Abstract
Many plant disease resistance (R) genes encode proteins predicted to have an N-terminal coiled-coil (CC) domain, a central nucleotide-binding site (NBS) domain and a C-terminal leucine-rich repeat (LRR) domain. These CC–NBS–LRR proteins recognize specific pathogen-derived products and initiate a resistance response that often includes a type of cell death known as the hypersensitive response (HR). Co-expression of the potato CC–NBS–LRR protein Rx and its elicitor, the PVX coat protein (CP), results in a rapid HR. Surprisingly, co-expression of the LRR and CC–NBS as separate domains also resulted in a CP-dependent HR. Likewise, the CC domain complemented a version of Rx lacking this domain (NBS– LRR). Correspondingly, the LRR domain interacted physically in planta with the CC–NBS, as did CC with NBS–LRR. Both interactions were disrupted in the presence of CP. However, the interaction between CC and NBS–LRR was dependent on a wild-type P-loop motif, whereas the interaction between CC–NBS and LRR was not. We propose that activation of Rx entails sequential disruption of at least two intramolecular interactions.
Keywords: disease resistance/hypersensitive response/NBS–LRR/PVX/R gene
Introduction
Plants have an innate immune system based on dominant resistance (R) genes in which disease resistance is elicited by the products of avirulence (Avr) genes from the pathogen (Flor, 1971). It is generally considered that the R proteins mediate elicitor recognition and activate downstream signalling responses leading to disease resistance. These responses are frequently associated with a type of programmed cell death termed the hypersensitive response (HR; Heath, 2000; Shirasu and Schulze-Lefert, 2000).
Most R genes are predicted to encode intracellular proteins with nucleotide-binding site and leucine-rich repeat (NBS–LRR) domains (Jones and Jones, 1997; Ellis et al., 2000; Young, 2000). The NBS of these proteins has an N-terminal sub-domain that contains consensus kinase 1a (P-loop), kinase 2 and kinase 3a motifs common to a large variety of nucleotide-binding proteins (Traut, 1994); we refer to this as the NB subdomain. The C-terminal part of the NBS, referred to as the ARC (apoptosis, R gene products and CED-4) subdomain, is conserved in plant NBS–LRR proteins as well as in several NBS-containing proteins involved in animal innate immunity and apoptosis (van der Biezen and Jones, 1998a; Aravind et al., 1999). In certain animal proteins, the NBS domain mediates oligomerization that ultimately results in activation of the N-terminal signalling domains (Srinivasula et al., 1998; Yang et al., 1998; Cain et al., 2000; Inohara et al., 2000).
The N-terminus of NBS–LRR proteins influences the requirement for downstream signalling components (Feys and Parker, 2000) and is either a Toll and interleukin receptor (TIR) domain or a loosely predicted coiled coil (CC) (Pan et al., 2000). The C-terminal LRR region is implicated, by genetic studies, in elicitor recognition specificity (Botella et al., 1998; McDowell et al., 1998; Meyers et al., 1998; Ellis et al., 1999; Noel et al., 1999; Bittner-Eddy et al., 2000; Dodds et al., 2001).
The candidate recognition domain of NBS–LRR proteins is the C-terminal LRR. It is the most variable region in closely related NBS–LRR proteins and is under selection to diverge (Meyers et al., 1998; Noel et al., 1999). Functional analysis of recombinant R proteins also indicates that recognition specificity resides in the LRR (Ellis et al., 1999). However, there is indirect evidence that the LRR may contribute to signalling as well as recognition (Warren et al., 1998; Banerjee et al., 2001).
In this paper, we address the function of a CC–NBS– LRR protein, Rx, that confers resistance to potato virus X (PVX) in Solanaceae (Bendahmane et al., 1999), and for which the elicitor is the PVX coat protein (CP; Bendahmane et al., 1995). Transient expression of wild-type Rx in the presence of the CP (Bendahmane et al., 1999) or of gain-of-function mutants (Bendahmane et al., 2002) results in a rapid HR that, like many disease resistance responses in plants, is dependent on the expression of a putative ubiquitin ligase-associated protein SGT1 (Austin et al., 2002; Azevedo et al., 2002; Peart et al., 2002).
We show that co-expression of the CC–NBS and LRR regions of Rx as separate molecules resulted in a CP-dependent HR. Similarly, co-expression of the CC domain with NBS–LRR resulted in a CP-dependent HR. These results suggested that a functional Rx protein was reconstituted through physical interactions between domains. Accordingly, co-immunoprecipitation experiments showed physical interactions between the same pairs of domains. We present evidence that there are at least two distinct interactions involving the CC and LRR domains of Rx that are disrupted in the presence of CP. In the intact Rx protein, these interactions are likely to be intramolecular. We propose that CP recognition initiates a sequence of conformational changes in Rx that involve disruption of these intramolecular interactions. A likely consequence of these conformational changes is the activation of Rx so that it is competent to initiate signalling that leads to disease resistance.
Results
The Rx CC–NBS and LRR regions can act in trans
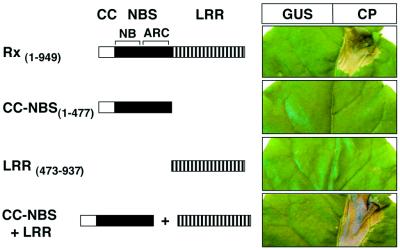
From a previous analysis of constitutive gain-of-function Rx mutants, we inferred that there are interactions between the domains of Rx (Bendahmane et al., 2002). To test this idea, we transiently expressed the CC–NBS and LRR domains as separate HA epitope-tagged constructs in Nicotiana benthamiana leaves (Figure 1). We expected that a cis interaction in the wild-type protein would also operate in trans so that the separated Rx domains would be active. The results (Figure 1) show that neither CC–NBS nor LRR could mediate an HR in the presence or absence of CP. However, co-expression of both proteins resulted in a CP-dependent HR (Figure 1). This complementation in trans indicates that both CC–NBS and LRR are required for Rx activity, but that they need not be encoded in the same polypeptide.
Fig. 1. Complementation of the Rx CC–NBS and LRR regions. Rx constructs are represented schematically. The CC region of Rx is represented by an open box. A black box represents the NBS domain (including the NB and ARC subdomains). The leucine-rich repeat (LRR) region is represented by bars. Versions of Rx were expressed via agroinfiltration (OD600 = 0.2 each) in N.benthamiana leaves from the Rx genomic promoter together with either 35S-GUS (left side of leaf) or 35S-CP (right side of leaf). Similar results were obtained when Rx fragments were expressed from the 35S promoter.
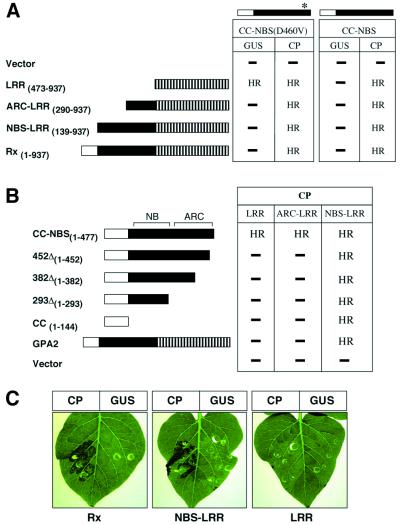
In a similar test, CC–NBS was derived from a mutant, Rx(D460V), that confers a CP-independent HR when transiently expressed (Bendahmane et al., 2002). We predicted that, if the LRR were required only for recognition of the CP elicitor, the CC–NBS from this mutant would produce an HR in the absence of the LRR; it did not (Figure 2A). However, co-expression of CC– NBS(D460V) and LRR (Figure 2A) resulted in a CP-independent HR. These data demonstrate that the LRR is required for activation of the signalling domains of Rx, concomitant with any roles it may have in recognition of the CP elicitor.
Fig. 2. Effect of cis elements on trans complementation. Rx constructs are schematically represented as in Figure 1. The CC–NBS(D460V) construct is identical to CC–NBS shown in Figure 1 except that residue D460 is mutated to V, indicated by an asterisk. (A) The CC– NBS(D460V) or CC–NBS polypeptides were co-expressed with the Rx derivatives indicated on the left, in conjunction with either 35S-GUS or 35S-CP via agroinfiltration in N.benthamiana leaves. (B) Rx derivatives indicated on the left were agroinfiltrated with CP plus either Rx LRR, ARC–LRR or NBS–LRR. The occurrence of a HR or not (–) is indicated. Similar results were obtained using either the Rx or the 35S promoter. No HR was observed with any combination in the absence of CP. (C) Potato leaves (rx genotype) were agroinfiltrated with pB1-Rx derivatives in conjunction with either 35S-CP or GUS, as indicated. Constructs were infiltrated at a concentration of agrobacterium of OD600 = 0.2 each.
To determine whether complementation in trans is affected by an overlap in the Rx domains, we transiently expressed CC–NBS and CC–NBS(D460V) with versions of Rx that contained the LRR with either all or part of the NBS. In both instances, the extended LRR proteins complemented CC–NBS and CC–NBS(D460V) to give a CP-dependent HR (Figure 2A). However, neither of the extended LRR proteins nor the full-length Rx produced a CP-independent HR when co-expressed with CC– NBS(D460V) (Figure 2A). The shortest extension of the LRR (ARC–LRR) was 183 amino acid residues extending through the ARC subdomain. Thus, the LRR can complement CC–NBS for a CP-dependent HR irrespective of whether the ARC subdomain is linked in cis. However, for activation of a CP-independent HR, the LRR must be free of the ARC subdomain.
Having established that overlapping fragments of CC–NBS and NBS–LRR could complement for a CP-dependent HR, we investigated whether the NBS domain could be functionally subdivided into its NB and ARC components by expression of deleted versions of CC–NBS (Figure 2B). None of these deleted constructs produced a CP-dependent HR when co-expressed with either LRR or ARC–LRR (Figure 2B). Therefore, the NB and ARC domains must be intact and in cis in order to be complemented by the LRR. However, it was not necessary for the intact NBS be linked to the CC because transient co-expression of Rx CC with NBS–LRR led to a CP-dependent HR (Figure 2B). These results also demonstrate that the CC domain is sufficient to complement NBS–LRR.
In a further characterization of Rx complementation, we co-expressed Rx domains with GPA2. GPA2 is an Rx paralogue from potato that confers nematode resistance. The encoded protein is 96% identical to Rx in the CC domain, but is more divergent in the LRR region (Bendahmane et al., 2000; van der Vossen et al., 2000). Co-expression of full-length GPA2 with either LRR or ARC–LRR of Rx did not lead to a CP-dependent HR. However, co-expression of GPA2 with Rx NBS–LRR resulted in a CP-dependent HR (Figure 2B). This result demonstrates that a CC domain can be provided by full-length CC–NBS–LRR protein. Our observation that Rx NBS–LRR produced a CP-dependent HR when expressed in rx genotype potato leaves can be explained in the same way (Figure 2C). Presumably, a CC domain was provided to NBS–LRR by full-length homologues of Rx and GPA2 that are present in the rx potato genome (Bendahmane et al., 1999).
The CC–NBS–LRR proteins encoded in the N.benthamiana genome, unlike those in the rx genotype potato, do not complement Rx NBS–LRR (Figure 2B). Therefore, it is likely that the ability to provide the CC function can be provided only by specific CC–NBS–LRR proteins. To investigate this specificity in the trans complementation, we used the Bs2 protein from pepper, which is one of the most similar NBS–LRR proteins to Rx outside of the potato (Tai et al., 1999). The NBS domains of Rx and Bs2 are 46% identical, and the CC domains are 29% identical, with 10 of the first 13 residues being identical. Bs2 NBS–LRR elicits a weak AvrBs2- dependant HR in N.benthamiana that is likely due to complementation by endogenous CC-containing proteins. This weak HR was enhanced by co-expression of the Bs2 CC, but not the Rx CC (Table I). Likewise, co-expression of Bs2 CC–NBS and LRR resulted in AvrBs2-dependent HR, but there was no complementation between analogous domains of Rx and Bs2 (Table I). Therefore, it is likely that the complementation in trans is dependent on a high degree of protein sequence specificity in the participating domains.
Table I. Specificity in Bs2 and Rx interactions.
| AvrBs2 | CP | ||
|---|---|---|---|
| Bs2 CC–NBS | Bs2 LRR | HR | – |
| Bs2 NBS–LRR | Bs2 CC | HR | – |
| Bs2 NBS–LRR | HR (weak) | – | |
| Rx CC–NBS | Rx LRR | – | HR |
| Rx NBS–LRR | Rx CC | – | HR |
| Rx CC–NBS | Bs2 LRR | – | – |
| Bs2 CC–NBS | Rx LRR | – | – |
| Rx NBS–LRR | Bs2 CC | – | – |
| Bs2 NBS–LRR | Rx CC | HR (weak) | – |
The Rx and Bs2 derivatives were transiently expressed in N.benthamiana and the production of a visible HR was assessed after 3 days. Bs2 constructs, CC–NBS, LRR, CC and NBS–LRR encode for residues 1–500, 489–905, 1–154 and 149–905, respectively.
The observation that separate Rx or Bs2 domains can complement in trans could be explained if they physically interact with each other. To investigate this possibility, we expressed Rx derivatives, either in the presence or absence of a CP elicitor, and assessed physical interactions by immunoprecipitation from plant extracts. The outcome of these assays is described in the following sections.
Physical interaction between domains of Rx
For analysis of physical interactions, various Rx derivatives were tagged with haemagglutinin (HA), myc (MT) and FLAG (FH) epitopes. The expression of the epitope-tagged proteins was confirmed by western blotting, and their function was verified in an HR assay, as described above (P.Moffett, unpublished data). However, we could not detect the epitope-tagged proteins if they were expressed with elicitor and in combinations that led to an HR. Presumably, there was cell death before the tagged proteins had accumulated to detectable levels. Therefore, to allow detection of the tagged proteins in the presence of elicitor and in combinations that are competent for downstream signalling, we exploited the observations that expression of SGT1 is required for resistance responses (Austin et al., 2002; Azevedo et al., 2002; Peart et al., 2002). Thus, where necessary, the analyses of physical interactions were carried out in SGT1-silenced N.benthamiana, in which the HR was suppressed.
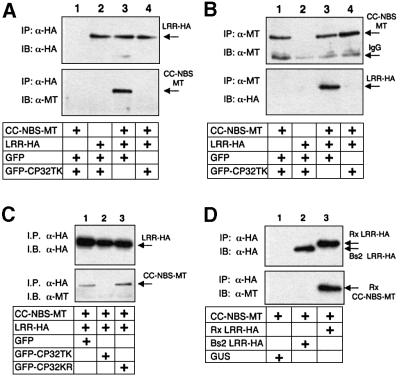
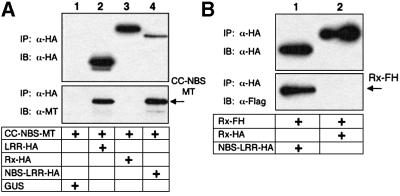
In our initial tests of Rx interactions, the elicitor was a deleted version (residues 32–142) of the PVX CP that was fused to GFP (GFP–CP32TK). GFP–CP32TK elicits an Rx-mediated HR to a similar degree as wild-type CP (P.Moffett, unpublished data). The equivalent derivative of a non-eliciting CP, GFP–CP32KR (Bendahmane et al., 1995), was expressed to similar levels as GFP–CP32TK (P.Moffett, unpublished data) and was used as a control. Results from these assays (Figure 3A and B) show that the domains of Rx interact in planta. Using anti-HA antibodies, CC–NBS-MT co-immunoprecipitated with LRR-HA (Figure 3A, lane 3) and, using anti-MT antibodies, the reciprocal interaction was confirmed (Figure 3B, lane 3). However, this interaction was abrogated in the presence of GFP–CP32TK (Figure 3A and B, lane 4). We can rule out that the loss of this interaction is due to protein degradation because LRR-HA and CC–NBS-MT were equally abundant either in the presence or absence of CP (Figure 3A and B, top panel). The interaction between LRR-HA and CC–NBS-MT was stable in the presence of the inactive elicitor GFP–CP32KR (Figure 3C). Therefore, it is likely that elicitor-mediated disruption of an LRR–CC–NBS interaction has a role in Rx-mediated resistance.
Fig. 3. Association of the Rx CC–NBS and LRR. (A) Nicotiana benthamiana plants were silenced for SGT1. Leaves were agroinfiltrated with 35S-driven constructs expressing Rx LRR-HA and CC–NBS-MT or GFP (OD600 = 0.2 each), as indicated. These were combined with either GFP or GFP–CP32 (OD600 = 0.1 each) such that the final OD600 = 0.5. Four days post-infiltration, protein extracts were subjected to immunoprecipitation with α-HA (3F10) antibody followed by immunoblotting with α-HA (3F10) antibody (top panel) or α-MT (9E10) antibody (lower panel). (B) Protein extracts were subjected to immunoprecipitation with α-MT (A-14) antibody followed by immunoblotting with α-MT (9E10) antibody (top panel) and α-HA (3F10) antibody (lower panel). (C) SGT1 silenced plants were infiltrated with CC–NBS-MT and LRR-HA (OD600 = 0.2 each) plus either GFP, GFP– CP32TK or GFP–CP32KR (OD600 = 0.1 each) as indicated. Protein extracts were subjected to immunoprecipitation with α-HA (3F10) antibody and samples immunoblotted with α-HA (3F10) antibody (top panel) and α-MT (A-14) antibody (lower panel). (D) Non- silenced N.benthamiana were agroinfiltrated with 35S-driven constructs expressing Rx CC–NBS-MT in combination with GUS, Bs2 LRR-HA, or Rx LRR-HA. Three days post-infiltration, protein extracts were subjected to immunoprecipitation with α-HA (3F10) antibody followed by immunoblotting with α-HA (3F10) antibody (top panel) and α-MT (A-14) antibody (lower panel).
We also showed that the CC–NBS and LRR domains interacted in plants in the absence of SGT1 silencing (Figure 3D, lane 3). In addition, the HA-tagged LRR of Bs2 failed to interact with Rx CC–NBS-MT (Figure 3D, lane 2), corresponding to the lack of HR in the bioassay (Table I). Therefore, the interaction of CC–NBS and LRR domains is specific and is not affected by silencing of SGT1.
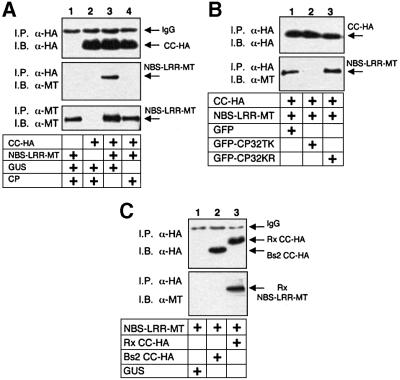
The observation that Rx CC complements NBS–LRR in an HR assay (Figure 2B) prompted us to investigate a physical interaction with this domain. The results, shown in Figure 4A, demonstrate that Rx NBS–LRR co-immunoprecipitated with CC in the absence, but not the presence, of CP (Figure 4A, lanes 3 and 4). As with the interaction of CC–NBS and LRR, the interaction of CC and NBS–LRR was unaffected by the non-eliciting GFP–CP32KR (Figure 4B) and Rx NBS–LRR did not interact with the Bs2 CC (Figure 4D). These results suggest that the CC domain is directly involved in a physical interaction with another domain of Rx.
Fig. 4. Association of the Rx CC and NBS–LRR. (A) SGT1 silenced leaves were agroinfiltrated with 35S-driven constructs expressing Rx CC-HA and Rx NBS–LRR-MT polypeptides or GFP (OD600 = 0.2 each). These were combined with either GFP or GFP–CPTK (OD600 = 0.1 each) such that the final OD600 was 0.5. Four days post-infiltration, protein extracts were subjected to immunoprecipitation with α-HA (3F10) antibody followed by immunoblotting with α-HA (3F10) antibody (top panel) and α-MT (A-14) antibody (middle panel). In the lower panel, protein extracts were subjected to immunoprecipitation with α-MT (9E10) antibody and subsequently immunoblotted with α-MT (A-14) antibody to demonstrate the presence of equal amounts of input NBS–LRR-MT protein. (B) SGT1 silenced plants were infiltrated with NBS–LRR-MT and CC-HA (OD600 = 0.2 each) plus GFP, GFP–CP32TK or GFP–CP32KR (OD600 = 0.1 each) as indicated. Protein extracts were subjected to immunoprecipitation with α-HA (3F10) antibody and samples immunoblotted with α-HA (3F10) antibody (top panel) and α-MT (A-14) antibody (lower panel). (C) Non- silenced N.benthamiana leaves were agroinfiltrated with 35S-driven constructs expressing Rx NBS–LRR-MT in combination with GUS, Bs2 CC-HA or Rx CC-HA. Three days post-infiltration, protein extracts were immunoprecipitated with α-HA (3F10) antibody and immunoblotted with α-HA (3F10) antibody (top panel) and α-MT (A-14) antibody (lower panel).
Separate CC and LRR interactions
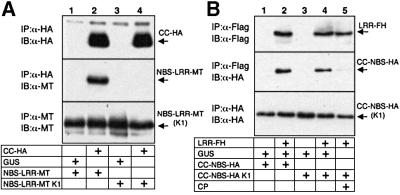
The data in Figures 3 and 4 could be explained if there is a single direct interaction between CC and LRR. However, a direct CC–LRR interaction is unlikely because these two domains do not interact when co-expressed in planta (P.Moffett, unpublished data). Furthermore, the CC and LRR interactions were differentially affected by a mutation in the NBS. Thus, mutations in the P-loop motif (G175A, K176A) of the NB subdomain (K1), which caused a loss of Rx-mediated HR (Bendahmane et al., 2002), also prevented the interaction of Rx CC-HA with NBS–LRR-MT (Figure 5A). In contrast, LRR-FH interacted equally well with both wild-type and the P-loop mutant CC–NBS-HA (Figure 5B). The presence of CP abrogated the interaction between LRR and CC–NBS K1 (Figure 5B) as with the wild-type domains in SGT1 silenced leaves (Figure 3A). Thus, the differential effects of the P-loop mutations demonstrate that the interactions involving the CC and LRR domains of Rx are not equivalent.
Fig. 5. Involvement of the NB domain in interdomain interactions. (A) Nicotiana benthamiana leaves were agroinfiltrated with 35S GUS (lanes 1 and 3) or 35S Rx CC-HA (lanes 2 and 4) along with wild-type Rx NBS–LRR-MT (lanes 1 and 2) or NBS–LRR-MT K1 (lanes 3 and 4). Two days post-infiltration, protein extracts were subjected to immunoprecipitation with α-HA (3F10) antibody and subsequently immunoblotted with α-HA (3F10) antibody (top panel) and α-MT (A-14) antibody (middle panel). In the lower panel, protein extracts were subjected to immunoprecipitation with α-MT (9E10) antibody and immunoblotted with α-MT (A-14) antibody to demonstrate the presence of equal amounts of input NBS–LRR-MT derivatives. (B) Nicotiana benthamiana leaves were agroinfiltrated with 35S GUS (lanes 1 and 3) or 35S Rx LRR-FH (lanes 2, 4 and 5) along with wild-type Rx CC–NBS-HA (lanes 1 and 2) or CC–NBS-HA K1 (lanes 3–5). These were combined with either GUS (lanes 1–4) or CP (lane 5). Two days post-infiltration, protein extracts were subjected to immunoprecipitation with α-FLAG (M2) antibody and subsequently immunoblotted with α-FLAG (M2) antibody (top panel) and α-HA (3F10) antibody (middle panel). In the lower panel, protein extracts were subjected to immunoprecipitation with α-HA (3F10) antibody and immunoblotted with α-HA (3F10) antibody to demonstrate the presence of equal amounts of input CC–NBS-HA derivatives.
Functional assays had indicated that the LRR interaction may be preferred in cis (Figure 2A). To address whether the physical interactions occur preferentially in cis, we tested the ability of CC–NBS to co-immunoprecipitate overlapping and non-overlapping forms of Rx. We predicted that if an interdomain interaction normally took place in cis then it would be excluded from interactions in trans.
The results, shown in Figure 6A, demonstrate that CC–NBS interacts with LRR and NBS–LRR, but not full-length Rx. Presumably, the LRR in full-length Rx interacts preferentially in cis with its own CC–NBS and is not available to interact in trans. In contrast, full-length Rx is competent to interact with NBS–LRR (Figure 6B). This interaction is likely mediated by the CC domain, as shown in Figure 4. However, there is no interaction between two full-length Rx molecules (Figure 6B) suggesting that trans interactions involving the CC domain take place only if one of the proteins lacks a CC domain. Therefore, we conclude that interdomain interactions normally take place within the same molecule.
Fig. 6. Effect of cis elements on trans interactions. (A) Nicotiana benthamiana leaves were agroinfiltrated with 35S-driven constructs expressing Rx CC–NBS-MT (all lanes) plus either GUS, Rx LRR-HA, Rx-HA or Rx NBS–LRR-HA. Two days post-infiltration, extracts were prepared and subjected to immunoprecipitation with α-HA antibody (3F10). Pellets were immunoblotted with α-HA (3F10) antibody (top panel) and α-MT (A-14) antibody (lower panel). (B) Nicotiana benthamiana leaves were agroinfiltrated with 35S-driven constructs expressing Rx-FH (both lanes) plus either Rx-HA or Rx NBS– LRR-HA. Two days post-infiltration, extracts were prepared and subjected to immunoprecipitation with α-HA antibody (3F10). Pellets were immunoblotted with with α-HA (3F10) antibody (top panel) and α-FLAG (M2) antibody (lower panel).
Discussion
The analysis of Rx described here involved biological and physical assays of different domains expressed either individually or in combinations. In the bioassays, the co-expression of CC–NBS with LRR, as well as CC with NBS–LRR, resulted in a CP-dependent HR (Figures 1 and 2). Correspondingly, the same pairs of Rx derivatives interacted physically in the absence, but not the presence, of CP (Figures 3 and 4). Therefore, the physical interactions in the absence of elicitor correlate with the reconstitution of Rx function when Rx domains were expressed separately. Therefore, it is likely that these interactions and their disruption are required for Rx-mediated resistance. Similar interactions may also take place in Bs2 because, like Rx, its domains can also act in trans in an HR bioassay (Table I). Additional support for interdomain interactions is suggested by the ability of the CC domain expressed in trans to affect the phenotype of full-length recombinant forms of Mi (Hwang et al., 2000). In other NBS–LRR proteins, intergenic recombination events often produce non-functional proteins (Ellis et al., 1999; Luck et al., 2000). This finding could be explained if heterologous domains in the recombinant proteins are unable to participate in the interdomain interactions.
From the differential effect of P-loop mutations, we conclude that there are at least two interactions involving CC and LRR domains of Rx (Figure 5). We have not formally ruled out the possibility that these interactions are indirect. However, if bridging molecules are involved, they are not limiting for the interactions and must be able to differentiate between Rx and Bs2 proteins. At present, we favour the possibility that these interdomain interactions are direct. We discuss below how these physical interactions could lead to the activation of Rx and other CC–NBS–LRR proteins. Animal NBS–LRR proteins involved in innate immune responses may also be activated in the same way (Inohara and Nunez, 2001).
Much of our discussion about the mechanism of Rx activation is focussed on intramolecular interactions because we did not find evidence for self-association of full-length molecules (Figure 6B). Our interpretations are based on the assumption that interactions between domains observed in trans are normally intramolecular. However, our demonstration that GPA2 can complement Rx NBS–LRR (Figure 2B) indicates that, in some cases, full-length NBS–LRR proteins could participate in trans interactions. For example, it is possible that trans interactions between alternatively spliced products of an NBS– LRR protein encoded at the N locus are required for resistance against tobacco mosaic virus (Dinesh-Kumar and Baker, 2000).
Activation of Rx
A general model for R protein-mediated disease resistance involves three loosely defined steps. First, the Avr gene product must be recognized, directly or indirectly, by the R gene product. This recognition process leads to the second step, in which the R protein is activated and becomes competent to signal to other components in the cell. These cellular components then mediate the third signalling step, ultimately leading to suppression of the pathogen. In this report, we have characterized changes to Rx that follow recognition of the elicitor and we propose that they are part of a mechanism for activating Rx.
An important feature of Rx activation involves the NBS/LRR interaction that could be inferred from the physical interaction of LRR with CC–NBS (Figure 3) and from the function complementation of CC–NBS constructs with LRR (Figure 2A). This interaction does not depend on P-loop function (Figure 6B), and is most likely between the LRR and the ARC subdomain of the NBS (Figure 2A). We conclude that activation of Rx involves disruption of the NBS/LRR interaction because the physical interaction between CC–NBS and LRR was not observed in the presence of CP (Figure 3). This disruption of the LRR interaction is independent of a functional NB subdomain (Figures 2A and 6B).
One interpretation of these data is that the LRR negatively regulates activation of Rx. In the presence of the elicitor, the disruption of the LRR interaction would lead to loss of the negative regulation. However, deletion of the LRR does not result in spontaneous activation of Rx (Figures 1 and 2) as would be expected if it were only a negative regulator. The requirement of the LRR for the elicitor-independent HR of the Rx(D460V) mutant (Figure 2) is also evidence for a positive role of this domain.
A positive role of the LRR is likely a general feature of NBS–LRR R proteins because plant genomes encode for many truncated R genes and splice variants lacking an LRR that do not cause spontaneous cell death (Lawrence et al., 1995; Anderson et al., 1997; Parker et al., 1997; Ayliffe et al., 1999; Dinesh-Kumar and Baker, 2000; Dinesh-Kumar et al., 2000; The Arabidopsis Genome Initiative, 2000). The properties of an LRR mutation have also been interpreted as indicators of a positive role for this domain in activation of downstream signalling (Warren et al., 1998).
Activation of Rx also involves an interaction of the CC within Rx. This second interaction is indicated by functional complementation and by physical interactions with CC domain constructs (Figures 2 and 4). Since the CC interaction is dependent on integrity of the P-loop (Figure 5A), we suggest that its disruption in the presence of elicitor may involve modulation of the nucleotide-binding status of the NBS. The trigger for this change to the NBS could be the elicitor-mediated change to the ARC/LRR interaction.
Other NBS-containing proteins provide precedent for this proposed mechanism of Rx activation. These proteins have an integrated nucleotide-binding pocket that is formed by the conserved NB subdomain together with a non-conserved carboxy-adjacent region that would be analogous to the ARC subdomain of Rx (Guenther et al., 1997; Jaroszewski et al., 2000). Protein–protein interactions with the NB-adjacent domain can influence nucleotide binding and indirectly, as a consequence of either nucleotide binding or hydrolysis, the conformation of the two subdomains of the NBS (Wang et al., 2001). These changes can, in turn, influence further protein– protein interactions involving the NBS and adjacent domains (Ogura and Wilkinson, 2001). In this way, the NBS acts as a molecular switch that can be regulated.
A specific example of conformational changes in NBS proteins is with the apoptosis regulator Apaf-1. Ligand binding to Apaf-1 causes a disruption of the interaction between the NBS and the C-terminus. This conformational change increases the nucleotide-binding affinity of the NBS domain, and allows assembly of a signalling complex at the Apaf-1 N-terminus (Jiang and Wang, 2000). In another example, the binding of ATP initially allows the NBS membrane fusion protein, NSF, to interact with another protein. Subsequently, ATP hydrolysis results in disassembly of the complex (May et al., 2001).
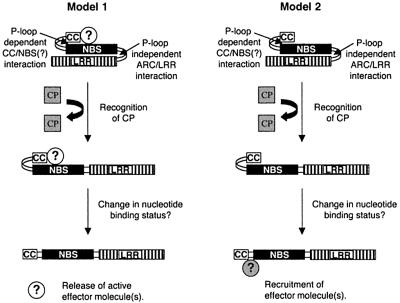
How could the CC domain conformational change lead to downstream signalling? It is unlikely to be simple release of the CC domain for interaction with downstream effector molecules because expression of the CC domain alone does not result in an HR (Figure 3A). At present, we favour two alternative, but not mutually exclusive, hypotheses. One of these is similar to the model proposed to account for the properties of recombinant forms of the Mi protein that confers nematode resistance (Hwang et al., 2000). According to this idea, the elicitor-mediated conformational changes allow the NBS–LRR protein to recruit or activate downstream signalling components. The second hypothesis is that the NBS–LRR protein binds a signalling factor in the absence of elicitor. The elicitor-induced conformational changes would allow release of this factor in an active form. Figure 7 shows the various steps that we propose for activation of Rx-mediated resistance based either on the recruitment or release of signalling factors.
Fig. 7. Model of Rx activation. A schematic representation of a possible mechanism for Rx activation is presented with two alternatives for effector molecule action. In both models, the coat protein (grey square) interacts transiently with the CC–NBS–LRR protein and initiates a two step conformational change. In the first step, the LRR/NBS interaction is disrupted, resulting in a change to the nucleotide binding status of the NBS. This change then results in disruption of an interaction of the CC. In model 1, an effector molecule(s) (white circles) is constitutively associated with the CC–NBS–LRR protein and is released in an active form upon dissociation of the CC. In model 2, the inactive effector molecule(s) (grey circle) is recruited to Rx upon dissociation of the CC domain.
Elicitor recognition
The most straightforward interpretation of R protein-mediated elicitor recognition invokes a direct intermolecular interaction. The elicitor would bind directly to the R protein and initiate the activation process discussed above. Consistent with this prediction, there is a direct interaction of the Pi-ta LRR with its elicitor (Jia et al., 2000). However, such straightforward binding may not be a general feature of elicitor recognition. For example, RPS2 may bind its elicitor as part of a complex (Leister and Katagiri, 2000). Moreover, although interactions between domains of Rx were readily detectable and were disrupted when the elicitor CP was present (Figure 3), we were unable to detect an interaction of CP with Rx or derivatives thereof (P.Moffett, unpublished data).
One explanation of these results is that Rx recognizes a host factor rather than the CP. Recognition would be triggered when the host factor interacts with or is modified by a virulence factor produced by the pathogen (van der Biezen and Jones, 1998b). Host factors may facilitate recognition, as suggested by analyses of LRR variants of RPS2 (Banerjee et al., 2001). However, we do not favour the idea that Rx recognizes a modified host factor because CP derivatives are elicitors even if they are not effective as virulence factors (Bendahmane et al., 1995). An alternative explanation of molecular recognition by Rx and other NBS–LRR proteins invokes a direct elicitor interaction that is either transient or unstable. According to this idea, the Rx protein would interact directly with the elicitor, but would be released as a consequence of the conformational changes that follow recognition (Figure 7).
Postscript
Despite the passage of several years since the first molecular cloning of NBS-LRR R proteins, there is very little definite understanding of how they mediate molecular recognition of pathogens and activate disease resistance signalling pathways. The results presented here provide direct evidence for conformational changes in an NBS–LRR protein and represent some of the first molecular events to be described that are associated with activation of disease resistance. In addition, the methods used here could be adapted and developed as tools for more detailed investigation of this process. For example, it may be possible to use variants of the immunoprecipitation assay with Rx domains to demonstrate the elusive elicitor–R protein interaction. It may be possible also to develop procedures based on this assay in a direct search for downstream signalling components that are either recruited or released following the elicitor-induced conformation change in Rx and other NBS–LRR proteins.
Materials and methods
Plasmid construction
The pBIN19-based binary vectors pB1-RxHA and pBIN61-RxHA, which drive expression of the Rx cDNA from the Rx and 35S promoters, respectively, have been described elsewhere (Bendahmane et al., 2002). To introduce six c-myc epitope tags at the C-terminus of Rx, the BamHI–SnaBI fragment of pCS2+MT (Rupp et al., 1994) was ligated into the BamHI/SmaI sites of pBIN61-RxHA to create pBIN61-Rx6MT. PCR was used to incorporate the sequence 5′-GGATCCGACTACAAAGACGATGATGACAAGCATCACCATCACCATCACTGACGAGCTTAAGCTGAGCTCGAATTC, encoding a FLAG epitope tag plus six histidine residues (FH tag), at the 5′ end of the Rx cDNA to create pBIN61-RxFH. All deletion versions of Rx and Bs2 were constructed by PCR using a 5′ primer, which incorporated a 5′ XbaI site followed (where necessary) by a Kozak consensus start codon (ACCACCATGG), and a 3′ primer, which incorporated a 3′ BamHI site into the cDNA. The residues encoded by each deletion construct are indicated in the text. PCR products were subsequently cloned into the XbaI/BamHI sites of pBIN61-RxHA, pBIN61-RxFH or pBIN61-Rx6MT, and subsequently transferred to the pB1 vector using the XbaI/SacI restriction sites. Introduction of inactivating point mutations in the P-loop domain of Rx is described elsewhere (Bendahmane et al., 2002).
For GFP fusions of the PVX coat protein, the GFP5 cDNA was amplified with primers GFP4SAL (5′-TTCTAGGTCGACATGAGTAAAGGAGAAGAAC-3′) and GFF3 (5′-TTATTTAAATTAACCCCCGGGTCCCTCGAGTCCACCGAGTTCGTCATGTTTGTATAG-3′). The resulting PCR product was ligated into the SmaI site of pBIN61 to create pBIN61-GFP6. Coding regions for residues 32–142 of PVX-CP TK and KR were PCR amplified from 35S-CP TK and 35S-CP KR (Bendahmane et al., 2000), using primers CP32 (5′-TATCATCTCGAGTTCACTATACCAGATGGGGAC-3′) and CP142 (5′-TATGAGCTCTTATGCGCTGTTGTTTGTTAACATCC-3′). PCR products were digested with XhoI and ligated into the XhoI/SmaI sites of pBIN61-GFP6 to create GFP–CP32TK and GFP–CP32KR. Throughout the text, CP refers to the TK version of the PVX coat protein unless otherwise indicated.
Agrobacterium-mediated transient expression (agroinfiltration)
Binary vectors were transformed into Agrobacterium tumefaciens strain C58C1 carrying the virulence plasmid pCH32. Agroinfiltrations were performed as described (Bendahmane et al., 2000). HR phenotypes generally presented at 24–48 h.
Immunoprecipitation and immunoblotting
Protein extracts from agroinfiltrated N.benthamiana leaves were prepared by grinding 1 g of leaf tissue in 2.5 ml of extraction buffer [25 mM Tris–HCl pH 7.5, 1 mM EDTA, 150 mM NaCl, 10% glycerol, 5 mM dithiothreitol (DTT)] in the presence of plant protease inhibitor cocktail (Sigma) and 2% polyvinylpolypyrrolidone. Extracts were spun at 12 Krcf at 4°C for 15 min, and 1 ml of extract was subsequently passed through a 5 ml G-25 Sephadex spin column (pre-equilibrated with extraction buffer). Immunoprecipitation was performed as follows: 400–800 µl of extract in a total volume of 1400 µl of extraction buffer plus 0.15% Nonidet P-40 (IP buffer) was pre-cleared with 50 µl of IgG agarose (Sigma) at 4°C for 30 min. Extracts were spun for 2 min at 12 Krcf and the supernatant was added to 25 µl of anti-HA (3F10) agarose beads (Roche), α-FLAG (M2, Sigma) agarose beads, α-myc (9E10) agarose beads (Santa Cruz) or 25 µl of protein G agarose plus 0.8 µg of anti-myc antibody A-14 (Santa Cruz). Extracts were incubated end over end at 4°C for 1 h, washed four times with IP buffer and the pellet was resuspended in 1× SDS–PAGE loading buffer. Immunoprecipitated samples were separated by SDS–PAGE and blotted to Immun-Blot PVDF membrane (Bio-Rad). Blots were pre-blocked with 5% skimmed milk powder in TBST and probed with 40–200 ng/ml of antibody in TBST. HA epitope tags were detected with 3F10 (Roche) and Myc tags were detected with either 9E10 or A-14 (Santa Cruz), followed by washing and incubation with an appropriate horseradish peroxidase (HRP)-conjugated secondary antibody. FLAG epitope tags were detected with HRP-conjugated M2 antibody (Sigma). Proteins were visualized with ECL-Plus (Amersham).
Virus-induced gene silencing (VIGS)
VIGS of the N.benthamiana homologue of SGT1 using a tobacco rattle virus vector is described elsewhere (Peart et al., 2002). Briefly, 14-day-old N.benthamiana plants were infiltrated with Agrobacterium carrying pBINTRA6 and pTV-SGT1 (Ratcliff et al., 2001), and plants were used for agroinfiltration experiments 3–4 weeks later.
Acknowledgments
Acknowledgements
We are grateful to Abdelhafid Bendahmane for biomaterials and discussions. We are also grateful to John Rathjen, Alan Herr and Scott Peck for helpful comments on the manuscript. The Bs2 and AvrBs2 cDNAs were generously provided by Brian Staskawicz. Recombinant viruses and transgenic plants were contained in greenhouses under DEFRA license PHL 161/4080. The work was supported by the Gatsby Charitable Foundation and the Biotechnology and Biological Sciences Research Council. P.M. is the recipient of an EMBO long term fellowship.
References
- Anderson P.A., Lawrence,G.J., Morrish,B.C., Ayliffe,M.A., Finnegan,E.J. and Ellis,J.G. (1997) Inactivation of the flax resistance gene M associated with loss of a repeated unit within the leucine-rich repeat coding region. Plant Cell, 9, 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L., Dixit,V.M. and Koonin,E.V. (1999) The domains of death: evolution of the apoptosis machinery. Trends Biochem. Sci., 24, 47–53. [DOI] [PubMed] [Google Scholar]
- Austin M.J., Muskett,P., Kahn,K., Feys,B.J., Jones,J.D.G. and Parker,J.E. (2002) Regulatory role of SGT1 in early R gene-mediated plant defenses. Science, 295, 2077–2080. [DOI] [PubMed] [Google Scholar]
- Ayliffe M.A., Frost,D.V., Finnegan,E.J., Lawrence,G.J., Anderson,P.A. and Ellis,J.G. (1999) Analysis of alternative transcripts of the flax L6 rust resistance gene. Plant J., 17, 287–292. [DOI] [PubMed] [Google Scholar]
- Azevedo C., Sadanandom,A., Kitagawa,K., Freialdenhoven,A., Shirasu,K. and Schulze-Lefert,P. (2002) The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science, 295, 2073–2076. [DOI] [PubMed] [Google Scholar]
- Banerjee D., Zhang,X.C. and Bent,A.F. (2001) The leucine-rich repeat domain can determine effective interaction between RPS2 and other host factors in Arabidopsis RPS2-mediated disease resistance. Genetics, 158, 439–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendahmane A., Köhm,B.A., Dedi,C. and Baulcombe,D.C. (1995) The coat protein of potato virus X is a strain-specific elicitor of Rx1-mediated virus resistance in potato. Plant J., 8, 933–941. [DOI] [PubMed] [Google Scholar]
- Bendahmane A., Kanyuka,K. and Baulcombe,D.C. (1999) The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell, 11, 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendahmane A., Querci,M., Kanyuka,K. and Baulcombe,D.C. (2000) Agrobacterium transient expression system as a tool for isolation of disease resistance genes: application to the Rx2 locus in potato. Plant J., 21, 73–81. [DOI] [PubMed] [Google Scholar]
- Bendahmane A., Farnham,G., Moffett,P. and Baulcombe,D.C. (2002) Constitutive gain-of-function mutants in a nucleotide binding site-leucine rich repeat protein encoded at the Rx locus of potato. Plant J., in press. [DOI] [PubMed] [Google Scholar]
- Bittner-Eddy P.D., Crute,I.R., Holub,E.B. and Beynon,J.L. (2000) RPP13 is a simple locus in Arabidopsis thaliana for alleles that specify downy mildew resistance to different avirulence determinants in Peronospora parasitica. Plant J., 21, 177–188. [DOI] [PubMed] [Google Scholar]
- Botella M.A., Parker,J.E., Frost,L.N., Bittner-Eddy,P.D., Beynon,J.L., Daniels,M.J., Holub,E.B. and Jones,J.D.G. (1998) Three genes of the Arabidopsis RPP1 complex resistance locus recognize distinct Peronospora parasitica avirulence determinants. Plant Cell, 10, 1847–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain K., Bratton,S.B., Langlais,C., Walker,G., Brown,D.G., Sun,X.M. and Cohen,G.M. (2000) Apaf-1 oligomerizes into biologically active similar to 700-kDa and inactive similar to 1.4-MDa apoptosome complexes. J. Biol. Chem., 275, 6067–6070. [DOI] [PubMed] [Google Scholar]
- Dinesh-Kumar S.P. and Baker,B.J. (2000) Alternatively spliced N resistance gene transcripts: their possible role in tobacco mosaic virus resistance. Proc. Natl Acad. Sci. USA, 97, 1908–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinesh-Kumar S.P., Tham,W.H. and Baker,B.J. (2000) Structure– function analysis of the tobacco mosaic virus resistance gene N. Proc. Natl Acad. Sci. USA, 97, 14789–14794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds P.N., Lawrence,G.J. and Ellis,J.G. (2001) Six amino acid changes confined to the leucine-rich repeat β-strand/β-turn motif determine the difference between the P and P2 rust resistance specificities in flax. Plant Cell, 13, 163–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J.G., Lawrence,G.J., Luck,J.E. and Dodds,P.N. (1999) Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell, 11, 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J., Dodds,P. and Pryor,T. (2000) Structure, function and evolution of plant disease resistance genes. Curr. Opin. Plant Biol., 3, 278–284. [DOI] [PubMed] [Google Scholar]
- Feys B.J. and Parker,J.E. (2000) Interplay of signaling pathways in plant disease resistance. Trends Genet., 16, 449–455. [DOI] [PubMed] [Google Scholar]
- Flor H.H. (1971) Current status of the gene-for-gene concept. Annu. Rev. Phytopathol., 9, 275–298. [Google Scholar]
- Guenther B., Onrust,R., Sali,A., Odonnell,M. and Kuriyan,J. (1997) Crystal structure of the δ subunit of the clamp-loader complex of E.coli DNA polymerase III. Cell, 91, 335–345. [DOI] [PubMed] [Google Scholar]
- Heath M.C. (2000) Hypersensitive response-related death. Plant Mol. Biol., 44, 321–334. [DOI] [PubMed] [Google Scholar]
- Hwang C.-F., Bhakta,A.V., Truesdell,G.M., Waclawa,M.P. and Williamson,V.M. (2000) Evidence for a role of the N terminus and leucine-rich repeat region of the Mi gene product in regulation of localized cell death. Plant Cell, 12, 1319–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inohara N. and Nunez,G. (2001) The NOD: a signaling module that regulates apoptosis and host defense against pathogens. Oncogene, 20, 6473–6481. [DOI] [PubMed] [Google Scholar]
- Inohara N., Koseki,T., Lin,J.M., del Peso,L., Lucas,P.C., Chen,F.F., Ogura,Y. and Nunez,G. (2000) An induced proximity model for NF-κB activation in the Nod1/RICK and RIP signaling pathways. J. Biol. Chem., 275, 27823–27831. [DOI] [PubMed] [Google Scholar]
- Jaroszewski L., Rychlewski,L., Reed,J.C. and Godzik,A. (2000) ATP-activated oligomerization as a mechanism for apoptosis regulation: fold and mechanism prediction for CED-4. Proteins, 39, 197–203. [DOI] [PubMed] [Google Scholar]
- Jia Y., McAdams,S.A., Bryan,G.T., Hershey,H.P. and Valent,B. (2000) Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J., 19, 4004–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X.J. and Wang,X.D. (2000) Cytochrome c promotes caspase-9 activation by inducing nucleotide binding to Apaf-1. J. Biol. Chem., 275, 31199–31203. [DOI] [PubMed] [Google Scholar]
- Jones D.A. and Jones,J.D.G. (1997) The role of leucine-rich repeat proteins in plant defences. Adv. Bot. Res., 24, 89–167. [Google Scholar]
- Lawrence G.J., Finnegan,E.J., Ayliffe,M.A. and Ellis,J.G. (1995) The L6 gene for flax rust resistance is related to the Arabidopsis bacterial resistance gene RPS2 and the tobacco viral resistance gene N. Plant Cell, 7, 1195–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister R.T. and Katagiri,F. (2000) A resistance gene product of the nucleotide binding site—leucine rich repeats class can form a complex with bacterial avirulence proteins in vivo. Plant J., 22, 345–354. [DOI] [PubMed] [Google Scholar]
- Luck J.E., Lawrence,G.J., Dodds,P., Shepherd,K.W. and Ellis,J.G. (2000) Regions outside of the leucine-rich repeats of flax rust resistance proteins play a role in specificity determination. Plant Cell, 12, 1367–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May A.P., Whiteheart,S.W. and Weis,W.I. (2001) Unraveling the mechanism of the vesicle transport ATPase NSF, the N-ethylmaleimide-sensitive factor. J. Biol. Chem., 276, 21991–21994. [DOI] [PubMed] [Google Scholar]
- McDowell J.M., Dhandaydham,M., Long,T.A., Aarts,M.G.M., Goff,S., Holub,E.B. and Dangl,J.L. (1998) Intragenic recombination and diversifying selection contribute to the evolution of downy mildew resistance at the RPP8 locus of Arabidopsis. Plant Cell, 10, 1861–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers B.C., Shen,K.A., Rohani,P., Gaut,B.S. and Michelmore,R.W. (1998) Receptor-like genes in the major resistance locus of lettuce are subject to divergent selection. Plant Cell, 10, 1833–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel L., Moores,T.L., van der Biezen,E.A., Parniske,M., Daniels,M.J., Parker,J.E. and Jones,J.D.G. (1999) Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell, 11, 2099–2111. [PMC free article] [PubMed] [Google Scholar]
- Ogura T. and Wilkinson,A.J. (2001) AAA(+) superfamily ATPases: common structure-diverse function. Genes Cells, 6, 575–597. [DOI] [PubMed] [Google Scholar]
- Pan Q.L., Wendel,J. and Fluhr,R. (2000) Divergent evolution of plant NBS–LRR resistance gene homologues in dicot and cereal genomes. J. Mol. Evol., 50, 203–213. [DOI] [PubMed] [Google Scholar]
- Parker J.E. et al. (1997) The Arabidopsis downy mildew resistance gene RPP5 shares similarity to the toll and interleukin-1 receptors with N and L6. Plant Cell, 9, 879–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart J.R. et al. (2002) The ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc. Natl Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff F., Martin-Hernandez,A.M. and Baulcombe,D.C. (2001) Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J., 25, 237–245. [DOI] [PubMed] [Google Scholar]
- Rupp R.A., Snider,L. and Weintraub,H. (1994) Xenopus embryos regulate the nuclear localization of XMyoD. Genes Dev., 8, 1311–1323. [DOI] [PubMed] [Google Scholar]
- Shirasu K. and Schulze-Lefert,P. (2000) Regulators of cell death in disease resistance. Plant Mol. Biol., 44, 371–385. [DOI] [PubMed] [Google Scholar]
- Srinivasula S.M., Ahmad,M., Fernandes-Alnemri,T. and Alnemri,E.S. (1998) Autoactivation of procaspase-9 by Apaf-1-mediated oligomerization. Mol. Cell, 1, 949–957. [DOI] [PubMed] [Google Scholar]
- Tai T.H., Dahlbeck,D., Clark,E.T., Gajiwala,P., Pasion,R., Whalen,M.C., Stall,R.E. and Staskawicz,B.J. (1999) Expression of the Bs2 pepper gene confers resistance to bacterial spot disease in tomato. Proc. Natl Acad. Sci. USA, 96, 14153–14158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiativ e (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Traut T.W. (1994) The functions and consensus motifs of nine types of peptide segments that form different types of nucleotide-binding sites. Eur. J. Biochem., 222, 9–19. [DOI] [PubMed] [Google Scholar]
- van der Biezen E.A. and Jones,J.D.G. (1998a) The NB-ARC domain: a novel signalling motif shared by plant resistance gene products and regulators of cell death in animals. Curr. Biol., 8, R226–R227. [DOI] [PubMed] [Google Scholar]
- van der Biezen E.A. and Jones,J.D.G. (1998b) Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem. Sci., 23, 454–456. [DOI] [PubMed] [Google Scholar]
- van der Vossen E.A.G., Rouppe van der Voort,J.N.A.M., Kanyuka,K., Bendahmane,A., Sandbrink,H., Baulcombe,D.C., Bakker,J., Stiekema,W.J. and Klein-Lankhorst,R.M. (2000) Homologues of a single resistance-gene cluster in potato confer resistance to distinct pathogens: a virus and a nematode. Plant J., 23, 567–576. [DOI] [PubMed] [Google Scholar]
- Wang J., Song,J.J., Seong,I.S., Franklin,M.C., Kamtekar,S., Eom,S.H. and Chung,C.H. (2001) Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU. Structure, 9, 1107–1116. [DOI] [PubMed] [Google Scholar]
- Warren R.F., Henk,A., Mowery,P., Holub,E. and Innes,R.W. (1998) A mutation within the leucine-rich repeat domain of the Arabidopsis disease resistance gene RPS5 partially suppresses multiple bacterial and downy mildew resistance genes. Plant Cell, 10, 1439–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Chang,H.Y. and Baltimore,D. (1998) Essential role of CED-4 oligomerization in CED-3 activation and apoptosis. Science, 281, 1355–1357. [DOI] [PubMed] [Google Scholar]
- Young N.D. (2000) The genetic architecture of resistance. Curr. Opin. Plant Biol., 3, 285–290. [DOI] [PubMed] [Google Scholar]