Abstract
Microbial transcription modulator NusG interacts with RNA polymerase and termination factor ρ, displaying striking functional homology to eukaryotic Spt5. The protein is also a translational regulator. We have determined crystal structures of Aquifex aeolicus NusG showing a modular design: an N-terminal RNP-like domain, a C-terminal element with a KOW sequence motif and a species-specific immunoglobulin-like fold. The structures reveal bona fide nucleic acid binding sites, and nucleic acid binding activities can be detected for NusG from three organisms and for the KOW element alone. A conserved KOW domain is defined as a new class of nucleic acid binding folds. This module is a close structural homolog of tudor protein–protein interaction motifs. Putative protein binding sites for the RNP and KOW domains can be deduced, which differ from the areas implicated in nucleic acid interactions. The results strongly argue that both protein and nucleic acid contacts are important for NusG’s functions and that the factor can act as an adaptor mediating indirect protein–nucleic acid associations.
Keywords: KOW domain/NusG/protein–nucleic acid interaction/regulation of transcription and translation/transcription termination and antitermination
Introduction
Transcription is mediated by conserved multi-subunit RNA polymerases (RNAP), which in bacteria exhibit an α2ββ′ω catalytic core (Cramer, 2002). During initiation, elongation and termination, the core is controlled by a complex network of biological signals, such as protein transcription factors, small molecule regulators, sequences on the template DNA or structures in the nascent RNA (Mooney et al., 1998). Among other proteins, four N-utilization substances, NusA, B, E [ribosomal (r-) protein S10] and G, are important elongation/termination modulators. Transcription regulation through these Nus factors has been intensively studied in the expression of genes from lambdoid phages (Greenblatt et al., 1998). Coliphage λ can activate delayed early gene expression by suppressing the effects of upstream terminators. While rudimentary antitermination can be evoked by a phage-encoded protein, N, alone, the host Nus factors are mandatory for optimal and long-range efficacy. λN and the Nus ensemble are recruited by RNAP upon encounter of RNA nut sequences, a linear 8–9 nucleotide (nt) segment (boxA) and a small hairpin (boxB). The Nus factors probably participate in antitermination during transcription of Escherichia coli rRNA (rrn) operons as well (Squires et al., 1993). Interestingly, a λ competitor, HK022, produces an N-like protein, Nun, which cooperates with the same group of host factors at nut loci to enhance transcription termination (Weisberg et al., 1999). Therefore, the mediating phage protein seems to guide the fundamental termination/antitermination decisions of RNAP, which are modulated in addition by the Nus factors.
Besides NusA, NusG is the only member of the Nus family that is essential for E.coli. Apart from antitermination, it is involved in other transcription control events. In some, it exerts similar effects as NusA, e.g. both factors increase the read-through of the attenuator upstream of the rpoB gene (Linn and Greenblatt, 1992). More often, the proteins act as antagonists: NusA prolongs pausing of RNAP at certain RNA hairpin structures (Schmidt and Chamberlin, 1984), thus enhancing ρ-independent termination (Schmidt and Chamberlin, 1987), while NusG reduces the dwell time of the enzyme at these sites (Burova et al., 1995). At the same time, NusA tends to decrease ρ-dependent termination, while NusG increases it (Burns et al., 1998; Pasman and von Hippel, 2000). The latter NusA effect may be explained by a correlation of RNAP and ribosome progression on their templates. The NusG effect probably rests in a direct interaction with termination factor ρ (Li et al., 1993; Pasman and von Hippel, 2000). Furthermore, NusG–RNAP contacts (Mason and Greenblatt, 1991; Li et al., 1992) have been observed in E.coli. Apart from simply facilitating the interaction of ρ with the transcription complex, NusG seems to prevent RNAP from backtracking or reverse backtracked states that are resistant to ρ.
A recent, surprising addition to the NusG activity profile has been its involvement in translational regulation. In NusG-depleted cells, polypeptide elongation rates for a β-galactosidase reporter have been found reduced with unchanged lacZ mRNA synthesis (Zellars and Squires, 1999).
Detailed structural information should prove useful for formulating hypotheses about NusG’s multiple roles. Herein we describe structures of NusG from Aquifex aeolicus (aaeNusG) in two crystal forms. The protein encompasses the entire E.coli homolog (ecoNusG) and in addition carries a unique insertion of ∼80 residues. In the common regions, aaeNusG and ecoNusG are >47% identical in sequence (Figure 1). Interestingly, the portions shared with ecoNusG show putative nucleic acid- and protein-binding elements. In combination with solution studies, the current structures advocate a functional role for NusG–nucleic acid interactions and suggest how the factor can contact other macromolecules.
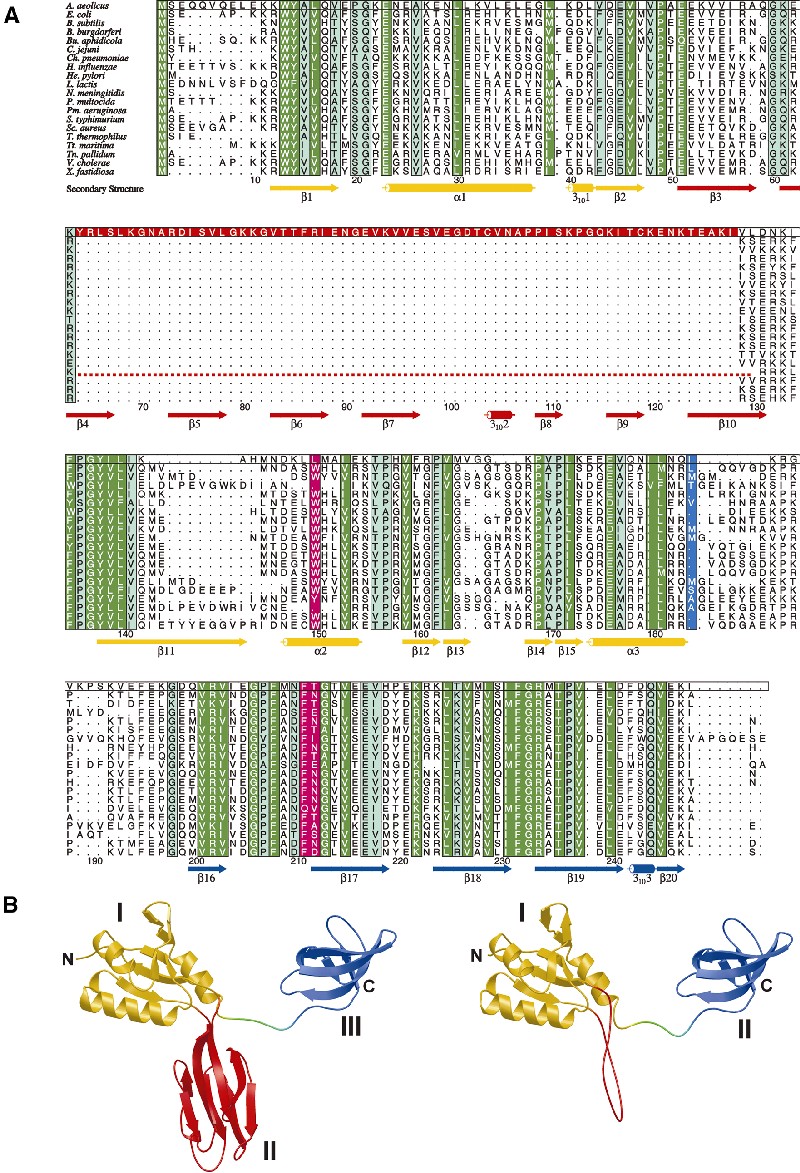
Fig. 1. (A) Alignment of 20 bacterial NusG sequences. Highly conserved residues, dark green; intermediately conserved residues, light green. The aaeNusG-specific insertion is shown in red, mutated residues in magenta (with a phenotype) and in light blue (no phenotype). An unrelated insertion in tmaNusG is indicated by a broken line. NusG from Thermus thermophilus carries 60 additional N-terminal residues compared with aaeNusG (data not shown). Other species, such as Streptomyces griseus and mycobacteria, also harbor N-terminal extensions of varying length (data not shown). The secondary structure elements corresponding to the aaeNusG crystal structures are indicated below the sequences, color coded by domains. Numbering is according to aaeNusG. (B) Domain organization in aaeNusG (left) and in a homology model of ecoNusG (right). Domains are in the same colors as in (A).
Results and discussion
Structure of aaeNusG
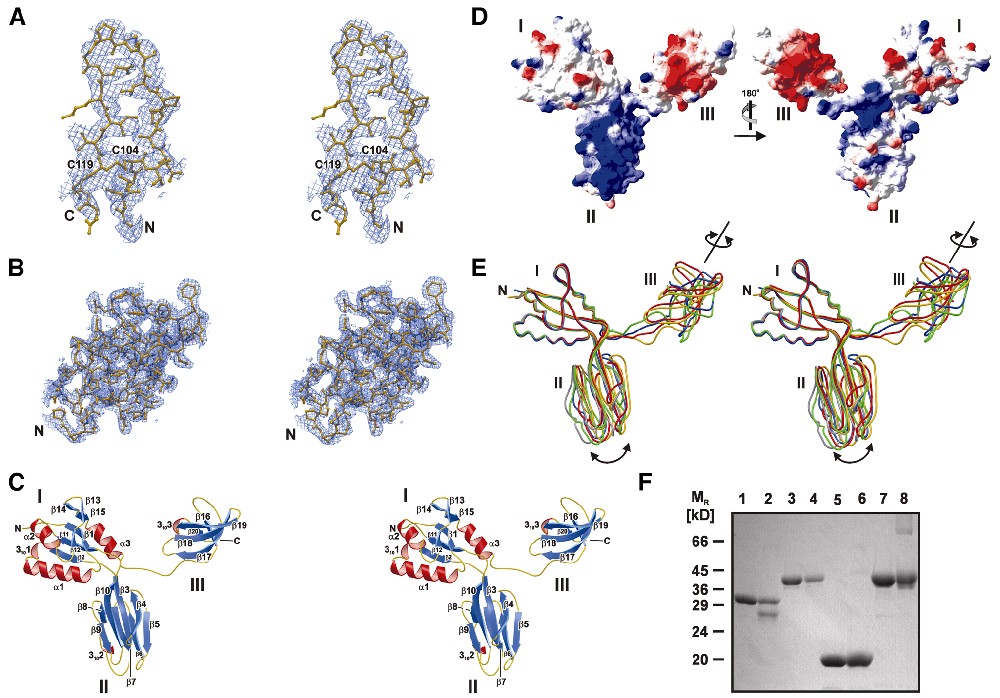
Overview. An aaeNusG structure in space group I222 was solved by an isomorphous replacement strategy and yielded a model for residues Q5–R186 (Table I; Figure 2A). No electron density was seen for 62 C-terminal residues, while a gel analysis of dissolved crystals showed the full-length molecule. A more densely packed structure in space group P21 could be solved by molecular replacement and clearly revealed the missing domain (Figure 2B). The four crystallographically independent molecules of the P21 structure contained all residues of aaeNusG except for 4–9 N-terminal amino acids and, in three cases, a flexible linker peptide (G187–E194). In the I222 and P21 crystal structures, 97.4 and 97%, respectively, of the residues displayed preferred φ/ψ conformations with 1.3 and 1.4%, respectively, in the disallowed regions. All residues with unusual main chain conformations were either very well defined in the electron densities or occupied flexible regions of the molecules.
Table I. Crystallographic data.
| Data collection | Form I |
Form II |
|||
|---|---|---|---|---|---|
| Native | KAu(CN)2 | HgCl2 | Ta6Br14 | Native | |
| Temperature (K) | 100 | 100 | 100 | 100 | 100 |
| Radiation source | Synchrotron | Rot. anode | Rot. anode | Rot. anode | Rot. anode |
| Wavelength (Å) | 0.95 | 1.5418 | 1.5418 | 1.5418 | 1.5418 |
| Space group | I222 | P21 | |||
| Unit cell (Å, °) | |||||
| a | 81.1 | 80.0 | 79.9 | 79.7 | 87.5 |
| b | 86.2 | 86.7 | 86.2 | 86.5 | 54.4 |
| c | 108.4 | 108.6 | 108.2 | 108.6 | 113.2 |
| β | 90.7 | ||||
| Resolution (Å) | 30.0–1.95 | 30.0–2.4 | 30.0–3.9 | 30.0–2.6 | 30.0–2.0 |
| Unique reflections | 27 602 | 13 654 | 2980 | 10 379 | 69 671 |
| Redundancy | 3.6 | 4.1 | 2.9 | 3.4 | 3.3 |
| Completeness (%) | 98.5 (98.4) | 90.3 (86.4) | 89.0 (88.9) | 87.4 (87.0) | 94.9 (79.2) |
| I/σ(I) | 30.0 (2.3) | 8.9 (1.4) | 8.9 (3.1) | 14.4 (1.3) | 8.9 (1.6) |
|
Rsyma (%) |
7.4 (50.4) |
10.1 (73.7) |
9.3 (15.0) |
4.7 (75.8) |
8.0 (40.6) |
| Phasing |
|
|
|
|
|
| Resolution (Å) | 30.0–2.4 | 30.0–2.4 | 30.0–3.9 | 30.0–2.7 | |
| Heavy atom sites | 3 | 3 | 2 | ||
| RCullisb | |||||
| Centrics | 0.73 | 0.79 | 0.94 | ||
| Acentrics | 0.75 | 0.82 | 0.96 | ||
| Phasing powerc | |||||
| Centrics | 1.15 | 0.82 | 0.31 | ||
| Acentrics | 1.55 | 1.18 | 0.54 | ||
| FOMd | |||||
| Before DMe | 0.34 | ||||
| After DM |
0.54 |
|
|
|
|
| Refinement |
|
|
|
|
|
| Resolution (Å) | 30.0–1.95 | 30.0–2.0 | |||
| Protein atoms | 1436 | 7640 | |||
| Water oxygens | 296 | 1078 | |||
| Rworkf (%) | 23.4 (28.3) | 25.1 (41.5) | |||
| Rfreef (%) | 25.9 (28.6) | 27.4 (43.9) | |||
| R.m.s.d. bond lengths (Å) | 0.006 | 0.008 | |||
| R.m.s.d. bond angles (°) | 1.42 | 1.51 | |||
| Protein B-factor (Å2) | 28.7 | 44.7 | |||
| Mean residual error (Å) | 0.14 | 0.21 | |||
| Preferred φ/ψ (%) | 97.4 | 97.0 | |||
aRsym(I) = (∑hkl∑i[|Ii(hkl) – <I(hkl)>|]/∑hkl∑i[Ii(hkl)]; Ii(hkl) is the intensity of the ith measurement of hkl; <I(hkl)> is the average value of hkl for all i measurements.
bRCullis = ∑hkl[||FPH ± FP| – |FH,calc||]/∑hkl[|FPH ± FP|]; (FPH + FP) if signs are opposite, (FPH – FP) if signs are equal.
cPhasing power = (∑n[|Fh|2]/∑n[|E|2])1/2; ∑n|E2| = lack of closure error = ∑n[|FPH|(obs) – |FPH|(calc)]2
dFOM = figure of merit = [|F(hkl)best|]/|F(hkl)|; F(hkl)best = ∑α[P(α)Fhkl(α)]/∑α[P(α)]
eDM is density modification (solvent flattening).
fRwork = ∑hkl[||Fobs| – k|Fcalc||]/∑hkl[|Fobs|]; Rfree = ∑hkl⊂T[||Fobs| – k|Fcalc||]/∑hkl⊂T[|Fobs|]; hkl⊂T is the test set.
Data for the last 0.1 Å are in parentheses.
Fig. 2. (A) MIRAS electron density of the I222 crystal form encompassing the disulfide bridge of domain II, superimposed on the final model. (B) 2Fo – Fc electron density for one domain III of the P21 crystal form. (C) Structural overview for a full-length aaeNusG molecule. Helices, red; β-strands, blue; coil, yellow. By default, all subsequent structures are displayed from the same point of view. (D) Two diametric views of the electrostatic surface potential of aaeNusG. The left panel is in the same orientation as (C); domains are indicated. Positive potential, blue; negative potential, red. (E) Superposition according to domain I of the four full-length P21 aaeNusG models [the molecule from (C) is in gold] and the I222 fragment (gray). Arrows indicate changes in positioning of domains II and III relative to I. (F) SDS gel analysis of glutaraldehyde-crosslinked NusG preparations. Lanes 1 and 2, aaeNusG, native, crosslinked; lanes 3 and 4, tmaNusG, native, crosslinked; lanes 5 and 6, ecoNusG, native, crosslinked; lanes 7 and 8, MJ0158 (positive dimeric control), native, crosslinked. Treatment of aaeNusG with glutaraldehyde leads to a faster migrating species.
aaeNusG features a modular structure of three domains (Figure 1B), which are arranged in a triangle with extensions ∼75 × 70 × 38 Å (Figure 2C). Domain I is a composite of residues Q5–V48 and P136–K185, interrupted by domain II (E51–F135). Two short linker segments (P49–A50 and K133–I134) connect these modules. Domain I is attached through a longer, unstructured linker (R186–V193) to domain III (E194–I248).
Domain I contains a four-stranded antiparallel β-sheet covered on one side by two α-helices and on the other by a single helix, which leads to the connection with domain III (Figure 2C). Strand β2 is preceded by a single 310-turn. The ‘fourth strand’ of the motif is composed of two half sides, β12 and β15, interrupted by a short β-hairpin, β13 and β14. Domain II is interspersed between strands β2 and β11. It consists of two sandwiched four-stranded antiparallel sheets. A short 310-helix (T103–A107) is found N-terminal of strand β8. C104 in this helix is linked through a disulfide bridge to C119 at the C-terminus of strand β9 (Figure 2A). While rare, disulfide bridges do occur in the reducing intracellular environments, in particular in hyperthermophiles (Vieille and Zeikus, 2001). Domain III is folded into a highly bent five-stranded antiparallel β-sheet. Residues F242–V245 again adopt a 310-conformation interspersed between strands β19 and β20. The four copies of domain III in the P21 structure are packed in different fashions against domains I and II of neighboring molecules.
Domain I shows a mixed electrostatic surface potential (Figure 2D). Domain II carries an extended positively charged patch covering strands β3, 4, 9 and 10. An area of negative surface potential is wrapped like a belt around domain III.
Domain arrangements. The five independent observations of aaeNusG allow separation of inherent structural tendencies from crystal packing effects. Individual domains superimpose extremely well: root-mean-square deviations (r.m.s.d.) are 0.6–1.4 Å for domain I (90 matching Cα), 0.4–0.8 Å for domain II (87 matching Cα) and 0.6–1.0 Å for domain III (55 matching Cα). After alignment of all molecules according to domain I (Figure 2E), the relative orientations of domains I and II appear rather independent of the environment and may, therefore, be maintained in solution. In contrast, two pairs of the third domain are seen to adopt two arrangements, distinguished by the azimuth around their long axes (∼110°). While in all cases domain III protrudes in the same direction from domain I/II, its flexible linker will probably allow it to swing out of the triangular plane in solution and, likewise, the portion missing in the I222 crystal structure suggests more than two possible conformations. Therefore, aaeNusG is composed of a relatively rigid part, domains I and II, and a flexible appendix, domain III.
Solution structure. The secondary structure composition for full-length aaeNusG was deduced from circular dichroism (CD) spectra. It agrees well with that calculated from the P21 crystal structure, suggesting that in solution the domains retain the observed folds (Table II). Furthermore, CD measurements of domain III in isolation indicate a fold similar to that in the crystalline state (Table II). Thus, the C-terminal region was presumably disordered as a rigid body in the I222 crystals and not partially unfolded due to marginal stability.
Table II. Secondary structure contents.
| Protein | Elements | Crystal structure (P21) (%) | Homology model (%) | CD (%) |
|---|---|---|---|---|
| aaeNusG, full length | Helices | 21 | – | 25 |
| β-structures | 53 | – | 46 | |
| aaeNusG, domain III | Helices | 6 | – | 14 |
| β-structures | 61 | – | 59 | |
| ecoNusG | Helices | – | 22 | 21 |
| β-structures | – | 39 | 42 |
Repeatedly, certain Nus factors have been inferred to form dimers. NusB from Mycobacterium tuberculosis exists as a dimer (Gopal et al., 2000) and one model for NusA activity invokes two of the molecules acting on the RNAP core (Bar-Nahum and Nudler, 2001). ecoNusG has been shown to be monomeric (Pasman and von Hippel, 2000). We conducted gel-filtration and velocity-sedimentation experiments for aaeNusG and NusG from Thermotoga maritima (tmaNusG), which suggested monomers in solution (data not shown). For aaeNusG, there are no conserved dimers among the contacting molecules in the crystals. aae, tma and ecoNusG were also treated with glutaraldehyde in order to covalently trap oligomers. On denaturing gels only monomers were detected (Figure 2F).
Homology model for ecoNusG. Based on the aaeNusG structure, a homology model was created for ecoNusG with the program SWISS-Model (http://www.expasy.ch). The folds of domains I and III were preserved in ecoNusG, while the domain II insertion was replaced by an extended loop (E.coli residues V44–G67; Figure 1B). This loop carried a large number of K and R residues, substituting for the positive potential on aaeNusG domain II. CD measurements on ecoNusG were in excellent agreement with the structural composition of the model (Table II). Therefore, the similarity of the folds (r.m.s.d. 0.27 Å for 163 Cα atoms) and the high degree of sequence identity warrant discussion of the information gathered for the E.coli protein in light of the present aaeNusG structures.
Nucleic acid and protein interaction modules in the structure of NusG. aaeNusG domain I showed similarity to the ribonucleoprotein (RNP) motif. The closest match for domain II was a portion of a T-cell antigen receptor Vα–Vβ structure (Housset et al., 1997). For domain III, there is a striking congruence with a region of r-protein L24 involved in rRNA interactions (Ban et al., 2000), as well as with the tudor domain of the human survival of motor neurons (SMN) protein (Selenko et al., 2001). Therefore, NusG seems to be a composite of both bona fide nucleic acid and protein interaction modules. Below we will derive possible modes of nucleic acid interactions for aaeNusG, demonstrate that NusGs from several organisms bind both DNA and RNA in solution, outline possible protein interaction sites and attempt to correlate mutational analyses with the derived binding surfaces.
Structural basis for NusG–nucleic acid interactions
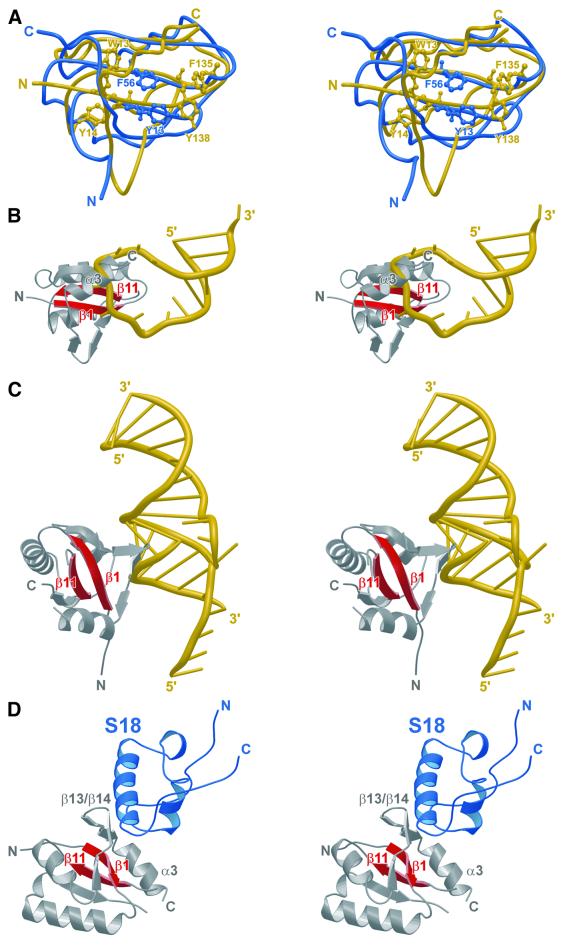
RNPs suggest nucleic acid interaction modes for domain I. RNPs contain a four-stranded antiparallel β-sheet with two α-helices on one side (Varani and Nagai, 1998). Normally, the open face of the sheet binds the RNA ligands. There are two short conserved sequence motifs on strands 1 and 3 of the modules, RNP2 and 1, respectively, with several aromatic residues to stack with the RNA bases. The central two β-strands of aaeNusG domain I also carry three aromatic residues: W13 and Y14 in β1, and Y138 in β11 (Figure 3A). In addition, F135 lies at the entrance to β11. Y14 is largely buried in the present conformation, while the other three residues are on the surface of the sheet. All these side chains are absolutely conserved (Figure 1A). However, RNA binding as, for example, in the spliceosomal U1A (Oubridge et al., 1994) is obstructed in aaeNusG by helix α3 (Figure 3B). V177, I180 and L181 on one side of α3 form a hydrophobic cluster with residues W13, L140 and L172 of the β-sheet and the neighboring I47 and P49. All these residues are well conserved. In addition, a salt bridge and a hydrogen bond link E174 of helix α3 to K142 of β11 and to W13 of β1, respectively. E174 and K142 are non-conserved side chains. Taken together, we assume that in the isolated state, blocking of the RNP sheet in NusG is a general feature of the family.
Fig. 3. (A) Superposition of aaeNusG domain I (yellow) on the U1A protein (blue). Relative to Figure 2C, the molecule was rotated 150° towards the viewer around the horizontal axis and 45° counterclockwise around the vertical axis. Aromatic residues in the central two β-strands are depicted in ball-and-stick. (B) Domain I binding mode for RNA based on the U1A–RNA structure. The RNA is seen to collide with helix α3. The orientation is the same as in (A). (C) Model for the domain I–RNA interaction based on the structure of the S6–rRNA complex. The protein is in gray with the central two β-strands in red. RNA is in gold. Relative to Figure 2C, the domain was rotated 90° counterclockwise both around the horizontal axis and the axis into the plane of the paper. (D) A model for a protein interaction mode in domain I (gray and red) based on the r-protein S6–S18 (blue) complex. The contact leaves the core of the domain I β-sheet available for nucleic acid interaction.
Helix α3 could act as a switch that prevents nucleic acid binding until an interaction between NusG and another component of the transcriptional apparatus removes the block. A similar mechanism of tunable RNA binding has recently been discovered for E.coli NusA (Mah, 2000). Such a mechanism could explain why nucleic acid contacts by ecoNusG have so far escaped detection.
A different mode of RNA binding by an RNP-like protein was seen in r-protein S6, which involves the β4 edge of the sheet (Schluenzen et al., 2000; Wimberly et al., 2000). After alignment with S6 (r.m.s.d. 1.8 Å for 36 Cα atoms), the β13/β14 hairpin of aaeNusG domain I pointed into the stacking junction of two RNA stems (Figure 3C). The β13/β14 region is well conserved and bordered by the positively charged K168 and K173 (Figure 1A), supporting its nucleic acid binding potential. Thus, two different nucleic acid binding modes can be hypothesized for domain I.
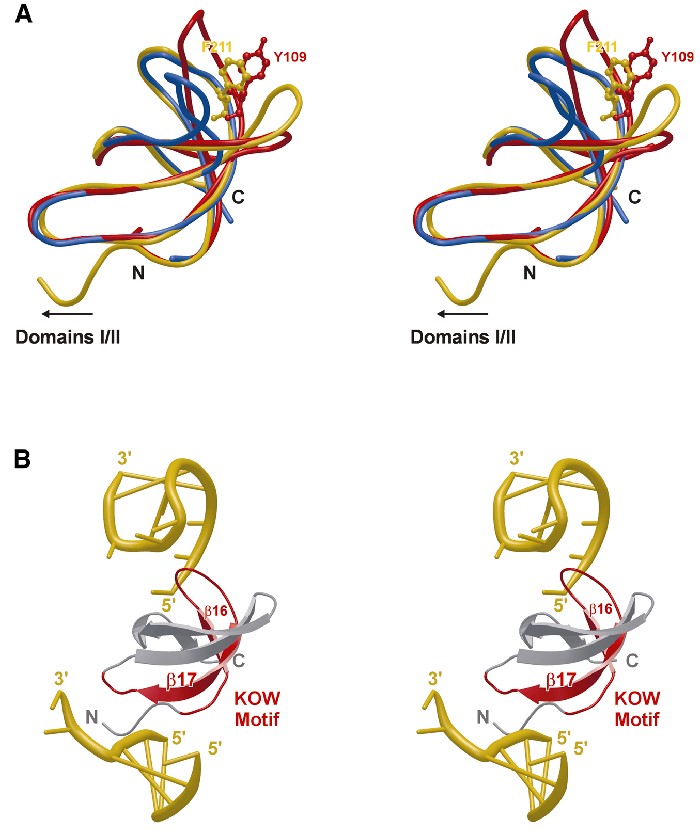
The domain III KOW motif is embedded in a structurally conserved RNA binding element. The region of domain III homologous to r-protein L24 encompasses a KOW sequence motif (E196–K222) (Kyrpides et al., 1996) constituting the first two strands, β16 and β17, of domain III and their adjacent loops. The motif has been implicated in RNA interactions since it was shared among three families of r-proteins (microbial L24 and eukaryotic eL26 and eL27) and the NusG factors. A comparison of domain III with L24 (r.m.s.d. 1.0 Å for 41 Cα atoms) showed the sequence embedded in a conserved fold (Figure 4A). The notion of the KOW sequence should, therefore, be expanded to that of a KOW domain as a separate nucleic acid interaction motif.
Fig. 4. (A) Superposition of aaeNusG domain III (gold), r-protein L24 (blue) and the SMN tudor domain (red). F211 of aaeNusG and Y109 of the tudor domain, presumably important for contacting other proteins, are shown in ball-and-stick. (B) Domain III in complex with RNA molecules according to the L24–rRNA structure. The sequence originally identified as the KOW element is shown in red.
Comparison with the L24–rRNA structure suggested that nucleic acids bind to domain III mainly through regions of the KOW sequence element (Figure 4B), i.e. the loop preceding strand β16 (K192–K197), the loop between β16 and β17 (E205–F208) and the C-terminal part of β17, plus the following loop (E217–K222). The first patch does not harbor highly conserved residues, but carries two positively charged K side chains, possibly to attract the negatively charged nucleic acid backbone. The second contact sequence is well conserved, encompassing the KOW-defining G206 residue. In concert with the neighboring P207, G206 allows strand reversal at this point and the close approach of protein and ligand backbones. The C-terminus of β17 is weakly conserved with H219 and K222 possibly maintaining electrostatic interactions and P220 important for the loop conformation.
Additional nucleic acid interactions through domain II? The lack of sequence homologs for domain II obviates the identification of conserved functional residues. However, the expanded positive patch could attract the negatively charged sugar–phosphate backbone of a nucleic acid. While expected to be non-specific, such interactions could mediate initial approach of the molecules and serve, for example, to span nucleic acids between domains I and III. Similar electrostatic interactions could be sponsored in ecoNusG by the positively charged loop replacing domain II.
NusG–nucleic acid interactions in solution
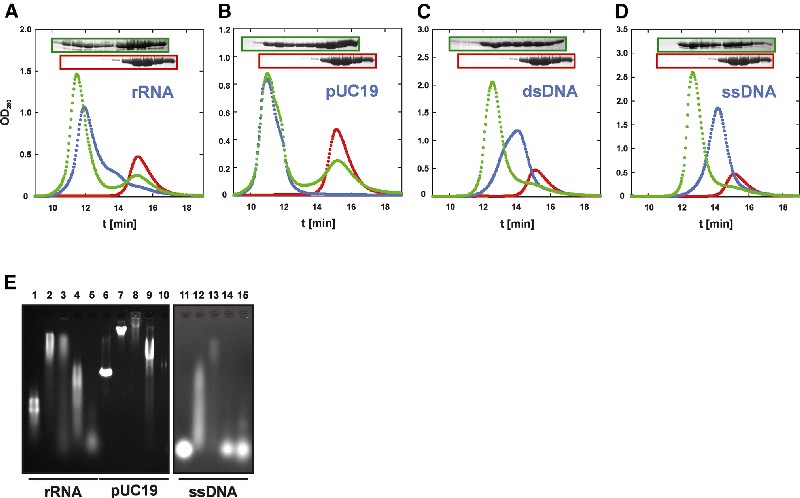
NusG factors from various organisms exhibit DNA- and RNA-binding activities. We tested whether aaeNusG binds nucleic acids by analytical gel-filtration chromatography. At pH 7.0 and 50 mM salt, a large portion of the protein co-eluted with the nucleic acids employed [16 and 23S rRNA, linearized pUC19 DNA, 64 bp double-stranded (ds) DNA and 64 nt single-stranded (ss) DNA; Figure 5A–D], well before the protein alone. The amount of bound protein was insensitive to pH changes between 7.0 and 9.0. It decreased but was still significant at 150 mM salt. Thus, aaeNusG displays nucleic acid binding capacities, which are at least partly mediated by electrostatic attraction via strongly basic residues (R or K). An electrostatic contribution can be most easily envisaged for the positive patch on domain II. Two proteins used as molecular weight standards without known nucleic acid binding capacities, carbonic anhydrase and chicken ovalbumin, did not co-migrate with the nucleic acids under the same conditions.
Fig. 5. (A–D) Gel-filtration assay. aaeNusG, red; nucleic acid, blue; complex, green. Nucleic acid types tested are indicated. SDS gel analyses are inserted and framed with the same color as their pertaining elution profile. (E) Agarose gel shifts. Lane 1, rRNA; lanes 2–5, aaeNusG, tmaNusG, ecoNusG and aaedomain III each with rRNA; lane 6, pUC19 (EcoRI linearized); lanes 7–10, aaeNusG, tmaNusG, ecoNusG and aaedomain III each with pUC19; lane 11, ssDNA 64mer; lanes 12–15, aaeNusG, tmaNusG, ecoNusG and aaedomain III each with ssDNA. Lanes 11–15 are from the same gel at higher exposure.
To confirm the results, similar mixtures of aaeNusG and nucleic acids were analyzed on agarose gels (Figure 5E). All nucleic acids tested were quantitatively shifted by aaeNusG. Only the complexes with the small nucleic acids penetrated 5% polyacrylamide gels, where results were consistent with the agarose gels. Taken together, aaeNusG exhibits general binding activities towards RNA as well as ds and ssDNA.
Previously, tmaNusG has been shown to bind nucleic acids cooperatively with a strong preference for dsDNA (Liao et al., 1996). As a control for our experimental set-up, we investigated tmaNusG as described for aaeNusG, and could verify significant binding to all nucleic acids tested. tmaNusG complexes with rRNA and ssDNA were similar in size as the aaeNusG complexes, but the complexes with plasmid DNA were larger and did not completely migrate into 1% agarose gels, presumably due to crosslinking (Figure 5E). Therefore, aaeNusG did not crosslink plasmid DNA to the same extent as tmaNusG.
Under the same conditions, we also detected binding of ecoNusG to both rRNA and plasmid DNA but not to the ss oligonucleotide in gel-shift experiments (Figure 5E). Therefore, some nucleic acid affinity seems to be a general feature of NusG proteins and does not require the presence of the extra domains of aaeNusG or tmaNusG.
With the KOW domain of aaeNusG alone, plasmid DNA was shifted slightly upward in gels, but staining with ethidium bromide was weak (Figure 5E). The rRNA fraction ran faster when KOW was present. We did not detect fragmentation of the rRNA in gel-filtration runs, which excludes the presence of RNases. The domain also bound weakly to the ssDNA 64mer. The different behavior compared with the full-length protein suggests that nucleic acid binding is implemented in several domains of aaeNusG.
In the gel-filtration experiments, the 256 nm absorbance for the aaeNusG–nucleic acid bands was higher than for the shifted protein plus the nucleic acids alone. A possible explanation would be the (partial) melting of the nucleic acids upon aaeNusG binding. For the 64 bp dsDNA, the protein-corrected absorbance of the complex reached ∼60% of the value for a completely ss sample (data not shown). Thus, aaeNusG’s affinity for ss nucleic acids can cause strand separation in duplex DNA. Consequently, electrostatics cannot be the sole basis for complex formation. Consistently, in gel-filtration experiments aaeNusG did not co-elute with dextran sulfate (MR ∼ 500 000), a strongly negatively charged polysaccharide lacking aromatic groups.
Apart from tmaNusG, NusG–nucleic acid interactions have so far not been described, in contrast with other Nus factors (Nodwell and Greenblatt, 1993). Under one experimental set-up, an interaction for ecoNusG with plasmid DNA was excluded (Mason and Greenblatt, 1991). Nevertheless, we regard the above results as significant: while the complexes were strongly diluted during the course of the gel-filtration experiments, sharp, well-separated peaks were observed, indicating stable complexation. The results were consistent in both chromatographic and electrophoretic experiments, and were in agreement with the nucleic acid binding folds of the crystal structures.
Species-specific differences in nucleic acid binding. NusG function does not seem to be absolutely conserved among bacteria. For example, Bacillus subtilis NusG is not essential for ρ-dependent termination or viability (Ingham et al., 1999). In line with these findings, the binding of ss nucleic acids by aaeNusG differs from the behavior of tmaNusG, which shows a preference for dsDNA (Liao et al., 1996), and of ecoNusG, which did not bind to ssDNA in our set-ups. Such distinctions may be related to the species-specific domain insertions and N-terminal appendices in certain NusG proteins. Nevertheless, the factors may generally be involved in rrn operon antitermination because boxA and boxB elements have been found in the leader and 16S/23S spacer regions of rrn operons in various bacterial and archaeal organisms (Berg et al., 1989), including A.aeolicus (T.Steiner and M.C.Wahl, unpublished data).
Previous observations could be explained by NusG– nucleic acid interactions. The exact pattern of transcript lengths at ρ-dependent termination sites is affected by ecoNusG in vitro (Nehrke et al., 1993). Lower NusG concentrations are necessary to elicit this effect than predicted based on the interaction efficiencies with ρ or RNAP. Furthermore, NusG alleviates the interference by DNA oligomers complementary to the RNA binding site of ρ, slows detachment of ρ from RNA transcripts and its stable association with the transcription complex requires the presence of RNA-bound ρ (Nehrke and Platt, 1994). All of the above effects could be based on an interaction of NusG with nucleic acids.
Structural basis for NusG–protein interactions
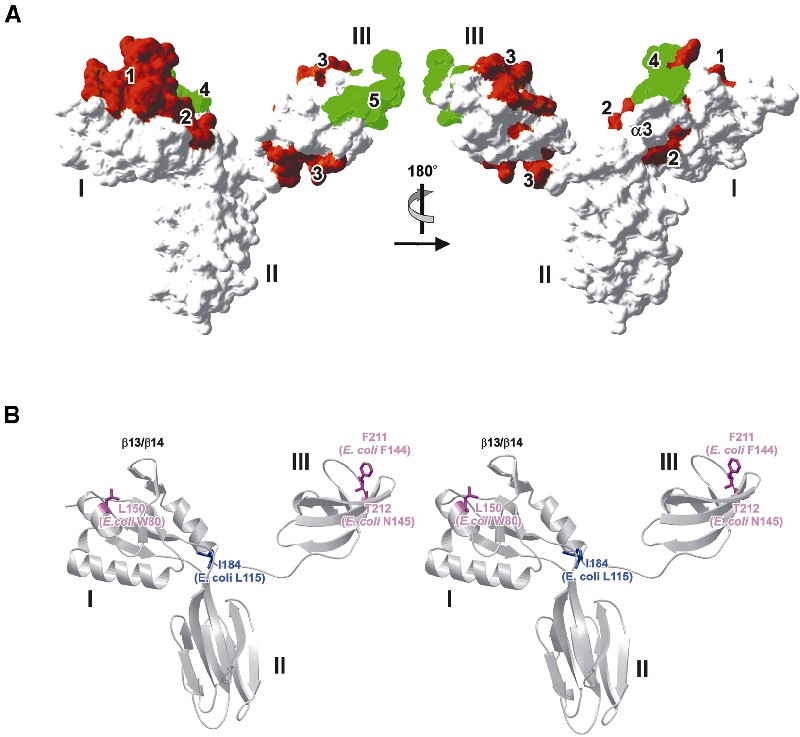
Model for protein interactions through domain I. Inside the 30S ribosomal subunit, the domain I homolog, S6, is in contact with S18. When S6 was overlaid on aaeNusG domain I, carrying S18 along in the process, the latter protein suggested an interaction site in the β13/14/15 region (Figure 3D). Like S6, NusG may concomitantly bind proteins and nucleic acids via its RNP-like domain. Interestingly, the attached protein does not occlude the nucleic acid binding sites deduced above (Figure 6A). Therefore, as an alternative or in addition to its function as a nucleic acid binding platform, domain I could be an anchor for other proteins.
Fig. 6. (A) Deduced nucleic acid (brown) and protein (green) interaction sites mapped onto the aaeNusG surface. The left panel is in the same orientation as Figure 2C. The three domains are labeled. Numbers identify potential interactions sites. 1, S6-like nucleic acid interaction site; 2, U1A-like nucleic acid interaction site; 3, nucleic acid interaction site mapped by L24–rRNA contacts; 4, S18-like protein interaction site; 5, tudor-like protein interaction site. If helix α3 of the RNP motif was removed upon U1A-like nucleic acid interaction, additional contact sites would become uncovered. Interaction sites that could not be deduced from the structures may also exist in domain II. (B) Stereo-ribbon plot of aaeNusG with the mutated residues in magenta (with phenotype) and light blue (no phenotype).
An immunoglobulin-like domain for protein contacts? Immunoglobulin folds are widespread eukaryotic β-sandwich motifs mediating protein–protein interactions (Bork et al., 1994). Protein contacts can take place through the connecting loops and the surfaces or sides of the sheets. Apart from the global resemblance, domain II is stabilized, like many immunoglobulin modules, by an inter-strand disulfide bridge. However, we could not assign the topology of aaeNusG domain II to a known class of immunoglobulin folds. Further work is required to decide which kind of macromolecule, if any, binds to this module.
The KOW domain is a close structural homolog of a known protein interaction motif. The tudor domain has been identified as a 10-fold repeat in the Drosophila protein tudor and subsequently in a large number of RNA binding proteins (Ponting, 1997). The NMR structure of the tudor domain of the human SMN protein (Selenko et al., 2001) exhibits a fold identical to the aaeNusG KOW element (r.m.s.d. 0.95 Å for 36 Cα atoms; Figure 4A), while lacking any significant sequence resemblance. In SMN, the tudor domain is employed for binding to the R/G-rich tails of Sm proteins (Friesen and Dreyfuss, 2000). According to chemical shift analyses (Selenko et al., 2001), the contact region corresponds in the KOW domain to the C-terminus of β16 (tudor A100–I101), the loop to β17 (tudor S103–C107), the very N-terminus of β17 (tudor Y109), the loop between β18 and β19 (tudor Y127–Y130) and the N-terminus of β19 (tudor G131–R133). Most of the contact residues are in flexible loops (Figure 4A). One absolutely conserved NusG residue, F211, aligns particularly well with Y109 of tudor. In addition, M235, T236 and P237 of NusG domain III align with tudor G131, N132 and R133. This latter stretch, plus the preceding loop between β18 and β19, is the most highly conserved area of the KOW domain (Figure 1A), hinting at a functional significance.
The protein binding site derived for the KOW domain is largely different from its deduced nucleic acid interaction surfaces. Therefore, proteins and nucleic acids could bind concomitantly to the domain (Figure 6A). The L24 KOW motif, NusG domain III and the SMN tudor domain are all highly negatively charged, which was thought to preclude nucleic acid interactions (Selenko et al., 2001). Nevertheless, for Haloarcula marismortui L24, rRNA binding has been unequivocally demonstrated (Ban et al., 2000). In general, many of the proteins from the H.marismortui ribosome are astonishingly acidic but still bind well to rRNA. Therefore, the negative charge does not, per se, disqualify RNA binding. It will be interesting to see whether certain tudor domains exhibit nucleic acid binding capacities apart from their protein binding potential or whether nucleic acid binding can functionally discriminate tudor and KOW domains.
The present analyses suggest that one or all of the aaeNusG domains could bind proteins and nucleic acids at the same time via different surfaces (Figure 6A). A similar conclusion was drawn for S1 and KH domains in tmaNusA (Worbs et al., 2001). These protein modules may therefore function as adaptors for indirect protein–nucleic acid interactions, which would explain their usefulness in multi-component systems like transcription and translation.
KOW domains may mediate RNAP and ribosome interactions. Spt5 is a eukaryotic transcription factor that exerts both positive and negative effects on RNAP II (Yamaguchi et al., 2001; Zorio and Bentley, 2001). Analogous to NusG, Spt5 binds to RNAP II and reduces pausing and transcript release by the enzyme at terminator sequences in cooperation with HIV1 Tat. In other systems, it can also increase RNAP II pausing, thus exhibiting the same dual activity as NusG. Furthermore, Spt5 contains four central repeats of the KOW motif and it has been shown that this region is required for interaction with RNAP II. Taken together, Spt5 seems to be a eukaryotic functional analog of NusG. The KOW elements, the only structural features linking Spt5 and NusG, could mediate some of the factor’s similar functions, in particular binding to RNAP.
Another KOW-containing protein has recently been discovered associated with ribosomes. This 21 kDa factor, YfjA, seems to be involved in 30S subunit maturation or translation initiation (Bylund et al., 1997). Because L24, eL26, eL27, NusG and YfjA show varying degrees of ribosome association, their other common characteristic, the KOW domain, could be responsible for this function. Again, both the RNA and the protein components of the ribosome would be possible interaction partners.
Mutational studies
Four mutations in ecoNusG (W80R, L115P, F144Y and N145D) have been described and their effects on Nun- and N-mediated termination/antitermination investigated (Burova et al., 1999). L115P had no observable phenotype; the other three point mutations destroyed Nun-mediated termination at the λnutR site but had no effect on termination mediated by λnutL. The nucleic acid sequences responsible for this difference lay outside the boxA–boxB regions. The mutations did not influence N-dependent antitermination, suggesting that NusG participates differently in the two phage systems.
ecoNusG W80, L115, F144 and N145 correspond to aaeNusG residues L150, I184, F211 and T212, respectively (Figure 6B). Residue L150 in helix α2 is partially solvent accessible and participates in a hydrophobic cluster with P162, M164 and P169. The interaction links α2 to strands β12, β13 and β14, which constitute the ‘irregular’ fourth strand of the RNP-like motif. The latter group of strands is the core element in the S6-like nucleic acid binding model for NusG. Thus, the involvement of specific nucleic acid sequences or their spacing in the mutational phenotypes and the current structures are consistent with the W80R mutation disturbing a nucleic acid binding site in NusG.
Residues F211 and T212 reside in the KOW domain at the N-terminus of β17, well accessible in a shallow groove. They are not among the residues suggested to contact nucleic acids and the conservative changes are unlikely to have a profound influence on the structure of the domain. Together with the congruence of F211 and Y109 of tudor (see above), the findings argue for a protein binding site on domain III encompassing the F211/T212 region.
Residue I184 is located in the flexible linker between the RNP and KOW domains and can be replaced without functional consequences.
Conclusions
In order to fulfill its broad spectrum of regulatory functions, NusG has to interact with various other components of the gene expression machinery. Consistently, the crystal structures of aaeNusG portray the protein as a multi-domain assembly. A flexible connection between two portions of the molecule allows for conformational changes and could support concomitant interaction of NusG with other factors far apart or at variant positions in the complexes. Two universal domains resemble known nucleic acid interaction motifs and a general affinity for nucleic acids exists in NusG proteins. Nucleic acid binding may be occluded in the isolated proteins and become uncovered only in functional complexes. At the same time, the aaeNusG domains harbor potential protein–protein interaction surfaces and may therefore serve as protein–nucleic acid adaptors. A task for the future will be to attribute ligand specificities to the separate folds.
Materials and methods
Cloning, expression and purification
The nusG gene from A.aeolicus was PCR amplified by standard procedures from total genomic DNA and inserted into pET22b(+) through NdeI/XhoI restriction sites. This and all other constructs were verified by sequencing of the promoter and insert regions. Expression of aaeNusG was achieved by electroporation of E.coli BL21(DE3) RIL cells, propagation of the crude transformation in 100 ml of LB medium (100 µg/ml ampicillin, 34 µg/ml chloramphenicol), inoculation of 6 l of selective LB medium with the overnight culture, growth to an OD600 of 0.8 and induction with 1 mM IPTG at 30°C for 3 h. Cells were pelleted and resuspended in buffer A (50 mM Tris–HCl pH 7.0, 2 mM DTT). The suspension was stored at –70°C until lysis with lysozyme and sonication.
The clarified lysate (30 min at 78 000 g) was treated for 10 min at 70°C and, after removal of the precipitate, for the same time at 90°C. All subsequent steps occurred at 4°C. The supernatant fraction was applied to an SP–Sepharose column equilibrated with buffer A, washed and eluted with a linear gradient to buffer A plus 1 M ammonium sulfate (buffer B). Relevant fractions were identified via SDS–PAGE, pooled and adjusted to 1 M ammonium sulfate. The pool was chromatographed on a butyl–Sepharose column with a gradient from buffer B to buffer A. aaeNusG fractions were identified as before, pooled, concentrated via 10 kDa cut-off membranes and the buffer exchanged to 10 mM Tris–HCl pH 7.0 and 2 mM DTT (buffer C). The eluate was concentrated to 1 mM, corresponding to ∼30 mg/ml, aliquoted, shock-frozen in liquid nitrogen and stored at –70°C.
The gene for tmaNusG was cloned into pET22b(+) using T.maritima total genomic DNA and NdeI/XhoI restriction sites. Expression was performed in the same way as for aaeNusG and the protein purified according to the procedure of Liao et al. (1996).
The econusG gene was cloned into pET22b(+) via NdeI/BamHI restriction sites employing E.coli total genomic DNA. Expression was performed as for aaeNusG in BL21(DE3) cells. The protein was purified in the same fashion as aaeNusG, omitting the heat steps.
A mutant of aaenusG, encompassing only domain III (residues K185–I248), was cloned from total A.aeolicus genomic DNA into pET22b(+) via NdeI/BamHI restriction sites, inserting an ATG start codon. Expression was achieved as for the wild-type protein. For purification of domain III, the same heat treatment and column chromatographic procedures as for the full-length protein were conducted. Unlike the wild type, domain III passed the SP–Sepharose column during the application and washing steps.
Analyses in solution
CD spectra were recorded with a J-715 spectropolarimeter (JASCO Corp., Tokyo, Japan), with protein samples at 0.2 mg/ml in buffer C. The spectra were interpreted as a mixture of helix, sheet and random coil structures with the program CDNN (Bohm et al., 1992).
To investigate the oligomeric states of the NusG preparations, crosslinking reactions were conducted with 1 mM glutaraldehyde in 20 mM Tris–HCl pH 7.0 and 150 mM NaCl at room temperature. The reactions proceeded for 15 min with the proteins at 0.02 mM. Samples were analyzed on 12% SDS gels. A positive control was conducted with Methanococcus jannaschii MJ0158, a known dimeric molecule (monomer MR ∼ 42 000).
Sedimentation velocity runs were performed with 400 µl of aaeNusG in PBS at 1.1 mg/ml [Beckman (Fullerton, CA) Optima XL-I analytical ultracentrifuge, Ti60 rotor, standard 12 mm Epon double cell]. Sedimentation was at 35 000 r.p.m. for 8 h at 20°C with PBS as the reference. The sedimentation behavior was analyzed with the program UltraScan 5.0 (http://www.ultrascan.uthscsa.edu).
Analytical gel-filtration analyses were undertaken with a Superdex-75 PC 3.2/30 size exclusion column (2.4 ml gel bed, column dimensions 3.2 × 300 mm) on a SMART™ FPLC system (Amersham-Pharmacia, Uppsala, Sweden). Thyroglobulin (MR 670 000), bovine γ-globulin (MR 158 000), bovine serum albumin (MR 66 000), chicken ovalbumin (MR 44 000), carbonic anhydrase (MR 29 000), equine myoglobin (MR 17 500) and vitamin B12 (MR 1300) served as size standards. Blue dextran (MR ∼ 2 000 000) and water were taken to represent the excluded volume (V0) and the total liquid phase volume (Vt), respectively. The apparent MR values of the proteins were extracted from plots of log(MR) versus Kd [Kd = (Ve – V0)/(Vt – V0); Ve are observed elution volumes].
Nucleic acid binding assays
Nucleic acid binding by the NusG preparations was tested via analytical gel-filtration analyses. For a standard run, 10 µl of a 1 mM NusG solution were mixed with 50 µl of running buffer and 5 µl of a 4 mg/ml nucleic acid solution [E.coli 16 and 23S rRNA (3.0 µM each), EcoRI-linearized pUC19 plasmid DNA (2.3 µM dsDNA), a synthetic 64 nt ssDNA fragment (0.2 mM ssDNA) or a synthetic 64 bp dsDNA (0.1 mM dsDNA)]. After incubation at room temperature for 1 h, 50 µl of the samples were injected. The eluates were fractionated and analyzed on 15% SDS–polyacrylamide gels. Standard runs were in 20 mM Tris–HCl pH 7.0 and 50 mM NaCl. The rRNA complexation was also tested in 20 mM Tris–HCl pH 7.5, 8.0, 8.5 and 9.0, with 50 mM NaCl in each case, and in 20 mM Tris–HCl pH 7.0 supplemented with 100 or 150 mM NaCl. For comparison, the nucleic acid and protein species were similarly analyzed alone in each buffer system. As negative controls, two proteins that were employed as MR references, carbonic anhydrase and chicken ovalbumin, were similarly incubated with the nucleic acids and analyzed. Dextran sulfate (MR ∼ 500 000) at the same concentration as the nucleic acids was used to check binding of NusG to a negatively charged polysaccharide that does not carry heterocyclic bases.
Mixtures of aaeNusG, tmaNusG, ecoNusG and aaeNusG domain III with nucleic acids, as used in gel-filtration experiments, were also analyzed on 1% agarose gels. Gels were run at 4°C for several hours in 1× TBE pH 7.1, and afterwards stained with ethidium bromide.
To detect changes in the absorption at 256 nm upon aaeNusG binding, we monitored gel-filtration runs of the 64 bp dsDNA alone or in complex with the protein in 20 mM Tris–HCl pH 7.0, 50 mM NaCl and of the DNA alone in 1 M NaOH. Similarly, the protein alone was analyzed in 20 mM Tris–HCl pH 7.0 and 50 mM NaCl. Samples were mixed as before and 8 µl were applied to the column in order to work in the linear response region of the detector.
Crystallization and data collection
Crystallizability of aaeNusG was checked with sparse matrix set-ups by sitting drop vapor diffusion and led to two crystallization conditions at 18°C (crystal form I: 0.1 M MES/Tris pH 5.8, 10% PEG 4000, 20% 2-propanol, 0.1 M NaCl; crystal form II: 0.1 M MES/Tris pH 8.2, 15% PEG 3000, 0.2 M NaCl). Both crystal forms could be frozen at 100 K in a liquid nitrogen stream by incubation with 15% dl-2,3-butanediol or 30% glycerol, respectively.
We collected a data set to 1.95 Å resolution from a form I crystal (space group I222) at beamline BW6 of the Deutsches Elektronensynchrotron (Hamburg, Germany; Table I). For heavy atom derivatization, crystals were incubated for several hours in mother liquor supplemented with KAu(CN)2 (100 mM), HgCl2 (2 mM) or Ta6Br14 (saturated) and derivatives measured at 100 K with CuKα X-radiation [Rigaku (Tokyo, Japan) RU200 rotating anode; MAR-Research (Hamburg, Germany) image plate]. Data reduction for the derivatives preserved their anomalous scattering contributions. A single form II specimen (space group P21) yielded data complete to 2.0 Å resolution with rotating anode X-rays (Table I).
Structure solution and refinement
The structure of the orthorhombic crystal form was solved by multiple isomorphous replacement with anomalous scattering with programs from the CCP4 collection (CCP4, 1994). Heavy atom positions were extracted from isomorphous difference Patterson maps (RSPS) and checked for consistency in difference Fourier maps (FFT). Heavy atom positions were refined and the data phased to 2.4 Å resolution with MLPHARE. Initial phases were improved by solvent flattening (DM; 74% solvent content). A molecular model encompassing residues Q5–R186 could be built into the resulting experimental electron density map with MAIN (Turk, 1996; Figure 2A). The model was refined at 1.95 Å resolution with CNS (Brunger et al., 1998) according to standard strategies (Table I). Final rounds of refinement were undertaken in REFMAC5 (CCP4, 1994) employing anisotropic corrections to the temperature factors of the individual domains.
Assuming four copies of the full-length protein per asymmetric unit, a solvent content of 47% was calculated for the P21 crystal form. Consistently, four of the I222 models could be positioned in the P21 cell with MOLREP. After initial refinement, additional density corresponding to four C-terminal domains was clearly visible in the maps and guided fitting of residues G187–I248. Refinement was completed in CNS and REFMAC5 as before. No NCS restraints/constraints were applied during the refinement.
All experimental data were included in the refinement processes and the free R-factors (5% of the observed reflections) were continuously monitored (Table I). The structures have been deposited with the Protein Data Bank [http://www.rscb.org/pdb; accession Nos 1M1H (I222) and 1M1G (P21)].
Acknowledgments
Acknowledgements
M.C.W. was supported by a postdoctoral fellowship from the Peter and Traudl Engelhorn-Stiftung. We thank Dr Georg Wiegand for help with the ultracentrifugation studies and the interpretation of the results, Elisabeth Weyher for help with recording and interpreting of the CD spectra, Drs Gleb Bourenkov, Hans D.Bartunik, Hans Brandstetter and Stefan Steinbacher for help with synchrotron data collection, and Dr Max Gottesman for helpful discussions and critical reading of the manuscript.
References
- Ban N., Nissen,P., Hansen,J., Moore,P.B. and Steitz,T.A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science, 289, 905–920. [DOI] [PubMed] [Google Scholar]
- Bar-Nahum G. and Nudler,E. (2001) Isolation and characterization of σ70-retaining transcription elongation complexes from Escherichia coli. Cell, 106, 443–451. [DOI] [PubMed] [Google Scholar]
- Berg K.L., Squires,C. and Squires,C.L. (1989) Ribosomal RNA operon anti-termination. Function of leader and spacer region box B–box A sequences and their conservation in diverse micro-organisms. J. Mol. Biol., 209, 345–358. [DOI] [PubMed] [Google Scholar]
- Bohm G., Muhr,R. and Jaenicke,R. (1992) Quantitative analysis of protein far UV circular dichroism spectra by neuronal networks. Protein Eng., 5, 191–195. [DOI] [PubMed] [Google Scholar]
- Bork P., Holm,L. and Sander,C. (1994) The immunoglobulin fold. Structural classification, sequence patterns and common core. J. Mol. Biol., 242, 309–320. [DOI] [PubMed] [Google Scholar]
- Brunger A.T. et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Burns C.M., Richardson,L.V. and Richardson,J.P. (1998) Combinatorial effects of NusA and NusG on transcription elongation and Rho-dependent termination in Escherichia coli. J. Mol. Biol., 278, 307–316. [DOI] [PubMed] [Google Scholar]
- Burova E., Hung,S.C., Sagitov,V., Stitt,B.L. and Gottesman,M.E. (1995) Escherichia coli NusG protein stimulates transcription elongation rates in vivo and in vitro. J. Bacteriol., 177, 1388–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burova E., Hung,S.C., Chen,J., Court,D.L., Zhou,J.G., Mogilnitskiy,G. and Gottesman,M.E. (1999) Escherichia coli nusG mutations that block transcription termination by coliphage HK022 Nun protein. Mol. Microbiol., 31, 1783–1793. [DOI] [PubMed] [Google Scholar]
- Bylund G.O., Persson,B.C., Lundberg,L.A. and Wikstrom,P.M. (1997) A novel ribosome-associated protein is important for efficient translation in Escherichia coli. J. Bacteriol., 179, 4567–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Cramer P. (2002) Multisubunit RNA polymerases. Curr. Opin. Struct. Biol., 12, 89–97. [DOI] [PubMed] [Google Scholar]
- Friesen W.J. and Dreyfuss,G. (2000) Specific sequences of the Sm and Sm-like (Lsm) proteins mediate their interaction with the spinal muscular atrophy disease gene product (SMN). J. Biol. Chem., 275, 26370–26375. [DOI] [PubMed] [Google Scholar]
- Gopal B., Haire,L.F., Cox,R.A., Jo Colston,M., Major,S., Brannigan,J.A., Smerdon,S.J. and Dodson,G. (2000) The crystal structure of NusB from Mycobacterium tuberculosis. Nat. Struct. Biol., 7, 475–478. [DOI] [PubMed] [Google Scholar]
- Greenblatt J., Mah,T.F., Legault,P., Mogridge,J., Li,J. and Kay,L.E. (1998) Structure and mechanism in transcriptional antitermination by the bacteriophage λ N protein. Cold Spring Harb. Symp. Quant. Biol., 63, 327–336. [DOI] [PubMed] [Google Scholar]
- Housset D., Mazza,G., Gregoire,C., Piras,C., Malissen,B. and Fontecilla-Camps,J.C. (1997) The three-dimensional structure of a T-cell antigen receptor VαVβ heterodimer reveals a novel arrangement of the Vβ domain. EMBO J., 16, 4205–4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham C.J., Dennis,J. and Furneaux,P.A. (1999) Autogenous regulation of transcription termination factor Rho and the requirement for Nus factors in Bacillus subtilis. Mol. Microbiol., 31, 651–663. [DOI] [PubMed] [Google Scholar]
- Kyrpides N.C., Woese,C.R. and Ouzounis,C.A. (1996) KOW: a novel motif linking a bacterial transcription factor with ribosomal proteins. Trends Biochem. Sci., 21, 425–426. [DOI] [PubMed] [Google Scholar]
- Li J., Horwitz,R., McCracken,S. and Greenblatt,J. (1992) NusG, a new Escherichia coli elongation factor involved in transcriptional antitermination by the N protein of phage λ. J. Biol. Chem., 267, 6012–6019. [PubMed] [Google Scholar]
- Li J., Mason,S.W. and Greenblatt,J. (1993) Elongation factor NusG interacts with termination factor rho to regulate termination and antitermination of transcription. Genes Dev., 7, 161–172. [DOI] [PubMed] [Google Scholar]
- Liao D., Lurz,R., Dobrinski,B. and Dennis,P.P. (1996) A NusG-like protein from Thermotoga maritima binds to DNA and RNA. J. Bacteriol., 178, 4089–4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linn T. and Greenblatt,J. (1992) The NusA and NusG proteins of Escherichia coli increase the in vitro readthrough frequency of a transcriptional attenuator preceding the gene for the β subunit of RNA polymerase. J. Biol. Chem., 267, 1449–1454. [PubMed] [Google Scholar]
- Mah T.-F., Kuznedelov,K., Mushegian,A., Severinov,K. and Greenblatt,J. (2000) The α-subunit of E.coli RNA polymerase activates RNA binding by NusA. Genes Dev., 14, 2664–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason S.W. and Greenblatt,J. (1991) Assembly of transcription elongation complexes containing the N protein of phage λ and the Escherichia coli elongation factors NusA, NusB, NusG, and S10. Genes Dev., 5, 1504–1512. [DOI] [PubMed] [Google Scholar]
- Mooney R.A., Artsimovitch,I. and Landick,R. (1998) Information processing by RNA polymerase: recognition of regulatory signals during RNA chain elongation. J. Bacteriol., 180, 3265–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehrke K.W. and Platt,T. (1994) A quaternary transcription termination complex. Reciprocal stabilization by Rho factor and NusG protein. J. Mol. Biol., 243, 830–839. [DOI] [PubMed] [Google Scholar]
- Nehrke K.W., Zalatan,F. and Platt,T. (1993) NusG alters rho-dependent termination of transcription in vitro independent of kinetic coupling. Gene Expr., 3, 119–133. [PMC free article] [PubMed] [Google Scholar]
- Nodwell J.R. and Greenblatt,J. (1993) Recognition of boxA antiterminator RNA by the E. coli antitermination factors NusB and ribosomal protein S10. Cell, 72, 261–268. [DOI] [PubMed] [Google Scholar]
- Oubridge C., Ito,N., Evans,P.R., Teo,C.H. and Nagai,K. (1994) Crystal structure at 1.92 Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature, 372, 432–438. [DOI] [PubMed] [Google Scholar]
- Pasman Z. and von Hippel,P.H. (2000) Regulation of rho-dependent transcription termination by NusG is specific to the Escherichia coli elongation complex. Biochemistry, 39, 5573–5585. [DOI] [PubMed] [Google Scholar]
- Ponting C.P. (1997) Tudor domains in proteins that interact with RNA. Trends Biochem. Sci., 22, 51–52. [DOI] [PubMed] [Google Scholar]
- Schluenzen F. et al. (2000) Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell, 102, 615–623. [DOI] [PubMed] [Google Scholar]
- Schmidt M.C. and Chamberlin,M.J. (1984) Amplification and isolation of Escherichia coli nusA protein and studies of its effects on in vitro RNA chain elongation. Biochemistry, 23, 197–203. [DOI] [PubMed] [Google Scholar]
- Schmidt M.C. and Chamberlin,M.J. (1987) nusA protein of Escherichia coli is an efficient transcription termination factor for certain terminator sites. J. Mol. Biol., 195, 809–818. [DOI] [PubMed] [Google Scholar]
- Selenko P., Sprangers,R., Stier,G., Buhler,D., Fischer,U. and Sattler,M. (2001) SMN tudor domain structure and its interaction with the Sm proteins. Nat. Struct. Biol., 8, 27–31. [DOI] [PubMed] [Google Scholar]
- Squires C.L., Greenblatt,J., Li,J. and Condon,C. (1993) Ribosomal RNA antitermination in vitro: requirement for Nus factors and one or more unidentified cellular components. Proc. Natl Acad. Sci. USA, 90, 970–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk D. (1996) An interactive software for density modifications, model building, structure refinement and analysis. In Bourne,P.E. and Watenpaugh,K. (eds), Proceedings from the 1996 Meeting of the International Union of Crystallography Macromolecular Computing School.
- Varani G. and Nagai,K. (1998) RNA recognition by RNP proteins during RNA processing. Annu. Rev. Biophys. Biomol. Struct., 27, 407–445. [DOI] [PubMed] [Google Scholar]
- Vieille C. and Zeikus,G.J. (2001) Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev., 65, 1–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg R.A., Gottesman,M.E., Hendrix,R.M. and Little,J.W. (1999) Family values in the age of genomics: comparative analyses of temperate bacteriophage HK022. Annu. Rev. Genet., 33, 565–602. [DOI] [PubMed] [Google Scholar]
- Wimberly B.T., Brodersen,D.E., Clemons,W.M., Morgan-Warren,R.J., Carter,A.P., Vonrhein,C., Hartsch,T. and Ramakrishnan,V. (2000) Structure of the 30S ribosomal subunit. Nature, 407, 327–339. [DOI] [PubMed] [Google Scholar]
- Worbs M., Bourenkov,G.P., Bartunik,H.D., Huber,R. and Wahl,M.C. (2001) An extended RNA binding surface through arrayed S1 and KH domains in transcription factor NusA. Mol. Cell, 7, 1177–1189. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y., Narita,T., Inukai,N., Wada,T. and Handa,H. (2001) SPT genes: key players in the regulation of transcription, chromatin structure and other cellular processes. J. Biochem., 129, 185–191. [DOI] [PubMed] [Google Scholar]
- Zellars M. and Squires,C.L. (1999) Antiterminator-dependent modulation of transcription elongation rates by NusB and NusG. Mol. Microbiol., 32, 1296–1304. [DOI] [PubMed] [Google Scholar]
- Zorio D.A. and Bentley,D.L. (2001) Transcription elongation: the ‘foggy’ is lifting. Curr. Biol., 11, R144–R146. [DOI] [PubMed] [Google Scholar]