Abstract
We recently demonstrated that the major decapping activity in mammalian cells involves DcpS, a scavenger pyrophosphatase that hydrolyzes the residual cap structure following 3′ to 5′ decay of an mRNA. The association of DcpS with 3′ to 5′ exonuclease exosome components suggests that these two activities are linked and there is a coupled exonucleolytic decay-dependent decapping pathway. We purified DcpS from mammalian cells and identified the cDNA encoding a novel 40 kDa protein possessing DcpS activity. Consistent with purified DcpS, the recombinant protein specifically hydrolyzed methylated cap analog but did not hydrolyze unmethylated cap analog nor did it function on intact capped RNA. Sequence alignments of DcpS from different organisms revealed the presence of a conserved hexapeptide, containing a histidine triad (HIT) sequence with three histidines separated by hydrophobic residues. Mutagenesis analysis revealed that the central histidine within the DcpS HIT motif is critical for decapping activity and defines the HIT motif as a new mRNA decapping domain, making DcpS the first member of the HIT family of proteins with a defined biological function.
Keywords: decapping/HIT motif/mammalian mRNA turnover
Introduction
Control of mRNA stability is a critical mechanism for the regulation of gene expression. All mRNAs have distinct half-lives, which are precisely regulated, and significant advances have recently been made in elucidating the regulation of eukaryotic mRNA stability (Guhaniyogi and Brewer, 2001; Wilusz et al., 2001). In the yeast Saccharomyces cerevisiae, the first step of the major mRNA turnover pathway involves removal of the poly(A) tail followed by Dcp1p-dependent decapping of the mRNA (Decker and Parker, 1993; Muhlrad and Parker, 1994). The exposed 5′ end undergoes subsequent degradation by the 5′ to 3′ exoribonuclease Xrn1p (Hsu and Stevens, 1993). An alternative, albeit less active, degradation pathway has also been identified in yeast that proceeds 3′ to 5′. This decay is carried out by a complex of proteins termed the exosome, which degrades the mRNA from the 3′ terminus following deadenylation (Mitchell et al., 1997; Anderson and Parker, 1998). The exosome was also recently shown to be the major component involved in the decay of mRNAs lacking a stop codon in a mechanism postulated to rid the cell of aberrant mRNAs that could encode abnormal proteins (Frischmeyer et al., 2002; van Hoof et al., 2002).
Analysis of the mammalian mRNA decay process suggests that the mRNA turnover machinery is conserved from yeast to mammals. Similarly to yeast, deadenylation appears to be the initial step in mammalian mRNA decay both in vivo and in vitro (Wilson and Treisman, 1988; Bernstein and Ross, 1989; Shyu et al., 1991; Couttet et al., 1997; Brewer, 1998; Ford et al., 1999; Wang et al., 1999; Wang and Kiledjian, 2001). Also similarly to yeast, both 5′ to 3′ (Wang and Kiledjian, 2001; Mukherjee et al., 2002) and 3′ to 5′ (Ross and Kobs, 1986; Brewer, 1998; Wang and Kiledjian, 2000, 2001; Chen et al., 2001; Mukherjee et al., 2002) exonucleolytic decay events have been detected in mammals. However, the relative contribution of the directional exonucleolytic decay pathways appears to be the opposite of that observed in yeast. The major pathway of mRNA decay in mammals proceeds through a coupled 3′ to 5′ exoribonucleolytic decay of the mRNA, followed by decapping by the DcpS decapping enzyme (Wang and Kiledjian, 2001). The minor pathway involves a decapping of the intact mRNA, which could potentially undergo 5′ to 3′ exonucleolytic decay.
Several lines of evidence support the premise that upon deadenylation, decay predominantly occurs from the 3′ end in mammals. First, c-myc mRNA, histone mRNA and generic RNA are all primarily degraded from the 3′ end in soluble or polysome-associated in vitro decay assays (Ross and Kobs, 1986; Brewer, 1998; Chen et al., 2001; Wang and Kiledjian, 2001; Mukherjee et al., 2002). Secondly, RNAs transfected into cells are predominantly degraded from the 3′ end even if the 5′ end lacks a cap structure to protect it (Wang and Kiledjian, 2001). Thirdly, using a circularization RT–PCR assay, Couttet et al. (1997) estimated that <3% of deadenylated mRNAs were decapped, suggesting that decapping of an intact mRNA followed by 5′ to 3′ decay is a minor pathway of mammalian mRNA decay. Consistent with this notion, a viral mRNA with an exposed 5′ end lacking a cap structure can remain stable in mammalian cells (Thoma et al., 2001), further implying that the 5′ to 3′ decay is a minor activity. Fourthly, the endogenous c-myc mRNA, within its natural mRNP context in cells, was shown to degrade primarily from the 3′ end (Wang and Kiledjian, 2001). These findings support the premise that the predominant decay pathway proceeds from the 3′ end in mammals.
The 3′ to 5′ exonuclease exosome complex plays an important role in RNA processing both in the nucleus and in the cytoplasm of eukaryotic cells (Decker, 1998; Butler, 2002). Numerous proteins have been shown to either directly or functionally associate with and regulate the exosome (Jacobs et al., 1998; van Hoof et al., 2000). Recently, the DcpS decapping enzyme was also shown to co-purify with exosomal components in a 300 kDa complex (Wang and Kiledjian, 2001). Therefore, the exosome complex is likely to be a target for modulation and may play an active role in the coordination of diverse processes of RNA metabolism.
Decapping plays a crucial role in mRNA turnover and decapping rates can vary greatly among different mRNAs (Muhlrad and Parker, 1994; Muhlrad et al., 1995). The yeast Dcp1p decapping enzyme is a pyrophosphatase capable of decapping the intact mRNA to release m7GDP. This activity preferentially hydrolyses methylated capped RNA >25 nucleotides long (LaGrandeur and Parker, 1998). A decapping activity that functions specifically on the residual cap structure to release m7GMP following 3′ to 5′ degradation of the mRNA body has also been identified (Nuss et al., 1975; Kumagai et al., 1992; Wang and Kiledjian, 2001). Its role is to complete the demise of the degraded mRNA and clear the cell of a potentially deleterious accumulation of cap analog, hence its name: scavenger decapping activity, DcpS (Wang and Kiledjian, 2001). We now report the identification of the gene encoding DcpS, which is a new member of the histidine triad (HIT) superfamily of pyrophosphatases. HIT proteins are nucleotide binding proteins that contain a conserved hexapeptide HIT motif, which consists of alternating histidines separated by hydrophobic amino acids (Seraphin, 1992). Although the biological function of HIT proteins is not known, they have been proposed to be regulators of dinucleotide (NpppN) signaling molecules (Kisselev et al., 1998). DcpS is the first HIT protein with a defined biological function and identifies the HIT motif as a new mRNA decapping domain.
Results
The scavenger decapping activity is carried out by a 40 kDa protein
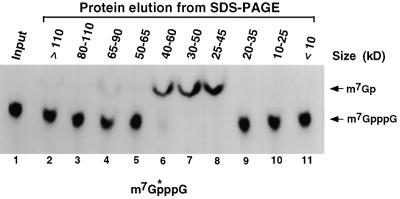
We recently reported that a major decapping activity in mammalian cell extract is carried out by a scavenger decapping enzyme, DcpS, which functions on the resulting cap structure following exonucleolytic decay (Wang and Kiledjian, 2001). DcpS can associate with the exosome in a 300 kDa complex, implying the existence of a coupled 3′ to 5′ degradation and decapping pathway. The DcpS activity hydrolyzed methylated cap analog and specifically released m7GMP, but did not function on intact mRNA or unmethylated cap analog. In an effort to purify the DcpS protein, we initially determined its monomer size. Gel filtration analysis indicated that the DcpS activity was 80 kDa (Wang and Kiledjian, 2001). However, it was unclear whether this size corresponded to a DcpS monomer or multimer. To determine the size of the DcpS activity, MEL S130 extract was resolved by SDS–PAGE. A sequential series of gel slices was excised from a lane containing the MEL S130 extract and proteins within the slices were eluted and renatured (see Materials and methods). The ability of the renatured protein to decap 32P-labeled cap structure containing the label at the first phosphate following the methylated guanosine (m7G*pppG) was tested. The reaction products were separated by polyethyleneimine (PEI) cellulose thin-layer chromatography (TLC) and developed in 0.45 M (NH4)2SO4. This buffer condition will separate the input cap structure from the resulting m7GMP. As shown in Figure 1, m7GMP generated by the DcpS decapping activity was detected in the proteins eluted from three overlapping fractions that all contained protein in the ∼40 kDa size range (lanes 6–8). Surprisingly, DcpS activity was not detected in the 80 kDa range (lanes 3 and 4), suggesting that the 80 kDa size detected by gel filtration in Wang and Kiledjian (2001) was most likely a consequence of dimerization, and the true size of DcpS was 40 kDa.
Fig. 1. DcpS activity elutes at the 40 kDa size range from an SDS–PAGE gel. MEL S130 extract (100 µg) was resolved by an SDS–PAGE and sequential gel slices were eluted and renatured as described (Rudolph and Lilie, 1996). The size and integrity of the protein in each sample were determined by rerunning an aliquot of the eluted protein on an SDS–PAGE gel and visualized by silver staining (data not shown). A sample of each eluted protein was tested for its ability to hydrolyze labeled cap analog and the products resolved by PEI TLC. The size range of the proteins is indicated at the top of each lane and migration of the standards visualized by ultraviolet shadowing is indicated on the left. Input, labeled cap analog is shown in lane 1. The schematic shown at the bottom represents the cap analog substrate used and the asterisk denotes the position of the labeled phosphate.
Purification of the DcpS protein and cloning of the corresponding gene
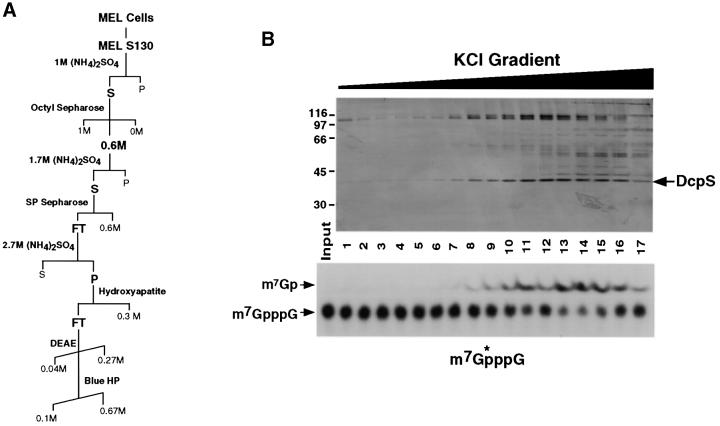
A biochemical purification was undertaken to identify the DcpS protein. MEL S130 extract was used as the starting point for the purification since DcpS activity is enriched in this fraction (Wang and Kiledjian, 2001). The purification scheme is shown in Figure 2A and described in detail in Materials and methods. The fractions eluted from the final Blue HP column are shown at the top of Figure 2B and the corresponding DcpS activity of each fraction is shown at the bottom. Only one band in the 40 kDa size range corresponded to the DcpS activity detected in the lower panel. This band was excised, the protein was eluted, renatured and tested in a decapping assay to confirm that it contained decapping activity (data not shown). Mass spectrometry analysis indicated that the protein corresponded to a predicted open reading frame encoded by an unknown human cDNA originally isolated from human CD34+ hematopoietic stem/progenitor cells (HSPC015; DDBJ/EMBL/GenBank accession No. AF077201) (Zhang et al., 2000). The clone encoded a predicted open reading frame of 337 amino acids with a calculated mass of 38.6 kDa. A search of the NCBI LocusLink database indicates that the gene is contained at human chromosome position 11q24.2.
Fig. 2. Purification of DcpS. (A) The purification scheme is outlined including the columns and fractions used. Elution gradients are indicated by diagonal lines. The fractions containing DcpS activity are in bold. (B) Protein eluted from the final Blue HP column purification of DcpS protein from 150 l of MEL cells is shown. Proteins were eluted with an increasing KCl gradient from 100 to 670 mM and either resolved by SDS–PAGE and visualized by silver staining (top), or used in a decapping assay and the products resolved by PEI TLC (bottom). Labeling is as described in the legend to Figure 1. The decapping assays were carried out in the presence of 1 µM cold cap analog to slow the reaction. The fraction numbers are indicated between the two panels. The candidate band, which corresponded with DcpS activity, is indicated. Protein size markers are shown on the left in kDa.
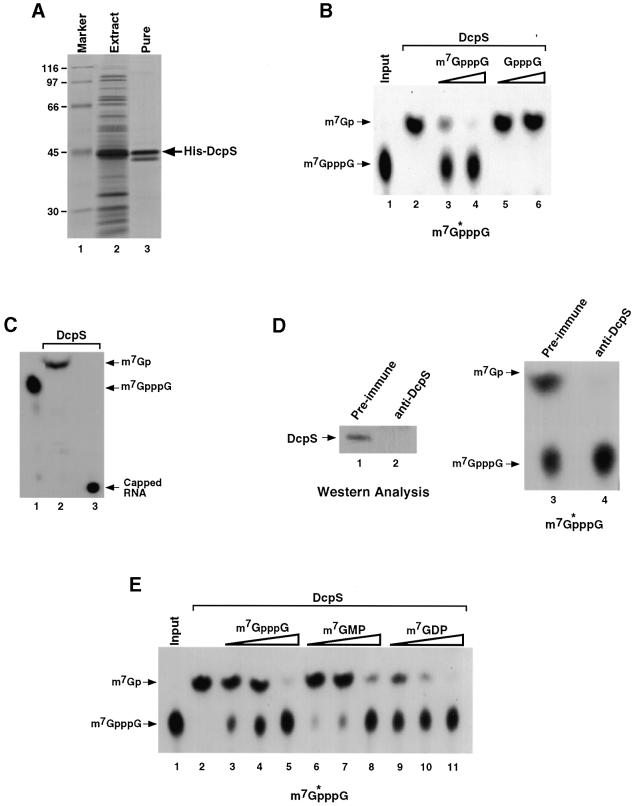
The HSPC015 cDNA was expressed as a His-tagged recombinant protein (Figure 3A) and used in a decapping assay with 32P-labeled cap analog as substrate to determine its decapping potential. The products were resolved by TLC and, as shown in Figure 3B, the recombinant protein expressed from the HSPC015 cDNA contained a decapping activity that releases m7GMP (lane 2) similar to DcpS activity from cytoplasmic extract. The decapping activity is sensitive to competition with unlabeled methylated cap analog (lanes 3 and 4) but minimally affected by the addition of unmethylated cap analog competitor (lanes 5 and 6), demonstrating substrate specificity for the N-7 methyl group in the decapping. As expected, this activity only functioned on the cap analog and did not utilize capped RNA as substrate (Figure 3C, lane 3). Furthermore, immunodepletion of human K562 cell S130 extract with polyclonal antisera directed against recombinant His- DcpS, which efficiently removed DcpS from the extract (Figure 3D, lanes 1 and 2), also abolished detectable DcpS decapping activity (lanes 3 and 4). Collectively, these data indicate that the HSPC015 cDNA encodes the DcpS protein, and it will be referred to as DcpS (DDBJ/EMBL/GenBank accession No. AF532613) hereafter.
Fig. 3. Recombinant DcpS is methyl-cap specific and does not hydrolyze capped RNA. (A) Total extract from E.coli BL21DE3 cells expressing His-tagged DcpS (lane 2) or purified recombinant protein (lane 3) resolved by SDS–PAGE and stained by Coomassie Blue are shown. The arrow denotes the His-tagged DcpS protein. Protein size markers are shown in lane 1 and the sizes indicated on the left. (B) His-tagged DcpS (100 ng) was tested for its ability to hydrolyze labeled cap analog (lanes 2–6). Lanes 3 and 4 contain 1 and 10 µM methylated cap analog competitor, while lanes 5 and 6 contain the same amount of unmethylated cap analog competitor. The substrate is shown at the bottom of the figure. (C) His-tagged DcpS was used to hydrolyze labeled cap analog (lane 2) or cap labeled RNA (lane 3). DcpS is unable to utilize the intact RNA as substrate. (D) Polyclonal antibody directed against recombinant His-DcpS was used to immunodeplete K562 S130 extract. A western blot of extract depleted with pre-immune or DcpS-specific antisera is shown in lanes 1 and 2, respectively. The products of a decapping assay using 10 µg of the respective depleted extract were resolved by PEI TLC (lanes 3 and 4). Migration of the standards and schematic of the substrate are indicated as above. Extract depleted of DcpS is devoid of scavenger decapping activity. (E) Decapping of labeled cap analog was carried out with His-tagged DcpS (100 ng) in the presence of the indicated competitors. Lanes 3, 6 and 9 contained 0.25 µM competitor. Lanes 4, 7 and 10 contained 1 µM competitor, and lanes 5, 8 and 11 contained 2.5 µM indicated competitor. The different levels of cap analog competitor susceptibility observed in (B) and (E) are due to the use of two separate preparations of recombinant DcpS for each panel.
The extent of decapping product inhibition of DcpS was also tested. Endogenous DcpS is inhibited by cap analog in extract (Wang and Kiledjian, 2001), as is recombinant DcpS (Figure 3A). However, the relative inhibition of the m7GMP product compared with the cap analog was not known. Decapping assays were carried out in the presence of increasing concentrations of m7GpppG, m7GMP and m7GDP. As shown in Figure 3E, the m7GMP product was able to compete for DcpS activity to a level comparable to that of m7GpppG cap analog (compare lanes 6–8 with 3–5). Surprisingly, we found that m7GDP was even more efficient at inhibiting DcpS activity than were either m7GMP or cap analog (lanes 9–11), but DcpS was unable to hydrolyze m7GDP (data not shown). These data indicate that DcpS is also competent in binding both m7GMP and m7GDP, and the binding to m7GDP is greater.
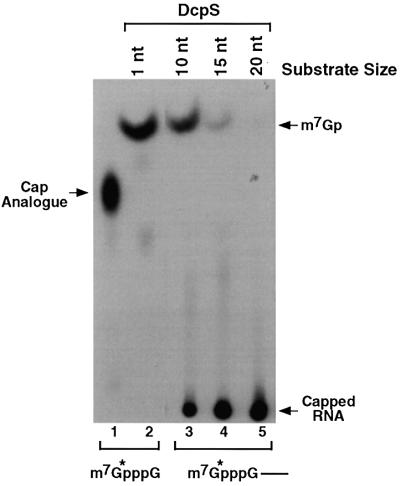
The ability of DcpS to hydrolyze the cap analog but not a 136 nucleotide capped RNA (Figure 3C) implied that there is a maximal RNA size limit that DcpS can utilize as substrate and decap. To determine the size range of RNA that DcpS can function on, RNase H cleavage was used to generate defined-length RNAs. Three oligonucleotides complementary to regions at the 5′ end of the polylinker (pcP) RNA were used to generate cap-labeled RNAs of 10, 15 and 20 nucleotides by RNase H cleavage (see Materials and methods). The RNAs were subsequently subjected to decapping by recombinant DcpS. As shown in Figure 4, DcpS was capable of decapping the cap analog (lane 2) as expected and was also able to hydrolyze the 10 nucleotide substrate (lane 3). However, the caps of the longer RNAs were not efficiently utilized as substrate and not cleaved (lanes 4 and 5). These data suggest that DcpS is capable of acting on an mRNA once it is degraded down to 10 nucleotides.
Fig. 4. DcpS preferentially hydrolyzes RNAs <10 nucleotides. Recombinant DcpS (500 ng) was used in decapping assays utilizing various length cap-labeled substrates. Labeled cap analog (lane 1), which contains one nucleotide following the m7G, was used as the substrate in lane 2. 5′-end-labeled oligonucleotides of 10, 15 and 20 bases in length were used in lanes 3, 4 and 5, respectively. The decapping products were resolved by PEI TLC. Labeling is as described in the legend to Figure 1.
Identification of DcpS homologs in S.cerevisiae
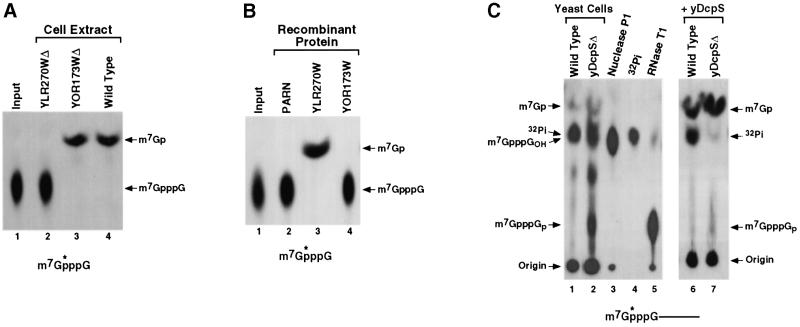
A DcpS decapping activity is present in yeast (Wang and Kiledjian, 2001), indicating that a gene encoding the yeast homolog of human DcpS would be contained in the yeast database. A sequence search of the S.cerevisiae genomic database identified two genes with unknown function that were potential homologs of DcpS. They were YLR270W and YOR173W, and the proteins encoded by the two genes are 30% identical (65% similar) and 34% identical (65% similar) to human DcpS, respectively. YLR270W is contained on yeast chromosome XII and YOR173W is on chromosome XV. To test whether either protein was responsible for the yeast DcpS activity, we obtained haploid yeast strains disrupted for either gene (ResGen; Invitrogen). Both yeast strains are viable, indicating that neither gene is essential, although the YLR270W strain had a slow growth phenotype. Decapping assays were carried out with extract derived from exponentially growing cells of each deletion strain to determine whether decapping was compromised in either cell. Labeled cap analog was used as substrate since it is cleaved by DcpS but is not hydrolyzed by the yeast Dcp1p decapping enzyme, which only functions on intact capped RNA (LaGrandeur and Parker, 1998). As shown in Figure 5A, extract from a wild-type yeast parental strain (BY4741) contains DcpS activity (lane 4), while extract derived from an isogenic strain with a disrupted YLR270W gene had no detectable decapping activity (lane 2). However, extract from a strain with a disrupted YOR173W gene contained wild-type levels of DcpS activity (lane 3). Therefore, the YLR270W gene product is required for the DcpS-mediated decapping in yeast, while the YOR173W gene product does not have an obvious contribution.
Fig. 5. The YLR270W gene encodes yeast DcpS (yDcpS). (A) Total yeast extract (50 µg) from the YLR270WΔ (lane 2), YOR173WΔ (lane 3) or wild-type cells (lane 4) was tested for the ability to hydrolyze labeled cap analog in a standard decay assay and the products resolved by PEI TLC. Extract from the YLR270WΔ strain lost the ability to hydrolyze cap analog (lane 2), while extract from the YOR173WΔ strain retains the ability to decap the same substrate. (B) The ability of His-tagged recombinant protein (500 ng) encoded by each yeast DcpS-like gene was used directly in an assay to determine their ability to cleave the labeled cap structure substrate. Consistent with the deletion strain data in (A) above, His-tagged protein expressed from the YLR270W gene was able to specifically hydrolyze the cap structure (lane 3), while protein expressed from YOR173W gene was not (lane 4). Purified His-tagged PARN was used as a negative control (lane 2). Labeling is as described in the legend to Figure 1. (C) Cap-labeled pcP RNA was electroporated into yeast cells and harvested 6 h post-electroporation and decapping products resolved by PEI TLC developed in 0.75 M LiCl to better resolve the lower spots. Decapping products from wild-type yeast cells (lane 1) and the yDcpSΔ strain (lane 2) are shown. The labeled cap analog standards were generated by treating cap-labeled pcP RNA with nuclease P1 (m7GpppGOH; lane 3) or with RNase T1 (m7GpppGp; lane 5). Migration of the 32Pi-free phosphate is shown in lane 4. Products shown in lanes 1 and 2 were treated with recombinant yDcpS and resolved by TLC as shown in lanes 6 and 7, respectively. DcpS will hydrolyze cap analog present in these lanes to m7GMP.
To directly determine whether the YLR270W gene product contains a pyrophosphatase activity, His-tagged recombinant protein encoded by the YLR270W gene was expressed in Escherichia coli and used in a decapping assay. As shown in Figure 5B, the recombinant protein contained a DcpS-like pyrophosphatase activity (lane 3), which released m7GMP, while His-tagged fusion protein from YOR173W (lane 4) did not nor did a control protein (lane 2). These data indicate that the YLR270W gene encodes the yeast DcpS (yDcpS) protein, while the function of a closely related protein encoded at YOR173W, which is 65% identical (90% similarity), remains unknown.
The significance of yDcpS for decapping in cells was tested by transfection of labeled RNA into yeast cells. RNA transfected into yeast cells is primarily degraded by the 3′ to 5′ decay pathway (Brown and Johnson, 2001). If yDcpS is involved in clearing the methylated cap analog following 3′ to 5′ decay, we would expect cap analog to accumulate in the deletion strain but not in the wild- type strain. 32P-cap-labeled RNA corresponding to the pcDNA3 pcP was transfected into either wild-type or yDcpSΔ strains. Post-electroporation (6 h), cells were lysed and decapping products detected by TLC resolved in 0.75 M LiCl. Both samples contained a product (Figure 5C, lanes 1 and 2) that co-migrated with labeled cap analog (lane 3). However, since free phosphate also co-migrates at this position with this buffer system (lane 4), the identity of the products could not be determined unambiguously. To this end, an aliquot of the samples used in lanes 1 and 2 was treated with recombinant yDcpS. yDcpS converts cap analog to m7GMP. yDcpS efficiently hydrolyzed the products from the yDcpSΔ strain in lane 2 to m7GMP (lane 7), indicating that cap analog accumulated in the deletion strain. However, as seen in lane 6, yDcpS had no effect on the product detected from the wild-type yeast cells shown in lane 1, demonstrating that the original major product was 32Pi, which was generated upon dephosphorylation of the m7GMP decapping product in the cells. Interestingly, two additional products are detected in lane 2 that are also hydrolyzed by DcpS and converted to m7GMP. The identity of the central product is unknown; however, the lower spot co-migrates with a cap analog containing a 3′ phosphate (lane 5), indicating that this spot probably corresponds to m7GpppGp. Furthermore, recombinant DcpS can efficiently hydrolyze cap analogs with either a 3′ hydroxyl or phosphate (data not shown). Collectively, these results provide evidence that yDcpS is involved in the decapping of cap analog following decay of the RNA both in cells and in extract.
DcpS contains a functional HIT pyrophosphatase motif
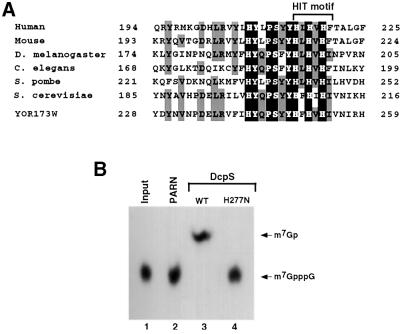
Sequence searches of the GenBank database using the DcpS protein sequence identified potential homologs in other organisms, indicating that DcpS is a conserved protein present in a variety of species. For example, overall protein identities between the human DcpS and putative DcpSs from the following species are: 89% with mouse, 42% with Drosophila melanogaster, 41% with Caenorhabditis elegans and 39% with Schizosaccharo myces pombe. Computer searches did not detect extensive obvious homology to other known proteins. A manual examination of the protein revealed the presence of a His-φ-His-φ-His-φ sequence, where φ denotes hydrophobic amino acids. These six amino acids are the central core of the HIT pyrophosphatase motif. Proteins with a HIT domain are nucleotide binding proteins that can hydrolyze a pyrophosphate bond (Guranowski et al., 1996; Guranowski, 2000). Other than the HIT hexapeptide, DcpS does not share any region of homology with other HIT-motif-containing proteins. An alignment of the HIT-motif region of putative DcpS proteins from a select number of organisms is shown in Figure 6A. A high degree of identity exists between the different DcpS proteins in the HIT region, suggesting a functional significance for this domain.
Fig. 6. The DcpS HIT motif and its significance for decapping. (A) An alignment of the HIT hexapeptide (boxed region) and adjacent amino acids from potential DcpS homologs from different organisms is shown. The alignment was generated using the ClustalW alignment program with the default parameters. The sequences shown are: human DcpS (NP_054745), mouse DcpS (BAB24138), D.melanogaster DcpS (XP_082208), C.elegans DcpS (NP_507876), S.pombe DcpS (NP_596194), S.cerevisiae DcpS YLR270W (NP_013372) and the DcpS-like yeast protein YOR173W (NP_014816). The amino acid positions are indicated on the left and right of the sequences. Amino acids that are identical at a particular position in all seven proteins are shaded in black. Amino acids that are identical in four or more proteins at a specific position are shaded in gray. The position of the HIT hexapeptide is denoted by the bracket. (B) The significance of the DcpS HIT motif for decapping was tested by substitution of the central histidine at amino acid 277 with asparagine (H277N). Recombinant His-tagged wild-type (lane 3) or mutant protein (lane 4) was tested for the ability to hydrolyze labeled cap analog using 500 ng of protein. His-tagged PARN was used as a negative control (lane 2). Substitution of the central His ablates decapping (lane 4).
To test the importance of the HIT motif within DcpS, the central histidine, which is critical for pyrophosphatase activity of other HIT proteins (Lima et al., 1997), was replaced with an asparagine (H277N) and the mutant human protein tested for pyrophosphatase activity. As shown in Figure 6B, this single amino acid substitution eliminated the decapping activity of DcpS (lane 4). These data confirm that DcpS contains a functional HIT pyrophosphatase domain and demonstrate the significance of the central histidine for decapping. Furthermore, they demonstrate that the detected activity is a consequence of the DcpS recombinant protein and not a co-purifying contaminant protein. Interestingly, the HIT-motif region is also highly conserved in the yeast YOR173W gene product (Figure 6A, bottom) despite the fact that it did not contain DcpS activity (Figure 5), demonstrating that the HIT motif is necessary, but not sufficient, for decapping.
Discussion
We present the purification of the major mammalian mRNA decapping enzyme DcpS and the identification of a novel cDNA (HSPC015) encoding the 40 kDa protein. Recombinant DcpS specifically hydrolyzed methylated cap analog or capped mRNA of ≤10 nucleotides. DcpS contains a functional HIT pyrophosphatase motif essential for its decapping activity. It is the first member of the HIT family of proteins with a defined biological function other than a general pyrophosphatase activity and identifies the HIT motif as a domain involved in mRNA decapping.
There are at least two distinct decapping activities in eukaryotes: one decaps the intact mRNA and triggers 5′ to 3′ exonucleolytic decay, while the second decaps the residual cap structure following 3′ to 5′ exonucleolytic decay of the mRNA. The decapping of intact RNA is carried out by the Dcp1p enzyme in yeast (Beelman et al., 1996; LaGrandeur and Parker, 1998) and by a protein containing Dcp1p-like activity in mammals (Wang et al., 2002). Both proteins hydrolyze the cap structure of an intact mRNA to release m7GDP. The scavenger decapping activity is carried out by DcpS in mammals and by its homolog, yDcpS, in yeast to complete the demise of an mRNA. The demonstration that m7GDP was a more efficient competitor of DcpS activity than were either cap analog or m7GMP (Figure 3B) suggests that a link between the two decapping activities could exist, where the product of the Dcp1-like decapping (m7GDP) may serve as a regulator of DcpS activity.
A decapping activity similar to DcpS was previously described and shown to function on capped RNAs shorter than seven nucleotides (Nuss et al., 1975; Nuss and Furuichi, 1977). It was postulated that this activity functioned as a scavenger to rid the cell of residual methyl cap analog following 3′ to 5′ decay of the mRNA (Nuss et al., 1975). A protein with a similar activity was also purified by Kumagai et al. (1992). Considering the results in Figure 3, where immunodepletion of DcpS eliminates this activity from cell extract, it is highly likely that the previously described decapping activities were carried out by DcpS.
It is intriguing that yeast contains two DcpS like proteins. These proteins are 65% identical (90% similar) to one another, both contain a HIT motif and they are ∼30% identical (65% similar) to the human DcpS protein. Although neither protein is essential for viability, one of these proteins, yDcpS (YLR270W), is capable of hydrolyzing the cap structure (Figure 5B) in extract as well as in cells (Figure 5C). The functional significance of the second protein encoded by YOR173W is not known. Curiously, we have not been able to detect a second gene similar to YOR173W encoding a DcpS-like protein in mammals or any of the other organisms listed in Figure 6. It will be interesting to determine the function of the YOR173W gene product and what its counterpart is in mammals.
The inability of DcpS to function on RNAs >10 nucleotides long suggests that DcpS is able to detect whether the mRNA is degraded or intact and specifically decap a structure that has a significant portion of the mRNA body removed. This hypothesis implies either that DcpS is unable to bind the cap structure when the RNA is intact (perhaps due to steric interference), or that it contains an RNA ‘sensing’ mechanism (perhaps a novel RNA-binding motif) and when it simultaneously detects the cap structure and mRNA its decapping activity is inhibited. We are currently testing these hypotheses.
The HIT motif that contains a conserved HIT sequence was originally discovered by sequence alignment (Seraphin, 1992). Proteins within the HIT family fall into two branches and proteins within each branch share substantial similarity to one another. One class is only found in animals and fungi, and constitutes the Fhit (fragile histidine triad) branch, while the second, Hint (HIT-nucleotide binding protein), is more ancestral and present in all phyla (Guranowski, 2000). Although a high degree of conservation exists among proteins within each branch, the HIT hexapeptide is the only region absolutely conserved between the two branches (Brenner et al., 1999). Structural analysis of representative HIT proteins from each of the two branches revealed that they form a homodimer with a 10-strand antiparallel β-sheet and two pyrophosphatase domains (Brenner et al., 1997; Pace et al., 1998). Although DcpS shares no obvious sequence homology with other HIT proteins, except for the HIT hexapeptide, its 80 kDa size determined by gel filtration (Wang and Kiledjian, 2001) implies that it too has the potential to form a homodimer. However, the specificity for methylated guanosine cap demonstrates that DcpS is distinct from the previously reported HIT protein dinucleotide triphosphatases, which can efficiently hydrolyze both methylated and unmethylated structures (Guranowski et al., 1996; Guranowski, 2000). DcpS and its homologs probably constitute a third branch within the HIT superfamily of proteins. The central histidine of a HIT motif is involved in the pyrophosphatase activity. It catalyzes a covalent nucleotidyl phosphohistidyl intermediate, which is utilized as a nucleophile for the α phosphate group of a dinucleotide triphosphate (Lima et al., 1997; Abend et al., 1999). Consistent with the significance of the central histidine in the pyrophosphatase activity, substitution mutagenesis of the central histidine in DcpS (H277N in Figure 6B) demonstrates that DcpS also contains a functional HIT motif.
Many of the HIT proteins identified appear to function on dinucleotide signal molecules linked by pyrophosphate bonds. For example, the Fhit protein preferentially hydrolyzes ApppA and this function is thought to contribute to tumor suppression. However, how the dinucleotide could potentially elicit a physiological consequence is unknown. Similarly, DcpS might be required for the elimination of the m7GpppN cap dinucleotide following mRNA decay, which could have at least two functional consequences. First, hydrolysis of the cap structure could minimize potential sequestration of eIF4E cap binding protein from the cap of translationally competent mRNA. Secondly, hydrolysis of the cap structure could be involved in the regulation of PARN-mediated deadenylation. Cap analog has been shown to inhibit PARN activity (Dehlin et al., 2000; Gao et al., 2000; Martinez et al., 2000). Therefore, one consequence of DcpS could be in a feedback mechanism to regulate PARN-mediated deadenylation. This could be a significant regulatory step since the initial step of decay is the removal of the poly(A) tail. Both of these potential regulatory roles are being addressed.
The association of DcpS with the mammalian exosome and the maximum size of capped oligonucleotide that can serve as a DcpS substrate suggest the following model for the 3′ to 5′ decay pathway in mammals. Following decay of an mRNA from the 3′ end by the exosome, the associated DcpS would be capable of cleaving the residual cap once the mRNA size is decreased to ≤10 nucleotides. The cap binding potential of DcpS, as well as its association with the exosome (Wang and Kiledjian, 2001), makes DcpS an attractive candidate to coordinate specific targeting of the exosome to capped mRNA. Future efforts will address any potential coordination of function between the exosome and DcpS in the regulation of mRNA decay.
Materials and methods
Plasmid constructs and recombinant protein purification
The pET28-hDcpS plasmid contains the human DcpS open reading frame derived by PCR amplification of K562 cell reverse transcribed mRNA. The 5′ primer (5′-AGGATCCCGCCTCCGCGGCAGCATG-3′) introduced a BamHI site and a 3′ primer (5′-TCTCGAGTCAGCTTTGC TGAGCCTCCTG-3′) introduced a XhoI site. The PCR products were cleaved with BamHI and XhoI, and inserted into the same sites of pET28a (Novagen, Madison, WI). The plasmids expressing yeast proteins encoded by YLR270W (pET28-yDcpS) and YOR173W (pET28-YOR173W) were PCR amplified from genomic DNA using primers that introduce BamHI and XhoI, and inserted into the same sites of pET28a as above. The primer set for yDcpS was 5′-AATTTGGATCCATGTCTCAACTGCCAACAG-3′ and 5′-CAGCCCTCGAGTTATTTAAAACCGTTCAC-3′. The primer set for YOR173W was 5′-TCGCGGGATCCATGAGTATAAAGGCAGAAAG-3′ and 5′-AGCGCCTCGAGTCAACCCAAATTGGTATTG-3′. The plasmid pET-hDcpS-H277N, which substitutes His277 with asparagine, was generated using the QuikChange mutagenesis system according to the manufacturer’s instructions (Stratagene, La Jolla, CA) with the following primers: 5′-CCCTCCTACTACCACCTGAATGTGCACTTCACCGCCCTG-3′ and 5′-CAGGGCGGTGAAGTGCACATTCAGGTGGTAGTA GGAGGG-3′. The plasmid expressing His-tagged PARN (pET30-PARN) was kindly provided by M.Wormington (University of Virginia) (Korner et al., 1998). Expression and purification of the His-tagged recombinant proteins His-yDcpS, His-YOR173W and His-PARN were carried out under native conditions according to the manufacturer’s suggested protocol (Novagen). Purification of His-hDcpS was carried out under denaturing conditions with 6 M urea according to the manufacturer’s instructions and the protein renatured by sequential dialysis against PBS to remove the urea. All PCR generated constructs were confirmed by sequencing.
Extract preparation
Murine erythroleukemia (MEL) cells were obtained from the National Culture Center (Minneapolis, MN). Cytosolic S130 extract was isolated as described in Rodgers et al. (2002). Yeast extracts from YLR173WΔ and YOR270WΔ deletion strains, or the parental BY4741 strain (purchased from ResGen Invitrogen Corporation, Huntsville, AL), were prepared as described previously (Zhang et al., 1999).
Protein isolation from SDS–PAGE
Proteins were eluted and renatured from an SDS–PAGE gel essentially as described in Rudolph and Lilie (1996) but with slight modifications. MEL S130 extract (100 µg) was resolved by 12.5% SDS–PAGE and, without staining, 1 cm gel slices were excised with a razor. Protein was electroeluted from the gel slice at 35–45 V in 1× TAE for 16 h at 4°C in a dialysis bag submerged in the same buffer. Protein was renatured by dialyzing overnight against renaturation buffer [50 mM Tris–HCl pH 8.5, 500 mM l-arginine, 5 mM EDTA, 0.4% (w/v) PEG3000, 5 mM reduced glutathione (Sigma, St Louis, MO) and 1 mM oxidized glutathione (Sigma)] and subsequently dialyzed against PBS with 10% glycerol and stored at –80°C.
Biochemical fractionation
S130 extract derived from 150 l of MEL cells was used as the starting point for DcpS purification. All purification steps were carried out at 4°C with ice-cold buffers. An ammonium sulfate cut was used to concentrate and enrich the S130 extract by the addition of solid (NH4)2SO4 to 25% saturation (1 M). After centrifugation at 4000 g for 20 min, the resulting supernatant was bound to a HiPrep Octyl–Sepharose column (Pharmacia) equilibrated in 1 M (NH4)2SO4 and 50 mM Na2HPO4 pH 7.5, and protein was eluted with 0.6 M (NH4)2SO4 and 50 mM Na2HPO4 pH 7.5. The extract was further enriched with the addition of solid (NH4)2SO4 to 40% saturation (1.7 M) and precipitated proteins were removed by centrifugation as above. The resulting supernatant was dialyzed against HG (20 mM HEPES pH 7.5, 10% glycerol) with 40 mM KCl (HG-40) and passed through an SP-Sepharose column (Pharmacia, Piscataway, NJ). Solid (NH4)2SO4 was added to the flow-through to achieve 60% saturation, and precipitated proteins were collected by centrifugation, dissolved in HAB (20 mM HEPES pH 7.5, 100 mM KCl, 1 mM DTT, 10% glycerol) and dialyzed against HAB with 10 mM Na2HPO4. The protein was subsequently bound to a hydroxyapatite column (Pharmacia) equilibrated in the same buffer. The flow-through was collected and dialyzed against HEG (20 mM HEPES pH 7.5, 1 mM EDTA, 10% glycerol) with 40 mM KCl (HEG-40), and loaded onto a DEAE column (Pharmacia) equilibrated in the same buffer. Bound protein was eluted with a 7 ml gradient from 40 to 270 mM KCl. Fractions (0.5 ml) were collected and tested for DcpS activity. Fractions containing activity were pooled, dialyzed against HEG-100 and loaded onto a Blue HP column (Pharmacia) equilibrated in the same buffer. Bound proteins were eluted with an 8.5 ml gradient from 100 to 670 mM KCl, 0.5 ml fractions collected and dialyzed against HG-40. An aliquot from each fraction was resolved by SDS–PAGE and protein visualized by silver staining, as previously described (Rodgers et al., 2002), as well as tested for DcpS activity. Mass spectrometry analysis of the DcpS candidate protein was carried out at the UMDNJ Mass Spectrometry Facility (Newark, NJ).
Generation of labeled RNA and labeled cap structure
Unlabeled, uncapped RNA corresponding to the pcDNA3 pcP was transcribed in vitro by T7 polymerase, according to the manufacturer’s instructions (Promega, Madison, WI), from a PCR-generated template using T7 and SP6 promoter primers. Cap labeling of the RNA was carried out with the vaccinia virus capping enzyme in the presence of [α-32P]GTP and S-adenosyl-methionine (SAM) to label the α-phosphate (relative to the terminal guanosine; m7G*pppN-), and gel purified by denaturing polyacrylamide gel, as described in Wang et al. (1999). Labeled cap structure containing a 3′ hydroxyl was generated by treating the cap-labeled RNA with nuclease P1 (Roche, Indianapolis, IN) to degrade the RNA leaving the cap structure intact, as described previously (Wang and Kiledjian, 2001). Labeled cap structure with a 3′ phosphate was similarly generated using 200 U of RNase T1 (Roche) for 1 h at 37°C. The samples were extracted once with an equal mixture of phenol:chloroform (1:1) and twice with chloroform, and the resulting supernatant containing the labeled cap analog was used as substrate for the decapping assays. Methylated cap analog and its derivatives m7GMP and m7GDP used as cold standards for TLC were purchased from Ambion.
Cap-labeled oligonucleotides of defined length in Figure 4 were generated with RNase H cleavage. Cap-labeled pcP RNA was generated by T7 polymerase and gel purified as above, annealed with either GCTCGGTACCAAGCT (T7/10), TCCGAGCTCGGTACC (T7/15) or GTGGATCCGAGCTCG (T7/20), and subjected to RNase H cleavage according to the manufacturer’s instructions (Promega). The antisense primers were designed to generate a 10 nucleotide (T7/10), a 15 nucleotide (T7/15) and a 20 nucleotide (T7/20) cap-labeled RNA. The resulting RNAs were extracted once with phenol:chloroform (1:1) and twice with chloroform. The supernatant containing the oligonucleotide was utilized as the substrate for the decapping assay.
Detection of decapping products
Proteins or fractions were incubated with the labeled cap structure in IVDA buffer (10 mM Tris pH 7.5, 100 mM KOAc, 2 mM MgOAc, 2 mM DTT, 10 mM creatine phosphate, 1 mM ATP, 0.4 mM GTP, 0.1 mM spermine) for 20–30 min at 37°C. The reactions were stopped and extracted once with an equal mixture of phenol:chloroform and twice with chloroform. An aliquot was applied to PEI–cellulose TLC plates (Sigma) that were pre-run in H2O and air dried prior to use, and the products were developed with 0.45 M (NH4)2SO4 at room temperature. The TLC in Figure 5C was developed in 0.75 M LiCl. The plates were dried and exposed to Kodak BioMax film overnight. Quantitations were carried out using a Molecular Dynamics PhosphorImager (Storm860) using ImageQuante-5 software.
RNA electroporation
RNA was electroporated into yeast cell as described previously (Searfoss et al., 2002) with slight modification. Cap-labeled RNA (2 × 107 c.p.m./2 µg) generated as above was electroporated into 200 µl of yeast spheroblasts (2 × 108 cells/ml) resuspended in 1 M sorbitol. Following discharge, the cells were washed twice with YPD containing 1 M sorbitol at room temperature, then resuspended in the same medium and incubated at 30°C with gentle swirling. Cells were harvested at the indicated times, washed twice in yeast spheroblast buffer A (Everett and Gallie, 1992; Searfoss et al., 2002) and lysed by freezing–thawing in liquid nitrogen. The supernatant was extracted twice with an equal mixture of phenol and chloroform, and the decapping products detected by TLC.
Generation of antibodies, immunodepletion and western analysis
Polyclonal antibody directed against recombinant His-DcpS was commercially generated in rabbits (Cocalico Biologicals, Reamstown, PA). Immunodepletions of 100 µg of K562 S130 extract were carried out with three sequential incubations of 30 µl of antibody bound to 20 µl of protein A beads in PBS/0.5% Tween-20, as described previously (Wang and Kiledjian, 2001). Primary antibody at 1:1000 dilution was used for western analysis and visualized by ECL using a horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (1:5000 dilution; Cappel, West Chester, PA).
Acknowledgments
Acknowledgements
We thank G.Qing, C.Lima, M.Steiger and R.Parker for helpful discussions. We also thank Z.Wang and A.Carr-Schmid for helpful discussions and critical reading of the manuscript. This work was supported by funds from the NIH (DK51611) to M.K.
References
- Abend A., Garrison,P.N., Barnes,L.D. and Frey,P.A. (1999) Stereochemical retention of the configuration in the action of Fhit on phosphorus-chiral substrates. Biochemistry, 38, 3668–3676. [DOI] [PubMed] [Google Scholar]
- Anderson J.S.J. and Parker,R.P. (1998) The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J., 17, 1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beelman C.A., Stevens,A., Caponigro,G., LaGrandeur,T.E., Hatfield,L., Fortner,D.M. and Parker,R. (1996) An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature, 382, 642–646. [DOI] [PubMed] [Google Scholar]
- Bernstein P. and Ross,J. (1989) Poly(A), poly(A) binding protein and the regulation of mRNA stability. Trends Biochem. Sci., 14, 373–377. [DOI] [PubMed] [Google Scholar]
- Brenner C., Garrison,P., Gilmour,J., Peisach,D., Ringe,D., Petsko,G.A. and Lowenstein,J.M. (1997) Crystal structures of HINT demonstrate that histidine triad proteins are GalT-related nucleotide-binding proteins. Nat. Struct. Biol., 4, 231–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner C., Bieganowski,P., Pace,H.C. and Huebner,K. (1999) The histidine triad superfamily of nucleotide-binding proteins. J. Cell Physiol., 181, 179–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer G. (1998) Characterization of c-myc 3′ to 5′ mRNA decay activities in an in vitro system. J. Biol. Chem., 273, 34770–34774. [DOI] [PubMed] [Google Scholar]
- Brown J.T. and Johnson,A.W. (2001) A cis-acting element known to block 3′ mRNA degradation enhances expression of polyA-minus mRNA in wild-type yeast cells and phenocopies a ski mutant. RNA, 7, 1566–1577. [PMC free article] [PubMed] [Google Scholar]
- Butler J.S. (2002) The yin and yang of the exosome. Trends Cell Biol., 12, 90–96. [DOI] [PubMed] [Google Scholar]
- Chen C.Y. et al. (2001) AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell, 107, 451–464. [DOI] [PubMed] [Google Scholar]
- Couttet P., Fromont-Racine,M., Steel,D., Pictet,R. and Grange,T. (1997) Messenger RNA deadenylylation precedes decapping in mammalian cells. Proc. Natl Acad. Sci. USA, 94, 5628–5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker C.J. (1998) The exosome: a versatile RNA processing machine. Curr. Biol., 8, 238–240. [DOI] [PubMed] [Google Scholar]
- Decker C.J. and Parker,R. (1993) A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev., 7, 1632–1643. [DOI] [PubMed] [Google Scholar]
- Dehlin E., Wormington,M., Korner,C.G. and Wahle,E. (2000) Cap-dependent deadenylation of mRNA. EMBO J., 19, 1079–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett J.G. and Gallie,D.R. (1992) RNA delivery in Saccharomyces cerevisiae using electroporation. Yeast, 8, 1007–1014. [DOI] [PubMed] [Google Scholar]
- Ford L.P., Watson,J., Keene,J.D. and Wilusz,J. (1999) ELAV proteins stabilize deadenylated intermediates in a novel in vitro mRNA deadenylation/degradation system. Genes Dev., 13, 188–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischmeyer P.A., van Hoof,A., O’Donnell,K., Guerrerio,A.L., Parker,R. and Dietz,H.C. (2002) An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science, 295, 2258–2261. [DOI] [PubMed] [Google Scholar]
- Gao M., Fritz,D.T., Ford,L.P. and Wilusz,J. (2000) Interaction between a poly(A)-specific ribonuclease and the 5′ cap influences mRNA deadenylation rates in vitro. Mol. Cell, 5, 479–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guhaniyogi J. and Brewer,G. (2001) Regulation of mRNA stability in mammalian cells. Gene, 265, 11–23. [DOI] [PubMed] [Google Scholar]
- Guranowski A. (2000) Specific and nonspecific enzymes involved in the catabolism of mononucleoside and dinucleoside polyphosphates. Pharmacol. Ther., 87, 117–139. [DOI] [PubMed] [Google Scholar]
- Guranowski A., Starzynska,E., Bojarska,E., Stepinski,J. and Darzynkiewicz,E. (1996) Dinucleoside 5′, 5′′′-P1, P3-triphosphate hydrolase from yellow lupin (Lupinus luteus) seeds: purification to homogeneity and hydrolysis of mRNA 5′-cap analogs. Protein Expr. Purif., 8, 416–422. [DOI] [PubMed] [Google Scholar]
- Hsu C.L. and Stevens,A. (1993) Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol. Cell. Biol., 13, 4826–4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs J.S., Anderson,A.R. and Parker,R.P. (1998) The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J., 17, 1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisselev L.L., Justesen,J., Wolfson,A.D. and Frolova,L.Y. (1998) Diadenosine oligophosphates (Ap(n)A), a novel class of signalling molecules? FEBS Lett., 427, 157–163. [DOI] [PubMed] [Google Scholar]
- Korner C.G., Wormington,M., Muckenthaler,M., Schneider,S., Dehlin,E. and Wahle,E. (1998) The deadenylating nuclease (DAN) is involved in poly(A) tail removal during the meiotic maturation of Xenopus oocytes. EMBO J., 17, 5427–5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai H., Kon,R., Hoshino,T., Aramaki,T., Nishikawa,M., Hirose,S. and Igarashi,K. (1992) Purification and properties of a decapping enzyme from rat liver cytosol. Biochim. Biophys. Acta, 1119, 45–51. [DOI] [PubMed] [Google Scholar]
- LaGrandeur T.E. and Parker,R. (1998) Isolation and characterization of Dcp1p, the yeast mRNA decapping enzyme. EMBO J., 17, 1487–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima C.D., Klein,M.G. and Hendrickson,W.A. (1997) Structure-based analysis of catalysis and substrate definition in the HIT protein family. Science, 278, 286–290. [DOI] [PubMed] [Google Scholar]
- Martinez J., Ren,Y.G., Thuresson,A.C., Hellman,U., Astrom,J. and Virtanen,A. (2000) A 54-kDa fragment of the poly(A)-specific ribonuclease is an oligomeric, processive, and cap-interacting poly(A)-specific 3′ exonuclease. J. Biol. Chem., 275, 24222–24230. [DOI] [PubMed] [Google Scholar]
- Mitchell P., Petfalski,E., Shevchenko,A., Mann,M. and Tollervey,D. (1997) The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell, 91, 457–466. [DOI] [PubMed] [Google Scholar]
- Muhlrad D. and Parker,R. (1994) Premature translational termination triggers mRNA decapping. Nature, 370, 578–581. [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Decker,C.J. and Parker,R. (1995) Turnover mechanisms of the stable yeast PGK1 mRNA. Mol. Cell. Biol., 15, 2145–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee D., Gao,M., O’Connor,J.P., Raijmakers,R., Pruijn,G., Lutz,C.S. and Wilusz,J. (2002) The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J., 21, 165–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuss D.L. and Furuichi,Y. (1977) Characterization of the m7G(5′)pppN-pyrophosphatase activity from HeLa cells. J. Biol. Chem., 252, 2815–2821. [PubMed] [Google Scholar]
- Nuss D.L., Furuichi,Y., Koch,G. and Shatkin,A.J. (1975) Detection in HeLa cell extracts of a 7-methyl guanosine specific enzyme activity that cleaves m7GpppNm. Cell, 6, 21–27. [DOI] [PubMed] [Google Scholar]
- Pace H.C. et al. (1998) Genetic, biochemical, and crystallographic characterization of Fhit–substrate complexes as the active signaling form of Fhit. Proc. Natl Acad. Sci. USA, 95, 5484–5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers N.D., Wang,Z. and Kiledjian,M. (2002) Characterization and purification of a mammalian endoribonuclease specific for the α-globin mRNA. J. Biol. Chem., 277, 2597–2604. [DOI] [PubMed] [Google Scholar]
- Ross J. and Kobs,G. (1986) H4 histone messenger RNA decay in cell-free extracts initiates at or near the 3′ terminus and proceeds 3′ to 5′. J. Mol. Biol., 188, 579–593. [DOI] [PubMed] [Google Scholar]
- Rudolph R. and Lilie,H. (1996) In vitro folding of inclusion body proteins. FASEB J., 10, 49–56. [PubMed] [Google Scholar]
- Searfoss A.M., Masison,D.C. and Wickner,R.B. (2002) Protein synthesis assayed by electroporation of mRNA in Saccharomyces cerevisiae. Methods Enzymol., 351, 631–639. [DOI] [PubMed] [Google Scholar]
- Seraphin B. (1992) The HIT protein family: a new family of proteins present in prokaryotes, yeast and mammals. DNA Seq., 3, 177–179. [DOI] [PubMed] [Google Scholar]
- Shyu A.B., Belasco,J.G. and Greenberg,M.E. (1991) Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev., 5, 221–231. [DOI] [PubMed] [Google Scholar]
- Thoma C. et al. (2001) Generation of stable mRNA fragments and translation of N-truncated proteins induced by antisense oligo deoxynucleotides. Mol. Cell, 8, 865–872. [DOI] [PubMed] [Google Scholar]
- van Hoof A., Lennertz,P. and Parker,R. (2000) Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol., 20, 441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A., Frischmeyer,P.A., Dietz,H.C. and Parker,R. (2002) Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science, 295, 2262–2264. [DOI] [PubMed] [Google Scholar]
- Wang Z. and Kiledjian,M. (2000) Identification of an erythroid-enriched endoribonuclease activity involved in specific mRNA cleavage. EMBO J., 19, 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. and Kiledjian,M. (2001) Functional link between the mammalian exosome and mRNA decapping. Cell, 107, 751–762. [DOI] [PubMed] [Google Scholar]
- Wang Z., Day,N., Trifillis,P. and Kiledjian,M. (1999) An mRNA stability complex functions with poly(A)-binding protein to stabilize mRNA in vitro. Mol. Cell. Biol., 19, 4552–4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Carr-Schmid,A., Jiao,X. and Kiledjian,M. (2002) The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc. Natl Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson T. and Treisman,R. (1988) Removal of poly(A) and consequent degradation of c-fos mRNA facilitated by 3′ AU-rich sequences. Nature, 336, 396–399. [DOI] [PubMed] [Google Scholar]
- Wilusz C.J., Wormington,M. and Peltz,S.W. (2001) The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol., 2, 237–246. [DOI] [PubMed] [Google Scholar]
- Zhang Q.H. et al. (2000) Cloning and functional analysis of cDNAs with open reading frames for 300 previously undefined genes expressed in CD34+ hematopoietic stem/progenitor cells. Genome Res., 10, 1546–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Williams,C.J., Wormington,M., Stevens,A. and Peltz,S.W. (1999) Monitoring mRNA decapping activity. Methods, 17, 46–51. [DOI] [PubMed] [Google Scholar]