Abstract
LTR-retrotransposons are abundant components of all eukaryotic genomes and appear to be key players in their evolution. They share with retroviruses a reverse transcription step during their replication cycle. To better understand the replication of retrotransposons as well as their similarities to and differences from retroviruses, we set up an in vitro model system to examine minus-strand cDNA synthesis of the yeast Ty1 LTR-retrotransposon. Results show that the 5′ and 3′ ends of Ty1 genomic RNA interact through 14 nucleotide 5′–3′ complementary sequences (CYC sequences). This 5′–3′ base pairing results in an efficient initiation of reverse transcription in vitro. Transposition of a marked Ty1 element and Ty1 cDNA synthesis in yeast rely on the ability of the CYC sequences to base pair. This 5′–3′ interaction is also supported by phylogenic analysis of all full-length Ty1 and Ty2 elements present in the Saccharomyces cerevisiae genome. These novel findings lead us to propose that circularization of the Ty1 genomic RNA controls initiation of reverse transcription and may limit reverse transcription of defective retroelements.
Keywords: cDNA synthesis/nucleocapsid/priming/RNA chaperone/RNA circularization
Introduction
Retrotransposons are mobile genetic elements, closely related to retroviruses, which replicate through a genomic single-stranded RNA intermediate that is converted into a double-stranded DNA by reverse transcriptase (RT). This process, called reverse transcription, takes place in a nucleoprotein particle containing the retrotransposon RT (Boeke and Stoye, 1997). Subsequently the newly formed DNA copy is integrated into the host genome. By means of this copy-and-paste mechanism, retrotransposons have efficiently invaded eukaryotic genomes. As a major portion of eukaryotic genomes they are thought to play a central role in their evolution. Retrotransposons have been involved in double-strand break repair (Teng et al., 1996; Yu and Gabriel, 1999) and are sources of insertional mutagenesis, homologous recombination and RT activity (Boeke and Stoye, 1997). The latter has been involved in pseudogene formation, gene transduction and exon shuffling, as well as intron loss when RT acts on cellular RNAs rather than on retrotransposon RNA (Fink, 1987; Derr and Strathern, 1993; Flavell et al., 1994; Moran et al., 1999; Esnault et al., 2000; Elrouby and Bureau, 2001). Therefore, understanding the mechanism of endogenous reverse transcription and the way it is controlled are major challenges. Long terminal repeat-containing retrotransposons (LTR-retrotransposons) share with oncoretroviruses a common organization of their genome. The close relationships between LTR-retrotransposons and retroviruses have led to the assumption that reverse transcription is identical in both retroelements. However, few studies have investigated reverse transcription at the molecular level in LTR-retrotransposons. The best-studied LTR-retrotransposons are Saccharomyces cerevisiae Ty1 and Ty3, and Schizosaccharomyces pombe Tf1. Initiation of reverse transcription, thought to be a strictly controlled step, has been studied in these three retrotransposons and striking differences from the retroviral modus operandi have been found. In retroviruses a specific cellular tRNA is annealed to an 18 nucleotide (nt) region called the primer binding site (PBS) near the 5′ end of genomic RNA. In Ty1 and Ty3 the primer for reverse transcription, initiator methionine tRNA [tRNA(iMet)], binds to canonical PBSs, which are much shorter (10 and 8 nt, respectively) than the 18 nt retroviral PBS. This has led to a search for additional contacts between tRNA(iMet) and genomic RNA that could account for the stability and specificity of the RT priming process. In these two elements a functional PBS was found split into at least three separate regions. In Ty1, tRNA(iMet) is annealed to boxes 0, 1 and 2.1 in the 5′ coding region of Ty1 RNA in addition to the canonical PBS (Figure 1) (Chapman et al., 1992; Keeney et al., 1995; Friant et al., 1996, 1998). In Ty3, tRNA(iMet) is annealed to sequences located at opposite ends of the genomic RNA, causing circularization of Ty3 RNA through a tRNA bridge (Gabus et al., 1998). Tf1 priming differs drastically from that of retroviruses, since the 5′ end of the genomic RNA folds back on itself, is cleaved and used as primer (Levin, 1995, 1996).
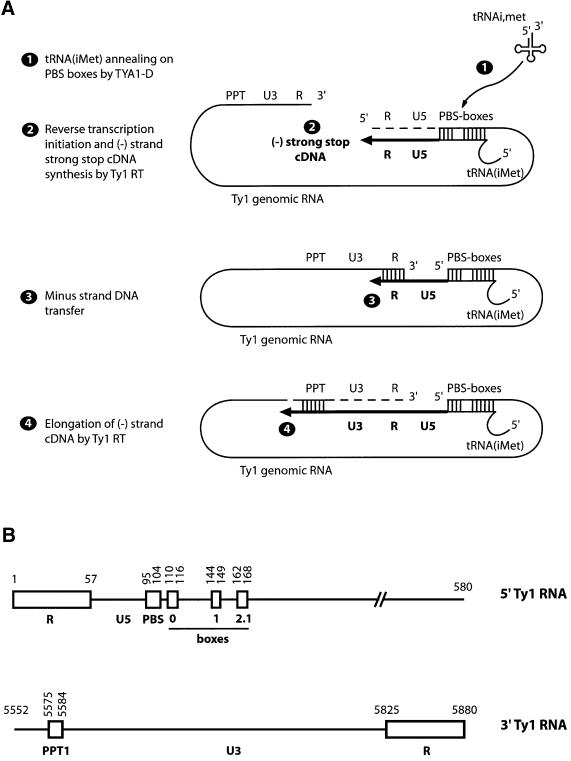
Fig. 1. Scheme of Ty1 reverse transcription. (A) Current model of Ty1 (–) strand cDNA synthesis. (1) Primer tRNA(iMet) is annealed to the primer binding site (PBS) and to boxes 0, 1 and 2.1 by the RNA-chaperone properties of the Ty1 Gag peptide, called TYA1-D. (2) The 3′ OH of tRNA(iMet) is elongated by Ty1 RT using Ty1 RNA as template (thin line) to generate minus-strand strong-stop cDNA (ss-cDNA, thick line). During the elongation process, RT RNase H degrades the RNA template (dotted line). (3) ss-cDNA is transferred to the 3′ end of Ty1 RNA genome (either intra- or intermolecular) by R sequences pairing and conducted by TYA1-D. (4) ss-cDNA is elongated by RT generating minus-strand cDNA product (st-cDNA). (B) WT RNAs used in the present study. Ty1 5′ RNA contains the 5′ repeat sequence R, U5 of the LTR, PBS and boxes 0, 1 and 2.1. Ty1 3′ RNA contains the polypurine tract (PPT1), U3 and 3′ R sequences of the LTR. Numbers indicate nucleotide positions with respect to the Ty1-H3 molecular clone.
Since the PBSs of Ty1 and Ty3 are multipartite, we wondered whether reverse transcription was strictly identical to the retroviral mode or might substantially diverge. To answer this question, we have set up an in vitro Ty1 model system to examine minus-strand cDNA synthesis. Surprisingly, this reconstituted system revealed that the 3′ region of Ty1 RNA greatly enhances the initiation efficiency, independently of minus-strand DNA transfer. This functional interaction requires base pairing between a 5′ Gag coding sequence of 14 nt and a complementary sequence located in the 3′ UTR. Furthermore, this 5′–3′ interaction is required in vivo for efficient transposition of a marked Ty1 element. The importance of the 5′–3′ RNA interaction is also strengthened by conservation or covariation in all full-length Ty1 and Ty2 elements present in the yeast genome. Based on these findings we propose that genomic RNA circularization, either directly, as in Ty1, or indirectly, as in Ty3, may be a common feature of reverse transcription in LTR-retrotransposons. This novel mechanism may limit reverse transcription of defective retroelements.
Results
A reconstituted in vitro Ty1 reverse transcription system
Ty1 cDNA synthesis occurs in a cytoplasmic nucleoprotein shell, called the virus-like particle (VLP), composed of Ty1 Gag (Garfinkel et al., 1985; Mellor et al., 1985), the dimeric RNA genome, primer tRNA(iMet), protease (PR), integrase and RT (Garfinkel et al., 1985; Mellor et al., 1985; Adams et al., 1987; Eichinger and Boeke, 1988; Chapman et al., 1992; Feng et al., 2000). Ty1 Gag is not processed further into matrix, capsid and nucleocapsid proteins (NCp) by PR, but rather the C-terminal 40 amino acids are cleaved by PR and then probably degraded (Merkulov et al., 1996). In a previous report, we showed that the C-terminal region of mature Ty1 Gag has in vitro nucleic acid chaperone properties similar to retroviral NCp (Cristofari et al., 2000). As a synthetic peptide called TYA1-D, this domain allows specific initiation of reverse transcription in vitro by annealing primer tRNA(iMet) to the PBS in physiological conditions and by inhibiting non-specific priming events (Cristofari et al., 2000). Retroviral NCps are nucleic acid chaperones that direct specific reverse transcription initiation and the two obligatory strand transfers required to generate the LTRs and complete proviral DNA by RT (Darlix et al., 2000).
By taking advantage of the recent characterization of both Ty1 RT and the Gag peptide with nucleic acid chaperone properties (Cristofari et al., 2000; Wilhelm et al., 2000), we reconstituted functional Ty1 nucleoprotein complexes to study minus-strand cDNA synthesis in vitro. The reconstituted Ty1 complexes comprise two in vitro generated RNAs (5′ and 3′ Ty1 RNA) mimicking the 5′ and 3′ regions of Ty1 genomic RNA, containing all sequences in 5′ (R, PBS and boxes 0, 1 and 2.1) or in 3′ (PPT1, R), respectively, thought to be required for transposition (Figure 1B) (Xu and Boeke, 1990). The Ty1 complexes also contain primer [32P]tRNA(iMet), TYA1-D peptide and recombinant Ty1 RT (Wilhelm et al., 2000).
Minus-strand strong-stop cDNA synthesis and transfer
5′ and 3′ Ty1 RNAs and 5′ 32P-labelled tRNA were first incubated with TYA1-D to form nucleoprotein complexes and to direct the annealing of tRNA to the multipartite PBS (PBS, box 0, box 1 and box 2.1; Figure 1A, step 1). Then Ty1 RT and dNTPs were added to start cDNA synthesis. All steps were carried out at 25°C, the optimal temperature for Ty1 transposition in yeast and for activity of the recombinant Ty1 RT.
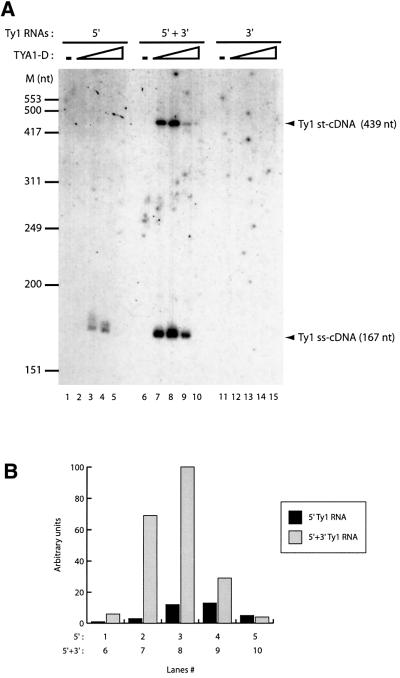
Two cDNA were synthesized (Figure 2A, lanes 6–10). The shorter one corresponds to the minus-strand strong-stop cDNA (ss-cDNA), i.e. the cDNA starting at the 3′ end of the tRNA and ending at the 5′ end of Ty1 5′ RNA (Figure 1A, step 2). The longer cDNA was of the expected size for the minus-strand transfer product (st-cDNA) (i.e. elongation of ss-cDNA after transfer to Ty1 3′ RNA; Figure 1A, steps 3 and 4). The maximal strand transfer efficiency was 40% st-cDNA of total cDNA synthesis (Figure 2A, lanes 7 and 8). In the absence of Ty1 3′ RNA, only ss-cDNA was detected (Figure 2A, lanes 1–5). A surprising observation was that Ty1 3′ RNA increased 10- to 20-fold the total level of ss-cDNA synthesis (Figure 2A, compare lanes 1–5 with 6–10, and B for quantification; see also Figure 6). In a control experiment with no Ty1 5′ RNA, no cDNA was detected (Figure 2A, lanes 11–15). DNA synthesis was strictly dependent on TYA1-D (Figure 2A, compare lanes 1 or 6 with 2–4 or 7–9, respectively). However, an excess of TYA1-D resulted in the inhibition of reverse transcription, as already observed with retroviral NCp (Guo et al., 1997; Lapadat-Tapolsky et al., 1997). Together, these two phenomena led to a sharp peak of cDNA synthesis. Therefore, several TYA1-D concentrations were always used to avoid missing the optimum (usually around 1:12 or 1:10 peptide to nucleotide ratio).
Fig. 2. In vitro synthesis of Ty1 minus-strand cDNA. (A) Ty1 nucleoprotein complexes containing 5′ 32P-labelled tRNA and Ty1 RNAs were incubated with Ty1 RT and dNTP. Ty1 5′ RNA alone (lanes 1–5), 5′ and 3′ RNAs (lanes 6–10) or 3′ RNA only (lanes 11–15) were used. After reverse transcription, nucleic acids were purified, analysed by 6% PAGE in denaturing conditions and the gel was autoradiographed. TYA1-D to nucleotide molar ratios were 0, 1:15, 1:12, 1:10 and 1:8. Arrowheads indicate minus-strand strong-stop cDNA (ss-cDNA, 167 nt) and strand transfer product (st-cDNA, 439 nt), covalently linked to [32P]tRNA(iMet). (B) Quantification of the gel shown in (A). Quantifications of lanes 1–5 are shown as black bars and those of lanes 6–10 as grey bars.
Fig. 6. Interaction between the ends of Ty1 RNA is required for efficient reverse transcription initiation. (A) ss-cDNA synthesis was performed with WT (lanes 1–9) or cyc5– (lanes 10–18) 5′ RNA, in the absence (lanes 1–3) or presence of WT (lanes 4–6 and 14–16) or cyc3– (lanes 7–9 and 16–18) 3′ RNA. TYA1-D to nucleotide molar ratios were 1:15, 1:12, 1:10 and 1:8. Ty1 RT RNase H(–) was used as before. (B) Quantification of the peak of ss-cDNA synthesis of the gel in (A) (lanes 2, 6, 10, 14 and 18, respectively).
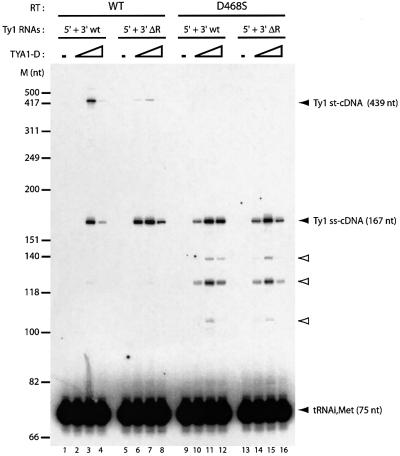
Since this model system was able to recapitulate the main steps of Ty1 minus-strand DNA synthesis in vitro, we examined the requirement for cis elements and trans-acting factors. For example, the 3′ R sequence and RT-associated RNase H are both necessary in retroviral strand transfers (Peliska and Benkovic, 1992; Allain et al., 1994). First, deletion of the 3′ R caused a drastic reduction of st-cDNA synthesis from 40% to <5% of strand transfer efficiency, whereas ss-cDNA level was intact (Figure 3, compare lanes 1–4 with 5–8). Next, Ty1 RT with the D468S mutation in the RNase H active site was used since it has a wild-type level of polymerase activity on homopolymeric template but no RNase H activity (Wilhelm et al., 2001). In vitro Ty1 RT D468S was able to synthesize ss-cDNA at similar levels to that of wild-type (WT) RT, whereas no st-cDNA was detected (Figure 3, compare lanes 1–4 with 9–12). Premature pauses before completion of ss-cDNA synthesis were observed with this mutant (Figure 3, lanes 9–16, white arrowheads), as already described for retroviral RNase H(–) RT (Dudding and Mizrahi, 1993). These results are in agreement with the effect of the D468S mutation on Ty1 cDNA synthesis in vivo (Uzun and Gabriel, 2001).
Fig. 3. Molecular requirements for minus-strand DNA transfer. Experiments were performed as in Figure 2. Ty1 5′ RNA was WT. Ty1 3′ RNA was either WT (lanes 1–4 and 9–12) or deleted of the repeated 3′ region (ΔR, lanes 5–8 and 13–16). Ty1 RT was either WT (lanes 1–8) or RNase H(–) (lanes 9–16). TYA1-D to nucleotide molar ratios were 0, 1:15, 1:10 and 1:8. Black arrowheads indicate strong-stop cDNA (ss-cDNA, 167 nt) and strand transfer product (st-cDNA, 439 nt), covalently linked to [32P]tRNA(iMet). Note that several premature stops before ss-cDNA completion were observed with the RT RNase H(–) mutant (white arrowheads).
The 3′ region of Ty1 RNA is required for efficient minus-strand ss-cDNA synthesis
As underlined above, Ty1 3′ RNA seemed to increase the total level of ss-cDNA synthesis (Figure 2). To determine whether strand transfer could be indirectly required for efficient ss-cDNA synthesis, we compared the level of ss-cDNA with the RNase H defective RT, in the presence or absence of Ty1 3′ RNA. ss-cDNA synthesis was increased by Ty1 3′ RNA, even when strand transfer was impaired due to the use of RT RNase H(–) (Figure 4A, compare lanes 1–5 with 6–10). The 5′ RNA PBS was also mutated to verify that no other initiation pathway was involved in the process. We used 5 nt point mutations preventing tRNA(iMet) pairing to PBS (Cristofari et al., 2000). Indeed, this mutation completely abolished ss-cDNA synthesis independently of added Ty1 3′ RNA (Figure 4A, compare lanes 1–10 with 11–20).
Fig. 4. The 3′ end of Ty1 RNA specifically enhances ss-cDNA synthesis. (A) Reverse transcription was performed in the presence of Ty1 RT RNase H(–) with WT (lanes 1–10) or pbs– (lanes 11–20) 5′ Ty1 RNA and with or without WT 3′ Ty1 RNA (lanes 1–5 and 11–15, or lanes 6–10 and 16–20, respectively). TYA1-D to nucleotide molar ratios were 0, 1:15, 1:12, 1:10 and 1:8. (B) Specificity of the Ty1 3′ RNA on ss-cDNA synthesis. Assays were performed with a constant amount of Ty1 5′ RNA (0.25 pmol) and a constant 1:10 TYA1-D protein to nucleotide molar ratio. An increasing 3′ to 5′ RNA molar ratio was used (0; 1; 2; 5). 3′ RNA was either Ty1 3′ RNA (lanes 1–4), HIV-1-derived 5′ RNA (nt 1–415) (lanes 5–8) or yeast poly(A)+ RNAs (lanes 9–12). Ty1 RT RNase H(–) was used in order to block strand transfer. The arrowhead indicates strong-stop cDNA covalently linked to [32P]tRNA(iMet) (ss-cDNA, 167 nt).
These findings suggest that the Ty1 3′ RNA contains a sequence required for efficient ss-cDNA synthesis independently of the other reverse transcription steps.
To assess the specificity of the Ty1 3′ RNA effect on ss-cDNA synthesis, Ty1 3′ RNA was replaced by heterologous RNA and reverse transcription was performed with RT RNase H(–) mutant to block strand transfer. Neither an in vitro-generated RNA encompassing nt 1–415 of HIV-1 RNA (Lapadat-Tapolsky et al., 1997), nor YH50 yeast poly(A)+ RNAs were able to increase the level of ss-cDNA synthesis as Ty1 3′ RNA did (Figure 4B). This indicated that Ty1 3′ RNA can specifically enhance ss-cDNA synthesis.
Direct interaction between the 5′ and 3′ ends of Ty1 genomic RNA
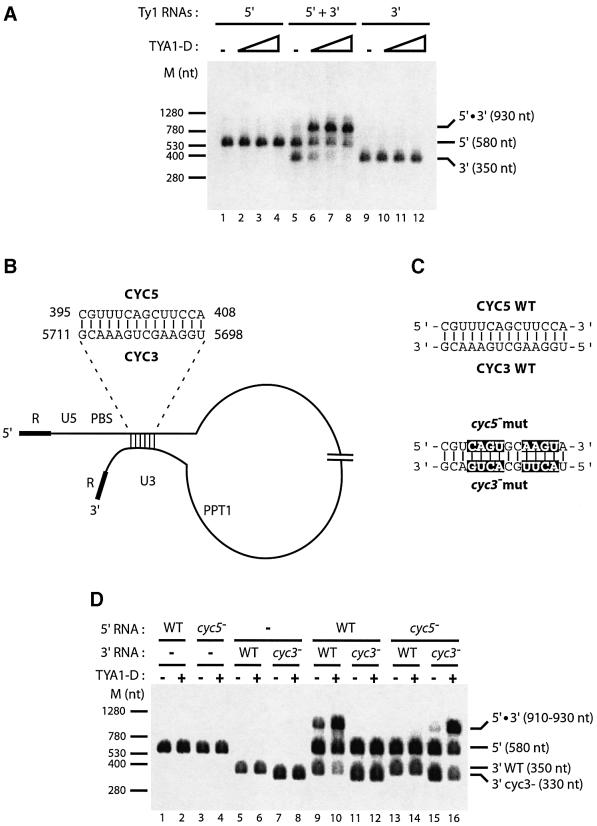
Since functional relationships appear to take place between the two ends of Ty1 genomic RNA, we wondered whether this could rely on direct physical interactions. To test this hypothesis, 5′ and 3′ Ty1 RNAs were incubated with TYA1-D. Following protein extraction, RNAs were analysed by electrophoresis in native conditions. With both Ty1 RNAs, a new RNA species with an apparent higher molecular weight of 930 nt was formed (Figure 5A, lanes 5–8). Conversely, incubation of 5′ RNA or 3′ RNA alone with TYA1-D did not generate this new RNA species (lanes 1–4 and 9–12). Interaction between 5′ and 3′ RNAs was strongly favoured by the TYA1-D RNA chaperone peptide (Figure 5A, compare lanes 5 and 8). When primer tRNA(iMet) was included in the assay, identical results were obtained (data not shown). This prompted us to perform a computer local alignment search for sequence complementarities at the 5′ and 3′ ends of Ty1 RNA. A 3′ sequence of 14 nt was discovered that perfectly matches a region downstream of the PBS, encompassing box 2.1 (Figure 5B). The 5′ and 3′ sequences were called CYC5 and CYC3, for cyclization in 5′ or in 3′, respectively. To examine whether these complementary sequences mediate the interaction between the 5′ and 3′ RNAs, we introduced point mutations in CYC5 to disrupt sequence complementarities. Compensatory point mutations were inserted in CYC3 to compensate for the CYC5 mutations (Figure 5C). Mutations in CYC5 or in CYC3 completely abolished RNA–RNA interactions (Figure 5D, compare lanes 9–10 with 11–14). In contrast, the use of both cyc5– and cyc3– mutations restored the 5′–3′ interaction (lanes 15–16). These results indicate that CYC5–CYC3 pairing mediates a direct molecular interaction between the ends of Ty1 RNA.
Fig. 5. Direct interaction between the 5′ and 3′ ends of Ty1 RNA. (A) The ends of Ty1 RNA can interact in vitro. Equimolar amounts of Ty1 5′ and 3′ RNAs were incubated with increasing amounts of TYA1-D, at nt molar ratios of 0, 1:20, 1:10 and 1:5. 5′ RNA alone (lanes 1–4), 5′ and 3′ RNAs (lanes 5–8) or 3′ RNA alone (lanes 9–12) were used. After incubation, nucleic acids were purified and analysed in native conditions by agarose gel electrophoresis, followed by EtBr staining. (B) Putative 5′–3′ interacting sequences. 5′ and 3′ sequences have been called CYC5 and CYC3, respectively. (C) Mutations introduced to destabilize putative CYC pairing. RNA mutants are cyc5– and cyc3–, respectively. (D) 5′–3′ interaction is mediated by CYC sequence pairing. Interaction between WT or mutant RNAs (cyc) was analysed as in (A) with a TYA1-D to nucleotide molar ratio of 1:10 (even lanes) or without TYA1-D (odd lanes). Note that mutant 3′ RNA is smaller than WT 3′ RNA because unlike WT it does not contain a poly(A) tail. Similar results were obtained when poly(A)– WT RNA was used (data not shown).
Base pairing between the 5′ and 3′ ends of Ty1 genomic RNA is required for efficient ss-cDNA synthesis in vitro
To investigate the requirement of CYC pairing for optimal ss-cDNA synthesis, the mutated 5′ and 3′ RNAs were used in the reconstituted reverse transcription system with RT RNase H(–) mutant to block strand transfer. Mutant cyc5– RNA with WT 3′ RNA or WT 5′ RNA with mutant cyc3– RNA were much less efficient than both 5′ and 3′ WT RNAs in promoting ss-cDNA synthesis (12 and 2% of the WT level, respectively; Figure 6A, compare lanes 4–6 with 7–9 and 13–15; Figure 6B for quantification). On the other hand, when both mutant RNAs were used, cDNA synthesis was restored to a level close to that of the wild type (Figure 6A, compare lanes 7–9, 13–15 and 16–18; Figure 6B for quantification). Thus CYC5–CYC3 base pairings are required for efficient ss-cDNA synthesis in vitro.
Ty1 3′ RNA enhances ss-cDNA synthesis by acting at the level of reverse transcription initiation
The Ty1 3′ RNA-mediated enhancement of ss-cDNA synthesis can take place at one (or more) of the following steps: tRNA(iMet) annealing, initiation, initiation-to-elongation transition or elongation. We have compared the annealing of tRNA(iMet) to the 5′ PBS with or without 3′ RNA. Results show that the level of tRNA(iMet) annealed to PBS was not increased by the 3′ RNA but was rather slightly decreased (Figure 7B, compare free tRNA(iMet) in lanes 5 and 10). Furthermore disrupting CYC pairing does not prevent tRNA(iMet) annealing to the PBS (see Supplementary figure 1 available at The EMBO Journal Online). This indicates that the 3′ RNA acts during a later step of ss-cDNA synthesis.
Fig. 7. The 3′ RNA acts at the level of reverse transcription initiation. (A and B) Annealing of tRNA(iMet) to 5′ RNA without (lanes 1–5) or with 3′ RNA (lanes 6–10). Experiments were as described in Figure 5, with an excess of [32P]tRNA(iMet). (A) and (B) are EtBr staining and autoradiography of the same gel, respectively. (C) Sensu stricto initiation of reverse transcription. Reverse transcription was performed as described in Figure 2, but an unlabelled tRNA(iMet) and solely [α-32P]dTTP were used instead of all four dNTPs. TYA1-D to nucleotide molar ratios were 1:30, 1:25, 1:20, 1:15, 1:12, 1:10 and 1:8. The arrowhead indicates [32P]dTMP covalently linked to tRNA(iMet) (76 nt). Labelled tRNA(iMet) and φX174 DNA HinfI markers (Promega) were used for size determination (not shown).
Next, we examined the influence of the 3′ RNA at the initiation step. To this end, we used an unlabelled tRNA(iMet) and solely [α-32P]dTTP instead of all four dNTPs to initiate reverse transcription. Since dTMP is the first nucleotide added by RT to tRNA(iMet), a 76 nt tRNA–[32P]dTMP product must arise upon addition of RT if initiation of reverse transcription occurs (scheme in Figure 7B). Indeed, we observed an initiation product with both 5′ and 3′ RNAs, whereas virtually no initiation product was detected with 5′ or 3′ RNA alone. Increasing the incubation time from 30 to 60 min did not increase initiation level. This suggests that tRNA(iMet)–5′ RNA–3′ RNA complexes are more competent for reverse transcription initiation than are tRNA(iMet)–Ty1 5′ RNA complexes.
CYC pairing is required for efficient cDNA synthesis in vivo and transposition
To analyse the role of the CYC sequences in vivo, we introduced a plasmid-borne Ty1 element under the control of the inducible GAL1 promoter (pGTy1-H3mHIS3AI) into a His– yeast strain. This element is genetically marked with a HIS3 reporter gene in the antisense orientation and interrupted by an artificial intron (AI) in the same orientation as Ty1 (Figure 8A). Thus the level of His+ revertants is an indicator of retrotransposition frequency since HIS3 expression only occurs after splicing, reverse transcription and integration (Curcio and Garfinkel, 1991). The YH50 strain and its isogenic rad52– counterpart AGY49 used here are spt3– to limit expression of endogenous Ty1s, which might complement in trans the HIS3-marked element (Boeke et al., 1986). Retrotrans position efficiencies in RAD52 and rad52– yeast were compared since cDNA incorporation into the yeast genome is known to occur by at least two pathways: one requires Ty1 integrase and a complete, double-stranded Ty1 DNA, and the second is dependent on homologous recombination (therefore on Rad52p) and could mediate integration of aberrant reverse transcripts (Sharon et al., 1994).
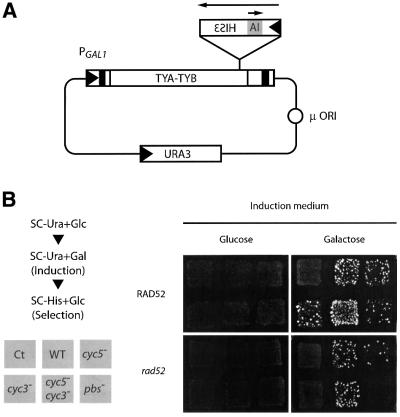
Fig. 8. Pairing of CYC5 and CYC3 sequences is required for Ty1 transposition. (A) Scheme of the Ty1 transposition assay. pGTy1-H3mHIS3AI plasmid contains the Ty1-H3 molecular clone, the expression of which is under the control of the inducible GAL1 promoter and marked with a HIS3 reporter gene in the antisense orientation. An artificial intron (AI) in the sense orientation has been inserted in the HIS3 gene. The plasmid also contains a URA3 gene to permit transformant selection and a high copy number origin of replication (µ ORI). (B) Effect of CYC mutations on Ty1 transposition. The WT plasmid or one of its mutant counterparts was transformed in a ura3– and his3– strain. Transformants were selected in synthetic medium containing glucose and lacking uracil (SC–Ura+Glc) and patched onto SC–Ura+ Glc plates. The position of each transformant is indicated in the grey squares. Transposition induction was performed by replica plating patches onto medium containing either glucose or galactose and lacking uracil (SC–Ura+Gal) at 30°C. Finally, transposition events were selected by replica plating colonies on medium containing glucose and lacking histidine (SC–His+Glc, plate shown). Strains were YH50 or its rad52 counterpart AGY49. Ct stands for plasmid pGmHIS3AI (see text).
Yeasts were transformed with pGTy1-H3mHIS3AI plasmid (WT Ty1) or with its mutated counterparts (cyc5–, cyc3– and double mutant cyc5–/cyc3–). Two negative controls included a pbs– mutant defective in the initiation of reverse transcription (Chapman et al., 1992) and the HIS3AI cassette alone under the GAL1 promoter as a control (pGmHIS3AI plasmid, Ct) of HIS3AI reverse transcription (Curcio and Garfinkel, 1991). The mutations chosen in vivo were those used in vitro. Since cyc5– and pbs– mutations were localized in the Gag coding sequence, silent mutations were chosen.
WT or mutated Ty1 elements were induced by growth on galactose and then replica-plated onto media lacking histidine. The level of His+ prototrophy was greatly decreased with single cyc5– and cyc3– mutations compared with WT (Figure 8B), but the number of His+ colonies was restored to a WT level when cyc5–/cyc3– compensatory mutations were inserted into the marked Ty1 element. The effect of the CYC mutations on transposition was qualitatively similar in RAD52 and rad52– strains. However, a number of cDNA insertions in the RAD52 strain did not depend on an intact PBS and therefore did not rely on accurate reverse transcription. In the RAD52 genetic background, the transposition rate of cyc5– or cyc3– mutants was similar to that of the pbs– mutant.
Transposition frequencies of the Ty1 mutants were quantified and compared with that of the WT element in the rad52– strain in which only integrase-dependent events are observed. Levels of cyc5– mutant and cyc3– mutant transposition were 5 and <1% that of WT Ty1, respectively (Table I). In contrast, the transposition level of the double cyc5–/cyc3– mutant was 33% higher than that of WT Ty1.
Table I. Transposition frequency of cyc and pbs mutants.
| Ty1 plasmid | Transposition frequency | Relative transposition frequency |
|---|---|---|
| WT | 3.2 (± 0.8) × 10–3 | 1.00 |
| Ct | <1.8 (± 0.4) × 10–5 | <0.01 |
| cyc5– | 1.5 (± 0.6) × 10–4 | 0.05 |
| cyc3– | <2.0 (± 0.2) × 10–5 | <0.01 |
| pbs– | 2.0 (± 2.2) × 10–5 | 0.01 |
| cyc5–/cyc3– | 4.3 (± 0.6) × 10–3 | 1.33 |
The transposition frequency was determined as the number of His+ cells/total number of cells. Transposition frequencies shown are the mean of three independent transformants and the standard deviation is given in parentheses. When no His+ cell was detected, we considered that it was less than one His+ cell. Relative transposition frequency is the ratio of the transposition frequency of mutant to that of WT Ty1.
These results strongly support the existence of a long-range interaction between CYC sequences in the native Ty1 genomic RNA and show that this interaction is required for Ty1 transposition.
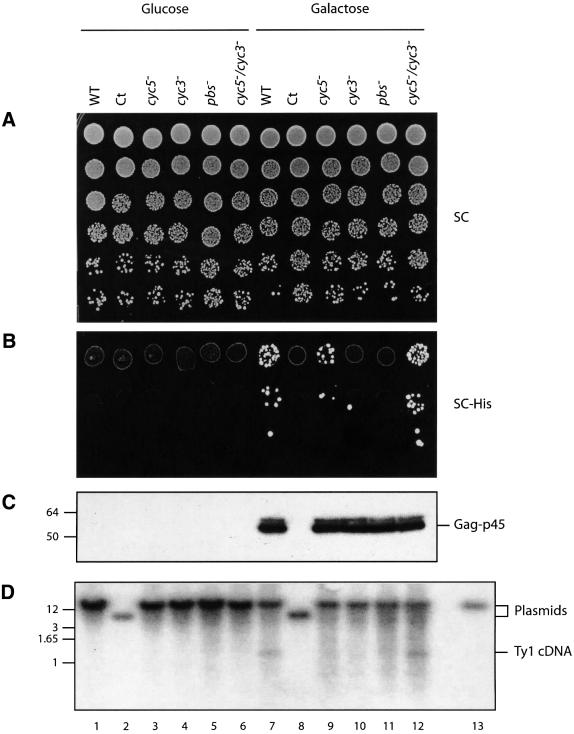
To further characterize the function of CYC pairing in vivo, we compared the transposition efficiency, and the steady-state levels of Ty1 Gag and cDNA in the rad52– strain at 22°C. To monitor transposition efficiency, serial dilutions of uninduced or induced cultures were plated on to non-selective medium (SC; Figure 9A) and selective medium (SC–His; Figure 9B). The expression of Ty1 Gag was strictly dependent on galactose induction (Figure 9C, compare lanes 1–6 with 7–12) and the steady-state level of Ty1 Gag was not influenced by cyc5–, cyc3–, pbs– or cyc5–/cyc3– mutations (Figure 9C, compare lanes 9–12 with 7). The silent mutations introduced into the Gag coding sequence (cyc5–, pbs– or cyc5–/cyc3–) had no effect on Gag expression. Ty1 cDNA synthesis was also strictly dependent on galactose induction (Figure 9D, compare lanes 1–6 with 7–12). However, Ty1 cDNA was no longer detectable in strains containing the Ty1 element with cyc5–, cyc3– or pbs– mutation, whereas similar levels of Ty1 cDNA were detected in WT and double mutant cyc5–/cyc3– Ty1 elements (Figure 9D, compare lanes 8–11 with 7 and 12). These observations show that CYC5–CYC3 pairing is required for Ty1 cDNA synthesis and transposition in vivo.
Fig. 9. Pairing of CYC5 and CYC3 sequences is required for Ty1 cDNA synthesis. (A and B) Transposition of WT or mutants of HIS3AI-marked Ty1 element in AGY49 cells at 22°C. Descending spots correspond to 5-fold dilutions. Transposition was assayed under repressing (lanes 1–6, glucose medium) and inducing (lanes 7–12, galactose medium) conditions by comparing growth on non-selective medium (A, SC) and on selective medium lacking histidine (B, SC–His). (C) Steady-state levels of Ty1 Gag protein as determined by immunoblotting of total protein extracts using anti-Gag antibodies. The same amount of total proteins was loaded in each lane. Molecular weights (left) are in kDa. (D) Steady-state levels of Ty1 cDNA as determined by Southern blot analysis of total DNA. The HIS3 probe hybridized to Ty1 cDNA (1.2 kbp) as well as to the pGTy1-H3mHIS3AI donor plasmid (14.5 kbp; all lanes except 2 and 7; lane 13 is a control with pGTy1-H3mHIS3AI alone). Molecular weights (left) are in kbp. Plasmid bands in lanes 2 and 7 are lower since they correspond to plasmid pGmHIS3AI (Ct) and the plasmid copy number increased when cells were grown in glucose rather than in galactose. Samples from the same cultures were used in experiments (A)–(D).
Functional interaction between the ends of Ty1 genomic RNA is supported by phylogenic conservation and covariation
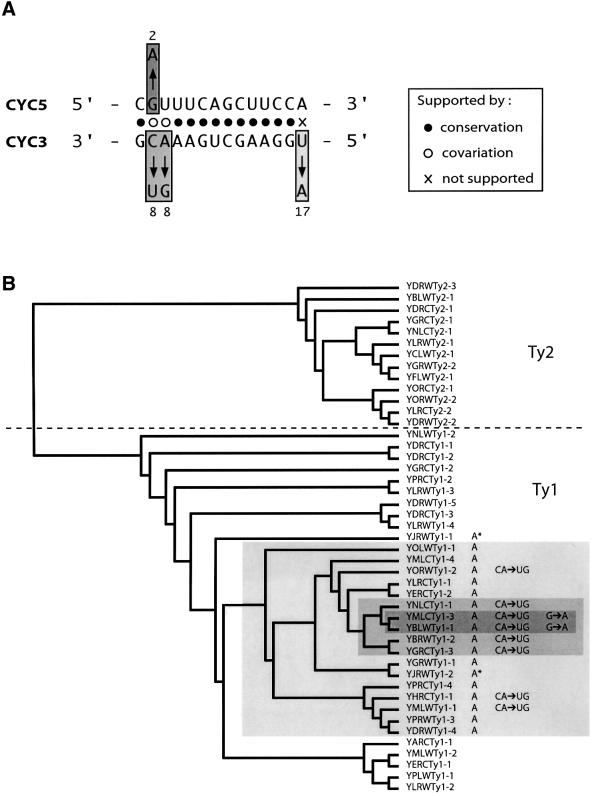
All full-length Ty1 and Ty2—a close relative of Ty1—elements present in the S.cerevisiae genome have been recovered (Kim et al., 1998). We aligned all full-length Ty1 (32 sequences) and Ty2 (13 sequences) elements and analysed conservation of the CYC5 and CYC3 sequences among them. CYC5 and CYC3 were found to be highly conserved in these Ty1/Ty2 elements, if the last base pair of the CYC interaction is excluded (Figure 10B). Indeed CYC5 sequences are strictly identical in 43 of 45 sequences. Only one of 13 positions (8%) in the CYC5 sequence alignment was affected by nucleotide variation, whereas 164 of 600 positions (27%) in the alignment of the first 600 nt of Ty1/Ty2 RNA showed at least one substitution. Similarly, CYC3 sequences were found to be strictly identical in 37 of 45 sequences. Only two of 13 positions (15%) in the CYC3 alignment were affected by nucleotide variation, whereas 94 of 371 positions (25%) in the alignment of the last 371 nt of Ty1/Ty2 RNA showed at least one substitution.
Fig. 10. Covariation of CYC sequences in all complete copies of Ty1 and Ty2 present in the S.cerevisiae genome. (A) Conservation and covariation of CYC sequences. Numbers indicate how many Ty1 elements contain the depicted changes. (B) Placement of CYC variants on an unrooted phylogenic tree based on the alignment of full Ty1–Ty2 sequences. Asterisks indicate mutations found in only one LTR, suggesting post-integration divergence. Note that CYC sequences are strictly conserved in all Ty2 elements, which are thought to form the most recent and active retrotransposon family in yeast (Kim et al., 1998).
Moreover, in cases of CYC sequence variation, base changes occur so as to maintain Watson–Crick or non-canonical G–U base pairs, often found in RNA secondary structures, without disturbing helix formation (Figure 10). Again the last base pair of the CYC sequences was not supported by covariation.
In conclusion, these analyses support the existence of an evolutionarily conserved 13 nt, long-range interaction bridging the 5′ and 3′ ends of Ty1 and Ty2 genomic RNA.
Discussion
To investigate possible differences of reverse transcription in retrotransposons and retroviruses, we have set up an in vitro model system for the yeast LTR-retrotransposon Ty1. Minus-strand DNA transfer is dependent on the R repeats and RT RNase H activity as shown for retroviruses (Peliska and Benkovic, 1992; Allain et al., 1994), which agrees with the analysis of newly made Ty1 cDNA in vivo (Uzun and Gabriel, 2001). However, in contrast to reverse transcription in retroviruses, the 3′ region of Ty1 genomic RNA is specifically involved in the initial stage of cDNA synthesis (Figures 2, 4 and 6). In fact, the 5′ and 3′ ends of Ty1 RNA were found to interact through complementary sequences called CYC5 and CYC3 (Figure 5). Mutations abrogating CYC pairing impaired efficient ss-cDNA synthesis, while compensatory mutations in CYC sequences restored efficient ss-cDNA synthesis in vitro (Figure 6). This base pairing is also required for Ty1 cDNA synthesis and transposition in vivo (Figures 8 and 9; Table I). These findings indicate that the 3′ region of Ty1 genomic RNA acts on reverse transcription initiation (Figure 7) through a long-range 5′–3′ CYC pairing, which is conserved in all complete Ty1 and Ty2 elements of the S.cerevisiae genome (Figure 10). Thus, CYC pairing most probably exists in native Ty1 genomic RNA and is of importance for Ty1 replication.
A structural model of the binary Ty1 5′ RNA–tRNA (iMet) complex has been proposed previously (Friant et al., 1996, 1998). In this model, the 3′ OH of tRNA(iMet) is embedded within a compact structure but how RT recognizes the primer 3′ end and extends it is poorly understood. Therefore, it is tempting to speculate that 5′–3′ pairing induces a switch between a closed Ty1 RNA– tRNA(iMet) structure, incompetent for initiation, to an open structure where the 3′ OH of tRNA(iMet) is accessible to RT and thus competent for reverse transcription initiation. This model could explain differences observed in vitro and in vivo between the cyc5– and the cyc3– mutant phenotypes. Additionally, in vitro ss-cDNA synthesis was increased 4- to 5-fold with the cyc5– mutant RNA alone compared with the 5′ WT RNA alone (Figure 6A, compare lanes 2 and 14, and B), indicating that the cyc5– mutation partially mimics the effect of the CYC sequences interaction in the absence of 3′ RNA. However, one cannot exclude that the 5′–3′ long-range interaction might be part of a more complex 3D RNA structure recognized by RT. Resolving the secondary structure of the 5′ RNA–3′ RNA–tRNA complex would be of great interest to discriminate between these two hypotheses and to determine whether other non-contiguous sequences are also involved in the formation or the stability of this ternary complex.
In yeast, retrotransposition is highly controlled since Ty1 RNA is very abundant (as much as 1% of total RNA in haploid cells) but the rate of transposition is only 10–7–10–5 per cell per generation. Post-translational maturation of VLPs by Ty1 protease is a key event that controls transposition (Curcio and Garfinkel, 1992). Maturation of Ty1 VLPs was also found to be associated with a conformational change of Ty1 dimeric RNA causing its stabilization (Feng et al., 2000). Based on these observations we propose a working model of Ty1 reverse transcription control. Dimeric RNA would first be encapsidated into immature Ty1 VLPs. Next, conversion of Gag-p49 precursor to its mature form Gag-p45 by Ty1 protease results in the formation of the 5′–3′ long-range CYC interactions by the Gag-p45 RNA chaperone. The change in RNA conformation then allows efficient priming of cDNA synthesis.
Interestingly, a putative interaction between the 5′ and 3′ ends of Ty1 and Ty2 was originally proposed in Warmington et al. (1985), giving rise to a circular RNA, which could facilitate minus-strand transfer. Our results do not exclude that the 5′–3′ long-range interaction might also play a role in other steps of the transposition process. However, the effect of cyc mutations on the early steps of reverse transcription has prevented us from looking for such additional roles.
The relationships between viral RNA folding and functions are hard to appreciate since viral genomes are often several kilobases long with a high level of genetic compaction that results in the overlapping of functions. The use of large RNAs in reconstituted in vitro systems has recently shed light on the important contribution of specific tertiary structures in biologically active RNA conformers, such as RNA circularization. For instance, in positive-strand RNA flaviviruses, an interaction between the 5′ and 3′ ends of the viral RNA is required for RNA synthesis in vitro and virus replication (You and Padmanabhan, 1999). Translational control of a viral RNA was found to occur by direct pairing between the 5′ and 3′ untranslated sequences resulting in the formation of a closed loop RNA (Guo et al., 2001). This loop mimics cap and poly(A) tail interaction usually mediated by the eukaryotic initiation factors 4E and 4G (eIF4E and eIF4G) and the poly(A) binding protein (PABP) (Wells et al., 1998), and allows uncapped, non-polyadenylated viral RNA to be translated and viral replication to occur (Guo et al., 2001).
We present here a remarkable long-distance base pairing between the CYC sequences resulting in the circularization of Ty1 genomic RNA (Figures 5, 8 and 10). This base pairing controls initiation of reverse transcription in vitro (Figures 2, 4, 6 and 7), and cDNA synthesis and transposition in vivo (Figures 8 and 9; Table I). In yeast Ty3 retrotransposon, tRNA(iMet) indirectly bridges the ends of the genomic RNA through a bipartite 5′–3′ PBS required for reverse transcription and transposition (Gabus et al., 1998).
In both cases, the Ty’s RNA chaperone proteins were found to promote direct or indirect bridging of Ty’s RNA ends and cDNA synthesis. This highlights the importance of RNA chaperone proteins in the folding of large RNAs, such as viral RNAs, into their active conformation(s) in physiological conditions. It suggests that these proteins regulate the functions of viral RNAs by means of trans-conformational processes (Cristofari and Darlix, 2002). This notion has recently been reinforced by the finding that the HIV-1 leader RNA exists in two alternative conformations functioning in either late or early phases of virus replication controlled by NCp7 (Berkhout et al., 2002).
The two active families of LTR-retrotransposons in S.cerevisiae, namely Ty1/Ty2 and Ty3, appear to require the 5′ and 3′ ends of their genomes to efficiently initiate reverse transcription. Furthermore, integration of reverse transcripts into the host genome is not strictly dependent on the integrity of cDNA extremities, but can occur by homologous recombination or gene conversion. Therefore aberrant cDNAs can potentially be incorporated into the host genome (Sharon et al., 1994; Nevo-Caspi and Kupiec, 1997). The present observations lead us to propose that circularization of Ty RNAs may have evolved to limit aberrant reverse transcription of defective elements in a cellular background where homologous recombination is highly efficient as in S.cerevisiae.
Materials and methods
Plasmid DNA
pGTy1-H3mHIS3AI and pGmHIS3AI were obtained from D.J.Garfinkel (Curcio and Garfinkel, 1991). Plasmid DNAs were constructed as described in Supplementary data.
RNAs
Recombinant RNAs were prepared by in vitro transcription using T7 RNA polymerase and plasmid DNA template digested by HindIII as reported in Cristofari et al. (2000). Yeast tRNA(iMet) was purified from S.cerevisiae and kindly provided by G.Keith (IBMC, Strasbourg, France). RNA concentration was determined by UV spectrometry (A260 nm).
Yeast tRNA(iMet) was dephosphorylated by CIP (Promega), 5′-labelled with [γ-32P]ATP by T4 kinase (Gibco) and gel purified. After purification, it was heated at 90°C and progressively cooled down at room temperature in 5 mM Tris–HCl pH 7 and 1 mM MgCl2 to allow proper folding.
Protein expression and purification
Highly pure Ty1 TYA1-D peptide was synthesized and purified as reported in Cristofari et al. (2000). The sequence of the TYA1-D peptide derives from the Ty1-H3 clone (GenBank accession No. M18706).
Recombinant WT and D468S RNase H(–) Ty1 RTs were produced in Escherichia coli and purified as described in Wilhelm et al. (2000, 2001).
Yeast strains
Yeast strain YH50 (MATα ho spt3-202 ura3-167 trp1Δ1 leu2-3 his3Δ200) and the isogenic rad52::LEU2 strain AGY49 were kindly provided by A.Gabriel (Teng et al., 1996). All strains are spt3, which eliminates endogenous Ty1 transcription and thus potential trans-complementation of plasmid-borne elements.
Formation of Ty1 nucleoprotein complexes
Ty1 5′ RNA (0.25 pmol), Ty1 3′ RNA (0.5 pmol), tRNA(iMet) (0.5 pmol) and TYA1-D, at the protein to nucleotide molar ratios indicated, were incubated for 10 min at 25°C in 10 µl of 20 mM Tris–HCl pH 7.5, 30 mM NaCl, 0.2 mM MgCl2, 5 mM dithiothreitol, 0.01 mM ZnCl2 and 5 U of RNasin (Promega). Nucleoprotein complexes were used for reverse transcription (see below) or reactions were stopped by 1% SDS and treated with proteinase K (2 µg) for 20 min at 25°C, and phenol–chloroform extracted. RNAs were separated in 1.3% agarose gel and visualized by ethidium bromide (EtBr) staining after migration to avoid structural changes induced by EtBr.
Reverse transcription
Once Ty1 nucleoprotein complexes were formed (see above), the reaction volume was increased to 25 µl by adding an excess (10 pmol) of Ty1 RT, dNTPs at 0.25 mM each, 30 mM NaCl and 3 mM MgCl2. Incubation was for 30 min at 25°C, after which the reaction was stopped by adding 0.5% SDS and 5 mM EDTA, and heating at 65°C for 3 min. Nucleic acids were purified by phenol–chloroform extraction and ethanol precipitated. Pellets were dissolved directly in formamide, denatured at 95°C for 1 min and analysed by 6% PAGE in 7 M urea.
Sequence alignments
All Ty1 and Ty2 sequences of the S.cerevisiae genome were retrieved from the GenBank database (NCBI) or Saccharomyces Genome Database (SDG) using coordinates described in Kim et al. (1998), and aligned with the ClustalX software (Jeanmougin et al., 1998). Local alignments to find potential interacting sequences were performed online using the Infobiogen LALIGN (Huang and Miller, 1991) server (http://www. infobiogen.fr) and were manually refined to take into account G–U base pairing.
Transposition assay
Saccharomyces cerevisiae strains containing Ty1 plasmids were first grown as patches at 30°C on SC–Ura+Glc plates for 2 days. Cells were replica plated on to SC–Ura+Gal to induce transposition or on to SC–Ura+Glc as a negative control. Induction was maintained for 4 days at 22°C or 2 days at 30°C. Cells were then replica plated on to SC–His+Glc and incubated at 30°C for 3 days to detect cells where Ty1 transposition has taken place. Experiments were repeated at least three times from three independent transformations. For quantitative assays, AGY49 cell patches from SC–Ura+Gal or SC–Ura+Glc plates grown at 22°C were scraped into 10 ml of water (dilution 1, OD 600 nm ∼0.25). Then 100 µl of dilution 1 were diluted in 10 ml of water (dilution 2). Fifty microlitres of dilution 1 were plated on selective SC–His+Glc to enumerate His+ revertants and 50 µl of dilution 2 were plated on YPD to enumerate total cells. The transposition efficiency was determined as the ratio of His+ prototrophs to the total number of cells. Quantifications were based on three independent transformations for each Ty1 construct.
Immunoblot and Southern blot analysis
Several independent AGY49 transformants were grown at 30°C in SC–Ura+Raf for 2 days (OD 600 nm ∼3). Cultures were diluted in 10 ml of SC–Ura+Glc or SC–Ura+Gal to an OD 600 nm of ∼0.5 and were grown for 36 h at 22°C. To determine qualitatively the transposition efficiency in each of these cultures, 5-fold serial dilutions were spotted as 10 µl drops on SC+Glc and on SC–His+Glc.
One millilitre of each culture was used to isolate total proteins by the post-alkaline extraction method (Kushnirov, 2000). One-tenth of each sample was resolved by 10% SDS–PAGE, visualized by Coomassie Blue staining and normalized by scanning the gel on a Fluor-S apparatus (Bio-Rad). The same amount of proteins (∼1/10 of each sample) was resolved by 10% SDS–PAGE, transferred on to PVDF membranes, and immunoblots were performed as described (Ausubel, 1995). A rabbit polyclonal antiserum raised against (His)6-tagged, recombinant TYA1-D [1/1000 dilution, serum PAS-3791-12 (Sigma; G.Cristofari and J.-L.Darlix, unpublished results)] was used as the primary antibody and a swine anti-rabbit polyclonal antiserum coupled to horseradish peroxidase (1/1000, Dako) was used as the secondary antibody. Westpico reagent (Pierce) was used for development.
The remaining cells were pelleted and total DNA was extracted by the glass-bead method (Ausubel, 1995). DNA was quantified by fluorometry (Hoefer DyNA Quant 200). Five micrograms of DNA was digested with NheI, RNase A treated and separated on a 0.7% agarose gel. DNA was transferred to a Hybond-N+ membrane with high-salt buffer and blotted as described (Ausubel, 1995). The blot was probed with a 32P-labelled HIS3 cDNA probe obtained by random priming and labelling (Invitrogen).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank A.Gabriel for providing strains and informations on RNase H mutant before publication and for critical reading of the manuscript; D.J.Garfinkel for strains, plasmids pGTy1-H3mHIS3AI and pGmHIS3AI and helpful discussions; G.Keith for the generous gift of yeast natural tRNA(iMet) and C.Daudé for excellent technical assistance. We are also grateful to P.A.Defossez, G.Fourel, E.Lebrun and A.Roborel de Climens for their advice on and help with yeast experiments. This work was supported by INSERM, ANRS, the French Ministry of Research and ARC (grant 9589 to F.-X.W.).
References
- Adams S.E., Mellor,J., Gull,K., Sim,R.B., Tuite,M.F., Kingsman,S.M. and Kingsman,A.J. (1987) The functions and relationships of Ty-VLP proteins in yeast reflect those of mammalian retroviral proteins. Cell, 49, 111–119. [DOI] [PubMed] [Google Scholar]
- Allain B., Lapadat-Tapolsky,M., Berlioz,C. and Darlix,J.L. (1994) Transactivation of the minus-strand DNA transfer by nucleocapsid protein during reverse transcription of the retroviral genome. EMBO J., 13, 973–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F.M. (1995) Short Protocols in Molecular Biology: A Compendium of Methods from Current Protocols in Molecular Biology. Wiley, New York, NY.
- Berkhout B., Ooms,M., Beerens,N., Huthoff,H., Southern,E. and Verhoef,K. (2002) In vitro evidence that the untranslated leader of the HIV-1 genome is an RNA checkpoint that regulates multiple functions through conformational changes. J. Biol. Chem., 277, 19967–19975. [DOI] [PubMed] [Google Scholar]
- Boeke J.D. and Stoye,J.P. (1997) Retrotransposons, endogenous retroviruses and the evolution of retroelements. In Cofin,J.M., Hughes,S.H. and Varmus,H.E. (eds), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 343–435. [PubMed]
- Boeke J.D., Styles,C.A. and Fink,G.R. (1986) Saccharomyces cerevisiae SPT3 gene is required for transposition and transpositional recombination of chromosomal Ty elements. Mol. Cell. Biol., 6, 3575–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman K.B., Bystrom,A.S. and Boeke,J.D. (1992) Initiator methionine tRNA is essential for Ty1 transposition in yeast. Proc. Natl Acad. Sci. USA, 89, 3236–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofari G. and Darlix,J.L. (2002) The ubiquitous nature of RNA chaperone proteins. Prog. Nucleic Acid Res. Mol. Biol., 72, 223–268. [DOI] [PubMed] [Google Scholar]
- Cristofari G., Ficheux,D. and Darlix,J.L. (2000) The GAG-like protein of the yeast Ty1 retrotransposon contains a nucleic acid chaperone domain analogous to retroviral nucleocapsid proteins. J. Biol. Chem., 275, 19210–19217. [DOI] [PubMed] [Google Scholar]
- Curcio M.J. and Garfinkel,D.J. (1991) Single-step selection for Ty1 element retrotransposition. Proc. Natl Acad. Sci. USA, 88, 936–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio M.J. and Garfinkel,D.J. (1992) Posttranslational control of Ty1 retrotransposition occurs at the level of protein processing. Mol. Cell. Biol., 12, 2813–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlix J.L., Cristofari,G., Rau,M., Pechoux,C., Berthoux,L. and Roques,B. (2000) Nucleocapsid protein of human immunodeficiency virus as a model protein with chaperoning functions and as a target for antiviral drugs. Adv. Pharmacol., 48, 345–372. [DOI] [PubMed] [Google Scholar]
- Derr L.K. and Strathern,J.N. (1993) A role for reverse transcripts in gene conversion. Nature, 361, 170–173. [DOI] [PubMed] [Google Scholar]
- Dudding L.R. and Mizrahi,V. (1993) Rapid kinetic analysis of a point mutant of HIV-1 reverse transcriptase lacking ribonuclease H activity. Biochemistry, 32, 6116–6120. [DOI] [PubMed] [Google Scholar]
- Eichinger D.J. and Boeke,J.D. (1988) The DNA intermediate in yeast Ty1 element transposition copurifies with virus-like particles: cell-free Ty1 transposition. Cell, 54, 955–966. [DOI] [PubMed] [Google Scholar]
- Elrouby N. and Bureau,T.E. (2001) A novel hybrid open reading frame formed by multiple cellular gene transductions by a plant long terminal repeat retroelement. J. Biol. Chem., 276, 41963–41968. [DOI] [PubMed] [Google Scholar]
- Esnault C., Maestre,J. and Heidmann,T. (2000) Human LINE retrotransposons generate processed pseudogenes. Nat. Genet., 24, 363–367. [DOI] [PubMed] [Google Scholar]
- Feng Y.X., Moore,S.P., Garfinkel,D.J. and Rein,A. (2000) The genomic RNA in Ty1 virus-like particles is dimeric. J. Virol., 74, 10819–10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink G.R. (1987) Pseudogenes in yeast? Cell, 49, 5–6. [DOI] [PubMed] [Google Scholar]
- Flavell A.J., Pearce,S.R. and Kumar,A. (1994) Plant transposable elements and the genome. Curr. Opin. Genet. Dev., 4, 838–844. [DOI] [PubMed] [Google Scholar]
- Friant S., Heyman,T., Wilhelm,M.L. and Wilhelm,F.X. (1996) Extended interactions between the primer tRNAi(Met) and genomic RNA of the yeast Ty1 retrotransposon. Nucleic Acids Res., 24, 441–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friant S., Heyman,T., Bystrom,A.S., Wilhelm,M. and Wilhelm,F.X. (1998) Interactions between Ty1 retrotransposon RNA and the T and D regions of the tRNA(iMet) primer are required for initiation of reverse transcription in vivo. Mol. Cell. Biol., 18, 799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabus C., Ficheux,D., Rau,M., Keith,G., Sandmeyer,S. and Darlix,J.L. (1998) The yeast Ty3 retrotransposon contains a 5′–3′ bipartite primer-binding site and encodes nucleocapsid protein NCp9 functionally homologous to HIV-1 NCp7. EMBO J., 17, 4873–4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfinkel D.J., Boeke,J.D. and Fink,G.R. (1985) Ty element transposition: reverse transcriptase and virus-like particles. Cell, 42, 507–517. [DOI] [PubMed] [Google Scholar]
- Guo J., Henderson,L.E., Bess,J., Kane,B. and Levin,J.G. (1997) Human immunodeficiency virus type 1 nucleocapsid protein promotes efficient strand transfer and specific viral DNA synthesis by inhibiting TAR-dependent self-priming from minus-strand strong-stop DNA. J. Virol., 71, 5178–5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Allen,E.M. and Miller,W.A. (2001) Base-pairing between untranslated regions facilitates translation of uncapped, nonpolyadenylated viral RNA. Mol. Cell, 7, 1103–1109. [DOI] [PubMed] [Google Scholar]
- Huang X.Q. and Miller,W. (1991) A time-efficient, linear-space local similarity algorithm. Adv. Appl. Math., 12, 337–357. [Google Scholar]
- Jeanmougin F., Thompson,J.D., Gouy,M., Higgins,D.G. and Gibson,T.J. (1998) Multiple sequence alignment with Clustal X. Trends Biochem. Sci., 23, 403–405. [DOI] [PubMed] [Google Scholar]
- Keeney J.B., Chapman,K.B., Lauermann,V., Voytas,D.F., Astrom,S.U., von Pawel-Rammingen,U., Bystrom,A. and Boeke,J.D. (1995) Multiple molecular determinants for retrotransposition in a primer tRNA. Mol. Cell. Biol., 15, 217–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.M., Vanguri,S., Boeke,J.D., Gabriel,A. and Voytas,D.F. (1998) Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res., 8, 464–478. [DOI] [PubMed] [Google Scholar]
- Kushnirov V.V. (2000) Rapid and reliable protein extraction from yeast. Yeast, 16, 857–860. [DOI] [PubMed] [Google Scholar]
- Lapadat-Tapolsky M., Gabus,C., Rau,M. and Darlix,J.L. (1997) Possible roles of HIV-1 nucleocapsid protein in the specificity of proviral DNA synthesis and in its variability. J. Mol. Biol., 268, 250–260. [DOI] [PubMed] [Google Scholar]
- Levin H.L. (1995) A novel mechanism of self-primed reverse transcription defines a new family of retroelements. Mol. Cell. Biol., 15, 3310–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin H.L. (1996) An unusual mechanism of self-primed reverse transcription requires the RNase H domain of reverse transcriptase to cleave an RNA duplex. Mol. Cell. Biol., 16, 5645–5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor J., Malim,M.H., Gull,K., Tuite,M.F., McCready,S., Dibbayawan,T., Kingsman,S.M. and Kingsman,A.J. (1985) Reverse transcriptase activity and Ty RNA are associated with virus-like particles in yeast. Nature, 318, 583–586. [DOI] [PubMed] [Google Scholar]
- Merkulov G.V., Swiderek,K.M., Brachmann,C.B. and Boeke,J.D. (1996) A critical proteolytic cleavage site near the C terminus of the yeast retrotransposon Ty1 Gag protein. J. Virol., 70, 5548–5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran J.V., DeBerardinis,R.J. and Kazazian,H.H.,Jr (1999) Exon shuffling by L1 retrotransposition. Science, 283, 1530–1534. [DOI] [PubMed] [Google Scholar]
- Nevo-Caspi Y. and Kupiec,M. (1997) cDNA-mediated Ty recombin ation can take place in the absence of plus-strand cDNA synthesis, but not in the absence of the integrase protein. Curr. Genet., 32, 32–40. [DOI] [PubMed] [Google Scholar]
- Peliska J.A. and Benkovic,S.J. (1992) Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science, 258, 1112–1118. [DOI] [PubMed] [Google Scholar]
- Sharon G., Burkett,T.J. and Garfinkel,D.J. (1994) Efficient homologous recombination of Ty1 element cDNA when integration is blocked. Mol. Cell. Biol., 14, 6540–6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S.C., Kim,B. and Gabriel,A. (1996) Retrotransposon reverse-transcriptase-mediated repair of chromosomal breaks. Nature, 383, 641–644. [DOI] [PubMed] [Google Scholar]
- Uzun O. and Gabriel,A. (2001) A Ty1 reverse transcriptase active-site aspartate mutation blocks transposition but not polymerization. J. Virol., 75, 6337–6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmington J.R., Waring,R.B., Newlon,C.S., Indge,K.J. and Oliver,S.G. (1985) Nucleotide sequence characterization of Ty1-17, a class II transposon from yeast. Nucleic Acids Res., 13, 6679–6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells S.E., Hillner,P.E., Vale,R.D. and Sachs,A.B. (1998) Circular ization of mRNA by eukaryotic translation initiation factors. Mol. Cell, 2, 135–140. [DOI] [PubMed] [Google Scholar]
- Wilhelm M., Boutabout,M. and Wilhelm,F.X. (2000) Expression of an active form of recombinant Ty1 reverse transcriptase in Escherichia coli: a fusion protein containing the C-terminal region of the Ty1 integrase linked to the reverse transcriptase-RNase H domain exhibits polymerase and RNase H activities. Biochem. J., 348, 337–342. [PMC free article] [PubMed] [Google Scholar]
- Wilhelm M., Uzun,O., Mules,E.H., Gabriel,A. and Wilhelm,F.X. (2001) Polypurine tract formation by Ty1 RNase H. J. Biol. Chem., 276, 47695–47701. [DOI] [PubMed] [Google Scholar]
- Xu H. and Boeke,J.D. (1990) Localization of sequences required in cis for yeast Ty1 element transposition near the long terminal repeats: analysis of mini-Ty1 elements. Mol. Cell. Biol., 10, 2695–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You S. and Padmanabhan,R. (1999) A novel in vitro replication system for Dengue virus. Initiation of RNA synthesis at the 3′-end of exogenous viral RNA templates requires 5′- and 3′-terminal complementary sequence motifs of the viral RNA. J. Biol. Chem., 274, 33714–33722. [DOI] [PubMed] [Google Scholar]
- Yu X. and Gabriel,A. (1999) Patching broken chromosomes with extranuclear cellular DNA. Mol. Cell, 4, 873–881. [DOI] [PubMed] [Google Scholar]