Abstract
Spitz (Spi) is the most prominent ligand of the Drosophila EGF receptor (DER). It is produced as an inactive membrane precursor which is retained in the endoplasmic reticulum (ER). To allow cleavage, Star transports Spi to the Golgi, where it undergoes cleavage by Rhomboid (Rho). Since some DER phenotypes are not mimicked by any of its known activating ligands, we identified an additional ligand by database searches, and termed it Keren (Krn). Krn is a functional homolog of Spi since it can rescue the spi mutant phenotype in a Rho- and Star-dependent manner. In contrast to Spi, however, Krn also possesses a Rho/Star-independent ability to undergo low-level cleavage and activate DER, as evident both in cell culture and in flies. The difference in basal activity correlates with the cellular localization of the two ligands. While Spi is retained in the ER, the retention of Krn is only partial. Examining Spi/Krn chimeric and deletion constructs implicates the Spi cytoplasmic domain in inhibiting its basal activity. Low-level activity of Krn calls for tightly regulated expression of the Krn precursor.
Keywords: EGF receptor/ligand processing/Rhomboid/Spitz/Star
Introduction
The Drosophilia EGF receptor (DER) pathway fulfills multiple roles during oogenesis, embryonic and post-embryonic development. While the receptor and its downstream signaling components are broadly expressed (Zak et al., 1990), activation of the pathway is tightly regulated in time and space, and any perturbations of this regulation have phenotypic consequences. This regulation is achieved by a battery of ligands, conferring tissue-specific activation, different strengths of activation, and finally by negative and positive feedback circuits (reviewed in Schweitzer and Shilo, 1997; Nilson and Schupbach, 1999; Freeman, 2000).
Three activating DER ligands have been identified to date. The weakest one is Vein, which is a secreted factor that resembles neuregulins in that both possess an Ig-C2 domain in addition to the EGF-like domain (Schnepp et al., 1996). Vein has a reduced capacity to activate MAPK in cells and embryos compared with the other two ligands, and a limited ability to induce ectopic expression of DER target genes (Schnepp et al., 1998; Golembo et al., 1999).
The other two ligands, Gurken (Grk) and Spitz (Spi), are TGFα homologs. Grk is expressed exclusively in the oocyte (Neuman-Silberberg and Schupbach, 1993). Early in oogenesis it establishes posterior cell fates, while later in oogenesis it initiates a signaling cascade, which determines the dorsal follicle cell fates (Gonzalez-Reyes et al., 1995; Roth et al., 1995). Spi, on the other hand, is used repeatedly during all stages (Rutledge et al., 1992). While broadly expressed, Spi is not active in its precursor, transmembrane form. Even when Spi is ectopically expressed at high levels, it does not activate the receptor (Schweitzer et al., 1995; Pickup and Banerjee, 1999). Only when cleaved does Spi become a potent ligand of DER, as can be assayed by the induction of target genes and the accumulation of activated MAP kinase (dpERK) (Schweitzer et al., 1995; Gabay et al., 1996, 1997). It has recently been shown that, similar to Spi, Grk undergoes cleavage as well (Ghiglione et al., 2002).
The mechanism of Spi processing has been examined extensively. Two proteins are required for Spi cleavage: Rhomboid (Rho) and Star. Their involvement in processing was implied by several experiments. First, secreted Spi can bypass the requirement for Rho or Star (Schweitzer et al., 1995). Secondly, they function in a non-autonomous manner to trigger DER activation in adjacent cells (Golembo et al., 1996; Guichard et al., 1999). Finally, experiments in Xenopus show that both proteins have to be expressed in the same cell as the Spi precursor to promote its processing (Bang and Kintner, 2000). Star is a broadly expressed type II transmembrane domain protein with no known homologs (Kolodkin et al., 1994). Rho is a seven transmembrane domain serine protease, belonging to a new family of proteases, which has homologs in vertebrates (Bier et al., 1990; Pascall and Brown, 1998; Urban et al., 2001). Recently, their specific roles in Spi cleavage have been understood. Spi is retained in the endoplasmic reticulum (ER). Star binds Spi and translocates it from its perinuclear location to the Golgi, where Rho is present. Spi is then cleaved by Rho and secreted (Lee et al., 2001; Tsruya et al., 2002).
During development, the expression of Rho is extremely dynamic (Bier et al., 1990) and is synonymous with activation of DER (Gabay et al., 1997). Con sequently, ectopic expression of Rho leads to concomitant induction of the pathway (Sturtevant et al., 1993; Golembo et al., 1996). Interestingly, in tissues where multiple cycles of DER induction are observed (e.g. the ovary and eye), rho itself becomes a target gene for the DER pathway (Sapir et al., 1998; Wasserman and Freeman, 1998).
In the embryo, removal of Spi or elements required for its cleavage, Rho and Star, gives rise to the typical embryonic ‘spi group’ phenotype (Mayer and Nusslein-Volhard, 1988). However, there are several instances where Rho is necessary, and yet the phenotypes observed for spi mutants are less severe. In the wing imaginal disc, the position of the future veins is dictated by localized expression of Rho (Sturtevant et al., 1993). Formation of veins is eliminated in rho mutants, and ectopic expression of Rho leads to induction of ectopic veins. Activated MAPK, as well as the expression of target genes like argos, kekkon and sprouty, are detected specifically in the future veins, at the site of Rho expression (Musacchio and Perrimon, 1996; Gabay et al., 1997; Reich et al., 1999). This is consistent with a high level of activation in these domains, resulting from cleavage of a DER ligand. However, when mutant clones for several null spi alleles were generated in the wing, no defects in vein formation were observed (Guichard et al., 1999; Nagaraj et al., 1999). This implicated an activity of another unknown DER ligand, capable of undergoing cleavage by Rho.
The presence of such a ligand was also indicated by analysis of mutant clones in the eye disc. In the eye, a second member of the Rho family, Roughoid, has a redundant role. Neither rho nor roughoid mutant backgrounds give rise to severe defects in photoreceptor cell induction, while in the double mutant only photoreceptor R8 differentiation is observed (Wasserman et al., 2000). The spi phenotypes are less severe than the rho/roughoid double-mutant phenotype. The correct spacing of R8 photoreceptor cells requires DER and is triggered by induction of Rho/Roughoid expression in the future R8 cells by Atonal (Baonza et al., 2001). Yet, in spi clones, spacing of the R8 cells is normal (Tio and Moses, 1997). Taken together, these results imply that in addition to Spi, another ligand that requires Rho for its processing may be utilized in these tissues.
We describe the identification of an additional DER ligand showing homology to TGFα and in particular to Spi, which we term Keren (Krn). Like Spi, this ligand is activated by cleavage regulated by Rho and Star. However, in contrast to Spi, the precursor form of Krn is also active upon overexpression. This activity represents low-level cleavage, which is not dependent upon Rho or Star. Chimeric and deletion constructs identify the cytoplasmic domains of Spi and Krn as the domains responsible for their different cleavage profiles. This is a result of different levels of retention in the ER. These results highlight the functional significance of Spi retention, and provide another facet to the tight regulation of ligand processing.
Results
A novel TGFα homolog
In order to identify additional DER ligand(s), we searched databases for new Spi homologs. Two expressed sequence tags (ESTs) representing the same gene were found to be highly homologous to Spi (LD34470 and LD34429), mapping to 74E/F. The complete sequence of both ESTs revealed an open reading frame of 217 amino acids (NCBI AF245387) (Figure 1A).
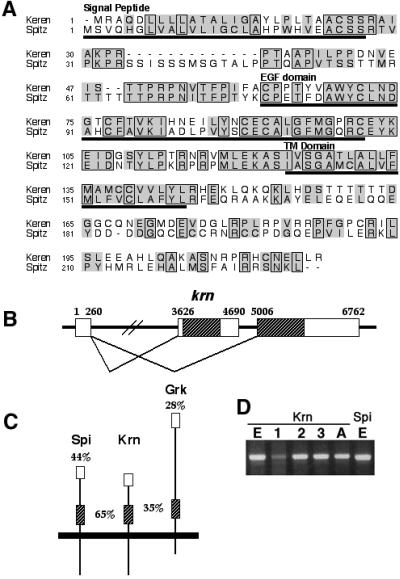
Fig. 1. Krn is highly homologous to Spi. (A) Alignment of the predicted amino acid sequence of Krn and Spi reveals a similar organization, and significant homology not only in the EGF domain, but also in several stretches of the extracellular region. On the other hand, the cytoplasmic domains of the two proteins are highly divergent. Boxes denote amino acid identity and shadows indicate conservative changes. (B) A schematic of the genomic organization of krn exons. According to the ESTs and RT–PCR, krn’s transcript is comprised of two exons, where the ORF is coded by the second exon. Additional ESTs show that the first exon can be alternatively spliced and joined to a different exon, encoding a different ORF with homology to novel vertebrate proteins. It seems that one promoter is regulating the expression of transcripts encoding two different proteins. The number of ESTs indicates that the transcript that does not include Krn is more prevalent. Thus, a low frequency of splicing may be one way to maintain low levels of the krn transcript. Open reading frames are hatched. (C) A schematic of the homologies of Spi and Grk to Krn. Overall amino acid identities, as well as the identities in the EGF domain (hatched), are shown. Krn is much more similar to Spi in sequence and overall organization. (D) RT–PCR (non-quantitative) of the krn transcript from embryos (E), first, second and third instar larvae (1, 2 and 3), and adults (A) shows the expected band of 0.6 kb. For comparison, see spi RT–PCR from embryos.
In the three activating DER ligands identified to date, the homology was restricted to the EGF domain. In contrast, the similarity between Spi and the new putative ligand, extending throughout the coding region, is 56%. The length and domain organization of both proteins are similar. Homologous stretches are concentrated mainly in the extracellular domain, with significant identity in the EGF domain (65%), while limited homology can be found in the cytoplasmic domain. Because of the similarity to Spi, we named this protein Keren, which is the Hebrew word for an antler, or a sharp object.
The ESTs of Krn are comprised of two exons, where the entire coding region is located in the second exon. Interestingly, the 5′ non-coding exon is also found in ESTs of a coding sequence located more 3′ to the gene encoding Krn (NCBI AY069849) (Figure 1B). This coding sequence is homologous to novel mouse and human proteins. The splicing of the 5′ non-coding exon to the TGFα transcript was confirmed by RT–PCR of RNAs extracted from adult flies and S2 cells (Figure 1D). According to the RT–PCR, Krn is expressed during the stages of embryonic and larval development, and in adults. We tried to examine the Krn expression pattern using RNA in situ hybridization and antibody stainings on wild-type embryos or imaginal discs, but the signal was below detection level.
Krn is a functional homolog of Spi
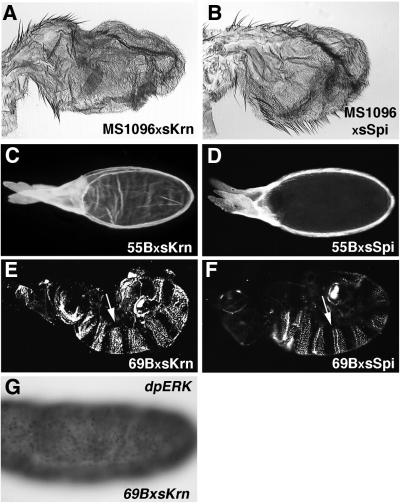
Spi is active only as a cleaved moiety (sSpi), which diffuses to neighboring cells and activates the receptor. Following this paradigm, we have overexpressed the secreted form of Krn (sKrn) in various tissues. Indeed, in all tissues, the phenotypes of sKrn resembled the ectopic phenotype of sSpi (compare left with right panels in Figure 2; see wild-type tissues in Figure 4). In the wing, MS1096-Gal4 driving sKrn gave rise to lethality, but several survivors had short bloated stumps for wings (Figure 2A). In the ovary, expression of sKrn in the anterior follicle cells using 55B-Gal4 resulted in the formation of extra dorsal appendages (Figure 2C). Ectopic sKrn driven by 69B-Gal4 in the embryonic ectodermal cells caused lethality, with the cuticle showing wider and square-edged denticle belts characteristic of DER hyperactivation (Figure 2E). We can look at the same embryos at the molecular level, by staining with the dpERK antibody, which detects the activated form of MAPK. It is evident that sKrn can activate MAPK wherever it is expressed in the embryo (Figure 2G), or in the wing disc (data not shown). These results show that sKrn can mimic sSpi activity and activate the DER pathway.
Fig. 2. Secreted Krn is biologically active. To follow the biological activity of sKrn, the protein was ectopically expressed in several tissues (left panels). The phenotypes obtained displayed striking similarity to those observed following ectopic expression of sSpi (right panels) and are all consistent with hyperactivation of DER. (A and B) Expression in the wing imaginal disc with the MS1096 driver is mostly lethal, and survivors have small, blistered wings with an excess of vein material. See wild-type wing in Figure 4A. (C and D) Expression in the follicle cells with the 55B driver gives rise to excess dorsal appendage material around the circumference of the egg, consistent with dorsalization of the egg. See wild-type egg in Figure 4E. (E and F) Expression in the embryo using the ubiquitous 69B driver generates cuticles where germ band retraction is defective, head structures are missing and the denticle bands are extended dorsally, terminating in a rectangular shape (arrows) instead of the typical wild-type trapezoidal shape. See wild-type cuticle in Figure 4G. (G) Staining of 69B/UAS-sKrn embryos with anti-dpERK antibodies reveals the ubiquitous activation of the DER signaling pathway. For wild-type staining see Figure 3A.
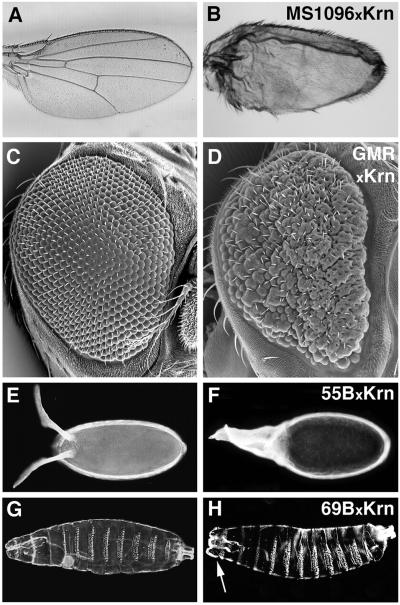
Fig. 4. Ectopic activity of membrane Krn. In contrast to the precursor of Spi, which does not induce ectopic phenotypes upon overexpression, the precursor of Krn gives rise to phenotypes that are consistent with DER hyperactivation in a variety of tissues, at a level that is significantly lower than that induced by sKrn. (A, C, E and G) The wild-type situation. (B) In the wing, Krn expression by MS1096 generated smaller, blistered wings. The photograph was taken at 1.6× magnification, compared with (A). (D) Rough, disorganized eyes were generated with the GMR driver. (F) Extended dorsal appendages were seen following induction by 55B. (H) Finally, in the embryo induction led to lethality. In contrast to the expression of sKrn, the denticle bands remained normal. However, the structures at the head region appeared wider, with a prevalent lobe on each side (arrow).
Having demonstrated the biological activity of sKrn, we wanted to determine whether its processing is regulated in a similar manner to Spi. Triggering of DER by Spi at stage 10 generates two prominent domains of activation, in the tracheal placodes and ventral ectoderm. These domains correspond to the sites of Rho expression in the tracheal placodes and midline, respectively. dpERK can be readily detected in these domains, while in spi mutant embryos no dpERK is observed at this stage (Gabay et al., 1997; Figure 3A). We examined the capacity of the Krn precursor to rescue spi mutant embryos. When Krn was ubiquitously expressed in the ectoderm of spi– embryos (using the 69B-Gal4 driver), complete rescue of the dpERK pattern was observed (Figure 3B). Induction of the pathway by Krn at the sites of Rho expression in the midline and tracheal placodes indicates that, like Spi, processing of Krn is dependent upon Rho. To test whether Krn cleavage requires Star, we expressed Krn in Star mutant embryos. No rescue of the phenotype was observed, as monitored by dpERK (Figure 3C). We thus conclude that, like Spi, processing of Krn is Rho and Star dependent.
Fig. 3. The capacity of Krn and Spi constructs to rescue spi or Star mutant embryos. (A) At stage 10, activation of the DER pathway generates a typical dpERK pattern in the tracheal placodes (TP) and ventral ectoderm (VE). No staining is observed at this stage in spi or Star mutant embryos. (B) Expression of Krn with the 69B driver in spi mutant embryos leads to complete rescue of the dpERK pattern in both tissues. This indicates that like Spi, cleavage of Krn is restricted to the domains of Rho expression. (C) Similar expression of Krn in Star mutant embryos did not lead to rescue, indicating that like Spi, Krn also requires Star for processing. (D) A construct of the Spi precursor in which the residues between the EGF and transmembrane domains have been deleted is unable to rescue spi mutant embryos, indicating that cleavage is necessary for the high level of DER activation. (E) A similar construct of the Krn precursor is unable to rescue spi mutants. (F) A chimeric precursor protein containing the Spi extracellular domain and the Krn intracellular domain is capable of undergoing processing and can rescue the spi mutant. (G) Similarly, a construct of the Spi precursor in which the cytoplasmic domain has been deleted can rescue spi.
Krn can also activate DER in a Rho- and Star-independent manner
While Spi is an extremely potent ligand in its secreted form, when expressed in the precursor form, even at high levels, it shows no ectopic activity. Following this paradigm, the rescue of spi mutant embryos by high levels of expression of Krn showed detectable levels of dpERK only at the tissues where Spi cleavage normally takes place.
To our surprise, in contrast to Spi, Krn was capable of eliciting phenotypes in a variety of tissues following overexpression. For example, in the wing, Krn overexpression gave rise to bloated wings, similar to those obtained upon moderate activation of DER with the λ-top construct (see Reich et al., 1999; Figure 4B). Over expression in the eye led to rough eyes and in the follicle cells to excess dorsal appendage material (Figure 4D and F). In the embryo, expression of Krn gave rise to embryonic lethality, with cuticles displaying expansion of head structures, but otherwise normal in appearance (Figures 4H and 6G).
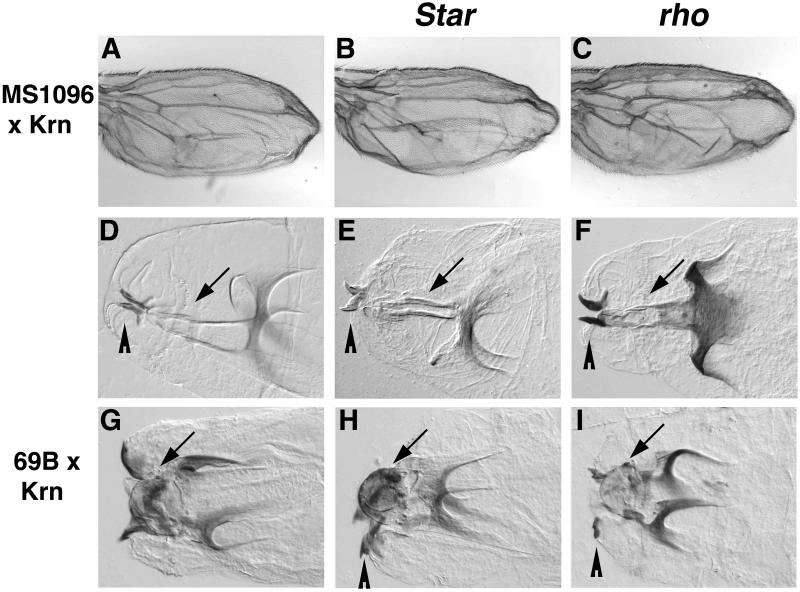
Fig. 6. The activation of DER by Krn is Star and Rho independent. (A) Expression of Krn in the wing disc by MS1096-Gal4 gave rise to a bloated wing phenotype. (B and C) This phenotype is not altered by halving the gene dosage of Star or rho. Using head structures as an assay for ectopic Krn activity in the embryo allowed us to test whether Star or Rho is necessary for generating this phenotype. (D) The mouth hooks (arrowhead) and H-piece (arrow) in a wild-type embryo. (E and F) The cephalopharyngeal skeleton is not disrupted in Star or rho mutant embryos. (G) Overexpression of Krn by 69B-Gal4 gave rise to a deformed cephalopharyngeal skeleton, where the mouth hooks are widely separated and the H-piece is displaced exteriorly, forming a lobe between the mouth hooks (arrow). (H and I) A similar phenotype was observed when Krn was expressed in Star or rho mutant embryos identified by the denticle belt defects, demonstrating that the low-level activity of Krn is independent of Star and Rho. Mouth hooks are indicated by an arrowhead.
These phenotypes indicated low levels of DER activation, not detected by dpERK antibody staining. Thus, uniform Krn expression in embryos did not expand the pattern of dpERK beyond the domains of Rho expression (Figure 3B). Following uniform Krn expression in the wing disc, the resulting phenotype reflected a broad and ubiquitous pattern of activation. Again, this activation did not expand the detectable dpERK pattern (data not shown).
The Krn overexpression phenotypes are consistent with uniform activation of DER. The eye phenotypes were tested for sensitivity to gene dosage of DER and its downstream components, to verify that they reflect DER activation. Indeed, the severity of the rough-eye phenotypes caused by Krn overexpression was reduced by halving the dosage of DER, sos or ras (Figure 5).
Fig. 5. Ectopic Krn activity activates the DER pathway. (A) Expression of Krn by GMR-Gal4 gave rise to rough eyes. (B) In a DER heterozygous background, the severity of the phenotype was reduced. A similar reduction was observed in sos heterozygous flies (C), or in ras heterozygous flies (D).
It was shown that high levels of Krn activity require Rho and Star. We wanted to test whether the low-level activity of Krn is also dependent on these proteins. The overexpression of Krn in the wing disc leads to a bloated wing phenotype that is not sensitive to a reduction in the dosage of either Star or rho (Figure 6A–C). This can also be assayed more rigorously by using the disorganization of head structures that can be identified in cuticle preparations of embryos in which Krn is ectopically expressed, where the mouth hooks are displaced and the H-piece is positioned on the outer surface (Figures 4H and 6G). Star or rho mutant embryos display cuticles that can be easily identified by virtue of reduced and often fused denticle belts. While the heads of these embryos are pointed, the position and arrangement of mouth hooks and cephalopharyngeal skeleton are normal (Figure 6D–F). Expres sion of Krn in Star or rho mutant embryos did not rescue the denticle defects, but was still capable of inducing the aberrant cephalopharyngeal skeleton phenotype (Figure 6G–I). These experiments demonstrate that the low-level activity of Krn can proceed in the absence of Star or Rho.
Two mechanisms may account for this lower level of Krn activity. One possibility is that it can activate DER as a transmembrane ligand, similar to what has been shown for several ligands (reviewed in Massagué and Pandiella, 1993). Alternatively, Krn may undergo low-level cleavage, and activate DER by residual levels of sKrn that are produced. The experiments below will distinguish between these alternatives.
Krn undergoes Rho- and Star-independent, low-level cleavage
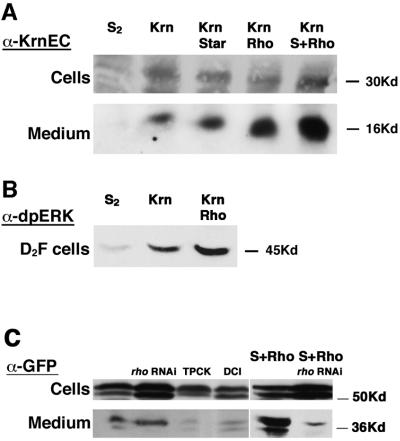
Cleavage of Spi and Krn can be tested in S2 cells. Spi expressed in these cells does not undergo cleavage (Schweitzer et al., 1995; Figure 8G). Only upon co-expression of Spi and Star, or of Spi, Star and Rho, is Spi cleaved and secreted to the medium (Lee et al., 2001; Tsruya et al., 2002). In contrast, expression of Krn in S2 cells resulted in the appearance of sKrn in the medium, as detected by an antibody directed against the extracellular domain of Krn (Figure 7A). sKrn accumulating in the medium is biologically active, as monitored by the capacity to trigger elevation of dpERK in S2 cells expressing DER (Figure 7B). We suggest that this cleavage of Krn in S2 cells represents the basal, low-level cleavage of Krn. Upon co-expression of Star or Rho with Krn, the amount of sKrn in the medium is markedly elevated. The highest levels of cleavage are reached when Krn is co-expressed with both Star and Rho (Figure 7A).
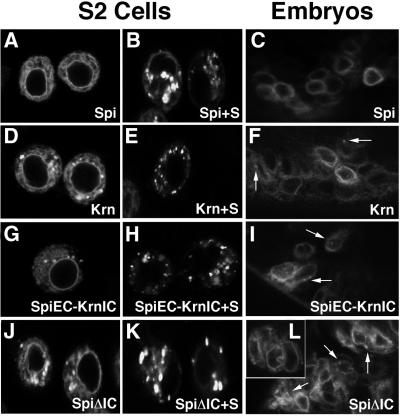
Fig. 8. Low-level cleavage of Krn in flies and in S2 cells. The chimeric and deletion constructs of Krn and Spi are illustrated. A summary of the high and low cleavage profile of each ligand is presented. All constructs, except those where the cleavage site has been truncated, were capable of rescuing the spi dpERK pattern, e.g. undergoing high-level cleavage (Figure 3). Low-level cleavage of the constructs was assayed in vivo by misexpression in the wing with the MS1096 driver. (A) Spi misexpression did not alter the wing. (B) Krn misexpression gave rise to smaller bloated wings. (C) Deletion construct of 16 amino acids that are essential for cleavage in Spi (comprising the sequence between the EGF and transmembrane domains) had no effect upon misexpression. (D) Deletion of this sequence in Krn abolished the capacity to induce the wing phenotype. This indicates that the low-level activation of Krn is dependent on cleavage. (E) A chimera containing the Spi extracellular domain and the Krn transmembrane and intracellular domains was capable of inducing the wing phenotype. (F) Deletion of the Spi intracellular domain allowed the protein to induce the wing phenotype. The wings in (B), (E) and (F) are shown at a magnification of 1.6× compared with (A), (C) and (D). (G) The upper panel shows sSpi in medium collected from cells transfected with different Spi constructs. Low levels of cleavage could be seen in cells transfected either with SpiΔIC alone or with SpiEC–KrnIC alone, indicating that the intracellular domain of Spi inhibits its low-level cleavage. SpiΔ16aa did not show cleavage under these conditions. The lower panel shows expression levels of each protein in cell extracts.
Fig. 7. Krn processing in S2 cells. (A) The upper panel shows comparable levels of Krn expression in stably transfected S2 cell extracts. These cells do not express detectable levels of Star and Rho. The lower panel shows sKrn in the medium collected from each transfection. When Krn was expressed alone, it was cleaved (consistent with the low-level activation observed in flies). When Rho, or Rho and Star were co-expressed with Krn, higher levels of sKrn were detected in the medium. This is likely to reflect the high-level cleavage of Krn, which is Rho and Star dependent. (B) sKrn in the medium was capable of triggering DER in S2 cells expressing the EGF receptor, as monitored by the accumulation of dpERK. The levels of MAPK activation were correlated to the levels of sKrn in the medium. (C) S2 cells were transiently transfected with Krn–GFP, and secreted protein was detected in the medium. Co-transfection of a rho RNAi construct did not reduce low-level cleavage, but dramatically reduced the cleavage in the presence of Star and Rho. Low-level cleavage was sensitive to the serine protease inhibitors TPCK and DCI (100 µM).
Elimation of possible residual Rho activity in S2 cells expressing Krn was carried out by co-expression of a rho RNAi construct. While this construct was capable of reducing the cleavage of Krn in the presence of Star and Rho, it had no effect on the low-level cleavage of Krn (Figure 7C). Cleavage of Spi facilitated by Rho was shown to be inhibited by serine protease inhibitors, such as TPCK and DCI (Urban et al., 2001). The low-level cleavage of Krn was also inhibited by these reagents (Figure 7C). This result is consistent with the involvement of other members of the Rho family in Krn cleavage, or with cleavage by other serine proteases. In view of the restricted expression of Rho family members, their engagement in ubiquitous cleavage of Krn is less likely.
An alternative approach to examine cleavage as the basis for the activity of Krn is to generate a construct in which the 16 amino acids (16aa) between the EGF and transmembrane domains were deleted. A similar deletion construct of Spi lost the capacity to undergo cleavage in Xenopus assays (Bang and Kintner, 2000), and in mammalian cells its cleavage was greatly compromised (Lee et al., 2001). The 16aa deletion constructs were generated for Krn and Spi, and tested in S2 cells (KrnΔ16aa and SpiΔ16aa). No accumulation of sSpi or sKrn was observed in the medium, when expressed alone or with Star (Figure 8G; data not shown) or upon co-expression of both Rho and Star (data not shown). When either construct was expressed in flies, no ectopic phenotypes were observed in the wing (Figure 8C and D). These results demonstrate that the biological activity of Krn relies on low-level cleavage, generating sKrn.
The two constructs were also tested for the capacity to rescue dpERK staining of spi mutant embryos, and no rescue was observed (Figure 3D and E). This result indicates that, in flies, these cleavage-impaired forms of Spi or Krn have no biological activity. Thus, high levels of DER activation depend on efficient generation of the cleaved ligands.
The intracellular domain controls the basal activity profile
What causes the difference in regulation of basal cleavage between Krn and Spi? The most obvious difference between the two ligands lies within the intracellular domains. We wanted to test whether switching these domains would lead to an alteration in basal cleavage. A chimeric protein was constructed, where the extracellular domain belonged to Spi and the transmembrane and cytoplasmic domains to Krn (SpiEC–KrnIC). The biological activity of the chimera was verified by its capacity to rescue the dpERK pattern in spi mutant embryos, demonstrating the ability to undergo regulated, high-level cleavage (Figure 3F). To test the low-level activity of the chimera, the wing phenotype was monitored following overexpression. Indeed, bloated wings were formed (Figure 8E). Furthermore, this chimera was able to undergo low-level cleavage in the absence of Star and Rho (Figure 8G), demonstrating that the control of Spi basal cleavage lies within the transmembrane or intracellular domains.
The above experiments highlighted the role of the Krn intracellular domain in controlling basal cleavage. It was not clear, however, if the cytoplasmic domain of Krn designates it for basal cleavage, thereby causing SpiEC– KrnIC to be cleaved in the same manner, or alternatively, whether this domain in Spi inhibits the same process. To distinguish between these possibilities, we have generated a Spi construct where the cytoplasmic domain was deleted, while the C-terminal leucine residue, which may be necessary for proper cellular trafficking, was retained (SpiΔIC). This construct behaved like the Spi–Krn chimera. It rescued spi mutant embryos (Figure 3G) and could trigger the typical bloated wing phenotype, consistent with basal-level cleavage (Figure 8F). In S2 cells, it was cleaved in the absence of Star and Rho, like Krn (Figure 8G). These results assign an inhibitory role for basal cleavage to the cytoplasmic, C-terminal domain of Spi.
Previous experiments have implicated the Spi cytoplasmic domain in repressing cleavage (Bang and Kintner, 2000). Specifically, the cytoplasmic domain of Spi was shown to be necessary for its ER retention, thus preventing it from reaching Rho and undergoing cleavage (Lee et al., 2001; Tsruya et al., 2002). This prompted us to ask what are the intracellular distributions of Krn and the above-mentioned constructs, which were able to undergo low levels of cleavage? We transfected S2 cells with a GFP-tagged version of each protein. Like Spi, Krn–GFP showed perinuclear localization, but unlike Spi, it was also detected in a punctate pattern consistent with limited exit from the ER (Figure 9D compared with A). The same distribution was seen with the chimera SpiEC–KrnIC– GFP (Figure 9G). The localization of these two GFP-tagged proteins mimicked the localization of SpiΔIC–GFP (Figure 9J; Tsruya et al., 2002). This indicates that the cytoplasmic domain of Krn is less potent in conferring ER retention, thus allowing some of the ligand molecules to proceed into the secretory pathway.
Fig. 9. Low-level cleavage in S2 cells and in embryos correlates with vesicular localization. Each of the proteins was constructed to contain GFP after its signal peptide. (A) Spi was shown to have perinuclear localization identified with the ER, in S2 cells. (B) Star was shown to transport Spi out of the ER. Spi is visualized in vesicles instead of the perinuclear ring. (C) After injection of the construct to embryos, Spi shows exclusive perinuclear localization. (D) Krn localization in S2 cells was predominantly perinuclear, but a punctate vesicular distribution, indicating exit from the ER, was also detected. (E) Star led to complete depletion of Krn from the ER. (F) In embryos, Krn also showed some vesicular localization (arrows), in addition to its perinuclear localization. (G) SpiEC–KrnIC showed similar cellular distribution as Krn, indicating that the intracellular domain is the key to ER retention levels. (H) Star caused complete transport of SpiEC–KrnIC from the ER. (I) In embryos, SpiEC–KrnIC mimicked the Krn pattern, showing some vesicular localization. (J) SpiΔIC showed perinuclear localization together with vesicular distribution, indicating that the intracellular domain of Spi inhibits it from exiting the ER. (K) SpiΔIC was depleted from the ER when co-expressed with Star. (L) Embryonic localization of SpiΔIC was similar to that of Krn.
If Krn can evade ER retention, what is the role of Star in Krn cleavage? Co-transfection of Star together with each of these constructs facilitated their efficient removal from the ER (Figure 9E, H and K). Star can thus recognize Krn and translocate it from the ER, in a similar manner as it transports Spi. Although Krn can ‘leak’ out from the ER in the absence of Star, full release from retention, leading to efficient cleavage, can occur only in the presence of Star.
We wanted to examine whether the intracellular distribution observed in S2 cells would also apply in embryos. Indeed, Krn–GFP showed a prominent perinuclear distribution with some vesicular localization (Figure 9F), as can also be seen in embryos injected with SpiEC–KrnIC–GFP or SpiΔIC–GFP (Figure 9I and L). The punctate distribution corroborates the localization seen in S2 cells. Furthermore, it highlights the role of the cytoplasmic domain of each ligand in retention versus partial evasion of retention in vivo. Another feature revealed in embryos was the higher stability of Krn compared with Spi. Unlike Spi–GFP, which is unstable, the GFP constructs of Krn, SpiEC–KrnIC or SpiΔIC were more stable, as evident by the higher percentage of injected embryos in which GFP can be readily detected.
Discussion
Krn structure and expression
The presence of another ligand was expected since in several tissues, such as the wing and eye imaginal discs, the phenotypes of spi mutants were not as severe as those of DER, and moreover, as those of rho mutants. Thus, a ligand that is cleaved by Rho was still missing. We searched EST databases for new Spi homologs, and identified Krn. No additional members were found beside Krn, suggesting that the final list of activating DER ligands will include four members. The protein sequence homology indicated that Krn has a significantly greater similarity to Spi than to the other DER ligands. This is evident in the overall homology, and in particular for the EGF domain of Krn, which is 65% identical to Spi. Indeed, just like sSpi, sKrn is a potent activator of DER. The activation capacity is likely to reside in the EGF domain itself, as was demonstrated by generation of chimeric ligands with different EGF domains (Schnepp et al., 1998).
We attempted to visualize Krn expression by RNA in situ hybridization and by using antibody staining, but did not detect a reliable endogenous signal. It therefore seems that in spite of the ability to detect krn transcripts by RT–PCR at all stages, they are present at very low levels. We have demonstrated that Krn undergoes low-level cleavage. This may account for the low levels of Krn expression. Since high levels of Krn can induce DER activation, tight control of expression is required, and appears to occur at the transcriptional level.
Cleavage of Krn versus Spi
A clearer understanding of Spi cleavage has been gained by studies in cells. Efficient cleavage of Spi occurred only in cells in which both Star and Rho were expressed. Spi is retained in the ER through its intracellular domain. Star binds Spi and translocates it from the ER to the Golgi, where Rho functions as a protease and cleaves Spi (Lee et al., 2001; Tsruya et al., 2002). We will address these steps in relation to Krn.
ER retention. Krn–GFP in Drosophila S2 cells showed partial release from retention, manifested by a vesicular distribution, indicating exit from the ER. The functional implication of this was observed through the ectopic phenotypes in various tissues. For instance, whereas ectopic Spi driven by ubiquitous GAL4 in embryos does not prevent hatching of larvae, ectopic Krn led to lethality. This highlights the importance of retention, as it allows expression of high amounts of Spi protein in the cell, yet controls its activity by preventing it from reaching further compartments where cleavage occurs.
The mechanism of Spi and Krn retention is not clear yet. Spi was also shown to be retained in a heterologous system of mammalian cells, implicating the action of conserved molecules or an intrinsic property of the protein. On one hand, association of the Spi cytoplasmic domain with an additional protein(s) could mediate retention. In that case, we would expect Krn to have lower affinity to this protein(s). On the other hand, the Spi C-terminus itself could have an intrinsic inhibitory capability through protein folding that sterically prohibits association to proteins, which would carry it further in the secretory pathway. In this case, we would expect Krn to posses a higher affinity to such chaperones, which would allow it to exit the ER without total dependence on Star.
Compared with Spi or Krn, the cytoplasmic domain of Grk, the third DER ligand with a transmembrane domain, is shorter (only 24 amino acids). Deletion of its cytoplasmic domain did not influence signaling by Grk (Queenan et al., 1999). Like Krn, overexpression of the full-length Grk protein causes ectopic wing phenotypes (Ghiglione et al., 2002). It would be interesting to see to what extent Grk is retained in the ER of S2 cells.
Low-level cleavage (Star and Rho independent). Expres sion of Krn in S2 cells allowed us to biochemically follow the mechanism of low-level cleavage, which was Star and Rho independent. What is the protease responsible for this cleavage? The sensitivity of Krn cleavage to inhibitors of serine proteases indicates that cleavage may be mediated by a protease of this family. Unlike Rho, which is expressed in a spatially and temporally regulated manner, we expect the protease to be ubiquitously expressed, since ectopic Krn caused abnormal phenotypes wherever it was expressed. This further elaborates the need for tight transcriptional control on Krn expression.
Co-expression of Star with Krn in S2 cells raises the amounts of secreted sKrn in the medium. This is a result of the efficient export of Krn from the ER by Star. Since higher levels of cleavage can be obtained by co-expressing both Rho and Star, it would seem to indicate that the protease involved in low-level cleavage is less efficient than Rho.
High-level cleavage (Star and Rho dependent). High-level cleavage of Krn was followed in embryos through the detection of dpERK. The activation profile followed the restricted expression of Rho, since Star is broadly expressed. Only in cell culture could Rho enhance cleavage of Krn without co-expression of Star (Figure 7). This probably occurs because Krn can ‘leak’ out of the ER, reach compartments where Rho is present and undergo cleavage by Rho independently of Star.
Sequence conservation within the transmembrane domains of the Rho protein suggested that the cleavage site of Spi would reside within the transmembrane domain. There is also evidence to indicate that Rho cleaves Spi within the membrane (Urban et al., 2001). The transmembrane regions of Spi and Krn are conserved, and show 50% sequence identity. The transmembrane domain of Krn may thus possess the same recognition sites as that of Spi.
In view of the two modes of Krn cleavage, where could it have biological roles? The capacity of Krn to undergo Rho- and Star-dependent high-level cleavage suggests that it could provide the missing ligand in tissues where the rho phenotype is more severe than that of spi. This includes the formation of veins in the wing, generation of correct R8 spacing and inhibition of apoptosis after the morphogenetic furrow in the eye disc. In the embryo, the spi phenotypes are similar to those of rho or Star mutants (Mayer and Nusslein-Volhard, 1988), indicating that Krn is not likely to be required at this stage.
Could the low-level cleavage of Krn also play a physiological role in activating DER? In the eye imaginal disc, generation of clones of DER mutants either anterior or posterior to the morphogenetic furrow was not possible, due to cell lethality (Xu and Rubin, 1993; Baonza et al., 2001). The known rho family genes are not expressed anterior to the furrow. It is thus possible that the low-level cleavage of Krn may provide the residual levels of secreted ligand necessary to trigger DER anterior to the furrow, and allow cell survival. The low levels of DER activation anterior to the furrow are consistent with the non-detectable levels of dpERK in this domain (Gabay et al., 1997). Sufficient levels of secreted ligand would need to be produced to elicit a biological response, in spite of the low level of krn expression. In the Krn misexpression assays presented in this work, high levels of expression were required to detect this low activity.
In order to examine the biological roles of Krn, we expressed krn double-stranded (ds) RNA. We focused our efforts in the eye and wing imaginal discs, where we expected Krn activity. Uniform expression of krn dsRNA in these tissues, even when combined with spi dsRNA, did not yield a noticeable phenotype. We examined the pattern of phospho-histone3 to follow cell cycle progression in the wing disc and anterior to the morphogenetic furrow in the eye disc, and did not detect abnormal phenotypes. The possible roles of Krn in these tissues may be confirmed only when mutations for krn become available and their phenotypes are tested, alone or in combination with mutants for spi.
In conclusion, we identify Krn, a new DER ligand, which is the functional homolog of Spi. Unlike Spi, Krn is capable of undergoing inefficient Star- and Rho-independent cleavage in flies and in cell culture. This is due to differences between the intracellular domains of Krn and Spi, which allow Krn to evade retention in the ER and reach further along in the secretory pathway. This calls for tight transcriptional control of Krn expression, in contrast to Spi, which can be ubiquitously and abundantly expressed.
Materials and methods
DNA constructs
UAS-Krn was produced by inserting the krn EST 34470 into pUAST through EcoRI and XhoI sites. UAS-sKrn was generated through PCR by inserting a stop codon after T113, and cloned by BglII–KpnI sites. UAS-KrnΔ16aa was produced by ligating two PCR fragments containing codons 1–107 and 124–217 through a ClaI site, and cloning in pUAST through EcoRI–XhoI sites. UAS-SpiΔ16aa was similarly generated, fusing the coding regions for 1–123 and 140–230. UAS-SpiEC–KrnIC was produced by similar restriction sites through fusion of two PCR fragments containing Spi codons 1–139 and Krn codons 124–217. UAS-SpiΔIC was generated by a PCR fragment containing codons 1–170, followed by the FLAG epitope (DYKDDDDK) and terminating with AAL residues. GFP-tagged Spi and SpiΔIC were described in Tsruya et al. (2002). GFP-tagged Krn and SpiEC–KrnIC constructs were made by ligating the GFP fragment preceded by the Spi signal peptide to each of the relevant constructs. In Krn, the fusion was at the position of I39, in SpiEC–KrnIC, at the position of S56 of the extracellular portion of Spi. UAS-rho RNAi was produced by cDNA bases 254–1027 (antisense orientation), followed by bases 86–1027 (sense orientation).
Antibody preparation and staining
Codons 4–114 of krn were inserted into the pRSET vector, using BamHI–HindIII sites. Antibodies to the purified protein were produced in rats. Anti-dpERK staining was performed with monoclonal antibody from Sigma at a dilution of 1/50. Jackson ImmunoResearch biotinylated anti-mouse antibodies were used, followed by amplification with the Vectastain ABC kit and Tyramide amplification (NEN). Rabbit anti-phospho-histone3 was from Upstate Biotechnology. For RNA in situ hybridization, RNA probes were prepared from the EST clone, using the SP6 promoter.
Flies
For ectopic expression, the following Gal4 drivers were used: MS1096, GMR, 55B and 69B. For testing rescue of spi mutants by Krn, spi0E92/CyO ftz-lac; 69B/69B flies were crossed to spi0E92, UAS-Krn/CyO ftz-lacZ or to spi0E92, UAS-Krn/CyO ftz-lacZ; UAS-Krn/UAS-Krn. spi mutant embryos were identified by the lack of anti-β-Gal staining. Similarly, for rescue of spi by the other constructs, the same driver line was crossed to lines carrying spi0E92 over CyO ftz-lacZ and the UAS-Krn construct on the X, second or third chromosomes. Rescue of Star mutants was tested by crossing the driver line SIIN23/CyO ftz-lacZ; 69B/69B to SIIN23/CyO ftz-lacZ flies with the UAS-Krn construct on the X or third chromosome. For ectopic Krn phenotypes in the background of spi or Star mutants, the same crosses were used and assayed by cuticle preparations. For ectopic Krn phenotypes in the background of rho mutants, the following cross was carried out: rhoΔ38, 69B/TM3 ftz-lacZ over UAS-Krn/UAS-Krn; rhoΔ38/TM3 ftz-lacZ. For genetic interactions in the eye, the following alleles were utilized: flb2G31, sosezH and rase1B. Transient injection of embryos has been described previously (Tsruya et al., 2002).
Cell culture
Schneider S2 cells were stably transfected with metallothionein promoter fusions to the coding regions of krn, Star or rho. Resistance to hygromycin was conferred by co-transfection with the pMK33 vector. Induction by Cu2+ was carried out for 48–72 h in serum-free medium, and the cells and medium were subsequently analyzed on western blots. Induction of DER was tested by incubating S2 cells constitutively expressing DER (D2F) with medium for 7–10 min, followed by centrifugation and lysis. The level of dpERK was analyzed by western blotting.
To examine the effect of rho RNAi, a UAS-rho RNAi construct was transiently co-transfected with UAS-krn–GFP and actin-Gal4 plasmids. Two days following transfection, cells were collected and incubated in serum-free medium for 3 h. The proteins secreted into the medium were TCA precipitated and resuspended in sample buffer. For the inhibitor studies, a similar transfection and collection protocol was used, but Krn–GFP-expressing cells were incubated in the presence of 100 µM Nα-tosyl-phe-chloromethyl ketone (TPCK) or diphenyleneiodonium chloride (DCI) (Calbiochem) for the 3 h of secretion to serum-free medium.
RT–PCR
For krn, the forward oligonucleotide was 5′-GCCAGCCAAAGCCAAGAATTC and the reverse 5′-CTCGTCCATGCCCTCGTTC. For spi, the following forward and reverse oligonucleotides were used: 5′-GCCCTAGCAGCGGACAACG and 5′-AGGCCAGGCAGACAAACAGC.
Acknowledgments
Acknowledgements
We thank A.Sapir for pointing out the Krn homology, M.Tugentman for producing the UAS-rho RNAi, members of the Shilo laboratory for discussions, and Z.Paroush and T.Volk for critical reading of the manuscript. This work was supported by the US–Israel binational fund to B.-Z.S., who is an incumbent of the Hilda and Cecil Lewis professorial chair in Molecular Genetics.
References
- Bang A.G. and Kintner,C. (2000) Rhomboid and Star facilitate presentation and processing of the Drosophila TGF-α homolog Spitz. Genes Dev., 14, 177–186. [PMC free article] [PubMed] [Google Scholar]
- Baonza A., Casci,T. and Freeman,M. (2001) A primary role for the epidermal growth factor receptor in ommatidial spacing in the Drosophila eye. Curr. Biol., 11, 396–404. [DOI] [PubMed] [Google Scholar]
- Bier E., Jan,L.Y. and Jan,Y.N. (1990) Rhomboid, a gene required for dorsoventral axis establishment and peripheral nervous system development in Drosophila melanogaster. Genes Dev., 4, 190–203. [DOI] [PubMed] [Google Scholar]
- Freeman M. (2000) Feedback control of intercellular signalling in development. Nature, 408, 313–319. [DOI] [PubMed] [Google Scholar]
- Gabay L., Scholz,H., Golembo,M., Klaes,A., Shilo,B.Z. and Klambt,C. (1996) EGF receptor signaling induces pointed P1 transcription and inactivates Yan protein in the Drosophila embryonic ventral ectoderm. Development, 122, 3355–3362. [DOI] [PubMed] [Google Scholar]
- Gabay L., Seger,R. and Shilo,B.Z. (1997) In situ activation pattern of Drosophila EGF receptor pathway during development. Science, 277, 1103–1106. [DOI] [PubMed] [Google Scholar]
- Ghiglione C., Bach,E.A., Paraiso,Y., Carraway,K.L., Noselli,S. and Perrimon,N. (2002) Mechanism of activation of the Drosophila EGF receptor by the TGFα ligand Gurken during oogenesis. Development, 129, 175–186. [DOI] [PubMed] [Google Scholar]
- Golembo M., Raz,E. and Shilo,B.Z. (1996) The Drosophila embryonic midline is the site of Spitz processing, and induces activation of the EGF receptor in the ventral ectoderm. Development, 122, 3363–3370. [DOI] [PubMed] [Google Scholar]
- Golembo M., Yarnitzky,T., Volk,T. and Shilo,B.Z. (1999) Vein expression is induced by the EGF receptor pathway to provide a positive feedback loop in patterning the Drosophila embryonic ventral ectoderm. Genes Dev., 13, 158–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Reyes A., Elliott,H. and St Johnston,D. (1995) Polarization of both major body axes in Drosophila by gurken–torpedo signalling. Nature, 375, 654–658. [DOI] [PubMed] [Google Scholar]
- Guichard A., Biehs,B., Sturtevant,M.A., Wickline,L., Chacko,J., Howard,K. and Bier,E. (1999) Rhomboid and Star interact synergistically to promote EGFR/MAPK signaling during Drosophila wing vein development. Development, 126, 2663–2676. [DOI] [PubMed] [Google Scholar]
- Kolodkin A.L., Pickup,A.T., Lin,D.M., Goodman,C.S. and Banerjee,U. (1994) Characterization of Star and its interactions with sevenless and EGF receptor during photoreceptor cell development in Drosophila. Development, 120, 1731–1745. [DOI] [PubMed] [Google Scholar]
- Lee J.R., Urban,S., Garvey,C.F. and Freeman,M. (2001) Regulated intracellular ligand transport and proteolysis control EGF signal activation in Drosophila. Cell, 107, 161–171. [DOI] [PubMed] [Google Scholar]
- Massagué J. and Pandiella,A. (1993) Membrane-anchored growth factors. Annu. Rev. Biochem., 62, 515–541. [DOI] [PubMed] [Google Scholar]
- Mayer U. and Nusslein-Volhard,C. (1988) A group of genes required for pattern formation in the ventral ectoderm of the Drosophila embryo. Genes Dev., 2, 1496–1511. [DOI] [PubMed] [Google Scholar]
- Musacchio M. and Perrimon,N. (1996) The Drosophila kekkon genes: novel members of both the leucine-rich repeat and immunoglobulin superfamilies expressed in the CNS. Dev. Biol., 178, 63–76. [DOI] [PubMed] [Google Scholar]
- Nagaraj R., Pickup,A.T., Howes,R., Moses,K., Freeman,M. and Banerjee,U. (1999) Role of the EGF receptor pathway in growth and patterning of the Drosophila wing through the regulation of vestigial. Development, 126, 975–985. [DOI] [PubMed] [Google Scholar]
- Neuman-Silberberg F.S. and Schupbach,T. (1993) The Drosophila dorsoventral patterning gene gurken produces a dorsally localized RNA and encodes a TGFα-like protein. Cell, 75, 165–174. [PubMed] [Google Scholar]
- Nilson L.A. and Schupbach,T. (1999) EGF receptor signaling in Drosophila oogenesis. Curr. Top. Dev. Biol., 44, 203–243. [DOI] [PubMed] [Google Scholar]
- Pascall J.C. and Brown,K.D. (1998) Characterization of a mammalian cDNA encoding a protein with high sequence similarity to the Drosophila regulatory protein Rhomboid. FEBS Lett., 429, 337–340. [DOI] [PubMed] [Google Scholar]
- Pickup A.T. and Banerjee,U. (1999) The role of star in the production of an activated ligand for the EGF receptor signaling pathway. Dev. Biol., 205, 254–259. [DOI] [PubMed] [Google Scholar]
- Queenan A.M., Barcelo,G., Van Buskirk,C. and Schupbach,T. (1999) The transmembrane region of Gurken is not required for biological activity, but is necessary for transport to the oocyte membrane in Drosophila. Mech. Dev., 89, 35–42. [DOI] [PubMed] [Google Scholar]
- Reich A., Sapir,A. and Shilo,B. (1999) Sprouty is a general inhibitor of receptor tyrosine kinase signaling. Development, 126, 4139–4147. [DOI] [PubMed] [Google Scholar]
- Roth S., Neuman-Silberberg,F.S., Barcelo,G. and Schupbach,T. (1995) Cornichon and the EGF receptor signaling process are necessary for both anterior–posterior and dorsal–ventral pattern formation in Drosophila. Cell, 81, 967–978. [DOI] [PubMed] [Google Scholar]
- Rutledge B.J., Zhang,K., Bier,E., Jan,Y.N. and Perrimon,N. (1992) The Drosophila spitz gene encodes a putative EGF-like growth factor involved in dorsal–ventral axis formation and neurogenesis. Genes Dev., 6, 1503–1517. [DOI] [PubMed] [Google Scholar]
- Sapir A., Schweitzer,R. and Shilo,B.Z. (1998) Sequential activation of the EGF receptor pathway during Drosophila oogenesis establishes the dorsoventral axis. Development, 125, 191–200. [DOI] [PubMed] [Google Scholar]
- Schnepp B., Grumbling,G., Donaldson,T. and Simcox,A. (1996) Vein is a novel component in the Drosophila epidermal growth factor receptor pathway with similarity to the neuregulins. Genes Dev., 10, 2302–2313. [DOI] [PubMed] [Google Scholar]
- Schnepp B., Donaldson,T., Grumbling,G., Ostrowski,S., Schweitzer,R., Shilo,B.Z. and Simcox,A. (1998) EGF domain swap converts a Drosophila EGF receptor activator into an inhibitor. Genes Dev., 12, 908–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer R. and Shilo,B.Z. (1997) A thousand and one roles for the Drosophila EGF receptor. Trends Genet., 13, 191–196. [DOI] [PubMed] [Google Scholar]
- Schweitzer R., Shaharabany,M., Seger,R. and Shilo,B.Z. (1995) Secreted Spitz triggers the DER signaling pathway and is a limiting component in embryonic ventral ectoderm determination. Genes Dev., 9, 1518–1529. [DOI] [PubMed] [Google Scholar]
- Sturtevant M.A., Roark,M. and Bier,E. (1993) The Drosophila rhomboid gene mediates the localized formation of wing veins and interacts genetically with components of the EGF-R signaling pathway. Genes Dev., 7, 961–973. [DOI] [PubMed] [Google Scholar]
- Tio M. and Moses,K. (1997) The Drosophila TGFα homolog Spitz acts in photoreceptor recruitment in the developing retina. Development, 124, 343–351. [DOI] [PubMed] [Google Scholar]
- Tsruya R., Schlesinger,A., Reich,A., Gabay,L., Sapir,A. and Shilo,B.Z. (2002) Intracellular trafficking by Star regulates cleavage of the Drosophila EGF receptor ligand Spitz. Genes Dev., 16, 222–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban S., Lee,J.R. and Freeman,M. (2001) Drosophila Rhomboid-1 defines a family of putative intramembrane serine proteases. Cell, 107, 173–182. [DOI] [PubMed] [Google Scholar]
- Wasserman J.D. and Freeman,M. (1998) An autoregulatory cascade of EGF receptor signaling patterns the Drosophila egg. Cell, 95, 355–364. [DOI] [PubMed] [Google Scholar]
- Wasserman J.D., Urban,S. and Freeman,M. (2000) A family of rhomboid-like genes: Drosophila rhomboid-1 and roughoid/rhomboid-3 cooperate to activate EGF receptor signaling. Genes Dev., 14, 1651–1663. [PMC free article] [PubMed] [Google Scholar]
- Xu T. and Rubin,G.M. (1993) Analysis of genetic mosaics in developing and adult Drosophila tissues. Development, 117, 1223–1237. [DOI] [PubMed] [Google Scholar]
- Zak N.B., Wides,R.J., Schejter,E.D., Raz,E. and Shilo,B.Z. (1990) Localization of the DER/flb protein in embryos: implications on the faint little ball lethal phenotype. Development, 109, 865–874. [DOI] [PubMed] [Google Scholar]