Deletion of the CArG box does not abolish FLC-mediated repression, suggesting that FLC also represses SOC1 expression by other more indirect mechanisms.
Materials and methods
Plant material and growth conditions
Wild type was the Landsberg erecta ecotype of A.thaliana. 35S::FLC plants and flc mutants were provided by R.Amasino (Michaels and Amasino, 1999, 2001). The fca-1 mutant was provided by M.Koornneef (University of Wageningen, Wageningen, The Netherlands). The 35S::CO and 35S::CO:GR plants were described previously and contain the co-2 tt4 mutations (Simon et al., 1996; Onouchi et al., 2000). We introduced SOC1::GUS reporter genes into 35S::CO, 35S::CO 35S::FLC, 35S::FLC and 35S::CO:GR plants by crossing. For most lines, homozygous plants were selected in the F3 generation. Flowering time was measured as described by Putterill et al. (1995). The numbers of rosette and cauline leaves on the main stem were scored and data are expressed as means ± SEM.
RNA isolation and northern analysis
RNA was extracted from whole seedlings grown on GM agar plates as described by Putterill et al. (1995). Tissue was harvested 5 h after dawn except where noted otherwise. Northern hybridization techniques were as in Suárez-López et al. (2001). The UBQ10-, FT- and β-TUB-specific probes have been described (Snustad et al., 1992; Wang et al., 1997; Samach et al., 2000). A SOC1-specific probe (nt +181 to 640 of the cDNA) was amplified by PCR from template pSOC1P (P.Reeves and G.Coupland, unpublished) using M13-20 and M13 Reverse primers (Stratagene). A GUS-specific probe was amplified by PCR from template pSLJ4K1 (Jones et al., 1992) using as primers GUS-2 and GUS-3. Images were visualized using a PhosphorImager (Molecular Dynamics) and band intensities quantified using ImageQuant software (Molecular Dynamics). Information on all primers cited in the text is provided in the Supplementary data available at The EMBO Journal Online.
Gel retardation assay
Gel retardations were performed as described by Hepworth et al. (1995) using as probe fragments of the SOC1 promoter whose 5′ overhangs were labelled by filling in with M-MLV reverse transcriptase (Life Technologies) in the presence of [α-32P]dCTP. A 20 µl gel retardation mixture was as described in Hepworth et al. (1995) plus 10 000 c.p.m. of radioactively labelled probe, the indicated competitor DNA and ∼120 ng of recombinant protein. After a 40 min incubation at room temperature, the samples were applied to an 8 or 12% polyacrylamide gel and run at 120 V. Images were visualized as above. Probes corresponding to all 300 bp fragments were amplified by PCR, digested with BamHI to give 5′-GATC overhangs, gel purified and labelled as above. The 30 bp fragment probes (SOC1 WT, SOC1 M1, SOC1 M2) were formed by annealing complementary oligonucleotides, which also generated 5′-GATC overhangs. Oligonucleotide sequences are provided in the Supplementary data.
Synthesis of His-tag CO and His-tag FLC proteins
Plasmids pET19b-CO and pET19b-FLC, which expressed His-tag CO and His-tag FLC proteins, respectively, were generated as described in the Supplementary data. Recombinant proteins were expressed in Escherichia coli BL21(DE3)pLysS and purified to apparent electrophoretic homogeneity over a cobalt resin column (Talon®; Clontech) according to the manufacturer’s instructions. The purified proteins were dialysed twice against PBS buffer at 4°C and stored at –20°C in 50% (v/v) glycerol.
Construction of SOC1::GUS reporter gene plasmids
SOC1::GUS reporter plasmids were derivatives of pGreen 0229, and the details of their construction are described in the Supplementary data.
Construction of mutant SOC1::GUS reporter gene plasmids
Site-directed mutagenesis was performed by PCR using the splice overlap extension procedure as described by Rowland and Segall (1998). To generate plasmid 1 kb ΔCArG SOC1::GUS, complementary reverse and forward oligonucleotides containing the desired mutations were used in separate PCRs with pSK208 as the template. Oligonucleotide sequences used to generate the mutations are provided in the Supplementary data.
GUS staining and activity measurement
For histochemical analysis of GUS expression, T2 or T3 seedlings were grown on GM agar. Homozygous T3 seedlings were used in most cases, but preliminary analyses were often performed in the T2, and the extreme late-flowering phenotype of 35S::FLC plants delayed the availability of T3 seedlings, making it necessary to use T2 material. Tissue was incubated in heptane for 5 min to remove the cuticle, air dried and then incubated in X-Gluc staining solution (Jefferson et al., 1987). Quantitative measurements of GUS activity in seedlings were determined based on the method of Jefferson et al. (1987). The protein concentration of samples was determined using a Bradford assay (Bio-Rad) according to the manufacturer’s instructions. GUS activity for each time point was determined in triplicate and expressed as relative fluorescence units (RFU) per microgram of protein. Data represent the means ± SEM.
Luciferase transient assay
The 1 kb SOC1 promoter was inserted upstream of the luciferase (LUC+; Promega) open reading frame and the nos poly(A) sequence. Details of the construct are provided in the Supplementary data. Thirty milligrams of 1 µm gold microcarriers were coated with both 1 kb SOC1::LUC and 35S::GFP according to the manufacturer’s instructions. A Biolistic PD-100/He particle delivery Gun (Bio-Rad) with Hepta adapter allowing the use of seven macrocarrier discs was used to deliver the microcarriers to the leaves of 15-day-old plants. Luciferase expression was detected after 24 h by spraying leaves with 1 mM luciferin, and detection of luminescence with a Hamamatsu Argus-50(20)/CA Imaging and Analysis system. Luminescence intensity was quantified using Hamamatsu HPD:CP software, and integrated density of the GFP images was analysed using Adobe PhotoShop and Scion Image software.
Transformation of plants and selection of homozygous lines
Wild-type plants were transformed with SOC1::GUS reporter gene constructs by floral dipping (Clough and Bent, 1998). The Agrobacterium strain used was C58C1 pGV101 pMP90. Basta®-resistant transformants were selected on soil by treatment of seedlings with the herbicide Challenge containing glufosinate-ammonium as the active ingredient (AgrEvo). Homozygous lines were selected in the T3 generation on GM agar plates that contained 12 mg/l phosphinothricin (Duchefa).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
The work was funded partly by an EC grant to G.C. within the REGIA programme. S.R.H. was supported by a post-doctoral fellowship from the NSERC (Canada) and an EMBO long-term fellowship; F.V. was supported by a FEBS Long-Term and EU Human Potential fellowship; D.R. received a BBSRC studentship. We thank R.Amasino and S.Michaels for providing 35S::FLC plants, as well as M.Yanofsky and D.Weigel for plasmids. We also thank Paula Suárez-López, Frederick Cremer and Owen Rowland for valuable comments.
References
- Alvarez-Buylla E.R., Liljegren,S.J., Pelaz,S., Gold,S.E., Burgeff,C., Ditta,G.S., Vergara-Silva,F. and Yanofsky,M.F. (2000) MADS-box gene evolution beyond flowers: expression in pollen, endosperm, guard cells, roots and trichomes. Plant J., 24, 457–466. [DOI] [PubMed] [Google Scholar]
- Araki T. (2001) Transition from vegetative to reproductive phase. Curr. Opin. Plant Biol., 4, 63–68. [DOI] [PubMed] [Google Scholar]
- Blázquez M.A. and Weigel,D. (2000) Integration of floral inductive signals in Arabidopsis. Nature, 404, 889–892. [DOI] [PubMed] [Google Scholar]
- Blázquez M.A., Soowal,L.N., Lee,I. and Weigel,D. (1997) LEAFY expression and flower initiation in Arabidopsis. Development, 124, 3835–3844. [DOI] [PubMed] [Google Scholar]
- Borden K.L.B. (1998) RING fingers and B-boxes: zinc-binding protein–protein interaction domains. Biochem. Cell Biol., 76, 351–358. [DOI] [PubMed] [Google Scholar]
- Borner R., Kampmann,G., Chandler,J., Gleißner,R., Wisman,E., Apel,K. and Melzer,S. (2000) A MADS domain gene involved in the transition to flowering in Arabidopsis. Plant J., 24, 591–599. [DOI] [PubMed] [Google Scholar]
- Clough S.J. and Bent,A.F. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J., 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Hartmann U., Höhmann,S., Nettesheim,K., Wisman,E., Saedler,H. and Huijser,P. (2000) Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. Plant J., 21, 351–360. [DOI] [PubMed] [Google Scholar]
- Hassler M. and Richmond,T.J. (2001) The B-box dominates SAP-1–SRF interactions in the structure of the ternary complex. EMBO J., 20, 3018–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth S.R., Ebisuzaki,L.K. and Segall,J. (1995) A 15-base-pair element activates the SPS4 gene midway through sporulation in Saccharomyces cerevisiae. Mol. Cell. Biol., 15, 3934–3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Mizukami,Y., Hu,Y. and Ma,H. (1993) Isolation and characterization of the binding sequences for the product of the Arabidopsis floral homeotic gene AGAMOUS. Nucleic Acids Res., 21, 4769–4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh,T.A. and Bevan,M.W. (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J., 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U., West,J., Lister,C., Michaels,S., Amasino,R. and Dean,C. (2000) Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science, 290, 344–347. [DOI] [PubMed] [Google Scholar]
- Jones J.D.J., Shlumukov,L., Carland,F., English,J., Scofield,S.R., Bishop,G.J. and Harrison,K. (1992) Effective vectors for transformation, expression of heterologous genes and assaying transposon excision in transgenic plants. Transgenic Res., 1, 285–297. [DOI] [PubMed] [Google Scholar]
- Kardailsky I., Shukla,V.K., Ahn,J.H., Dagenais,N., Christensen,S.K., Nguyen,J.T., Chory,J., Harrison,M.J. and Weigel,D. (1999) Activation tagging of the floral inducer FT. Science, 286, 1962–1965. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y., Kaya,H., Goto,K., Iwabuchi,M. and Araki,T. (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science, 286, 1960–1962. [DOI] [PubMed] [Google Scholar]
- Koornneef M., Hanhart,C.J. and van der Veen,J.H. (1991) A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet., 229, 57–66. [DOI] [PubMed] [Google Scholar]
- Koornneef M., Blankestijn-de Vries,H., Hanhart,C., Soppe,W. and Peeters,T. (1994) The phenotype of some late-flowering mutants is enhanced by a locus on chromosome 5 that is not effective in the Landsberg erecta wild-type. Plant J., 6, 911–919. [Google Scholar]
- Kurup S., Jones,H.D. and Holdsworth,M.J. (2000) Interactions of the developmental regulator ABI3 with proteins identified from developing Arabidopsis seeds. Plant J., 21, 143–155. [DOI] [PubMed] [Google Scholar]
- Lee H., Suh,S.-S., Park,E., Cho,E., Ahn,J.H., Kim,S.-G., Lee,J.S., Kwon,Y.M. and Lee,I. (2000) The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev., 14, 2366–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S.D. and Amasino,R.M. (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell, 11, 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S.D. and Amasino,R.M. (2001) Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell, 13, 935–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onouchi H., Igeño,M.I., Périlleux,C., Graves,K. and Coupland,G. (2000) Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell, 12, 885–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnueli L., Gutfinger,T., Hareven,D., Ben-Naim,O., Ron,N., Adir,N. and Lifschitz,E. (2001) Tomato SP-interacting proteins define a conserved signaling system that regulates shoot architecture and flowering. Plant Cell, 13, 2687–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill J., Robson,F., Lee,K., Simon,R. and Coupland,G. (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell, 80, 847–857. [DOI] [PubMed] [Google Scholar]
- Ratcliffe O.J., Nadzan,G.C., Reuber,T.L. and Riechmann,J.L. (2001) Regulation of flowering in Arabidopsis by an FLC homologue. Plant Physiol., 126, 122–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves P.H. and Coupland,G. (2000) Response of plant development to environment: control of flowering by daylength and temperature. Curr. Opin. Plant Biol., 3, 37–42. [DOI] [PubMed] [Google Scholar]
- Riechmann J.L. and Meyerowitz,E.M. (1997) MADS domain proteins in plant development. Biol. Chem., 378, 1079–1101. [PubMed] [Google Scholar]
- Robson F., Costa,M.M.R., Hepworth,S., Vizir,I., Piñeiro,M., Reeves,P.H., Putterill,J. and Coupland,G. (2001) Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J., 28, 619–631. [DOI] [PubMed] [Google Scholar]
- Rouse D.T., Sheldon,C.C., Bagnall,D.J., Peacock,W.J. and Dennis,E.S. (2002) FLC, a repressor of flowering, is regulated by genes in different inductive pathways. Plant J., 29, 183–191. [DOI] [PubMed] [Google Scholar]
- Rowland O. and Segall,J. (1998) A hydrophobic segment within the 81-amino-acid domain of TFIIIA from Saccharomyces cerevisiae is essential for its transcription factor activity. Mol. Cell. Biol., 18, 420–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A., Onouchi,H., Gold,S.E., Ditta,G.S., Schwarz-Sommer,Z., Yanofsky,M.F. and Coupland,G. (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science, 288, 1613–1616. [DOI] [PubMed] [Google Scholar]
- Sanda S.L. and Amasino,R. (1996) Interaction of FLC and late-flowering mutations in Arabidopsis thaliana. Mol. Gen. Genet., 251, 69–74. [DOI] [PubMed] [Google Scholar]
- Scortecci K.C., Michaels,S.D. and Amasino,R.M. (2001) Identification of a MADS-box gene, FLOWERING LOCUS M, that represses flowering. Plant J., 26, 229–236. [DOI] [PubMed] [Google Scholar]
- Sheldon C.C., Burn,J.E., Perez,P.P., Metzger,J., Edwards,J.A., Peacock,W.J. and Dennis,E.S. (1999) The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell, 11, 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon C.C., Rouse,D.T., Finnegan,E.J., Peacock,W.J. and Dennis,E.S. (2000) The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC) Proc. Natl Acad. Sci. USA, 97, 3753–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi H., Okada,K. and Shimura,Y. (1993) Nucleotide sequences recognized by the AGAMOUS MADS domain of Arabidopsis thaliana in vitro. Plant J., 4, 385–398. [DOI] [PubMed] [Google Scholar]
- Shore P. and Sharrocks,A.D. (1995) The MADS-box family of transcription factors. Eur. J. Biochem., 229, 1–13. [DOI] [PubMed] [Google Scholar]
- Simon R., Igeño,I.M. and Coupland,G. (1996) Activation of floral meristem identify genes in Arabidopsis. Nature, 384, 59–62. [DOI] [PubMed] [Google Scholar]
- Simpson G.G., Gendall,A.R. and Dean,C. (1999) When to switch to flowering. Annu. Rev. Cell Dev. Biol., 15, 519–550. [DOI] [PubMed] [Google Scholar]
- Snustad D.P., Haas,N.A., Kopczak,S.D. and Silflow,C.D. (1992) The small genome of Arabidopsis contains at least nine expressed β-tubulin genes. Plant Cell, 4, 549–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-López P., Wheatley,K., Robson,F., Onouchi,H., Valverde,F. and Coupland,G. (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature, 410, 1116–1119. [DOI] [PubMed] [Google Scholar]
- Wang Z.Y., Kenigsbuch,D., Sun,L., Harel,E., Ong,M.S. and Tobin,E.M. (1997) A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell, 9, 491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]