Abstract
Cells respond to a wide variety of stresses through the transcriptional activation of genes that harbour stress elements within their promoters. While many of these elements are shared by genes encoding proteins representative of all subcellular compartments, cells can also respond to stresses that are specific to individual organelles, such as the endoplasmic reticulum un folded protein response. Here we report on the discovery and characterization of a mitochondrial stress response in mammalian cells. We find that the accumulation of unfolded protein within the mitochondrial matrix results in the transcriptional upregulation of nuclear genes encoding mitochondrial stress proteins such as chaperonin 60, chaperonin 10, mtDnaJ and ClpP, but not those encoding stress proteins of the endoplasmic reticulum. Analysis of the chaperonin 60/10 bidirectional promoter identified a CHOP element as the mitochondrial stress response element. Dominant-negative mutant forms of CHOP and overexpression of CHOP revealed that this transcription factor, in association with C/EBPβ, regulates expression of mitochondrial stress genes in response to the accumulation of unfolded proteins.
Keywords: gene regulation/mitochondria/molecular chaperones/stress
Introduction
Exposure of cells to stress results in protein unfolding and the impairment of essential cellular processes. Such stress leads to the transcriptional activation of genes that harbour stress response elements within their promoters. The heat shock element (HSE) is found in promoters of genes encoding proteins representative of all subcellular compartments (Morimoto, 1998), enabling cells to respond to global stresses by the increased synthesis of heat shock proteins and other molecular chaperones involved in repair (Hartl, 1996). However, cells can also respond to stresses that are specific to individual organelles. For example, the endoplasmic reticulum (ER) unfolded protein response (UPR) is a well-documented example wherein a wide range of genes encoding proteins involved in the maintenance of ER function are upregulated (Travers et al., 2000). The accumulation of unfolded proteins in the ER of yeast activates a transmembrane kinase/nuclease, Ire1p (Cox et al., 1993; Mori et al., 1993; Shamu and Walter, 1996), which leads ultimately to the production of Hac1p, a bZIP transcription factor that activates the transcription of a set of UPR target genes (reviewed by Chapman et al., 1998).
In mammalian cells, UPR is more complex and diverse (reviewed by Ma and Hendershot, 2001), having a partly parallel Ire1 pathway (Yoshida et al., 2001; Calfon et al., 2002) and a unique system largely dependent on PERK and ATF6, not found in yeast (Harding et al., 2000; Ye et al., 2000; Yoshida et al., 2000). One of the genes upregulated by UPR is the CHOP gene encoding a bZIP transcription factor CHOP [C/EBP homology protein, also called GADD 153 (growth arrest DNA damage 153)] that dimerizes with members of the C/EBP (CCAAT/enhancer-binding protein) family of transcription factors (Ron and Habener, 1992; Fawcett et al., 1996). The CHOP gene has been shown previously to be upregulated by a wide variety of cellular stress responses such as growth arrest and DNA damage (Fornace et al., 1989; Schmitt-Ney and Habener, 2000), amino acid and glucose deprivation (Carlson et al., 1993; Marten et al., 1994; Bruhat et al., 1997), hypoxia (Price and Calderwood, 1992), or exposure of cells to chemicals such as reducing agents (Chen et al., 1992), tunicamycin (Ubeda et al., 1996) and calcium ionophores (Bartlett et al., 1992).
CHOP expression is regulated by a number of transcriptional and translational mechanisms (Jousse et al., 2001) and there is increasing evidence that CHOP is implicated in apoptosis (reviewed by Spear and Ng, 2001). Surprisingly, only three CHOP-inducible genes have so far been identified (Wang et al., 1998; Sok et al., 1999).
The mitochondrial matrix contains its own set of molecular chaperones involved in the subsequent folding of newly imported proteins, and also for the folding of some of the 13 polypeptides encoded by mtDNA (reviewed by Ryan et al., 1997a). These molecular chaperones include chaperonin 60 (Cpn60/Hsp60), chaperonin 10 (Cpn10/Hsp10), mtHsp70, mtGrpE and mtDnaJ, which are homologues of Escherichia coli GroEL, GroES, DnaK, GrpE and DnaJ, respectively. We therefore reasoned that an additional UPR pathway might also exist in a cell, which senses and responds to a mitochondrial specific stress.
We have reported previously that deletion of mtDNA from mammalian cells induced a stress response by stimulating the transcription of the nuclear genes encoding Cpn60 and Cpn10 (Martinus et al., 1996). Stress-inducible chaperones from non-mitochondrial compartments, such as the Hsp70 isoforms BiP (ER) and Hsp72 (cytosol), were not upregulated. However, so far a mitochondrial stress response (MSR), whereby accumulation of unfolded proteins within the mitochondrial compartment results in the selective upregulation of target genes encoding mitochondrial stress proteins, has not been described. Here, we report on the discovery of a MSR in mammalian cells and identify the transcription factors responsible for target gene activation as CHOP and C/EBPβ. We identify four more CHOP target genes, all encoding mitochondrial stress proteins. These are the Cpn60 and Cpn10 genes, which are linked by a bidirectional promoter (Ryan et al., 1997b), the gene encoding the mitochondrial isoform of DnaJ (mtDnaJ), and the ClpP gene, encoding the catalytic subunit of the ATP-dependent mitochondrial protease.
Results
Unfolded proteins in mitochondria induce nuclear genes encoding mitochondrial stress proteins
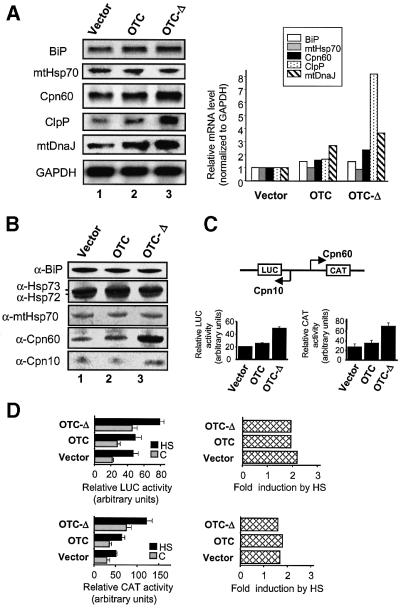
We created a model for the accumulation of protein aggregates within the mitochondrial matrix by making a mutant form of the mitochondrial matrix protein, ornithine transcarbamylase (OTC), by deletion of amino acids 30–114, encompassing the carbamyl phosphate-binding domain (McDowall et al., 1990; OTC-Δ, Figure 1A). This protein is expressed only in liver and small intestine and is absent from transformed cells grown in culture. Transfection of COS-7 cells with cDNA encoding the precursor forms of wild-type or mutant OTC followed by pulse labelling and immunoprecipitation with anti-OTC antibodies showed that both were proteolytically processed by removal of the presequence in a membrane potential (Δψ)-dependent manner, a precondition for protein import (Figure 1B). Specific localization of both OTC and OTC-Δ to mitochondria in transfected cells was confirmed by immunofluorescence using antibodies to OTC (Figure 1C), and by subcellular fractionation of transfected cells, radiolabelled 36 h after transfection (Figure 1D). Detergent extraction of mitochondria isolated from transfected cells showed that, whereas wild-type processed OTC was in the soluble fraction, processed OTC-Δ was insoluble (Figure 1E). The precursor forms of both wild-type and mutant OTC were found to be associated with the outer surface of the mitochondria and were also in the insoluble fraction, as shown by proteinase K digestion of mitochondria (data not shown). The mitochondrial molecular chaperones Cpn60 and Cpn10 have been shown previously to recognize OTC folding intermediates as substrates (Hartman et al., 1992). We therefore investigated the stress response of cells expressing the mutant, aggregated form of OTC. Total RNA was isolated from cells transfected with either vector alone, vector encoding OTC or vector encoding OTC-Δ, and subjected to northern blot analysis. A selective increase in the level of transcripts for the mitochondrial chaperones Cpn60, mtDnaJ, and the mitochondrial protease ClpP, in cells overexpressing OTC-Δ was observed (Figure 2A). Transcripts for other mitochondrial proteins, Cpn10 (see data below) and the Lon protease but not ClpX (J.Wang, unpublished data) were also upregulated, whereas transcripts encoding non-mitochondrial chaperones, such as those encoding the ER proteins, BiP, Grp94, protein disulfide isomerase, calnexin and calreticulin (Figures 2A and 6C) were not significantly affected by the accumulation of unfolded proteins. The increased levels of transcripts for Cpn60 and Cpn10 were accompanied by increased levels of protein expression in response to transfection with OTC-Δ, as shown by western blot analysis (Figure 2B), whereas levels of the ER chaperone BiP, the cytosolic chaperones Hsp73 and Hsp72 and mitochondrial Hsp70 were unchanged. We conclude from these data that the accumulation of protein aggregates in mitochondria produces a mitochondrial stress protein response characterized by the upregulation of nuclear genes encoding mitochondrial molecular chaperones and proteases.
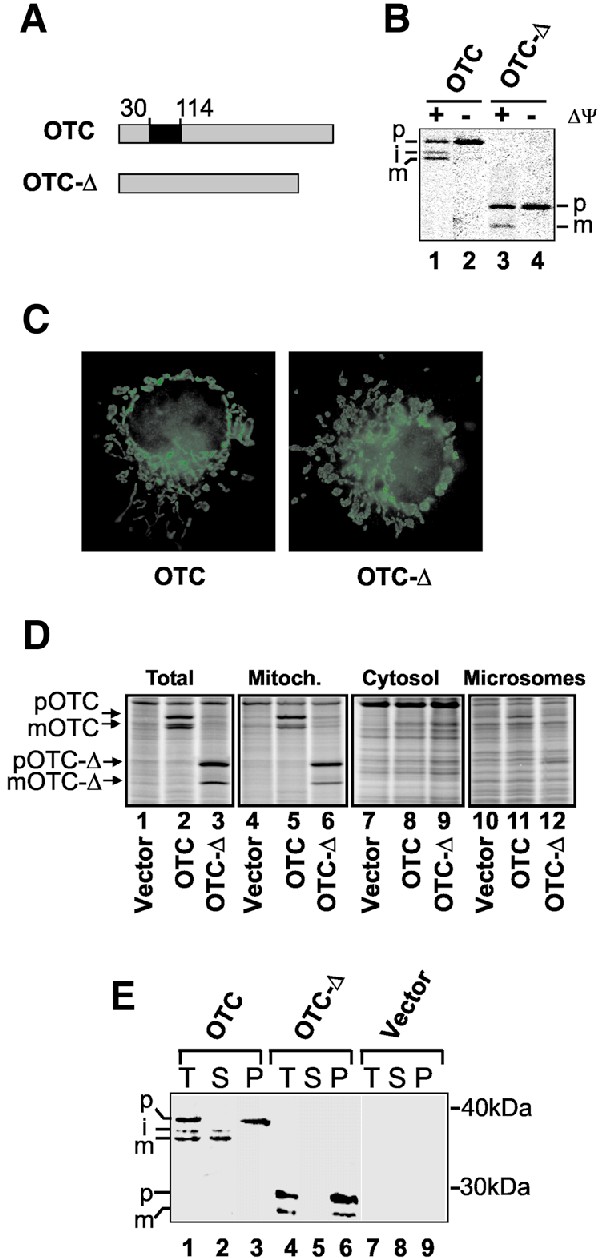
Fig. 1. Establishment of a mitochondrial UPR. (A) Diagrammatic representation of the wild-type OTC and deletion mutant (OTC-Δ) from which the carbamyl phosphate-binding domain (amino acids 30–114 of mature OTC) has been removed. (B) OTC and OTC-Δ are imported into mitochondria in vivo. COS-7 cells transfected with OTC (lanes 1 and 2) or OTC-Δ (lanes 3 and 4) were labelled with [35S]methionine 36 h after transfection in the presence or absence of rhodamine 6G to disrupt the mitochondrial inner membrane potential (Δψ). Cell extracts were immunoprecipitated with antibodies specific for OTC and subjected to SDS–PAGE and PhosphorImager analysis (p, precursor; i, processing intermediate; m, mature). (C) OTC-Δ localizes to mitochondria. Immunofluorescence microscopy of cells transfected with OTC or OTC-Δ was employed using antibodies against OTC. (D) OTC and OTC-Δ are associated with the mitochondrial fraction in radiolabelled cells. Cells were transfected with vector alone or vector expressing OTC or OTC-Δ and, 32 h later, radiolabelled for 4 h. Cells were harvested and subjected to subcellular fractionation, SDS–PAGE and PhosphorImager analysis. Radiolabelled forms of precursor OTC/OTC-Δ and matrix-processed OTC/OTC-Δ are indicated. (E) OTC-Δ forms aggregates in mitochondria. Cells expressing OTC or OTC-Δ were harvested 36 h post-transfection. The total mitochondrial fraction (T) was separated into 0.5% (w/v) Triton X-100-soluble (S) and -insoluble (P) fractions, and subjected to SDS–PAGE followed by western blot analysis using OTC antibodies.
Fig. 2. Transcriptional activation of genes by unfolded proteins in mitochondria. (A) Northern blot analysis of total RNA isolated from COS-7 cells transfected with vector OTC or OTC-Δ constructs (left panel). The blot was hybridized sequentially with radiolabelled Cpn60, Cpn10, ClpP, BiP, mtHsp70, mtDnaJ and control GAPDH cDNA probes. The change in transcript levels normalized against GAPDH levels was quantitated relative to that found in cells transfected with vector alone (right panel). (B) Increase in the levels of chaperones within mitochondria. Immunoblot analysis of various stress proteins from equal amounts of whole-cell lysates from COS-7 cells transfected with vector, OTC or OTC-Δ. The blot was sequentially probed with antibodies against Cpn60, Cpn10, mtHsp70, Hsp72/73 and BiP. (C) Activation of the Cpn60/10 bidirectional promoter by mitochondrial stress. A reporter construct was used whereby the Cpn10 side of the bidirectional promoter regulates LUC expression and the Cpn60 side regulates CAT expression (top panel). COS-7 cells co-transfected with vector, OTC or OTC-Δ, Cpn60/10 reporter plasmid and β-galactosidase plasmid were assayed 36 h after transfection for LUC and CAT activities and normalized against β-galactosidase activity. Data represent the mean ± SEM from experiments performed in triplicate. (D) Activation of Cpn60/10 promoter by heat shock and mitochondrial stress is additive. COS-7 cells were co-transfected with vector, OTC or OTC-Δ, the Cpn60/10 reporter construct, and β-galactosidase plasmid. After 12 h, cells were split into two plates and 6 h later one set was heat shocked (HS, black bars) at 43°C for 1 h and the others left at 37°C (C, grey bars). Cell extracts were assayed after 16 h for LUC and CAT activities relative to β-galactosidase activity (left side). The fold activation due to HS compared with controls of the Cpn60/10 promoter in either orientation (Cpn60/CAT or Cpn10/LUC) is shown (right side).
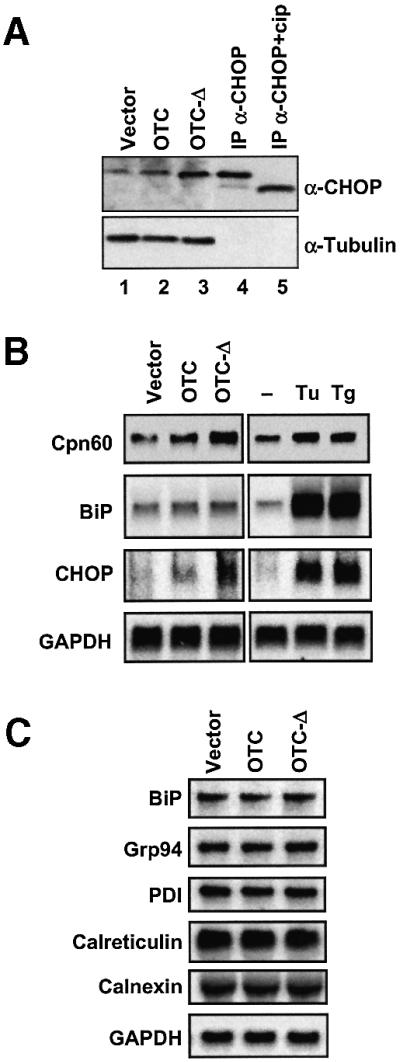
Fig. 6. CHOP induction and specificity of the MSR. (A) CHOP expression in response to mitochondrial stress. Extracts from cells transfected with vector, wild-type OTC or mutant OTC-Δ constructs were subjected to western blotting transfers and probed with antibodies against CHOP (α-CHOP) and tubulin (α-tubulin) as control. Extracts of cells transfected with wild-type CHOP cDNA were subjected to immunoprecipitation with antibodies against CHOP (IP α-CHOP) and half of the immunoprecipitate was treated with calf intestinal phosphatase (IP α-CHOP + cip) followed by the western blot analysis probed with antibodies against CHOP. (B) Specificity of MSR and the ER UPR. RNA was isolated from cells transfected with vector, wild-type OTC, OTC-Δ or with agents that trigger the ER stress response, tunicamycin (Tu) and thapsigargin (Tg) and subjected to northern blot analysis. The blot was sequentially hybridized with radiolabelled Cpn60, BiP, CHOP and GAPDH cDNA probes. (C) MSR does not affect expression of genes for ER stress proteins. RNA isolated from cells transfected with vector, wild-type OTC or OTC-Δ was subjected to northern blot analysis using probes corresponding to transcripts encoding ER proteins as well as GAPDH.
Mammalian Cpn60 and Cpn10 genes share a single, bidirectional promoter (Ryan et al., 1997b). This promoter was linked at either end to chloramphenicol acetyl transferase (CAT) and luciferase (LUC) reporter cDNAs (Figure 2C). When co-transfected into COS-7 cells expressing either wild-type or mutant OTC protein, there was a 2- to 3-fold induction of both reporter genes (Figure 2C), showing that the accumulation of unfolded proteins resulted in the activation of the promoter of the nuclear genes encoding mitochondrial Cpn60 and Cpn10. The level of induction due to MSR was similar to that produced by heat shock (Figure 2D); however, the transcriptional activation of the Cpn60/10 promoter by mitochondrial stress was not through the HSE present in the promoter. The effects of mitochondrial stress induced by OTC-Δ and heat shock were additive. Indeed, the fold induction due to heat shock was the same in cells transfected with OTC-Δ as in cells transfected with vector alone or wild-type OTC (Figure 2D), and deletion of the HSE had no effect on transcriptional activation by OTC-Δ (see Figure 4A).
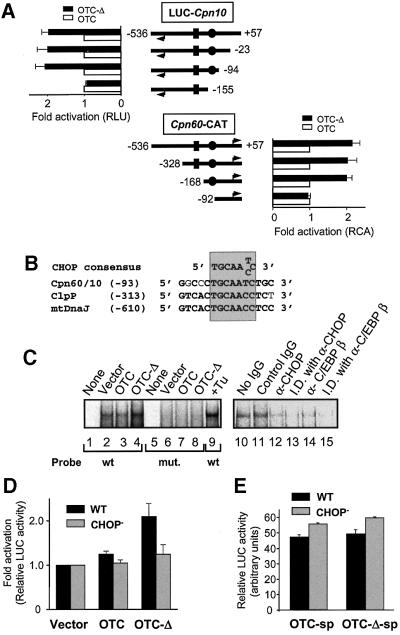
Fig. 4. Identification of the MSR element in the Cpn60/10 promoter. (A) Promoter deletion analysis was carried out by making deletions from either the Cpn60 side (top panels) or Cpn10 side (bottom panels) of the Cpn60/10 bidirectional promoter. Deletions are shown as distances (bp) from the Cpn60 transcription start site. Cpn60 and Cpn10 transcription start sites are indicated by arrowheads. The relative position of the HSE is indicated by rectangles while circles indicate the CHOP element. The fold activation of the promoter construct in cells expressing OTC-Δ (solid bars) compared with those expressing wild-type OTC (open bars) is shown as relative luciferase (RLU) or CAT (RCA) activity. Data represents the mean ± SEM from experiments performed in triplicate. (B) Nucleotide sequence alignment of the CHOP consensus element in the Cpn60/10 and the putative ClpP and mtDnaJ promoters. Bold letters show identical nucleotides and numbers refer to distance from transcription initiation site of Cpn60 or the first base of the initiation codon for ClpP and mtDnaJ. The CHOP consensus binding site is shaded. (C) EMSA of nuclear extracts from COS-7 cells transfected with OTC constructs. A probe corresponding to a 30 bp Cpn60/10 promoter fragment that contains the CHOP-binding element (wt) and a mutant probe (mut.) containing an altered CHOP site were incubated with nuclear extracts isolated from cells transfected with various constructs or incubated with tunicamycin (Tu) as indicated and subjected to PAGE and PhosphorImager analysis. Immunocompetition and immunodepletion assays were performed using nuclear extracts of COS-7 cells transfected with OTC-Δ. Nuclear extracts were incubated with no IgG (lane 10), control (anti-β-galactosidase) IgG (lane 11), or with anti-CHOP or C/EBPβ antibodies (lanes 12 and 14), before adding radiolabelled wild-type probe. In addition, nuclear extracts were first immunodepleted (I.D.) of CHOP or C/EBPβ using antibodies attached to protein A–Sepharose followed by incubation with radiolabelled wild-type probe (lanes 13 and 15). (D) The CHOP site in the Cpn60/10 promoter is involved in the MSR response. The effect of introduction of a mutated CHOP element (CHOP–) on the Cpn60/10 promoter activity was determined by co-transfecting cells with OTC constructs (vector, OTC, OTC-Δ) and wild-type (WT, black bars) or mutant (CHOP–, grey bars) Cpn60/10 promoter constructs. Promoter activity was measured 36 h after transfection and is based on measurement of LUC expression (mean ± SEM). (E) The effect of expressing wild-type (OTC-sp) or mutant (OTC-Δ-sp) OTC lacking the mitochondrial signal peptide (sp) was measured by co-transfecting cells with OTC constructs and with wild-type (WT, black bars) or mutant (CHOP–, grey bars) Cpn60/10 promoter reporter constructs. Promoter activity was measured 36 h after transfection and is based on measurement of LUC expression (mean ± SEM).
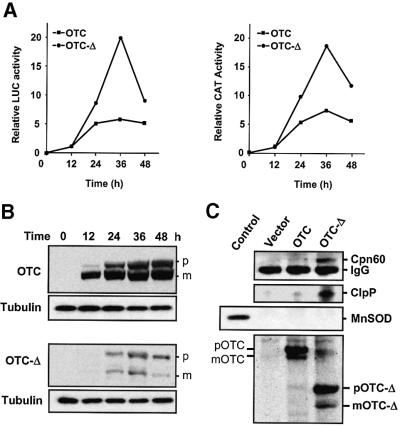
The MSR is transient and correlates with the level of aggregated protein
Time-course experiments showed that the upregulation was transient for both Cpn10 and Cpn60 promoter activities (Figure 3A). This induction is closely correlated to the steady-state level of processed, aggregated protein present in mitochondria as measured by western blot analysis using antibodies specific to OTC, while analysis of tubulin was used to control loading (Figure 3B). The peak promoter activity was maximal 36 h post-transfection and dropped markedly between 36 and 48 h. The maximum amount of mutant protein found in cells also occurred at 36 h post-transfection (Figure 3B). In contrast, the accumulation of wild-type, soluble OTC continued in a linear manner and was accompanied by only a slightly increased stimulation of promoter activity, despite the very large amounts of protein that accumulated in the mitochondria (Figure 3A and B). The persistence of mutant protein in transfected cells was relatively short lived, due to a decrease in its half-life as shown by a comparison of pulse labelling of newly synthesized protein using [35S]methionine with that of the steady-state level of protein revealed by western blot analysis. Pulse–chase experiments showed that, whereas mature wild-type OTC is a stable protein, the processed form of the deletion mutant is not (data not shown). We sought to investigate whether a direct association of stress-induced proteins could be observed with the unstable mutant OTC. Co-immunoprecipitation analysis revealed that both Cpn60 and ClpP were stably associated with the OTC deletion mutant, but not with wild-type OTC. The endogenous mitochondrial matrix protein manganese superoxide dismutase (MnSOD) was not found in the immunoprecipitates, indicating that non-specific interactions of matrix proteins with OTC-Δ were not occurring. The presence of OTC and OTC-Δ in the immunoprecipitates was also confirmed (Figure 3C, bottom panel) and no Cpn60, ClpP or OTC was immunoprecipitated from cells transfected with vector alone (Figure 3C, top panel). The increased turnover of the mutant OTC protein observed is therefore likely to be due to the increased expression of mitochondrial proteases such as ClpP.
Fig. 3. MSR is transient and correlates with the level of aggregated protein in mitochondria. (A) Cpn60 and Cpn10 promoter activation is transient. COS-7 cells were co-transfected with the Cpn60/10 promoter construct along with constructs expressing OTC or OTC-Δ and β-galactosidase plasmid. Cells were harvested at the indicated times and CAT and LUC activity relative to β-galactosidase activity was measured. (B) Changes in the steady-state levels of wild-type OTC and mutant OTC after transfection of cells. Equal amounts of protein from whole-cell extracts were subjected to SDS–PAGE, blotted and probed with an anti-OTC antibody, and anti-tubulin antibodies as a loading control. p, precursor of OTC; m, mature form of OTC. (C) Cpn60 and ClpP associate with mutant OTC-Δ. Mitochondrial proteins from COS-7 cells transfected with vector or OTC constructs were translabelled with [35S]methionine, immunoprecipitated with OTC antibodies and subjected to western blot analysis. The blot was probed with Cpn60 and ClpP antibodies (upper panels) and MnSOD antibodies as a control (middle panel; control on left side). The immunoprecipitated 35S- labelled wild-type OTC and OTC-Δ were visualized by PhosphorImager analysis of the filter (bottom panel).
Identification of the MSR element
In order to identify the promoter region involved in MSR, deletion analysis of the Cpn60/10 bidirectional promoter was performed, and reporter activities were measured in cells transfected with OTC-Δ relative to those transfected with OTC (Figure 4A). Upregulation of LUC expression by OTC-Δ was observed when 94 bp, but not 155 bp, were deleted upstream of the Cpn60 transcription start site (Figure 4A, upper panel). Likewise, upregulation of CAT expression by OTC-Δ was seen when 168 bp, but not 92 bp, were retained upstream of the Cpn60 transcription start site (Figure 4A, lower panel). Deletion of the HSE found at bases –207 to –226 (rectangle) did not impair the MSR (Figure 4A, lower panel; compare constructs beginning at –328 and –168). The MSR responsive element was therefore found to lie between bases –94 and –155. Scanning of this 62 bp region revealed that it contained a site (TGCAATC) that could potentially be recognized by the transcription factor CHOP (GADD153) (Fornace et al., 1989; Ubeda et al., 1996) and is 99 bp upstream of the Cpn60 transcription start site (Ryan et al., 1997b). Since ClpP and mtDnaJ mRNA levels were also upregulated by mitochondrial stress, we searched for CHOP-binding elements within their promoter regions. This revealed that ClpP (AC 011491) and mtDnaJ (AC 012676) promoters both possessed a CHOP-binding element. Moreover, this element encompassed a 15 bp region that is highly conserved in all three promoters (Figure 4B). To confirm that CHOP could bind to this promoter region, electrophoretic mobility shift assays (EMSA) were employed. A 30 bp probe encompassing the CHOP site was incubated with nuclear extracts from cells transfected with mutant OTC or from cells transfected with vector alone or wild-type OTC. As can be seen (Figure 4C, lanes 2–4), an increase in the level of a high molecular weight complex is observed with incubation of nuclear extracts from cells expressing OTC-Δ. This increase was not obtained with a probe containing a mutated CHOP site (Figure 4C, lanes 6–8). As a control, nuclear extracts from cells treated with tunicamycin were used in the EMSA, since this reagent induces the expression of CHOP (Yoshida et al., 2000) and leads to an increase in the level of the high molecular weight complex (Figure 4C, lane 9). We then assessed whether the complex observed was due to binding of CHOP or its partner C/EBPβ to this promoter region. Nuclear extracts from cells subjected to MSR using OTC-Δ were isolated and either mixed with monoclonal antibodies against CHOP or C/EBPβ (Figure 4C, lanes 12 and 14), compared with no IgG (lane 10) or control (anti-β-galactosidase) IgG (lane 11), or subjected to immuno-depletion experiments using either of the antibodies absorbed to protein A–Sepharose (lanes 13 and 15), prior to conducting the EMSA. As can be seen, the complex was depleted in immunocompeted extracts and totally removed in immunodepleted nuclear extracts, suggesting that the band shift was dependent on binding of CHOP and C/EBPβ to this region of DNA. Furthermore, mutation of this Cpn60/10 CHOP-binding site from TGCAATC to TGTCGCC within the 594 bp promoter resulted in an ablation of the MSR (Figure 4D). Transfection of cells with OTC lacking the mitochondrial signal peptide (OTC-sp and OTC-Δ-sp), which prevented their import into mitochondria, showed that the mutant protein did not activate the mitochondrial UPR (Figure 4E), despite being expressed at levels equivalent to those of OTC and OTC-Δ (data not shown), confirming that the response was obtained only if mutant protein accumulated inside mitochondria.
MSR is regulated by CHOP and C/EBPβ
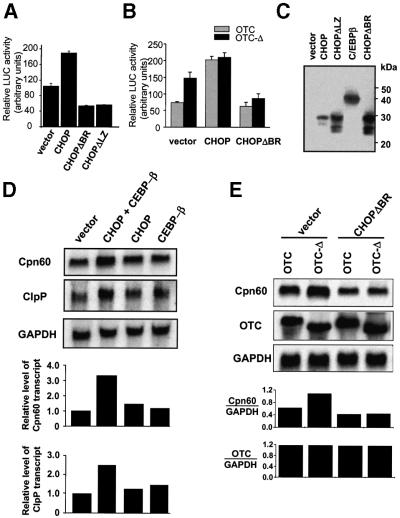
To directly confirm that CHOP and C/EBPβ can activate the Cpn60/10 promoter, cells were co-transfected with the Cpn60/10 promoter reporter construct along with constructs expressing CHOP and C/EBPβ (Figure 5A). Over-expression of CHOP/CEBPβ resulted in an ∼2-fold increase in promoter activity, compared with that obtained in transfections with vector alone. In contrast, cells overexpressing mutant CHOP, which either lacked the basic DNA-binding domain (CHOPΔBR) or the leucine zipper domain (CHOPΔLZ), did not activate promoter activity. Indeed, the level of Cpn60/10 promoter activity in cells expressing the mutant forms of CHOP was approximately half of that obtained with control cells transfected with vector alone (Figure 5A). This suggests that at least part of the constitutive activity of the Cpn60/10 gene is dependent on CHOP, an observation which agrees with earlier findings that CHOP is constitutively expressed at low levels under normal conditions and is induced in response to various forms of stress (Carlson et al., 1993; Wang et al., 1996; see also Figure 6A).
Fig. 5. Effect of CHOP expression on Cpn60/10 promoter activity. (A) Overexpression of CHOP and C/EBPβ activates the Cpn60/10 promoter. COS-7 cells were co-transfected with the Cpn60/10 promoter reporter construct and cDNA encoding C/EBPβ, as well as either vector alone, wild-type CHOP cDNA or CHOP constructs lacking either the basic DNA-binding region (CHOPΔBR) or the leucine zipper region (CHOPΔLZ). Relative LUC activity was measured 36 h after transfection. Results represent the mean of transfections performed in triplicate ± SEM. (B) CHOP-dependent induction of MSR. The effect of a dominant-negative mutant of CHOP (CHOPΔBR) on the MSR was measured by co-transfecting cells with the Cpn60/10 promoter reporter construct, OTC constructs (OTC, grey bars; OTC-Δ, black bars), cDNA encoding C/EBPβ and either vector alone, wild-type CHOP or CHOPΔBR cDNA. Relative LUC activities were measured in cell extracts 36 h after transfection and represent the mean of transfections performed in triplicate ± SEM. (C) Level of expression of transcription vectors. The expression of CHOP constructs and C/EBPβ by transfected cells (30 µg of total protein/lane) is shown by western blot analysis using specific antibodies. (D) Overexpression of CHOP and C/EBPβ increases the level of Cpn60 and ClpP transcripts. COS-7 cells were transfected with cDNA encoding CHOP and C/EBPβ and total RNA extracted from cells subjected to northern blot analysis. The blot was sequentially hybridized with radiolabelled Cpn60, ClpP and GAPDH cDNA probes (top panels). Blots were quantitated relative to GAPDH by PhosphorImager analysis (lower panels) with the ratio of transcripts from cells transfected with vector alone being set at 1.0. (E) Induction of Cpn60 transcripts by MSR is ablated by a dominant-negative form of CHOP. The effect of transfection of COS-7 cells with vector alone or dominant-negative mutant CHOPΔBR on Cpn60 mRNA levels was measured by northern blot analysis from cells co-transfected with wild-type OTC and OTC-Δ. The levels of wild-type and mutant OTC transcripts were also determined as a control (upper panels). Bands were quantitated by PhosphorImager analysis relative to the levels of GAPDH (lower panels).
Dominant-negative experiments were then carried out to determine whether CHOP produced in response to mitochondrial stress is responsible for the induction of the Cpn60/10 promoter. Overexpression of wild-type CHOP and C/EBPβ in cells co-transfected with wild-type OTC stimulated the Cpn60/10 promoter-LUC reporter gene to the same level as cells transfected with OTC-Δ (Figure 5B). In contrast, overexpression of C/EBPβ and a CHOP mutant lacking the basic DNA-binding domain (CHOPΔBR) impaired the activation of the Cpn60/10 promoter reporter construct in cells expressing OTC-Δ (Figure 5B). Western blot analysis was performed as a control to verify that CHOP, C/EBPβ and the CHOP mutants were indeed being expressed (Figure 5C). CHOP and C/EBPβ were also able to activate transcription of MSR responsive genes, Cpn60 and ClpP (Figure 5D). Northern blot analysis revealed that the overexpression of both CHOP and C/EBPβ led to an ∼3- and 2.5-fold increase in Cpn60 and ClpP mRNA levels, respectively, whereas expression of either CHOP or C/EBPβ alone had no appreciable effect on Cpn60 and ClpP transcription. Conversely, MSR induced in cells transfected with the construct expressing OTC-Δ was ablated by co-transfection of C/EBPβ and the CHOPΔBR mutant cDNA, as determined by northern blot analysis of Cpn60 transcripts (Figure 5E). Indeed, the mutant form of CHOP reduced the levels of Cpn60 transcripts below that found in control cells transfected with vector alone (Figure 5E, lower panel), in a manner similar to the effect obtained for the Cpn60 promoter-LUC construct (Figure 5A). Confirm ation that expression of OTC was the same in experiments with vector control and mutant CHOP was determined by northern blot analysis and compared with the expression of control GAPDH (Figure 5E, middle and lower panels).
Levels of CHOP are upregulated by MSR
The steady-state level of CHOP was increased in cells undergoing mitochondrial stress. Western blot analysis of extracts of cells transfected with vector alone, or vector expressing OTC, showed the presence of low levels of CHOP, which was increased in cells expressing the mutant OTC-Δ (Figure 6A, lanes 1–3). To control for protein loading, blots were also probed with the marker protein tubulin (Figure 6A, lower panel). These results provide further confirmation that protein aggregates in mitochondria lead to an increase in CHOP expression. Virtually all the CHOP found in cells, including unstressed cells, seems to be in a phosphorylated form, since treatment with calf intestinal alkaline phosphatase of CHOP immunoprecipitated from cells transfected with CHOP cDNA resulted in the formation of a faster migrating species of CHOP in SDS–PAGE (Figure 6A, compare lanes 4 and 5).
Since both the ER and the mitochondrial UPR result in the activation of CHOP transcription, we sought to determine whether each of these stress responses is organelle specific. Cells treated with tunicamycin or thapsigargin, both of which trigger the ER stress response (Li et al., 2000; Yoshida et al., 2000), showed strong upregulation of BiP transcription but had only a marginal effect on Cpn60 transcription (Figure 6B). Indeed, an experiment to determine the dose response of tunicamycin treatment on cells transfected with reporter constructs under the control of either BiP or Cpn60/10 promoters showed that, whereas 1 µg/ml tunicamycin produced a maximum 1.4-fold activation for the Cpn60/10 promoter, this same concentration stimulated the BiP promoter 4-fold (data not shown). The activation of the BiP gene has been shown previously to be through a pathway involving the proteolytic release of the transcription factor ATF6 from the ER membrane and its translocation to the nucleus, where it induces a number of factors, including CHOP (Fawcett et al., 1996; Yoshida et al., 2000). Indeed, tunicamycin and thapsigargin have a much more pronounced effect on increasing the levels of CHOP transcripts than does the expression of OTC-Δ and yet, had only a marginal effect on Cpn60 induction. Conversely, northern blot analysis and quantitation data normalized to the levels of GAPDH revealed the accumulation of mutant OTC-Δ protein in mitochondria induced Cpn60 transcription (Figure 6B), but had no effect on the induction of BiP, Grp94, protein disulfide isomerase (PDI), calreticulin or calnexin, all of which are induced by the ER UPR (Figure 6C). Thus, we conclude that the ER unfolded protein stress response and the MSR are organelle specific.
Discussion
We report on the discovery of a mitochondrial specific stress response, or MSR. By introducing a mutation in the mitochondrial targeted preprotein, OTC, the imported protein was prevented from folding properly in the matrix, leading to the induction of nuclear genes encoding molecular chaperones Cpn60 and Cpn10, mtDnaJ, and the protease, ClpP (Figures 1 and 2). Cpn60 and Cpn10 have been shown previously to be upregulated in ρ° cells (Martinus et al., 1996) and by stress such as heat shock (Naylor et al., 1996; Ryan et al., 1997b), whilst mtHsp70 was not induced by heat shock. The effect of stress on mammalian mtDnaJ and ClpP has not been described previously. The upregulation of mtDnaJ, but not of mtHsp70, is unexpected, although mtDnaJ contains a CHOP element whereas mtHsp70 does not, indicating that mtDnaJ may act independently of mtHsp70 in protein folding or in concert with another mtHsp70 isoform. The level of induction of Cpn60 and Cpn10 was very similar to that obtained in response to heat shock (Ryan et al., 1997b), but the activation of the MSR was not through the heat shock response, as deletion of the HSE did not affect the induction caused by the accumulation of unfolded protein (Figure 4). Further, the pattern of proteins induced by heat shock is quite distinct from that induced by MSR.
The finding that the MSR involves the transcription factors CHOP and C/EBPβ provides another example of the participation of CHOP in stress pathways. Thus, CHOP is also induced by the ER UPR, although the role of CHOP in this response is still uncertain (Spear and Ng, 2001). The fact that CHOP is upregulated in many different, but specific, cellular stress responses suggests that additional factors must play a role in determining specificity involving CHOP. Since CHOP acts in association with other transcription factors, this specificity may reside in either the nature of the proteins with which CHOP associates, such as C/EBPβ (Friedman and McKnight, 1990; Ron and Habener, 1992; Fawcett et al., 1996; Friedman, 1996), ATF3 (Chen et al., 1992) and AP-1 (Ubeda et al., 1999), or in the nature of the signalling pathway that is activated in response to various forms of stress. Since the accumulation of unfolded proteins in mitochondria increases the levels of CHOP in cells, this raises questions about the mechanism of sensing of mitochondrial stress and mitochondrial–nuclear communication that ultimately leads to activation of target genes.
Most of the genes that regulate the function of mitochondria are nuclear located. It is therefore axiomatic that there must be effective communication between mitochondria and the nucleus. Evidence for such communication comes from the substantial changes in gene expression profiles in ρ° yeast cells that have had their mtDNA removed (Traven et al., 2001). The change in nuclear gene expression in response to alterations in mitochondrial function has been termed retrograde regulation (Parikh et al., 1987; Liao and Butow, 1993). One example of this is the upregulation of a number of genes encoding peroxisomal enzymes which mitigate the loss of tricarboxylic acid cycle activity in respiration-deficient mitochondria (Epstein et al., 2001). In yeast cells, this form of regulation is through a number of signalling pathways, the best described of which is via the helix– loop–helix leucine zipper transcription factors, Rtg 1 and Rtg 3, which bind to an upstream element in target genes (Jia et al., 1997) and which become activated by the cytoplasmic factor, Rtg 2 (Sekito et al., 2000). In mammalian cells, a communication pathway exists to regulate nuclear genes in response to loss of mtDNA via Ca2+ signalling of a c-Jun N-terminal kinase pathway (Biswas et al., 1999). The form of communication between mitochondria and the nucleus in response to MSR is unknown. In the UPR, sensing of stress appears to be via binding of BiP to unfolded proteins (Dorner et al., 1992). An analogous mechanism may exist in mitochondria through the association of Cpn60 with mutant proteins (Figure 3). An alternative model may involve the release from mitochondria of peptides generated by proteases upregulated by MSR, and these may stimulate a signalling pathway leading to transcriptional activation of the CHOP gene (Young et al., 2001).
Materials and methods
Plasmid construction, transfection and promoter analysis
Wild-type OTC was cloned into the mammalian expression vector pCAGGS (Niwa et al., 1991). For construction of the deletion mutant pOTCΔ(30–114) (OTC-Δ), an internal 255 bp BglII fragment was removed from the wild-type OTC. Equivalent constructs lacking the N-terminal mitochondrial signal peptide (OTC-sp and OTC-Δ-sp) were constructed by PCR by deleting amino acids 1–32 from the pOTC constructs. The pCAT/LUC-Cpn60/10 promoter vector has been described previously (Ryan et al., 1997b). Serial deletion constructs of the Cpn60/10 promoter were obtained by PCR by introducing flanking HindIII sites (Cpn10 side) or PstI sites (Cpn60 side) and cloning the resulting fragments into the pCAT/LUC vector (Figure 2B). The CHOPΔ-Cpn60/10 promoter construct was obtained by PCR overlap extension (Ho et al., 1989), using mutagenic primers that altered the CHOP element from TGCAATC to TGTCGCC (5′-CGCCCCGGCCCTGTCGCCTGCATGCGA-3′, and complementary strand). The pcDNA1-CHOP, pcDNA1-CHOPΔLZ and pEF-BOS-C/EBPβ expression constructs were gifts from Drs David Ron and Masataka Mori. The pcDNA1-CHOPΔBR expression construct was made by removing amino acid residues R102–K122 (Ubeda et al., 1996).
COS-7 cells were cultured in DME/10% fetal calf serum and transfected at 70% confluence using Lipofectamine (Gibco-BRL) and 10 µg of plasmid DNA. For heat shock experiments, transfected cells were split, with one set of cells subjected to heat shock (43°C) for 1 h and the other left at 37°C. All cells were harvested 16 h later for assay. For promoter deletion analysis, transfected cells were incubated at 37°C for 36 h, lysed and assayed for LUC and CAT activity (Promega and Roche, respectively). Transfection efficiencies were normalized on the basis of co-transfection with a plasmid expressing β-galactosidase (Li et al., 2000). To induce UPR, cells were treated with 2 µg/ml tunicamycin or 300 nM thapsigargin for 10 h.
Mitochondrial fractionation and OTC import in vivo
Mitochondria were isolated from COS-7 cells according to Yang et al. (1997), and soluble proteins were separated from aggregated proteins by extraction with 0.5% (w/v) Triton X-100 in phosphate-buffered saline for 5 min on ice. The whole lysate, supernatant and pellet fractions were subjected to 12% SDS–PAGE. For the in vivo OTC import assay, the mitochondrial membrane potential was disrupted with 2.1 µM rhodamine 6G chloride (Sigma), and cells were labelled with l-[35S]methionine/cysteine (New Life Science) 36 h after transfection, for 30 min before immunoprecipitation.
Immunochemical and northern blot analysis
Antibodies against OTC, Cpn60 and ClpP were made in rabbits using recombinant proteins expressed in E.coli. For western blot analyses, protein extracts were prepared from cells lysed with buffer containing 1% (v/v) Triton X-100, 0.1% (w/v) SDS and 0.5% (w/v) sodium deoxycholate, separated by SDS–PAGE and transferred to nitrocellulose filters using a semi-dry method. Quenched membranes were probed with antibodies and analysed with enhanced chemiluminescence reagent (Amersham Pharmacia Biotech) with subsequent exposure on X-Omat film (Kodak). Immunoprecipitation experiments were performed on mitochondria lysed with buffer containing 0.5% NP-40, using rabbit antibodies against OTC or monoclonal antibodies against CHOP. For immunolocalization of protein constructs, cells were grown on microscope coverslips in 6-well plates and 36 h after transfection were probed with antibodies against OTC and fluorescein-labelled secondary anti-rabbit antibodies. For northern blotting, RNA was extracted from cultures using TriZol Reagent (Life Technologies Inc.). Total RNA (20 µg) was separated on 1% (w/v) agarose, 6% (v/v) formaldehyde gels and transferred to Hybond-XL membrane (Amersham Pharmacia Biotech). Blots hybridized with cDNA probes labelled by the Megaprime DNA labelling system in Rapid-Hyb buffer (Amersham Pharmacia Biotech) were scanned with a PhosphorImager (Molecular Dynamics) and data quantified using IMAGE-QUANT software.
Electrophoretic mobility shift assays
Complementary oligonucleotides encompassing the desired binding sites were synthesized (wild type, 5′-GCGTCGCATGCAGATTGCAGGG CCGGG-3′, and complementary strand; mutant, 5′-GCGTCGCATGC AGGCGACAGGGCCGGG-3′, and complementary strand). All duplex olignonucleotides were annealed by mixing in a 1:1 molar ratio in 10 mM Tris–HCl pH 7.5, 1 mM EDTA, 50 mM KCl, heating to 100°C for 5 min and cooling to 25°C. The 3 bp overhangs were filled with [32P]dCTP using Klenow enzyme. The annealed labelled products were purified by passage through Nutrap Probe purification columns (Stratagene). A 10 000 c.p.m. aliquot of the probe was incubated for 30 min at 25°C with an aliquot of COS-7 nuclear extract (Andrews and Faller, 1991) containing 10 µg of protein and 2 µg of dI–dC (Pharmacia) in 4% (v/v) glycerol, 2 mM MgCl2, 0.5 mM EDTA, 0.5 mM dithiothreitol, 50 mM NaCl, 10 mM Tris–HCl pH 9.0, 300 µg/ml bovine serum albumin in a total volume of 20 µl. The reaction mixtures were resolved by electrophoresis on a 5% polyacrylamide gel and dried gels were analysed by phosphoimaging. Antibody competition assays used monoclonal antibodies against CHOP and C/EBPβ (Santa Cruz) or control β-galactosidase antibodies (1–2 µg) which were added to the nuclear extract and left for 30 min on ice before addition of the labelled oligonucleotides. Immunodepletion of nuclear extracts was carried out by pre-incubating the extracts with protein A–Sepharose beads containing the monoclonal antibodies before EMSA.
Acknowledgments
Acknowledgements
Complementary DNA clones for CHOP and CHOPΔLZ, C/EBPβ, ClpP and hTid-1 (mtDnaJ) were gifts from D.Ron, M.Mori, P.Bross and K.Munger, respectively. We thank J.Myazaki for the pCAGGS vector, J.Pereira for assistance in the production of the OTC deletion mutant and A.Giraud for antiserum against Cpn10. This work was supported by grants from the Australian Research Council and the Australian National Health and Medical Research Council.
References
- Andrews N.C. and Faller,D.V. (1991) A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res., 19, 2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett J.D., Luethy,J.D., Carlson,S.G., Sollott,S.J. and Holbrook,N.J. (1992) Calcium ionophore A23187 induces expression of the growth arrest and DNA damage inducible CCAAT/enhancer-binding protein (C/EBP)-related gene, gadd153. Ca2+ increases transcriptional activity and mRNA stability. J. Biol. Chem., 267, 20465–20470. [PubMed] [Google Scholar]
- Biswas G., Adebanjo,O.A., Freedman,B.D., Anandatheerthavarada,H.K., Vijayasarathy,C., Zaidi,M., Kotlikoff,M. and Avadhani,N.G. (1999) Retrograde Ca2+ signaling in C2C12 skeletal myocytes in response to mitochondrial genetic and metabolic stress: a novel mode of inter-organelle crosstalk. EMBO J., 18, 522–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruhat A., Jousse,C., Wang,X.Z., Ron,D., Ferrara,M. and Fafournoux,P. (1997) Amino acid limitation induces expression of CHOP, a CCAAT/enhancer binding protein-related gene, at both tran scriptional and post-transcriptional levels. J. Biol. Chem., 272, 17588–17593. [DOI] [PubMed] [Google Scholar]
- Calfon M., Zeng,H., Urano,F., Till,J.H., Hubbard,S.R., Harding,H.P., Clark,S.G. and Ron,D. (2002) IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature, 415, 92–96. [DOI] [PubMed] [Google Scholar]
- Carlson S.G., Fawcett,T.W., Bartlett,J.D., Bernier,M. and Holbrook,N.J. (1993) Regulation of the C/EBP-related gene gadd153 by glucose deprivation. Mol. Cell. Biol., 13, 4736–4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman R., Sidrauski,C. and Walter,P. (1998) Intracellular signaling from the endoplasmic reticulum to the nucleus. Annu. Rev. Cell Dev. Biol., 14, 459–485. [DOI] [PubMed] [Google Scholar]
- Chen Q., Yu,K., Holbrook,N.J. and Stevens,J.L. (1992) Activation of the growth arrest and DNA damage-inducible gene gadd153 by nephrotoxic cysteine conjugates and dithiothreitol. J. Biol. Chem., 267, 8207–8212. [PubMed] [Google Scholar]
- Cox J.S., Shamu,C.E. and Walter,P. (1993) Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell, 73, 1197–1206. [DOI] [PubMed] [Google Scholar]
- Dorner A.J., Wasley,L.C. and Kaufman,R.J. (1992) Overexpression of GRP78 mitigates stress induction of glucose regulated proteins and blocks secretion of selective proteins in Chinese hamster ovary cells. EMBO J., 11, 1563–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein C.B., Waddle,J.A., Hale,W.,IV, Dave,V., Thornton,J., Macatee,T.L., Garner,H.R. and Butow,R.A. (2001) Genome-wide responses to mitochondrial dysfunction. Mol. Biol. Cell, 12, 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett T.W., Eastman,H.B., Martindale,J.L. and Holbrook,N.J. (1996) Physical and functional association between GADD153 and CCAAT/enhancer-binding protein β during cellular stress. J. Biol. Chem., 271, 14285–14289. [DOI] [PubMed] [Google Scholar]
- Fornace A.J. Jr, Nebert,D.W., Hollander,M.C., Luethy,J.D., Papathanasiou,M., Fargnoli,J. and Holbrook,N.J. (1989) Mammalian genes coordinately regulated by growth arrest signals and DNA-damaging agents. Mol. Cell. Biol., 9, 4196–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman A.D. (1996) GADD153/CHOP, a DNA damage-inducible protein, reduced CAAT/enhancer binding protein activities and increased apoptosis in 32D c13 myeloid cells. Cancer Res., 56, 3250–3256. [PubMed] [Google Scholar]
- Friedman A.D. and McKnight,S.L. (1990) Identification of two polypeptide segments of CCAAT/enhancer-binding protein required for transcriptional activation of the serum albumin gene. Genes Dev., 4, 1416–1426. [DOI] [PubMed] [Google Scholar]
- Harding H.P., Novoa,I.I., Zhang,Y., Zeng,H., Wek,R., Schapira,M. and Ron,D. (2000) Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell, 6, 1099–1108. [DOI] [PubMed] [Google Scholar]
- Hartl F.U. (1996) Molecular chaperones in cellular protein folding. Nature, 381, 571–579. [DOI] [PubMed] [Google Scholar]
- Hartman D.J., Hoogenraad,N.J., Condron,R. and Hoj,P.B. (1992) Identification of a mammalian 10-kDa heat shock protein, a mitochondrial chaperonin 10 homologue essential for assisted folding of trimeric ornithine transcarbamoylase in vitro. Proc. Natl Acad. Sci. USA, 89, 3394–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S.N., Hunt,H.D., Horton,R.M., Pullen,J.K. and Pease,L.R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene, 77, 51–59. [DOI] [PubMed] [Google Scholar]
- Jia Y., Rothermel,B., Thornton,J. and Butow,R.A. (1997) A basic helix–loop–helix–leucine zipper transcription complex in yeast functions in a signaling pathway from mitochondria to the nucleus. Mol. Cell. Biol., 17, 1110–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousse C., Bruhat,A., Carraro,V., Urano,F., Ferrara,M., Ron,D. and Fafournoux,P. (2001) Inhibition of CHOP translation by a peptide encoded by an open reading frame localized in the Chop 5′ UTR. Nucleic Acids Res., 29, 4341–4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Baumeister,P., Roy,B., Phan,T., Foti,D., Luo,S. and Lee,A.S. (2000) ATF6 as a transcription activator of the endoplasmic reticulum stress element: thapsigargin stress-induced changes and synergistic interactions with NF-Y and YY1. Mol. Cell. Biol., 20, 5096–5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X. and Butow,R.A. (1993) RTG1 and RTG2: two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell, 72, 61–71. [DOI] [PubMed] [Google Scholar]
- Ma Y. and Hendershot,L.M. (2001) The unfolding tale of the unfolded protein response. Cell, 107, 827–830. [DOI] [PubMed] [Google Scholar]
- Marten N.W., Burke,E.J., Hayden,J.M. and Straus,D.S. (1994) Effect of amino acid limitation on the expression of 19 genes in rat hepatoma cells. FASEB J., 8, 538–544. [DOI] [PubMed] [Google Scholar]
- Martinus R.D., Garth,G.P., Webster,T.L., Cartwright,P., Naylor,D.J., Hoj,P.B. and Hoogenraad,N.J. (1996) Selective induction of mitochondrial chaperones in response to loss of the mitochondrial genome. Eur. J. Biochem., 240, 98–103. [DOI] [PubMed] [Google Scholar]
- McDowall S., van Heeswijck,R. and Hoogenraad,N. (1990) Site-directed mutagenesis of Arg60 and Cys271 in ornithine transcarbamylase from rat liver. Protein Eng., 4, 73–77. [DOI] [PubMed] [Google Scholar]
- Mori K., Ma,W., Gething,M.J. and Sambrook,J. (1993) A trans membrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell, 74, 743–756. [DOI] [PubMed] [Google Scholar]
- Morimoto R.I. (1998) Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones and negative regulators. Genes Dev., 12, 3788–3796. [DOI] [PubMed] [Google Scholar]
- Naylor D.J., Hoogenraad,N.J., and Høj,P.B. (1996) Isolation and characterization of a cDNA encoding rat mitochondrial GrpE, a stress-inducible nucleotide-exchange factor of ubiquitous appearance in mammalian organs. FEBS Lett., 396, 181–188. [DOI] [PubMed] [Google Scholar]
- Niwa H., Yamamura,K. and Miyazaki,J. (1991) Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene, 108, 193–199. [DOI] [PubMed] [Google Scholar]
- Parikh V.S., Morgan,M.M., Scott,R., Clements,L.S. and Butow,R.A. (1987) The mitochondrial genotype can influence nuclear gene expression in yeast. Science, 235, 576–580. [DOI] [PubMed] [Google Scholar]
- Price B.D. and Calderwood,S.K. (1992) Gadd45 and Gadd153 messenger RNA levels are increased during hypoxia and after exposure of cells to agents which elevate the levels of the glucose-regulated proteins. Cancer Res., 52, 3814–3817. [PubMed] [Google Scholar]
- Ron D. and Habener,J.F. (1992) CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Genes Dev., 6, 439–453. [DOI] [PubMed] [Google Scholar]
- Ryan M.T., Naylor,D.J., Hoj,P.B., Clark,M.S. and Hoogenraad,N.J. (1997a) The role of molecular chaperones in mitochondrial protein import and folding. Int. Rev. Cytol., 174, 127–193. [DOI] [PubMed] [Google Scholar]
- Ryan M.T., Herd,S.M., Sberna,G., Samuel,M.M., Hoogenraad,N.J. and Hoj,P.B. (1997b) The genes encoding mammalian chaperonin 60 and chaperonin 10 are linked head-to-head and share a bidirectional promoter. Gene, 196, 9–17. [DOI] [PubMed] [Google Scholar]
- Schmitt-Ney M. and Habener,J.F. (2000) CHOP/GADD153 gene expression response to cellular stresses inhibited by prior exposure to ultraviolet light wavelength band C (UVC). Inhibitory sequence mediating the UVC response localized to exon 1. J. Biol. Chem., 275, 40839–40845. [DOI] [PubMed] [Google Scholar]
- Sekito T., Thornton,J. and Butow,R.A. (2000) Mitochondria-to-nuclear signaling is regulated by the subcellular localization of the transcription factors Rtg1p and Rtg3p. Mol. Biol. Cell, 11, 2103–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamu C.E. and Walter,P. (1996) Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J., 15, 3028–3039. [PMC free article] [PubMed] [Google Scholar]
- Sok J., Wang,X.Z., Batchvarova,N., Kuroda,M., Harding,H. and Ron,D. (1999) CHOP-dependent stress-inducible expression of a novel form of carbonic anhydrase VI. Mol. Cell. Biol., 19, 495–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear E. and Ng,D.T. (2001) The unfolded protein response: no longer just a special teams player. Traffic, 2, 515–523. [DOI] [PubMed] [Google Scholar]
- Traven A., Wong,J.M., Xu,D., Sopta,M. and Ingles,C.J. (2001) Interorganellar communication. Altered nuclear gene expression profiles in a yeast mitochondrial DNA mutant. J. Biol. Chem., 276, 4020–4027. [DOI] [PubMed] [Google Scholar]
- Travers K.J., Patil,C.K., Wodicka,L., Lockhart,D.J., Weissman,J.S. and Walter,P. (2000) Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell, 101, 249–258. [DOI] [PubMed] [Google Scholar]
- Ubeda M., Wang,X.Z., Zinszner,H., Wu,I., Habener,J.F. and Ron,D. (1996) Stress-induced binding of the transcriptional factor CHOP to a novel DNA control element. Mol. Cell. Biol., 16, 1479–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda M., Vallejo,M. and Habener,J.F. (1999) CHOP enhancement of gene transcription by interactions with Jun/Fos AP-1 complex proteins. Mol. Cell. Biol., 19, 7589–7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.Z. et al. (1996) Signals from the stressed endoplasmic reticulum induce C/EBP-homologous protein (CHOP/GADD153). Mol. Cell. Biol., 16, 4273–4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.Z., Kuroda,B., Sok,J., Batchvarova,N., Kimmel,R., Chung,P., Zinszner,H. and Ron,D. (1998) Identification of novel stress-induced genes downstream of chop. EMBO J., 17, 3619–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Liu,X., Bhalla,K., Kim,C.N., Ibrado,A.M., Cai,J., Peng,T.I., Jones,D.P. and Wang,X. (1997) Prevention of apoptosis by Bcl-2: release of cytochrome c from mitochondria blocked. Science, 275, 1129–1132. [DOI] [PubMed] [Google Scholar]
- Ye J., Rawson,R.B., Komuro,R., Chen,X., Dave,U.P., Prywes,R., Brown,M.S. and Goldstein,J.L. (2000) ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell, 6, 1355–1364. [DOI] [PubMed] [Google Scholar]
- Yoshida H., Okada,T., Haze,K., Yanagi,H., Yura,T., Negishi,M. and Mori,K. (2000) ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol. Cell. Biol., 20, 6755–6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M., Matsui,T., Yamamoto,A., Okada,T. and Mori,K. (2001) XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell, 107, 881–891. [DOI] [PubMed] [Google Scholar]
- Young L., Leonhard,K., Tatsuta,T., Trowsdale,J. and Langer,T. (2001) Role of the ABC transporter Mdl1 in peptide export from mitochondria. Science, 291, 2135–2138. [DOI] [PubMed] [Google Scholar]