Abstract
3-phosphoinositide dependent protein kinase-1 (PDK1) plays a key role in regulating signalling pathways by activating AGC kinases such as PKB/Akt and S6K. Here we describe the 2.0 Å crystal structure of the PDK1 kinase domain in complex with ATP. The structure defines the hydrophobic pocket termed the ‘PIF-pocket’, which plays a key role in mediating the interaction and phosphorylation of certain substrates such as S6K1. Phosphorylation of S6K1 at its C-terminal PIF-pocket-interacting motif promotes the binding of S6K1 with PDK1. In the PDK1 structure, this pocket is occupied by a crystallographic contact with another molecule of PDK1. Interestingly, close to the PIF-pocket in PDK1, there is an ordered sulfate ion, interacting tightly with four surrounding side chains. The roles of these residues were investigated through a combination of site-directed mutagenesis and kinetic studies, the results of which confirm that this region of PDK1 represents a phosphate-dependent docking site. We discuss the possibility that an analogous phosphate-binding regulatory motif may participate in the activation of other AGC kinases. Furthermore, the structure of PDK1 provides a scaffold for the design of specific PDK1 inhibitors.
Keywords: AGC protein kinases/crystal structure/PDK1/PDK1-interacting fragment/phosphatidylinositol 3,4,5-trisphosphate
Introduction
3-phosphoinositide dependent protein kinase-1 (PDK1) is a key protein kinase, regulating the activity of a group of related protein kinases through phosphorylation. These kinases include isoforms of protein kinase B (PKB; also known as Akt; Brazil and Hemmings, 2001; Scheid and Woodgett, 2001), p70 ribosomal S6 kinase (S6K; Volarevic and Thomas, 2001), p90 ribosomal S6 kinase (RSK; Frodin and Gammeltoft, 1999) and the serum- and glucocorticoid-induced protein kinase (SGK; Lang and Cohen, 2001). These enzymes are stimulated by hormones and growth factors, and phosphorylate regulatory proteins mediating the various physiological effects of these agonists.
PDK1 possesses an N-terminal kinase catalytic domain and a C-terminal pleckstrin homology (PH) domain (Alessi et al., 1997; Stephens et al., 1998). PDK1 activates its substrates by phosphorylating these kinases at their activation loop (reviewed in Toker and Newton, 2000; Alessi, 2001). The phosphorylation of PKB by PDK1 is dependent upon prior activation of the phosphoinositide 3-kinase (PI-3-kinase) and the production of the se cond messenger, phosphatidylinositol 3,4,5-trisphosphate [PtdIns(3,4,5)P3], which binds to the PH domains of PDK1 and PKB. PtdIns(3,4,5)P3 does not activate either PKB or PDK1, but instead recruits and co-localizes these enzymes at the plasma membrane, allowing PDK1 to activate PKB. Unlike PKB, the other PDK1 substrates described thus far do not interact with PtdIns(3,4,5)P3 nor is the rate at which they are phosphorylated by PDK1 further enhanced by the binding of PDK1 to PtdIns(3,4,5)P3. Instead the ability of PDK1 to phosphorylate S6K, SGK and RSK is promoted by phosphorylation of these enzymes at a residue located C-terminal to the kinase catalytic domain in a region known as the hydrophobic motif (Alessi et al., 1997; Pullen et al., 1998; Kobayashi and Cohen, 1999). The kinases that phosphorylate the hydrophobic motif of S6K and SGK are unknown, but as the phosphorylation of this residue in vivo is dependent on PI-3-kinase activation, the hydrophobic motif kinases and/or the hydrophobic motif phosphatases may be regulated by PtdIns(3,4,5)P3. In the case of RSK isoforms, phosphorylation by the ERK1/ERK2 MAP kinases induces the phosphorylation of the hydrophobic motif (reviewed in Frodin and Gammeltoft, 1999).
PDK1 belongs to the same subfamily of protein kinases as its substrates, termed the AGC protein kinases, related to the cAMP dependent protein kinase (PKA), cGMP dependent protein kinase and protein kinase C (PKC). PKA is the only AGC kinase whose crystal structure has been solved (Taylor et al., 1992). Like all protein kinases, its catalytic core possesses an N-terminal lobe consisting mainly of β-sheet and a predominantly α-helical C- terminal lobe (Taylor et al., 1992; Husen and Kuriyan, 2002). The ATP-binding site is located in between the two lobes (Knighton et al., 1991; Johnson et al., 2001). At the very end of the C-terminus, PKA possesses an extended loop that terminates in the sequence FXXF, which resembles the first part of the hydrophobic motif phosphorylation site of S6K and SGK (FXXFS/TY) in which the Ser/Thr is the phosphorylated residue (Biondi et al., 2000). In the structure of PKA, the FXXF motif is buried in a hydrophobic pocket in the small lobe of the PKA catalytic domain (Knighton et al., 1991) and mutation of either Phe residue drastically reduces PKA activity towards a peptide substrate (Etchebehere et al., 1997; Batkin et al., 2000). Unlike other AGC kinases, PDK1 does not possess a hydrophobic motif C-terminal to its catalytic domain. However, there is evidence that PDK1 possesses a hydrophobic pocket in the small lobe of its catalytic domain similar to that in PKA. We have demonstrated biochemically that the interaction of PDK1 with four of its substrates [S6K1, SGK1, PKCζ and PKC related kinase-2 (PRK2)] is reduced or abolished by mutation of residues predicted to form part of this pocket (Balendran et al., 2000; Biondi et al., 2000). Furthermore, mutation of a central residue in the predicted pocket, Leu155, prevented PDK1 from phosphorylating and activating S6K1 and SGK1 without affecting its ability to phosphorylate either PKB or a short peptide substrate that encompasses the activation loop of PKB (T308tide; Biondi et al., 2000). The hydrophobic pocket on the kinase domain of PDK1 has been termed the ‘PIF-pocket’ after the name of the first AGC-kinase hydrophobic motif-containing peptide (PDK1-interacting fragment) that was found to bind PDK1 (Balendran et al., 1999a). It has been suggested that the PIF-pocket in PDK1 functions as a docking site, enabling PDK1 to interact with some of its physiological substrates. Furthermore, there is evidence that phosphorylation of the hydrophobic motif of S6K1, SGK and RSK2 (Balendran et al., 1999b; Frodin et al., 2000; Biondi et al., 2001) promotes the interaction of these enzymes with PDK1. These findings suggest that the PIF-pocket on PDK1 could contain a phosphate-binding site promoting the binding of PDK1 to a subset of substrates (S6K, SGK and RSK) once these enzymes have been phosphorylated at their hydrophobic motif. This would result in a physiological phosphate-dependent interaction. In addition there is evidence that occupancy of the PIF-pocket activates PDK1, because peptides that encompass the hydrophobic motif of PRK2 (Biondi et al., 2000) and RSK (Frodin et al., 2000) induce a 4- to 6-fold activation of PDK1.
Here, we describe the first crystal structure of the kinase domain of PDK1 at 2 Å resolution. The structure defines the PIF-pocket and reveals an adjacent phosphate-binding site. Furthermore, we performed structure-based mutagenesis and biochemical analysis which support the existence of such a phosphate-binding site, which could mediate the phosphate-dependent docking interaction with substrates such as S6K and SGK. Finally, we have used a novel algorithm to define the conformational state of PDK1 relative to all the reported structures of PKA, which shows that while PDK1 has all the signs of being in an active form in the crystal, its overall conformation is in between an ‘open’ and ‘closed’ state.
Results and discussion
Overall structure
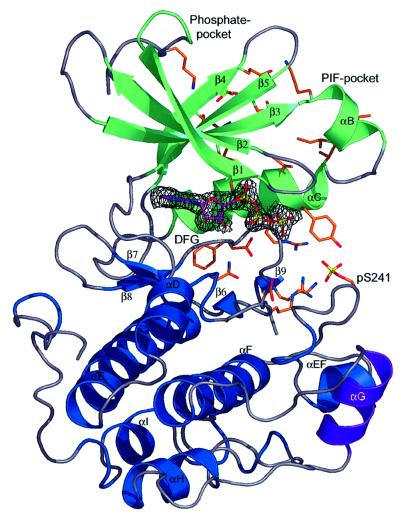
The structure of the catalytic domain of PDK1 was solved by molecular replacement and refined to an R-factor of 0.19 (Rfree = 0.22). PDK1 assumes the classic bilobal kinase fold (Figure 1) and is similar to the only other AGC kinase structure solved, that of PKA [root mean square deviation (r.m.s.d.) of 1.0 Å on Cα atoms with Protein Data Bank (PDB) entry 1STC (Prade et al., 1997)]. The form of PDK1 that was crystallized comprised residues 51–359. The tip of the activation loop (residues 233–236) is disordered, as observed in other kinase structures (Johnson et al., 1996). The N-terminus (residues 51–70), which is pointing into a large void generated by the crystallographic symmetry, is also disordered. In contrast, the N-terminal extension to the kinase domain of PKA assumes an amphipathic α-helix (termed αA-helix), and packs against the kinase core (Knighton et al., 1991). The cluster of hydrophobic residues that mediates this interaction in PKA is not present in PDK1, suggesting that the N-terminus of PDK1 could have a different function from that of PKA. Interestingly, it has recently been shown that the N-terminus of PDK1 (residues 1–50) interacts with Ral guanine nucleotide exchange factors (Tian et al., 2002). Thus, this region may assume a unique conformation in PDK1, which is not defined by the structure described here.
Fig. 1. Overview of the PDK1 structure.The PDK1 kinase domain backbone is shown in a ribbon representation, with the secondary structure elements for residues 74–163 coloured green, and for residues 164–358 coloured blue. Helix αG, encompassing residues 287–295 (which makes a crystal contact to a symmetry related PDK1 molecule, Figure 2), is coloured purple. Key residues discussed in the text are shown as a stick model with carbons coloured orange. ATP is shown as a stick model with carbon atoms coloured purple. A simulated annealing omit map for the ATP molecule is shown in black, contoured at 3σ. The phosphoserine (pS241) and the sulfate discussed in the text are also shown.
PDK1 was crystallized in the presence of ATP, but in the absence of any divalent cations. In the early stages of the refinement, well-defined density for the entire ATP molecule could be observed. ATP adopts a different conformation from that observed in other kinase–ATP complexes (Figure 1). Perhaps due to the absence of divalent cations, the generally observed kink between the β and γ phosphate, caused by the interaction with such an ion, is not seen in the PDK1 structure.
It is known that PDK1 can phosphorylate itself on residue Ser241 in the activation loop and that this phosphorylation is required for PDK1 activity (Casamayor et al., 1999). Indeed, we observed density for a phosphate attached to this residue (Figure 1), and extensive interactions are observed between this phosphoserine and residues from the C-terminal lobe and αC-helix (Figure 1). The interaction between Ser241 and the C-terminal lobe is similar to the equivalent interactions in PKA, but as discussed below the binding to the αC-helix differs.
The PIF-pocket
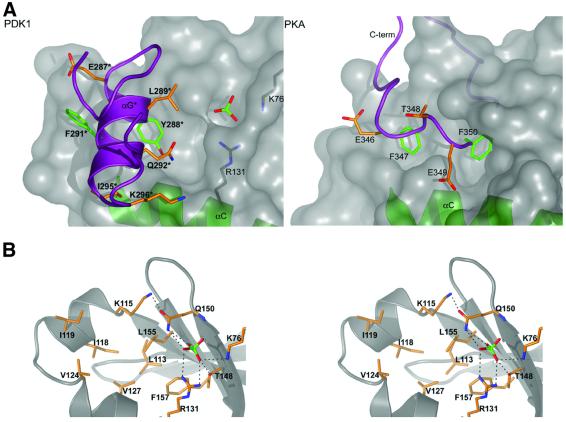
As outlined in the Introduction, PDK1 was postulated to possess a pocket (the ‘PIF-pocket’) in the small lobe of its catalytic domain, required for the binding of PDK1 to the hydrophobic motif of its substrates (Biondi et al., 2000). The PDK1 structure described here indeed reveals such a pocket, and shows that it lies in a location similar to the FXXF-binding pocket in PKA (Figure 2). PDK1 residues Lys115, Ile118, Ile119 on the αB-helix (Figure 2), Val124, Val127 on the αC-helix and Leu155 on β-sheet 5 form an ∼5 Å deep pocket. Previous work has shown that mutation of Leu155 to Glu abolishes the ability of PDK1 to interact with a peptide that encompasses the hydrophobic motif of PRK2 (PIFtide; Biondi et al., 2000) as well as with S6K1, SGK1, PKCζ and PRK2 (Balendran et al., 2000; Biondi et al., 2000). In addition, mutation of Lys115, Ile119, Glu150 and Leu155 to Ala, reduced the affinity of PDK1 for PIFtide ∼10-fold, but did not significantly diminish the ability of PDK1 to phosphorylate and activate S6K1 and SGK1 (Biondi et al., 2001). These results are in agreement with the crystal structure of the PIF-pocket, since Leu155 is located at the centre and the other residues line the wall of the pocket (Figure 2). Interestingly, in our structure, the PIF-pocket is occupied by helix αG of a symmetry-related molecule (Figure 2). Tyr288 and Phe291 make hydrophobic contacts in this pocket with almost all pocket-lining residues, remarkably reminiscent of the interactions of the phenylalanines in the FXXF motif in PKA and their hydrophobic docking site in the equivalent region of the kinase domain (Figure 2). In addition, residues Glu287, Gln292, Ile295 and Lys296 on the symmetry related loop also form contacts with residues lining the PIF-pocket. In total, 244 Å2 of accessible surface are buried by this contact. The significance of this interaction is not clear as an oligomerization event for PDK1 has not been demonstrated in solution previously. Indeed, both the isolated catalytic domain of PDK1, which was crystallized, and full-length PDK1 migrate in gel filtration chromatography as apparent monomeric species (data not shown).
Fig. 2. The PIF-pocket. (A) A surface representation of the putative PIF-binding pocket is shown and compared with the pocket interacting with the C-terminal FXXF motif in PKA. For PDK1, the αG-helix of a symmetry-related molecule is shown as a purple coil, in PKA the C-terminus is shown as a purple coil. Aromatic amino acids buried in the pocket are shown as sticks with green carbons, further side chains interacting with the pocket are shown with orange carbons. Helix αC is shown as a green ribbon in both PDK1 and PKA. In PDK1, the ordered sulfate ion and basic residues interacting with it are also shown. The asterisk indicates residues from the symmetry related molecule. (B) A stereo image of the residues lining the PIF/ phosphate-pockets is shown. The PDK1 backbone is shown as a grey ribbon. Side chains are shown with carbon atoms coloured orange. Hydrogen bonds to the sulfate ion are shown as black dotted lines.
The phosphate pocket
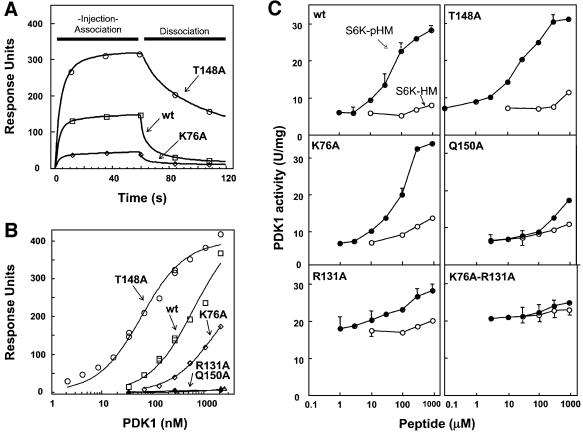
As outlined in the Introduction, substrates of PDK1, such as S6K1, interact with the PIF-pocket of PDK1 with higher affinity when they are phosphorylated at their hydrophobic motif. This suggested that a regulatory phosphate-binding site may be located close to the PIF-pocket. During refinement of the PDK1 structure, it became clear that next to the PIF-pocket another small pocket was present, occupied by a tetrahedral oxy-anion (Figure 2). As 2.0 M sulfate was present in the crystallization conditions, this was assigned as a sulfate ion. The ion interacts with four residues lining the pocket, namely Lys76, Arg131, Thr148 and Gln150. Because of its close proximity to the PIF-pocket (∼5 Å) it is possible that this sulfate-occupied pocket could represent the binding site for the phosphate on the phosphopeptide. To investigate this further, we mutated Lys76, Arg131, Thr148 and Gln150 to Ala, in order to verify the contribution of each of these residues in enabling PDK1 to interact with a peptide encompassing the hydrophobic motif of S6K1, in which the residue equivalent to Thr412 was phosphorylated (termed S6K-pHM). A quantitative surface plasmon resonance based binding assay (Figure 4A) showed that wild type PDK1 interacted with S6K-pHM, with a Kd of 0.6 µM, but not detectably with the non-phosphorylated form of this peptide (S6K-HM). The PDK1[R131A] and PDK1[Q150A] mutants did not interact detectably with S6K-pHM in this assay (Figure 4B), confirming that the interactions these residues make in the PDK1 structure are of key importance. The PDK1[K76A] mutant interacted with 3-fold lower affinity (Kd = 1.7 µM) with S6K-pHM. The PDK1[T148A] mutant, however, possessed ∼10-fold higher (Kd = 0.06 µM) affinity for S6K-pHM than wild type PDK1. Moreover, the dissociation of PDK1[T148A] from S6K-pHM is markedly slower than that of wild type PDK1 or PDK1[K76A] (Figure 4A). These findings are unexpected as Thr148 is within hydrogen bonding distance of the sulfate ion (Figure 2), but indicate that this residue may play a role in enabling the dissociation of PDK1 from S6K-pHM.
Fig. 4. PDK1 binding and activation studies. Binding and activation of wild type and mutant forms of PDK1 to a phosphopeptide derived from the hydrophobic motif of S6K1. The binding of the wild type (wt) PDK1 and indicated mutants to a phosphopeptide comprising the hydrophobic motif of S6K1 (S6K-pHM: SESANQVFLGFT*YVAPSV, where T* indicates phospho-threonine) was analysed by surface plasmon resonance as described in Materials and methods. (A) The sensor chip SA was coated with 12 RU of the biotinylated S6K-pHM peptide and the binding was analysed following injection of 270 nM wild type PDK1, PDK1[T148A] and PDK1[K76A]. No detectable binding to S6K-pHM was observed using PDK1[R131A] or PDK1[Q150] (data not shown). (B) As in (A) except that binding was analysed over a range of PDK1 concentrations (2–2150 nM). The response level at the steady-state binding is plotted versus the log of the PDK1 concentration. The estimated Kd was obtained by fitting the data to the formula [m1 × m0/(m0 + m2)] using Kaleidagraph software. Kd for wild type PDK1 equals 642 ± 131 nM, PDK1[T148A] Kd = 64 ± 7 nM and PDK1[K76A] Kd = 1744 ± 167 nM. No detectable binding of PDK1 to the non-phosphorylated S6K-HM peptide (SESANQVFLGFTYVAPSV) was detected with wild type PDK1 or any of the mutants (data not shown). (C) Activation of the indicated forms of PDK1 by S6K-pHM and S6K-HM. PDK1 activity was measured using the peptide substrate (T308tide) in the presence of the indicated concentrations of S6K-pHM (closed circles) and S6K-HM (open circles) as described in Materials and methods. Assays were performed in triplicate and similar results obtained in two separate experiments. The results are the average ± SD for a single experiment.
The binding of PDK1 to PIFtide stimulates up to 4-fold the rate at which PDK1 phosphorylates a small peptide that encompasses the activation loop motif of PKB (termed T308tide; Biondi et al., 2000), indicating that occupancy of the PIF-pocket of PDK1 activates the enzyme. Similarly, the binding of a phosphopeptide corresponding to the hydrophobic motif of RSK stimulated PDK1 activity 6-fold (Frodin et al., 2000). We have now also found that the binding of S6K-pHM to wild type PDK1 induces a maximal 5-fold activation, with a half-maximal activation occurring at a concentration of ∼50 µM S6K-pHM (Figure 4C). We next assayed the specific activities of PDK1[K76A], PDK1[R131A], PDK1[T148A] and PDK1[Q150A] mutants in the absence or presence of increasing concentrations of S6K-pHM (Figure 4C). The PDK1[K76A] mutant possessed the same specific activity towards T308tide in the absence of S6K-pHM as wild type PDK1, but an ∼3-fold higher concentration of S6K-pHM was required to half-maximally activate PDK1[K76A], consistent with the reduced affinity of this form of PDK1 for S6K-pHM (Figure 4A and B). The PDK1[R131A] mutant possessed a 3-fold higher specific activity towards Thr308tide in the absence of S6K-pHM (Figure 4C), as has been observed previ ously with certain other PIF-pocket mutants of PDK1 (PDK1[K115A] and PDK1[L155E]; Biondi et al., 2000). However, in accordance with the inability of PDK1[R131A] to bind S6K-pHM in the BiaCore assay (Figure 4B), it was not activated significantly by concentrations of S6K-pHM below 0.1 mM and its activity was only increased moderately by the addition of high concentrations (0.3 and 1 mM) of S6K-pHM (Figure 4C). Interestingly, our findings suggest that Arg131 has a fine regulatory role, participating both in the inhibition of basal PDK1 activity (since mutation to Ala increases basal activity) and in the activation process, upon occupancy of the phosphate pocket. The activity of a mutant of PDK1 in which both Lys76 and Arg131 were changed to Ala was activated even less significantly by these high concentrations of S6K-pHM. The PDK1[T148A] and PDK1 [Q150A] mutants possessed similar specific activity towards T308tide as wild type PDK1 in the absence of S6K-pHM. In the presence of S6K-pHM, the PDK1 [T148A] mutant was activated similarly compared with wild type PDK1. Consistent with the inability of PDK1[Q150A] to interact with S6K-pHM, this mutant of PDK1 was not significantly activated by concentrations of S6K-pHM below 0.1 mM, but at a peptide concentration of 0.3 and 1 mM, a 2- to 3-fold activation was observed (Figure 4).
At very high peptide concentrations (0.3–1 mM), the non-phosphorylated S6K-HM peptide induced a small (<2-fold) activation of PDK1 (Figure 4C). Interestingly, despite the PDK1[K76A] and PDK1[R131A] mutants being markedly less able to interact with the phosphorylated S6K-pHM peptide than wild type PDK1, high concentrations of the S6K-HM peptide activated PDK1[K76A] and PDK1[R131A] to a similar extent as wild type PDK1, indicating that the ability of these mutants to interact weakly with the S6K-HM peptide was not affected.
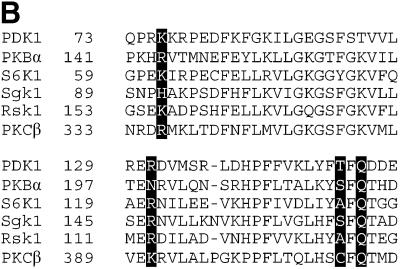
We evaluated the sequence conservation in the phosphate pocket of the insulin/growth factor-activated AGC family kinases (PKBα, S6K1, SGK1 and RSK1). Sequence alignments indicate that this pocket is conserved amongst these kinases (Figure 5B). The most conserved residue is Gln150, which is found in all of these AGC kinases, and the residue equivalent to Lys76 is always a basic residue (Figure 5B). Arg131 is conserved in S6K1, SGK1, RSK1 and PKCβ, but not in PKBα, PKBβ or PKBγ, where it is an Asn or Ser. Thr148 is conserved in PKBα and SGK1, but is an Ala in S6K1/RSK1 and a Cys in PKCβ. Interestingly, we have found that the Thr148Ala mutation in PDK1 did not disrupt the phosphate pocket (Figure 4). As PKBα, S6K1, SGK1 and RSK1 need to be phosphorylated at their hydrophobic motif in order to be maximally activated, it is tempting to speculate that the C-terminal hydrophobic motifs of these enzymes, when phosphorylated, bind to their own PIF/phosphate pockets, thereby generating a network of interactions similar to that of PDK1. Consistent with PKA not possessing a phosphate pocket, Lys76 and Gln150 are not conserved in PKA (Figure 3), and indeed such a pocket is not observed in the PKA structure (Figure 2).
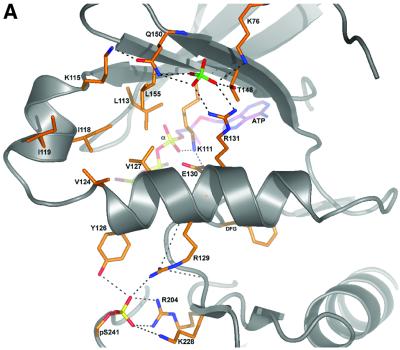
Fig. 5. Interactions of regulatory phosphates with the αC-helix. (A) The PDK1 backbone is shown as a ribbon, with helix αC in the centre of the view. Key residues are shown as sticks with carbons coloured orange. The sulfate ion and the phosphate on the activation loop are also shown. A stick model of ATP is shown with carbons coloured purple. Hydrogen bonds are shown as black dotted lines. (B) Alignment of the amino acid sequence forming part of the phosphate pocket on PDK1 with the equivalent region of the indicated AGC kinases. The residues corresponding to Lys76, Arg131, Thr148 and Gln150 of PDK1 are indicated in bold.
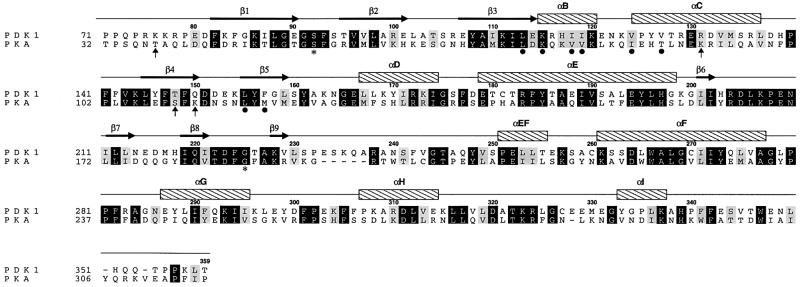
Fig. 3. Structure-based sequence alignment. The sequences of PKA and PDK1 are aligned according to a structural superposition performed in WHAT IF (Vriend, 1990). Sequence numbering is according to PDK1. β-strands (arrows) and α-helices (bars) are shown for the PDK1 structure according to a DSSP (Kabsch and Sander, 1983) secondary structure assignment, and labelled consistent with the secondary structure element names proposed for PKA (Taylor and Radzioandzelm, 1994). Residues lining the PIF-pocket are indicated with a black dot. Residues hydrogen bonding the sulfate ion are indicated by arrows. The PDK1 residues equivalent to Ser53 and Gly186 in PKA are labelled with an asterisk.
In the case of conventional PKC, phosphorylation of the hydrophobic motif plays a role in stabilizing these enzymes rather than triggering their activation (Bornancin and Parker, 1997; Edwards and Newton, 1997). Recent studies indicate that PDK1 also interacts with isolated C-terminal fragment of PKCβ with an order of magnitude higher affinity after this is phosphorylated at the hydrophobic motif (Gao et al., 2001). Interestingly, however, PDK1 can only interact with full-length PKC when it is not phosphorylated at its hydrophobic motif and phosphorylation of this motif was shown to somehow mask this region, preventing it from interacting with PDK1 (Gao et al., 2001). It is likely that the hydrophobic motif of conventional PKC isoforms in its dephosphorylated state will be available for interaction with PDK1, but that following its phosphorylation, it will interact more strongly with the equivalent PIF/phosphate-binding pocket in its own kinase domain, thereby becoming inaccessible for binding to PDK1 (Gao et al., 2001). Our results support the existence of a phosphate-binding site in PKC isoforms that could account for the higher affinity binding of the phosphorylated C-terminal hydrophobic motif to its own pocket. The alignment in Figure 5A predicts that Arg336, Lys391, Cys409 and Gln411 in PKCβ will comprise the hydrophobic motif phosphate-binding site in the PKCβ kinase catalytic domain. Thus, binding of AGC kinases to PDK1 could depend not only on the sequence surrounding the hydrophobic motif and its phosphorylation status, but also on the availability of this sequence for interaction. The structure of PDK1 does not provide a major insight into the explanation as to why PDK1 appears to interact more strongly with the hydrophobic motif of PRK2 (PIF) compared with the equivalent fragments of S6K1 and PKB phosphorylated at their hydrophobic motif (Biondi et al., 2001). Co-crystallization of PDK1 bound to substrates or peptides encompassing hydrophobic motifs could shed light into other contacts between PDK1 and its substrates.
The αC-helix
The PDK1 structure shows that, as in other protein kinases (Johnson et al., 2001; Husen and Kuriyan, 2002), the αC-helix (residues 124–136) is a key signal integration motif in the kinase core. One turn of the PDK1 αC-helix (residues 129–131, Figures 3 and 5) links together the N-terminal lobe, the C-terminal lobe and the active site. Arg129 points towards the activation loop and forms two hydrogen bonds with the phosphorylated Ser241, whereas Arg131 forms two hydrogen bonds with the sulfate in the phosphate pocket (Figure 5). Glu130 coordinates Lys111, which forms a hydrogen bond with the α-phosphate of the bound ATP. This interaction is conserved in all protein kinases and was shown to be crucial for activation (Johnson et al., 2001; Husen and Kuriyan, 2002). An additional residue, Tyr126, forms a third hydrogen bond with the phosphorylated Ser241. Val124 and Val127 on the αC-helix are involved in formation of the PIF-pocket (Figure 5). The αC-helix provides a structural link between the putative phosphopeptide-binding pocket and the phosphoserine in the activation loop. The fact that R131A has higher basal activity than wild type PDK1 may indicate that this residue plays a tuning role in the PDK1 structure, not only participating in the activation of PDK1 in the presence of a phosphate ion, but also in keeping the equilibrium of the enzyme towards an inactive conformation in the absence of S6K-pHM. To our knowledge this is the first report of a kinase structure in which the αC-helix is positioned by two regulatory phosphate-binding sites on either side of the helix (Figure 5). This provides a possible sensor mechanism for linking the phosphorylation-state of the activation loop and the phos phopeptide binding event in the PIF-pocket to PDK1 activity. Our findings could provide an explanation for the observation that inactive dephosphorylated PKCβ can interact with PDK1, whilst phosphorylated PKCβ does not (Gao et al., 2001). This would be possible since in the absence of activation-loop phosphorylation, the αC-helix may not be fixed in a position that enables the formation of the hydrophobic and phosphate pocket that permits the binding of the hydrophobic motif to its own kinase domain. Following phosphorylation of the activation loop by PDK1 and autophosphorylation of the hydrophobic motif, the PKCβ hydrophobic and phosphate pocket would be stabilized as in the PDK1 crystal structure.
Activation state
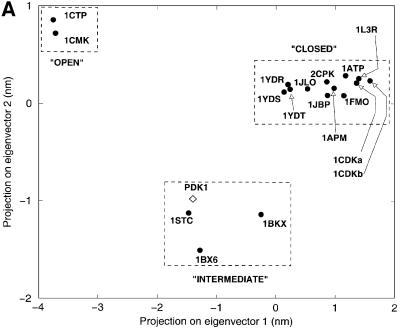
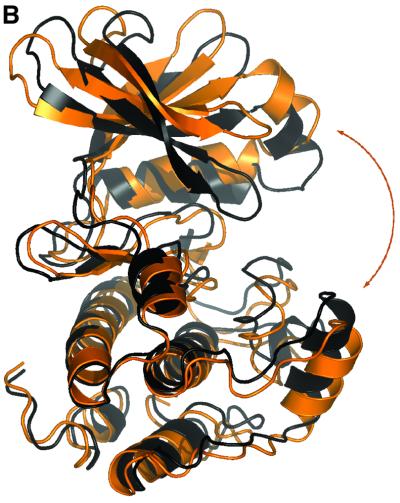
All structures of PKA solved to date show a phosphorylated T-loop and are therefore assumed to be in an active state. In addition to the unphosphorylated versus phosphorylated states of PKA, there appear to be two main conformational states possible for the latter (Zheng et al., 1993; Johnson et al., 2001). In the active, closed conformation, all residues are positioned to facilitate phosphoryl transfer. In contrast, the inactive, open conformation is seen in the absence of a nucleotide, and differs from the closed conformation by conformational changes of the N- and C-terminal domains with respect to each other. In addition, three ‘intermediate’ structures were described from PKA, having either adenosine (PDB entry 1BKX; Narayana et al., 1997) or the inhibitors staurosporine (PDB entry 1STC; Prade et al., 1997) and balanol (PDB entry 1BX6; Narayana et al., 1999) in the ATP-binding site. Taylor et al. (1992) have described a method to distinguish between the active and inactive conformations, based on three distances: His87–pThr197 (αC-helix positioning), Ser53–Gly186 (opening of the glycine-rich loop) and Glu170–Tyr330 (C-terminal tail distance to active site; Johnson et al., 2001). In PDK1, only one of these distances, the opening state of the glycine-rich loop, can be measured due to sequence conservation (Figure 3). This distance is 12.4 Å, similar to a PKA intermediate conformation [this distance in PKA is 14.2 Å for the open, 11.8 Å for intermediate and 10.0 Å for the closed conformation (Johnson et al., 2001)]. To allow a more direct comparison of the PDK1 structure with the available PKA structures, we have analysed the conformational state of PDK1 in detail using a novel approach, which involves a principal component analysis [also called ‘essential dynamics’ (Amadei et al., 1993)] of the crystallographic coordinates. In short, this involves the construction of a covariance matrix containing the correlations between atomic shifts (with respect to an average structure) in the ensemble of all available PKA crystal structures. Diagonalization of this matrix gives eigen vector/eigenvalue sets, which describe concerted shifts of atoms (eigenvectors) together with the corresponding mean square fluctuation of the structures (eigenvalues). This approach allows a condensed description of PKA conformational states using only a few degrees of freedom, as shown previously for a range of other proteins (van Aalten et al., 1997, 2000; de Groot et al., 1998). Diagonalization of a covariance matrix built from the backbone atoms of residues 37–196, 198–283 and 286–305 results in a set of eigenvectors that describe concerted motions of the PKA backbone. In Figure 6A, all PKA structures are projected on a subspace spanned by the first two eigenvectors (i.e. those with the two largest eigenvalues). It appears that the PKA structures cluster in three main areas along the first eigenvector. On the left of the average structure (which by definition has a projection of 0.0 on all eigenvectors) are the structures that are known to be in the ‘open’ conformation (Figure 6A). Around the average structure lie the structures that have been shown to be in an ‘intermediate’ conformation (complexes with the inhibitors staurosporine, balanol and adenosine). More to the right of the average structure are the PKA structures that are known to be in the ‘closed’ conformation. Thus, we have captured the conformational state of PKA in a single variable, the translation along the first eigenvector. This is further clarified by investigation of the atomic shifts described by this eigenvector in Cartesian space (Figure 6B). A hinge-bending motion is observed between the N- and C-terminal lobes, opening and closing the active site. It is now possible to directly compare the PDK1 conformational state by projecting the structure (backbone atoms only) onto the PKA eigenvectors. Figure 6A shows that the conformation of PDK1 is close to the PKA structures that are in an ‘intermediate’ conformation, consistent with the other structural analyses described above.
Fig. 6. Essential dynamics. (A) Projection of all available PKA crystal structures (labelled dots) and the PDK1 structure (diamond) onto the first two eigenvectors (the ones with the two largest eigenvalues) calculated from the PKA structures. (B) Graphic representation of the motion along the first eigenvector, generated by projecting two structures at –4 nm (black) and +4 nm (orange).
Conclusions
We have reported the structure of the PDK1 catalytic domain, which, although similar to PKA, has revealed important features that increase our understanding of the mechanism by which PDK1 is regulated. The structure, together with mutational analyses, defines a phosphopeptide-binding pocket, consisting of a separate hydrophobic PIF-pocket and a phosphate-binding site, which mediates the interaction of PDK1 with the phosphorylated hydrophobic motif of S6K. This is consistent with the previous hypothesis that phosphorylation of S6K and SGK (Biondi et al., 2001) as well as RSK (Frodin et al., 2000) at their FXXFS/T hydrophobic motif is the trigger for their interaction and phosphorylation by PDK1. In this mechanism, the PIF-pocket would physiologically only interact with the Phe residues when the Ser/Thr residue is phosphorylated. Furthermore, as the phosphate pocket is conserved in other AGC kinases, the structural features and network of interaction of the phosphate pocket with the αC-helix on PDK1 could provide insight into the mode of activation of other AGC kinases.
A significant number of human cancers, such as those in which the PTEN tumour suppressor is mutated, possess markedly elevated PtdIns(3,4,5)P3 levels resulting in constitutive activation of PKB and S6K, which are likely to be major contributors to the proliferation and enhanced survival of these tumour cells (Leslie and Downes, 2002). Inhibitors of PDK1 could prove effective for the treatment of these types of cancer by suppressing PKB and S6K activity. The structure of the catalytic domain of PDK1 provides a molecular framework to develop inhibitors that target either the ATP-binding site or the PIF-binding pocket. Both types of drug would hinder PDK1 function in vivo, but the advantage of the latter class of drugs is that they may be more specific than ATP-competitive inhibitors and therefore less likely to inhibit the activity of other protein kinases in cells.
Materials and methods
Materials
Mammalian and insect cell culture reagents were from Life Technologies. SensorChips SA were from BiaCore AB (Stevenage, UK). Glutathione–Sepharose, as well as pre-packed HiTrap Q HP and Hiload Superdex 200 prep grade columns were from Amersham Biosciences. Dialysis cassettes were from the Slide-A-Lyzer series (Pierce). Ni-NTA–agarose was from Qiagen. Disposable ultrafiltration devices (polyethersulfone membranes) were from Vivascience. Crystallization research tools (primary screens, additive screens and crystallization plates) were from Hampton Research. Peptides were synthesized by Dr G.Blomberg (University of Bristol, UK).
General methods
Molecular biology techniques were performed using standard protocols. Site-directed mutagenesis was performed using a QuikChange kit (Stratagene) following instructions provided by the manufacturer. DNA constructs used for transfection were purified from bacteria using Qiagen plasmid Mega kit according to the manufacturer’s protocol, and their sequence verified. Human kidney embryonic 293 cells were cultured on 10 cm diameter dishes in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum.
Buffers
Low-salt buffer: 25 mM Tris–HCl pH 7.5, 150 mM NaCl; High-salt buffer: 25 mM Tris–HCl pH 7.5, 500 mM NaCl. Lysis buffer: 25 mM Tris–HCl pH 7.5, 150 mM NaCl 0.07% β-mercaptoethanol, 1 mM benzamidine and 20 µg/ml PMSF. Buffer A: 50 mM Tris–HCl pH 7.5, 1 mM EGTA, 1 mM EDTA, 1% (by mass) Triton X-100, 1 mM sodium orthovanadate, 50 mM sodium fluoride, 5 mM sodium pyrophosphate, 0.27 M sucrose, 1 µM microcystin-LR, 0.1% (by vol.) β-mercaptoethanol and ‘complete’ proteinase inhibitor cocktail (one tablet per 50 ml; Roche). Buffer B: 50 mM Tris–HCl pH 7.5, 0.1 mM EGTA, 10 mM β-mercaptoethanol and 0.27 M sucrose.
Expression, purification and characterization of the kinase domain of PDK1
A cDNA encoding for human PDK1 amino acid residues 51–359, with a stop codon inserted at position 360, was amplified by PCR using full-length human PDK1 cDNA in the pCMV5 vector (Alessi et al., 1997) as a template. The PCR strategy employed incorporates at the 5′ end of the gene an initiating methionine, a His6 tag followed by a PreScission protease recognition sequence prior to the residue equivalent to Met51 of PDK1 (GGATCCTATAAATATGGCACATCATCATCATCATCATCTGGAAGTTCTGTTCCAGGGGCCCATGGACGGCACTGCAGCCGAGCCTCGG). The 3′ primer applied in this reaction was: 5′-GGA TCCTCAGGTGAGCTTCGGAGGCGTCTGCTGGTG-3′. The resulting PCR product was ligated into pCR 2.1 TOPO vector (Invitrogen) and then subcloned as a BamHI–BamHI fragment into pFastbac1 vector (Life Technologies) for baculovirus protein expression. The resulting construct was then used to generate recombinant baculovirus using the Bac-to-Bac system (Life Technologies) following the manufacturer’s protocol. The resulting baculoviruses were used to infect Sf21 cells at 1.5 × 106/ml. The infected cells were harvested by centrifugation 72 h post-infection. Cell pellets corresponding to 7 l of culture were resuspended in 900 ml of Lysis buffer and cells lysed in a nitrogen cavitation chamber. Cell debris was then pelleted by centrifugation, the supernatant made 0.5 M NaCl by addition of 4 M NaCl and then incubated with Ni-NTA–agarose (10 ml resin) for 1 h. The resin was then washed 10 × with 40 ml of Lysis buffer containing 0.5 M NaCl and then placed in a disposable Econo-Pac column (Bio-Rad), where the resin was further washed with 700 ml of High-salt buffer and then with 100 ml of Low-salt buffer, both supplemented with 10 mM imidazole. Elution was performed with 200 mM imidazole in High-salt buffer and 2 ml fractions were collected. Fractions containing protein were pooled, diluted to 200 mM NaCl with 25 mM Tris–HCl pH 7.5, and loaded onto a 5 ml Hi-trap Q sepharose column. The flow-through from this step, containing PDK1, was concentrated to 4 ml and then chromatographed on a 16/60 Superdex 200 gel filtration column using an AKTA Explorer system (Amersham Biosciences) equilibrated with High-salt buffer with the addition of 1 mM dithiothreitol (DTT). PDK1 eluted in a large symmetric peak at the expected size for a monomer. The PDK1-containing peak was again pooled, concentrated and incubated with 300 µg of glutathione S-transferase (GST)– PreScission protease (expression construct kindly provided by John Heath, University of Birmingham, UK) on ice for 4 h. In order to eliminate the cleaved His-tag sequences as well as any remaining uncleaved His-PDK1 and the GST–PreScission protease, the mixture was incubated with a mixture of 200 µl of glutathione–Sepharose and 200 µl of Ni-NTA–agarose resin for 15 min and the PDK1 protein that did not bind was collected. The resulting protein consists of PDK1 (51–359) preceded by a Gly-Pro at the N-terminus. The protein at this stage of the purification was apparently homogeneous as revealed by a single band after electrophoresis of 20 µg of protein on an SDS–polyacrylamide gel and staining with Coomassie Blue R250 (data not shown). Electrospray mass spectrometry revealed a main peak mass close to the expected size of this fragment of PDK1. The specific activity of PDK1 (51–359) towards the peptide T308tide and its activation in the presence of PIFtide were identical to wild type full-length PDK1 (Biondi et al., 2000), and tryptic peptide mass finger printing indicated that PDK1 was quantitatively phosphorylated at Ser241 (data not shown). In BiaCore experiments, the steady-state binding of PDK1 (51–359) to the peptide PIFtide was similar to that of the His-tag PDK1 (51–556) protein characterized previously (Balendran et al., 1999a).
Crystallization and data collection
The PDK1 (51–359) protein was concentrated to a final concentration of 8.5 mg/ml (as determined by a Bradford assay using bovine serum albumin as a standard). The sitting drop vapour diffusion method was used for producing crystals. Sitting drops were formed by mixing 1 µl of protein solution + 0.2 µl of EDTA (100 mM) with 1 µl of a mother liquor solution (0.1 M Tris–HCl pH 8.5, 2.0 M ammonium sulfate, 16.6 mM ATP). Hexagonal crystals (Table I) of PDK1 were grown at 20°C from a mother liquor containing 0.1 M Tris–HCl pH 8.5, 2.0 M ammonium sulfate, 16.6 mM ATP. Crystals appeared after 1 day, growing to 0.05 × 0.05 × 0.2 mm over 20 days. Crystals were frozen in a nitrogen gas stream after being soaked in 0.075 M Tris pH 8.5, 1.5 M ammonium sulfate and 25% glycerol.
Table I. Details of data collection and structure refinement for the PDK1 kinase domaina.
| Wavelength (Å) | 0.933 |
| Space group | P3221 |
| Unit cell (Å) | a = 123.01, b = 123.01, c = 47.62 |
| Resolution (Å) | 25–2.0 (2.07–2.0) |
| Observed reflections | 77 315 |
| Unique reflections | 27 643 |
| Redundancy | 2.8 (2.5) |
| Completeness (%) | 98.0 (93.5) |
| Rmerge | 0.091 (0.454) |
| I/σI | 7.3 (2.0) |
| Rfree reflections | 579 |
| Rcryst | 0.195 |
| Rfree | 0.222 |
| Number of groups | |
| Protein residues | 71–359 |
| Water | 200 |
| ATP | 1 |
| SO4 | 5 |
| Glycerol | 8 |
| Wilson B (Å2) | 22.4 |
| <B> Protein | 25.6 |
| <B> Water | 35.7 |
| <B> ATP | 38.8 |
| R.m.s.d. from ideal geometry | |
| Bond lengths (Å) | 0.005 |
| Bond angles (°) | 1.34 |
| Main chain B (Å2) | 1.5 |
aValues in parentheses are for the highest resolution shell. All measured data were included in structure refinement.
Expression and purification of wild type and mutant forms of GST–PDK1
Wild type PDK1 (Alessi et al., 1997), PDK1[R76A], PDK1[R131A], PDK1[R76A, R131A], PDK1[T148A] and PDK1[Q150A] in the pEBG2T vector were used to express the wild type and indicated mutants of PDK1 fused through their N-terminus to GST. The GST fusion proteins were expressed in human embryonic kidney 293 cells. For the expression of each construct, twenty 10 cm diameter dishes of 293 cells were cultured and each dish transfected with 10 µg of the pEBG-2T construct, using a modified calcium phosphate method. Thirty-six hours post-transfection, the cells were lysed in 0.6 ml of ice-cold buffer A, the lysates pooled, centrifuged at 4°C for 10 min at 13 000 g and the GST fusion proteins were purified by affinity chromatography on glutath ione–Sepharose and eluted in buffer B supplemented with 20 mM glutathione as described previously (Alessi et al., 1997). Typically between 1 and 2 mg of each GST fusion protein were obtained and each protein was >75% pure, as judged by SDS–PAGE (data not shown).
PDK1 catalytic activity measurements
The ability of wild type and mutant PDK1 to phosphorylate the synthetic peptide T308tide (KTFCGTPEYLAPEVRR; Biondi et al., 2000) was carried out in 30 µl assays containing 100 ng of wild type or mutant PDK1, 50 mM Tris–HCl pH 7.5, 0.1% β-mercaptoethanol, 10 mM MgCl2, 100 µM [γ-32P]ATP (200 c.p.m./pmol), 0.5 µM microcystin-LR, 1 mM T308tide in the presence or absence of the indicated concentrations of the S6K-pHM peptide [SESANQVFLGFT(P)YVAPSV] or S6K-HM peptide (SESANQVFLGFTYVAPSV). After incubation for 10 min at 30°C, 25 µl of the resultant mixture were spotted onto P81 phospho cellulose paper (2 × 2 cm) and the papers washed and analysed as described previously for assays of MAP kinase. Control assays were carried out in parallel in which either PDK1 or peptide substrate was omitted; these values were always <5% of the activity measured in the presence of these reagents. One unit of PDK1 activity was defined as that amount required to catalyse the phosphorylation of 1 nmol of the T308tide in 1 min.
BiaCore analysis
Binding was analysed in a BiaCore 3000 system (BiaCore AB). Biotinylated S6K-pHM [Biotin-C12-SESANQVFLGFT(P)YVAPSV] or the non-phosphorylated form of this peptide S6K-HM was bound to a streptavidin-coated Sensor chip (SA) (12 response units, RU). A 30 µl aliquot of wild type or the indicated mutant GST–PDK1 was injected at a flow rate of 30 µl/min, in buffer HBS-P [10 mM HEPES pH 7.4, 0.15 M NaCl, 0.005% (by vol.) polysorbate-20] supplemented with 1 mM DTT. Specific interactions between S6K-pHM and PDK1 proteins were obtained in the concentration range of 2–2150 nM PDK1. Steady-state binding was determined at each concentration. Dissociation of PDK1 from the phosphopeptide was monitored over a 1 min period. Regeneration of the sensor chip surface was performed with 10 µl injections of 0.05% SDS. As previously found for PDK1 binding to PIFtide (Biondi et al., 2000), the interaction data obtained using BiaCore did not fit a simple 1:1 interaction model. For all PDK1 constructs tested, the off rates for S6Kp-HM were high in comparison to those of PIFtide binding with the time taken for 50% dissociation to occur for S6K-pHM being 30 s compared with 1000 s for PIFtide. This could account for the reduced ability of S6K-pHM to displace binding from PIFtide (Biondi et al., 2000).
Data collection, structure solution and refinement
Data on PDK1 crystals were collected at the European Synchrotron Radiation Facility (Grenoble, France) beamline ID14-EH1, using an ADSC Q4 CCD detector. The temperature of the crystals was maintained at 100 K using a nitrogen cryostream. Data were processed using the HKL package (Otwinowski and Minor, 1997), and statistics are shown in Table I.
The structure of PDK1 was solved by molecular replacement with AMoRe (Navaza, 1994) using the structure of PKA in complex with an inhibitory peptide as a search model (PDB entry 1YDB), against 8–4 Å data. A single, well separated solution was found with an R-factor of 0.479 (correlation coefficient = 0.428). The structure was automatically built using warpNtrace (Perrakis et al., 1999), which found 262 of a possible 309 residues, giving an initial protein model with R = 0.293 (Rfree = 0.318) after simulated annealing in CNS (Brünger et al., 1998). Iterative protein building in O (Jones et al., 1991) together with refinement in CNS, which included incorporation of a model for ATP, the phosphoserine in the activation loop, solvent molecules and a key sulfate molecule, resulted in a final model with R = 0.195 (Rfree = 0.222). No electron density was observed for residues 51–70 (the N-terminus of the construct) and 233–236 (the tip of the activation loop).
All figures were made with PyMOL (http://www.pymol.org).
PDB coordinates
The coordinates and structure factors have been deposited with the PDB (entry 1h1w).
Acknowledgments
Acknowledgements
We thank the European Synchrotron Radiation Facility, Grenoble, for the time at beamline ID14-EH1, as well as Agnieszka Kieloch for assistance with tissue culture, Felicity Newell for baculovirus production, Nick Morrice for mass spectrometry analysis and the DNA Sequencing Service, School of Life Sciences, University of Dundee, for DNA sequencing. We also thank Jennifer Gallop and Evangelis Dioletis for their help in early stages of this work. D.K. is supported by an MRC Predoctoral Fellowship, C.C.T. by a BBSRC CASE studentship, D.M.F.v.A. by a Wellcome Trust Career Development Research Fellowship, D.R.A. by the Medical Research Council (UK), Diabetes UK, Association for International Cancer Research, and the pharmaceutical companies supporting the Division of Signal Transduction Therapy unit in Dundee (AstraZeneca, Boehringer Ingelheim, GlaxoSmithKline, Novo-Nordisk, Pfizer).
References
- Alessi D.R. (2001) Discovery of PDK1, one of the missing links in insulin signal transduction. Biochem. Soc. Trans., 29, 1–14. [DOI] [PubMed] [Google Scholar]
- Alessi D.R. et al. (1997) 3-phosphoinositide-dependent protein kinase-1 (PDK1): structural and functional homology with the Drosophila DSTPK61 kinase. Curr. Biol., 7, 776–789. [DOI] [PubMed] [Google Scholar]
- Amadei A., Linssen,A.B.M. and Berendsen,H.J.C. (1993) Essential dynamics of proteins. Proteins, 17, 412–425. [DOI] [PubMed] [Google Scholar]
- Balendran A., Casamayor,A., Deak,M., Paterson,A., Gaffney,P., Currie,R., Downes,C.P. and Alessi,D.R. (1999a) Pdk1 acquires PDK2 activity in the presence of a synthetic peptide derived from the carboxyl terminus of PRK2. Curr. Biol., 9, 393–404. [DOI] [PubMed] [Google Scholar]
- Balendran A., Currie,R., Armstrong,C.G., Avruch,J. and Alessi,D.R. (1999b) Evidence that 3-phosphoinositide-dependent protein kinase-1 mediates phosphorylation of p70 S6 kinase in vivo at Thr-412 as well as Thr-252. J. Biol. Chem., 274, 37400–37406. [DOI] [PubMed] [Google Scholar]
- Balendran A., Biondi,R.M., Cheung,P.C.F., Casamayor,A., Deak,M. and Alessi,D.R. (2000) A 3-phosphoinositide-dependent protein kinase-1 (PDK1) docking site is required for the phosphorylation of protein kinase Cζ (PKCζ) and PKC-related kinase 2 by PDK1. J. Biol. Chem., 275, 20806–20813. [DOI] [PubMed] [Google Scholar]
- Batkin M., Schvartz,I. and Shaltiel,S. (2000) Snapping of the carboxyl terminal tail of the catalytic subunit of PKA onto its core: Characterization of the sites by mutagenesis. Biochemistry, 39, 5366–5373. [DOI] [PubMed] [Google Scholar]
- Biondi R.M., Cheung,P.C.F., Casamayor,A., Deak,M., Currie,R.A. and Alessi,D.R. (2000) Identification of a pocket in the PDK1 kinase domain that interacts with PIF and the C-terminal residues of PKA. EMBO J., 19, 979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biondi R.M., Kieloch,A., Currie,R.A., Deak,M. and Alessi,D.R. (2001) The PIF-binding pocket in PDK1 is essential for activation of S6K and SGK, but not PKB. EMBO J., 20, 4380–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornancin F. and Parker,P.J. (1997) Phosphorylation of protein kinase C-ζ on serine 657 controls the accumulation of active enzyme and contributes to its phosphatase-resistant state. J. Biol. Chem., 272, 3544–3549. [DOI] [PubMed] [Google Scholar]
- Brazil D.P. and Hemmings,B.A. (2001) Ten years of protein kinase B signalling: a hard Akt to follow. Trends Biochem. Sci., 26, 657–664. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: A new software system for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Casamayor A., Morrice,N.A. and Alessi,D.R. (1999) Protein phosphorylation of Ser-241 is essential for the activity of 3-phosphoinositide-dependent protein kinase-1: identification of five sites of phosphorylation in vivo. Biochem. J., 342, 287–292. [PMC free article] [PubMed] [Google Scholar]
- de Groot B.L., Hayward,S., van Aalten,D.M.F., Amadei,A. and Berendsen,H.J.C. (1998) Domain motions in bacteriophage T4 lysozyme: A comparison between molecular dynamics and crystallographic data. Proteins, 31, 116–127. [DOI] [PubMed] [Google Scholar]
- Edwards A.S. and Newton,A.C. (1997) Phosphorylation at conserved carboxyl-terminal hydrophobic motif regulates the catalytic and regulatory domains of protein kinase c. J. Biol. Chem., 272, 18382–18390. [DOI] [PubMed] [Google Scholar]
- Etchebehere L.C., Van Bemmelen,M.X.P., Anjard,C., Traincard,F., Assemat,K., Reymond,C. and Veron,M. (1997) The catalytic subunit of Dictyostelium cAMP-dependent protein kinase—role of the N-terminal domain and of the C-terminal residues in catalytic activity and stability. Eur. J. Biochem., 248, 820–826. [DOI] [PubMed] [Google Scholar]
- Frodin M. and Gammeltoft,S. (1999) Role and regulation of 90 kDa ribosomal S6 kinase (RSK) in signal transduction. Mol. Cell. Endocrinol., 151, 65–77. [DOI] [PubMed] [Google Scholar]
- Frodin M., Jensen,C.J., Merienne,K. and Gammeltoft,S. (2000) A phosphoserine-regulated docking site in the protein kinase RSK2 that recruits and activates PDK1. EMBO J., 19, 2924–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao T.Y., Toker,A. and Newton,A.C. (2001) The carboxyl terminus of protein kinase C provides a switch to regulate its interaction with the phosphoinositide-dependent kinase, PDK1. J. Biol. Chem., 276, 19588–19596. [DOI] [PubMed] [Google Scholar]
- Husen M. and Kuriyan,J. (2002) The conformational plasticity of protein kinases. Cell, 109, 275–282. [DOI] [PubMed] [Google Scholar]
- Johnson D.A., Akamine,P., Radzio-Andzelm,E., Madhusudan and Taylor,S.S. (2001) Dynamics of cAMP-dependent protein kinase. Chem. Rev., 101, 2243–2270. [DOI] [PubMed] [Google Scholar]
- Johnson L.N., Noble,M.E.M. and Owen,D.J. (1996) Active and inactive protein kinases: Structural basis for regulation. Cell, 85, 149–158. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kabsch W. and Sander,C. (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers, 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- Knighton D.R., Zheng,J.H., Teneyck,L.F., Ashford,V.A., Xuong,N.H., Taylor,S.S. and Sowadski,J.M. (1991) Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science, 253, 407–414. [DOI] [PubMed] [Google Scholar]
- Kobayashi T. and Cohen,P. (1999) Activation of serum- and glucocorticoid-regulated protein kinase by agonists that activate phosphatidylinositide 3-kinase is mediated by 3-phosphoinositide-dependent protein kinase-1 (PDK1) and PDK2. Biochem. J., 339, 319–328. [PMC free article] [PubMed] [Google Scholar]
- Lang F. and Cohen,P. (2001) Regulation and physiological roles of serum- and glucocorticoid-induced protein kinase isoforms. Science, 108, RE17. [DOI] [PubMed] [Google Scholar]
- Leslie N.R. and Downes,C.P. (2002) PTEN: The down side of PI 3-kinase signalling. Cell. Signal., 14, 285–295. [DOI] [PubMed] [Google Scholar]
- Narayana N., Cox,S., Xuong,N.H., Ten Eyck,L.F. and Taylor,S.S. (1997) A binary complex of the catalytic subunit of cAMP-dependent protein kinase and adenosine further defines conformational flexibility. Structure, 5, 921–935. [DOI] [PubMed] [Google Scholar]
- Narayana N., Diller,T.C., Koide,K., Bunnage,M.E., Nicolaou,K.C., Brunton,L.L., Xuong,N.H., Ten Eyck,L.F. and Taylor,S.S. (1999) Crystal structure of the potent natural product inhibitor balanol in complex with the catalytic subunit of cAMP-dependent protein kinase. Biochemistry, 38, 2367–2376. [DOI] [PubMed] [Google Scholar]
- Navaza J. (1994) AMoRe: an automated package for molecular replacement. Acta Crystallogr. A, 50, 157–163. [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Perrakis A., Morris,R. and Lamzin,V.S. (1999) Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol., 6, 458–463. [DOI] [PubMed] [Google Scholar]
- Prade L., Engh,R.A., Girod,A., Kinzel,V., Huber,R. and Bossemeyer,D. (1997) Staurosporine-induced conformational changes of cAMP-dependent protein kinase catalytic subunit explain inhibitory potential. Structure, 5, 1627–1637. [DOI] [PubMed] [Google Scholar]
- Pullen N., Dennis,P.B. Andjelkovic,M., Dufner,A., Kozma,S.C., Hemmings,B.A. and Thomas,G. (1998) Phosphorylation and activation of p70S6k by PDK1. Science, 279, 707–710. [DOI] [PubMed] [Google Scholar]
- Scheid M.P. and Woodgett,J.R. (2001) PVB/Akt: Functional insights from genetic models. Nat. Rev. Mol. Cell Biol., 2, 760–768. [DOI] [PubMed] [Google Scholar]
- Stephens L. et al. (1998) Protein kinase B kinases that mediate phosphatidylinositol 3,4,5-trisphosphate-dependent activation of protein kinase B. Science, 279, 710–714. [DOI] [PubMed] [Google Scholar]
- Taylor S.S. and Radzioandzelm,E. (1994) Three protein-kinase structures define a common motif. Structure, 2, 345–355. [DOI] [PubMed] [Google Scholar]
- Taylor S.S., Knighton,D.R., Zheng,J.H., Teneyck,L.F. and Sowadski, J.M. (1992) Structural framework for the protein-kinase family. Annu. Rev. Cell Biol., 8, 429–462. [DOI] [PubMed] [Google Scholar]
- Tian X.J., Rusanescu,G., Hou,W.M., Schaffhausen,B. and Feig,L.A. (2002) PDK1 mediates growth factor-induced Ral-GEF activation by a kinase-independent mechanism. EMBO J., 21, 1327–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toker A. and Newton,A.C. (2000) Cellular signaling: Pivoting around PDK1. Cell, 103, 185–188. [DOI] [PubMed] [Google Scholar]
- van Aalten D.M.F., Conn,D.A., de Groot,B.L., Berendsen,H.J.C., Findlay,J.B.C. and Amadei,A. (1997) Protein dynamics derived from clusters of crystal structures. Biophys. J., 73, 2891–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Aalten D.M.F., Chong,C.R. and Joshua-Tor,L. (2000) Crystal structure of carboxypeptidase A complexed with D-cysteine at 1.75 Å: inhibitor-induced conformational changes. Biochemistry, 39, 10082–10089. [DOI] [PubMed] [Google Scholar]
- Volarevic S. and Thomas,G. (2001) Role of S6 phosphorylation and S6 kinase in cell growth. Prog. Nucleic Acid Res. Mol. Biol., 65, 101–127. [DOI] [PubMed] [Google Scholar]
- Vriend G. (1990) WHAT IF: a molecular modeling and drug design program. J. Mol. Graph., 8, 52–56. [DOI] [PubMed] [Google Scholar]
- Zheng J.H., Knighton,D.R., Xuong,N.H., Taylor,S.S., Sowadski,J.M. and Teneyck,L.F. (1993) Crystal structures of the myristylated catalytic subunit of cAMP-dependent protein kinase reveal open and closed conformations. Protein Sci., 2, 1559–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]