Abstract
Mammalian chromatin remodeling complexes are involved in both activation and repression of transcription. Here, we show that NoRC, a SNF2h- containing nucleolar chromatin remodeling complex, represses ribosomal gene transcription. NoRC-mediated rDNA silencing was alleviated by trichostatin A, indicating that histone deacetylation is causally involved in silencing. Chromatin immunoprecipitation experiments demonstrate that overexpression of TIP5, the large subunit of NoRC, mediates deacetylation of nucleosomes in the vicinity of the rDNA promoter. Protein–protein interaction assays reveal association of TIP5 with the histone deacetylase HDAC1 in vivo and in vitro. Deletion of the C-terminal PHD finger and bromodomain abolishes the interaction of TIP5 and HDAC1, and abrogates transcriptional repression. The results suggest that NoRC silences the rDNA locus by targeting the SIN3 corepressor complex to the rDNA promoter, thereby establishing a repressed chromatin structure.
Keywords: histone modification/RNA polymerase I/silencing/TIP5/transcription
Introduction
The nucleolus, the site of ribosome synthesis, is the most striking example of a correlation of specialized transcription and nuclear compartmentalization. In the mouse diploid genome, there are ∼400 ribosomal genes spread over a number of different chromosomes. The overall rDNA transcriptional activity of a given cell depends on the demand for protein synthesis and hence on metabolic activity. In eukaryotic cells, two types of ribosomal chromatin exist: one that contains nucleosomes and represents the inactive gene copies, and one that lacks nucleosomes and corresponds to the transcribed genes. The two states of chromatin are maintained independently of the transcriptional activity and are stably propagated through the cell cycle (Conconi et al., 1989). The mechanisms that maintain the ratio of active versus silent rDNA repeats are poorly understood, but different histone modifications have been demonstrated to be involved in the establishment of a euchromatic or heterochromatic gene structure (Strahl and Allis, 2000).
Previous in vitro studies revealed that transcription by RNA polymerase I (Pol I) on chromatin templates requires ATP-dependent nucleosome remodeling (Längst et al., 1997, 1998). In a search for remodeling complex(es) that have the potential to alter the chromatin structure of the rDNA promoter in vitro and in vivo, we have identified NoRC (nucleolar remodeling complex), a complex containing a >200 kDa subunit, TIP5 (TTF-I interacting protein #5), and SNF2h, the mammalian homolog of the ATPase ISWI (Strohner et al., 2001). NoRC induces nucleosome sliding in an ATP- and histone H4 tail-dependent fashion. This suggests that NoRC may alter the position of nucleosomes at the rDNA locus and/or modify the N-terminal tails of the core histones, creating a biochemical code that regulates transcription of chromatin templates.
The core histone tails contain a variety of covalent modifications, including acetylation, phosphorylation, methylation and ubiquitylation. Increasing evidence suggests that the different histone modifications are functionally interconnected. In general, hyperacetylated regions of chromatin frequently contain active transcription units, whereas hypoacetylated chromatin is transcriptionally silent. Transcriptional coactivators, such as Gcn5, p300/CBP, PCAF, TAFII250 and ACTR, acetylate the tails of histones H3 and H4, affect the mobility of nucleosomes and facilitate the access of specific DNA-binding proteins to nucleosomal DNA (Brown and Robinson, 2000; Sterner and Berger, 2000; Strahl and Allis, 2000; Turner, 2000; Roth et al., 2001). Likewise, histone deacetylases (HDACs) are active components of transcriptional corepressor complexes. In mammals, HDAC1 and HDAC2 are found in two distinct complexes, SIN3 and NuRD (Ayer, 1999; Ahringer, 2000). Both complexes are large molecular assemblies comprised of multiple components. The emerging view is that these complexes have specific developmental roles, rather than being required for general cellular functions (Pazin and Kadonaga, 1997; Kuzmichev and Reinberg, 2001).
The functions of the different members of mammalian ISWI/SNF2h-containing chromatin remodeling complexes are just beginning to be elucidated. Biochemical and molecular biological studies revealed that ISWI/SNF2h-containing complexes may play both a positive and a negative role in transcription regulation. In vitro, dNURF, ACF and hRSF were initially found to promote access of transcription factors to chromatin and activate transcription from chromatin templates (Ito et al., 1996; Mizuguchi et al., 1997; LeRoy et al., 1998; Kingston and Narlikar, 1999). However, the in vivo functions of individual remodeling machines are just beginning to be uncovered. Some complexes of the ISWI/SNF2h subfamily repress transcription in vivo (Goldmark et al., 2000; Fazzio et al., 2001). The WSTF–ISWI chromatin remodeling complex WICH is targeted to pericentromeric heterochromatin, suggesting that it may play a role in either the establishment or maintenance of silent chromatin (Bozhenok et al., 2002). Moreover, the fact that, in Drosophila, ISWI does not co-localize with RNA polymerase II indicates that the majority of ISWI is associated with inactive chromatin (Deuring et al., 2000). This suggests that ISWI/SNF2h complexes may serve a function in resetting the chromatin structure to a repressed ground state (Varga-Weisz, 2001).
The primary structure of TIP5 reveals the presence of several conserved sequence motifs, including a tandem PHD (plant homeodomain) finger and bromodomain. Tandem arrangement of PHD fingers and bromodomain motifs has defined a growing family of transcriptional corepressors, including KAP-1, TIF1α and TIF1γ (Le Douarin et al., 1995; Venturini et al., 1999; Schultz et al., 2001). All members of this protein family have been shown to repress transcription when tethered to DNA. This suggests that the PHD finger and bromodomain form a cooperative unit that plays a role in the establishment and/or maintenance of heterochromatin and transcriptional silencing. The finding that remodeling complexes can also cause previously exposed sites to become occluded (for a review, see Tyler and Kadonaga, 1999) prompted us to investigate the effect of NoRC on cellular rRNA synthesis. We found that overexpression of TIP5 inhibits Pol I transcription by recruiting a deacetylase complex containing HDAC1, mSin3A and RbAp46 to rDNA. The results suggest that targeted recruitment of the SIN3 corepressor complex leads to the deacetylation of promoter-bound nucleosomes and the formation of a chromatin structure that is incompatible with transcription initiation.
Results
Overexpression of TIP5 inhibits Pol I transcription in vivo
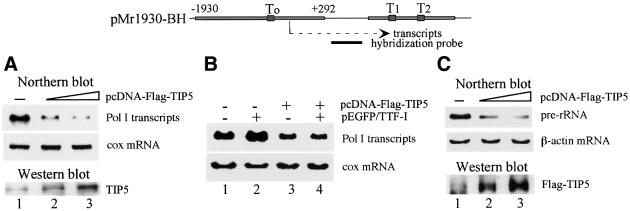
Previous in vitro data have revealed that ATP-dependent remodeling of nucleosomes at the Pol I transcription start site is required for transcription activation on preassembled chromatin templates (Längst et al., 1997, 1998). On the other hand, ISWI is a transcriptional repressor in yeast. To examine the effect of NoRC on Pol I transcription in vivo, we co-transfected NIH 3T3 cells with a ribosomal minigene reporter (pMr1930-BH) and increasing amounts of an expression vector (pcDNA-Flag-TIP5) encoding Flag-tagged TIP5, the large subunit of NoRC. Expression of TIP5 was visualized on immunoblots, and transcripts from the reporter plasmid were monitored on northern blots. Surprisingly, increasing amounts of TIP5 repressed transcription from the Pol I reporter (Figure 1A), whereas expression of a luciferase reporter plasmid was not affected (data not shown). The fact that overexpression of TIP5 abrogates transcription of the rDNA reporter plasmid suggests that NoRC acts as a repressor of Pol I transcription.
Fig. 1. Overexpression of TIP5 inhibits Pol I transcription in vivo. (A) TIP5 represses Pol I transcription in vivo. NIH 3T3 cells were transiently transfected with 2.5 µg of Pol I reporter (pMr1930-BH) and 2 µg (lane 2) or 4 µg (lane 3) of pcDNA-Flag-TIP5. Total RNA was extracted and Pol I transcripts were monitored on northern blots. As an internal control, the amount of cytochrome c oxidase (cox) mRNA was determined. Overexpression of TIP5 was visualized on western blots using α-TIP5 antibodies. A schematic representation of the Pol I reporter and the hybridization probe used in northern blots is shown above. (B) TIP5 counteracts transcription activation of TTF-I in vivo. NIH 3T3 cells were transfected with 2.5 µg of Pol I reporter, 2 µg of pEGFP/TTF-I and 4 µg of pcDNA-Flag-TIP5 as indicated. Total RNA was extracted and transcripts from the Pol I reporter were monitored on northern blots. To normalize for variations of RNA loading, the blot was also hybridized with a probe complementary to cox mRNA. (C) Overexpression of TIP5 inhibits cellular 45S pre-rRNA synthesis. 293T cells were transiently transfected with pcDNA-Flag-TIP5, and RNA was analyzed on northern blots using a riboprobe complementary to nucleotides 1–155 of human pre-rRNA. The blots were subsequently reprobed for actin mRNA. Overexpression of TIP5 was visualized on western blots using α-Flag antibodies.
Remodeling at the rDNA promoter is triggered by binding of the transcription termination factor TTF-I to the promoter-proximal terminator T0. As TIP5 interacts with TTF-I, we wondered whether TIP5 would synergize or antagonize TTF-I function in Pol I transcription. Con sistent with in vitro results demonstrating transcription activation by TTF-I on chromatin templates, co-transfection of an expression vector encoding EGFP-tagged TTF-I enhanced transcription from the Pol I reporter plasmid (Figure 1B, lane 2). Overexpression of TIP5, on the other hand, counteracted transcriptional activation by TTF-I (lane 4).
To examine whether NoRC affects transcription of the endogenous rDNA genes in a manner similar to that of the transfected reporter plasmid, we assayed the effect of overexpression of TIP5 on cellular 45S pre-rRNA synthesis. For this, 293T cells, which can be more efficiently transfected than NIH 3T3 cells, were used. In the experiment shown in Figure 1C, >90% of the 293T cells were transfected with pcDNA-Flag-TIP5 and the level of 45S pre-rRNA was monitored on northern blots. Consistent with NoRC repressing Pol I transcription, overexpression of TIP5 drastically decreased cellular pre-rRNA synthesis (Figure 1C, lanes 2 and 3). Thus, repression of Pol I transcription by overexpression of TIP5 is not limited to transiently transfected rDNA reporter genes: the endogenous rDNA genes are affected in a similar way.
Transcriptional repression by TIP5 is mediated by histone deacetylation
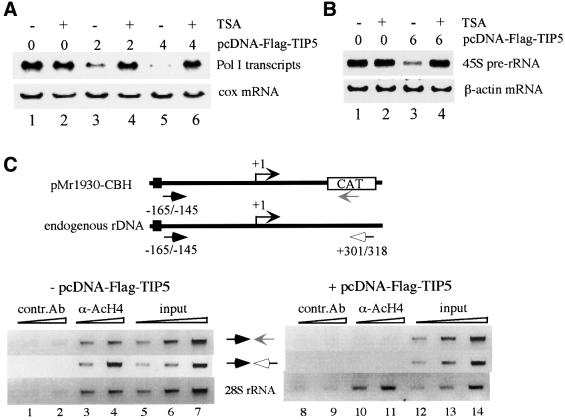
The C-terminus of TIP5 contains a tandem PHD finger and bromodomain, a motif that has defined an emerging family of transcriptional repressors. The PHD finger and bromodomain have been shown to form a cooperative unit that targets HDAC complexes to specific gene promoters in vivo (Le Douarin et al., 1995; Venturini et al., 1999; Schultz et al., 2001). Histone deacetylation is linked with transcriptionally silent chromatin states (for reviews, see Ng and Bird, 2000; Jenuwein and Allis, 2001). To gain insight into the mechanism by which NoRC represses Pol I transcription, we investigated whether deacetylation of histones may be causally involved in rDNA silencing. First, we assayed the effect of the deacetylase inhibitor trichostatin A (TSA) on TIP5-mediated transcriptional repression. Consistent with previous data (Muth et al., 2001), transcription of the Pol I reporter plasmid was not affected by TSA treatment (Figure 2A, lanes 1 and 2). Strikingly, repression by TIP5 was alleviated in the presence of TSA (lanes 3–6), indicating that TIP5-mediated inhibition of rDNA transcription is brought about by histone deacetylation. Again, overexpression of TIP5 repressed transcription of both the transfected reporter plasmid and the endogenous rDNA genes, and inhibition of endogenous Pol I transcription was overcome by TSA treatment (Figure 2B, lanes 3 and 4).
Fig. 2. Transcription repression by TIP5 is mediated by histone deacetylation. (A) TSA counteracts TIP5-mediated repression of Pol I transcription. NIH 3T3 cells were co-transfected with 2.5 µg of Pol I reporter (pMr1930-BH) and 2 µg (lanes 3 and 4) or 4 µg (lanes 5 and 6) of pcDNA-Flag-TIP5. Where indicated, TSA (33 nM) was added 24 h after transfection, and cells were cultured for a further 24 h. Transcripts from the Pol I reporter and endogenous cox gene were monitored on northern blots. (B) TIP5-mediated repression of endogenous rDNA transcription is alleviated by TSA. 293T cells were transfected with 6 µg of pcDNA-Flag-TIP5 and cultured in the absence or presence of 33 nM TSA as indicated. 45S pre-rRNA synthesis was monitored on northern blots. (C) ChIP assay. NIH 3T3 cells were co-transfected with 2.5 µg of Pol I reporter (pMr1930-CBH) in the absence (lanes 1–7) or presence (lanes 8–14) of 4 µg of pcDNA-Flag-TIP5. After formaldehyde cross-linking, cells were sonicated and soluble chromatin was immunoprecipitated with anti-AcH4 (lanes 3, 4, 10 and 11) or anti-myc antibodies (lanes 1, 2, 8 and 9). Different amounts of precipitated DNA were amplified by PCR. Primers were used that amplify either the promoter of the reporter plasmid or the endogenous rDNA (as indicated in the scheme above) or part of the 28S rRNA coding region (lower). Quantitative PCR analysis was also performed before immunoprecipitation to ensure equal transfection efficiencies (input).
NoRC-dependent sliding of nucleosomes requires the tail of histone H4 (Strohner et al., 2001). As histones are the major target of TSA-dependent HDACs, we studied whether overexpression of TIP5 causes deacetylation of histone H4 at the rDNA promoter. To test this, chromatin immunoprecipitation (ChIP) experiments were carried out. NIH 3T3 cells were transfected with the Pol I reporter plasmid pMr1930-CBH (Santoro and Grummt, 2001) in the absence or presence of pcDNA-Flag-TIP5, fixed with formaldehyde, and soluble chromatin was immunoprecipitated with either antibodies against acetylated histone H4 (α-AcH4) or control antibodies (α-myc). To monitor histone acetylation at the reporter plasmid, precipitated DNA was amplified with a pair of primers that hybridize to rDNA promoter sequences (–165/–145) and the CAT marker gene. Co-precipitated endogenous rDNA was analyzed by PCR using primers that amplify either the rDNA promoter or the 28S rRNA coding region (Figure 2C). In the absence of exogenous TIP5, antibodies against α-AcH4 co-precipitated the promoter of the Pol I reporter gene, the endogenous rDNA promoter and the 28S RNA coding region (Figure 2C, lanes 3 and 4). This demonstrates that a significant fraction of nucleosomes associated with exogenous and endogenous rDNA was acetylated at histone H4. Overexpression of TIP5 reduced the amount of exogenous and endogenous promoter DNA that was immunoprecipitated with α-AcH4 antibodies to background levels (Figure 2C, lanes 10 and 11). This demonstrates that transcriptional repression by TIP5 correlates with histone deacetylation. Strikingly, the acetylation status of histones associated with the 28S RNA coding region remained unaffected by overexpression of TIP5 (Figure 2C, lower, lanes 10 and 11), indicating that TIP5-mediated histone deacetylation is restricted to the 5′-end of the ribosomal genes and does not spread into the transcribed region.
TIP5 interacts with HDAC1 in vivo and in vitro
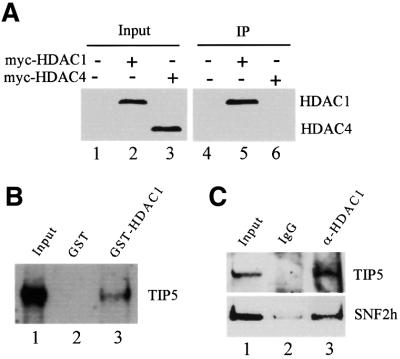
The observation that overexpression of TIP5 results in deacetylation of histones at the ribosomal gene promoter and TIP5-mediated transcriptional inhibition can be overcome by TSA suggests that NoRC represses Pol I transcription by recruiting HDAC(s) to the rDNA locus. To gain insight into which HDAC(s) may deacetylate nucleosomes at the rDNA promoter, we co-transfected 293T cells with Flag-tagged TIP5 and myc-tagged HDAC1 or HDAC4. After immunoprecipitation with anti-Flag antibodies, the interaction of TIP5 and HDAC1 and HDAC4 was monitored on immunoblots using α-myc antibodies. As shown in Figure 3A, a significant amount of HDAC1 was associated with TIP5 (lane 5). In contrast, no interaction of TIP5 and HDAC4 was observed (lane 6). Thus, TIP5 appears to specifically interact with class I but not class II HDACs.
Fig. 3. TIP5 interacts with HDAC1 in vitro and in vivo. (A) HDAC1 but not HDAC4 co-immunoprecipitates with TIP5. pcDNA-myc-HDAC1 or pcDNA-myc-HDAC4 was co-transfected with pcDNA-Flag-TIP5 into 293T cells. Cells were lysed and NoRC was precipitated with anti-Flag antibodies. Co-immunoprecipitated HDAC1 and HDAC4 were monitored on western blots with α-myc antibodies. The amount of HDAC1 and HDAC4 present in 5% of the cell lysates is shown in lanes 1–3. (B) TIP5 interacts with HDAC1. A total of 100 µl of a partially purified nuclear extract (DEAE-280 fraction) were applied to 10 µl of glutathione–Sepharose beads containing equivalent amounts of immobilized GST (lane 2) or GST–HDAC1 (lane 3). After washing with buffer AM-300, bound proteins (50%) were analyzed on western blots using anti-TIP5 antibodies. Ten percent of input of the DEAE-280 fraction is shown in lane 1. (C) NoRC interacts with HDAC1 in vivo. Pre-immune serum (lane 2) or anti-HDAC1 antibodies (lane 3) were bound to protein G–agarose, and 10 µl of beads were incubated with 100 µl of DEAE-280 fraction at 4°C for 4 h. Captured NoRC complex was analyzed on western blots using anti-TIP5 and SNF2h antibodies. Ten percent of the DEAE-280 fraction (lane 1) and 50% of bead-bound proteins are shown in lanes 2 and 3.
To examine whether cellular NoRC, rather than TIP5 alone, interacts with HDAC1, GST pull-down experiments were performed. Immobilized GST and GST–HDAC1 were incubated with a NoRC-containing protein fraction (DEAE-280; Schnapp and Grummt, 1996), and captured proteins were analyzed on immunoblots. Consistent with the results above, TIP5 was retained by GST–HDAC1 but not GST alone (Figure 3B). To verify an in vivo interaction of NoRC and HDAC1, we precipitated HDAC1 from the DEAE-280 fraction and monitored co-precipitated TIP5 and SNF2h. As shown in Figure 3C, both proteins were precipitated with anti-HDAC1 antibodies but not pre-immune IgGs (lanes 2 and 3). This result demonstrates a physical association of NoRC with HDAC1 in vivo.
The PHD-finger/bromodomain motif of TIP5 interacts with HDAC1
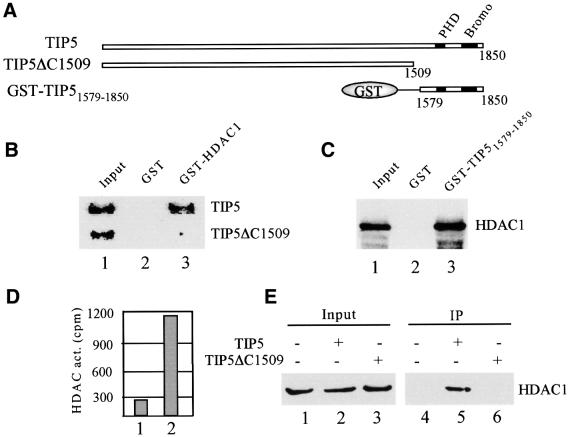
The tandem arrangement of PHD finger and bromodomain has been implicated in transcriptional repression by the KAP-1 corepressor (Schultz et al., 2001 and references therein). Therefore, we tested whether this C-terminal motif mediates the interaction of TIP5 and HDAC1. We compared binding of HDAC1 with full-length TIP5 and TIP5ΔC1509, a C-terminal deletion mutant lacking the PHD finger and bromodomain. As shown in Figure 4B, full-length TIP5 but not TIP5ΔC1509 interacted with immobilized GST–HDAC1, underscoring the involvement of the C-terminus in the interaction of TIP5 and HDAC1. Moreover, a GST fusion protein containing amino acids 1579–1850 (GST–TIP51579–1850) efficiently captured 35S-labeled HDAC1 (Figure 4C), demonstrating that the C-terminal fragment harboring the PHD finger and bromodomain is sufficient for the interaction of TIP5 and HDAC1. If the immobilized fusion protein was incubated with mouse nuclear extracts, HDAC activity was found to be retained by GST–TIP51579–1850 but not GST alone (Figure 4D).
Fig. 4. The C-terminal part of TIP5 interacts with HDAC1. (A) Schematic representation of TIP5, TIP5ΔC1509 and GST–TIP51579–1850. The position of the PHD finger and bromodomain is indicated. (B) HDAC1 interacts with the C-terminal region of TIP5. A total of 10 µl of immobilized GST (lane 2) or GST–HDAC1 (lane 3) were incubated with 5 µl of 35S-labeled TIP5 and TIP5ΔC1509 in 200 µl of buffer AM-100. Bound proteins (50%) were analyzed by electrophoresis and autoradiography. Ten percent of input proteins are shown in lane 1. (C) The C-terminal part of TIP5 including the PHD finger and bromodomain interacts with HDAC1. A total of 10 µl of bead-bound GST (lane 2) or GST–TIP51579–1850 (lane 3) were incubated with 5 µl of 35S-labeled HDAC1, and captured HDAC1 was analyzed by 10% SDS–PAGE and autoradiography. Ten percent of input is shown in lane 1. (D) The C-terminal part of TIP5 recruits HDAC activity. Bead-bound GST and GST–TIP51579–1850 were incubated with 200 µl of nuclear extracts for 4 h at 4°C. Washed beads were assayed for the release of [3H]acetate (c.p.m.) from 3H-acetylated histones. (E) Interaction of TIP5 and HDAC1 in vivo. 293T cells were co-transfected with 4 µg of pcDNA-myc-HDAC1 and 8 µg of pcDNA-Flag-TIP5 or pcDNA-Flag-TIP5ΔC1509. TIP5-containing protein complexes were precipitated with α-Flag antibodies, and the presence of HDAC1 in the immunoprecipitates was analyzed on western blots with α-myc antibodies. Five percent of input is shown in lanes 1–3.
The interaction of HDAC1 and the C-terminal part of TIP5 was verified by co-immunoprecipitation experiments. HDAC1 and either Flag-tagged TIP5 or TIP5ΔC1509 were overexpressed in 293T cells and precipitated with anti-Flag antibodies, and the immunoprecipitates were probed for HDAC1. As shown in Figure 4E, full-length TIP5 but not TIP5ΔC1509 was found to be associated with HDAC1 (lanes 5 and 6). Thus, the PHD finger and bromodomain mediate the interaction of TIP5 and HDAC1 in vivo and in vitro.
NoRC recruits the SIN3 corepressor complex to the rDNA promoter
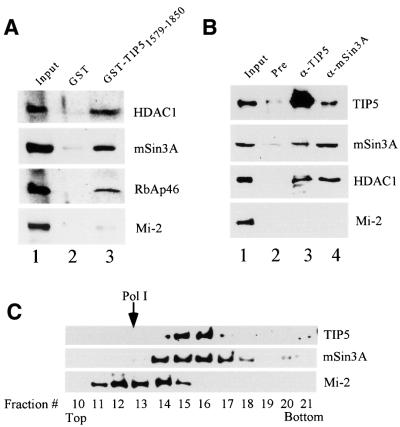
Macromolecular complexes with HDAC activity have been implicated in chromatin dynamics and transcriptional silencing. In mammals, two major HDAC1/2-containing corepressor complexes have been biochemically characterized: SIN3 and NuRD. Both corepressor complexes share four polypeptides, HDAC1, HDAC2, RbAp46 and RbAp48 (Ayer, 1999; Ahringer, 2000), but differ in the composition of other subunits. The different subunit composition of the SIN3 and NuRD complexes suggests that they participate in distinct biological functions. To analyze whether NoRC preferentially interacts with SIN3 or NuRD, GST–TIP51579–1850 was incubated with a fractionated nuclear extract (DEAE-280 fraction), and captured proteins were analyzed on western blots. As shown in Figure 5A, HDAC1, RbAp46 and mSin3A, but not Mi-2, were found to be associated with the GST– TIP51579–1850 fusion protein, indicating that NoRC interacts with the SIN3 HDAC complex.
Fig. 5. NoRC interacts with the SIN3 deacetylase complex. (A) TIP5 interacts with the SIN3 complex in vitro. A total of 100 µl of a DEAE-280 fraction were incubated with 10 µl of immobilized GST (lane 2) or GST–TIP51579–1850 (lane 3). After washing with buffer AM-300, bound proteins were analyzed on western blots using α-HDAC1, α-mSin3A, α-RbAp46 and α-Mi-2 antibodies. Ten percent of input is shown in lane 1. (B) In vivo association of NoRC with the SIN3 complex. A total of 100 µl of a DEAE-280 fraction were incubated with pre-immune serum (lane 2), α-TIP5 (lane 3) and α-mSin3A antibodies (lane 4) bound to protein G–agarose. After washing with buffer AM-200, 50% of bead-bound proteins were analyzed on western blots using antibodies against HDAC1, TIP5, mSin3A and Mi-2. Ten percent of input proteins are shown in lane 1. (C) Co-sedimentation of NoRC with cellular SIN3. A total of 250 µl of a DEAE-280 fraction were centrifuged through a 12.5–30% glycerol gradient. Fractions (150 µl) were precipitated with trichloroacetic acid and analyzed on immunoblots using α-TIP5, α-Sin3A and α-Mi-2 antibodies. The position of Pol I (∼600 kDa) is marked by an arrow.
The interaction of TIP5 and the SIN3 corepressor was also demonstrated by immunoprecipitation experiments (Figure 5B). The DEAE-280 fraction was incubated with immobilized control IgGs (pre), α-TIP5 and α-mSin3A antibodies, respectively, and bead-bound TIP5, mSin3A, HDAC1 and Mi-2 were monitored on immunoblots. Consistent with the results above, HDAC1 was co-immunoprecipitated with both α-TIP5 and α-mSin3A antibodies, demonstrating a physical association of the NoRC and the SIN3 complex. On the other hand, Mi-2, a unique subunit of the NuRD complex, was not detected in the immunoprecipitates. Thus, NoRC targets the SIN3 complex but not the NuRD corepressor complex.
Given that NoRC interacts with the SIN3 complex, a subpopulation of both cellular complexes should be associated in cell extracts. To test this, we fractionated whole-cell extracts on DEAE–Sepharose and by glycerol gradient centrifugation, and probed the presence of TIP5 and mSin3A on western blots (Figure 5C). Consistent with previous gel filtration data (Strohner et al., 2001), TIP5 was detected in fractions corresponding to a molecular weight >800 kDa. It is noteworthy that the TIP5-containing fractions (14–17) also contain mSin3A, indicating that at least part of the cellular SIN3 complex is associated with NoRC in a large multiprotein complex. Mi-2-containing protein complexes, on the other hand, did not co-sediment with TIP5, underscoring the association of NoRC with the SIN3 complex but not the NuRD complex.
Deletion of the C-terminal PHD-finger/bromodomain motif converts TIP5 into a transcriptional activator
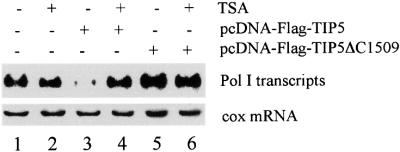
In order to evaluate the role of TIP5-mediated recruitment of HDAC1 in the repression of rDNA transcription, we chose to disrupt interactions of TIP5 and HDAC1 by overexpressing the deletion mutant TIP5ΔC1509 in NIH 3T3 cells and monitored transcription of the co-transfected Pol I reporter plasmid in the absence and presence of TSA (Figure 6). Treatment with TSA did not affect transcription of the Pol I reporter plasmid (lanes 1 and 2). Consistent with the results in Figure 2, full-length TIP5 repressed transcription and repression was alleviated by TSA (lanes 3 and 4). Surprisingly, overexpression of TIP5ΔC1509 activated Pol I transcription. A 3- to 4-fold stimulation of the reporter construct was observed at optimal concentrations of pcDNA-TIP5ΔC1509 (cf. lanes 1 and 5). Thus, C-terminally truncated TIP5 lacking the PHD finger and bromodomain augments rDNA transcription via a dominant-negative mechanism.
Fig. 6. Overexpression of a TIP5 C-terminal deletion mutant enhances rDNA transcription. NIH 3T3 cells were transfected with 2.5 µg of Pol I reporter (pMr1930-BH) and 4 µg of either pcDNA-Flag-TIP5 or pcDNA-Flag-TIP5ΔC1509. Where indicated, cells were treated with TSA (33 nM) for 24 h. Pol I transcripts and cox mRNA were visualized on northern blots.
Discussion
In this study, we investigated the role of NoRC, a SNF2h-containing nucleolar remodeling complex, in ribosomal gene transcription. Previous experiments have demonstrated that Pol I transcription on chromatin templates requires ATP-dependent remodeling of nucleosomes at the rDNA promoter triggered by TTF-I (Längst et al., 1997, 1998). Relatives of NoRC, i.e. dNURF, ACF and hRSF, activate transcription from chromatin templates in vitro (Ito et al., 1996; Mizuguchi et al., 1997; LeRoy et al., 1998). Like other members of ISWI/SNF2-containing complexes, NoRC induces nucleosome sliding in an ATP- and histone H4 tail-dependent fashion (Strohner et al., 2001). Moreover, the large subunit of NoRC, TIP5, interacts with TTF-I. This suggested a stepwise model of Pol I transcription activation, including recruitment of NoRC to the rDNA promoter, translational repositioning of nucleosomes and pre-initiation complex formation. Surprisingly, we found that overexpression of TIP5 repressed ribosomal gene transcription in a concentration-dependent manner, indicating that NoRC exerts a repressive rather than a stimulatory effect on rDNA transcription.
The in vivo functions of mammalian chromatin remodeling complexes are just beginning to be elucidated. Although the role of the central ATPase subunit is well established, very little is known about the role of the remaining subunits. In principle, they can modulate the activity of the ATPase subunit or target the remodeling complex to specific promoters. The current view is that activators that bind to their recognition sites in chromatin recruit a remodeling complex to the promoter, which, in turn, facilitates the binding of other factors that cannot bind directly to chromatin. Different members of SNF2-like nucleosome remodeling complexes have been shown to affect chromatin dynamics and transcription in diverse ways. Most in vitro studies revealed an activating effect of remodeling machines on transcription, indicating that chromatin remodeling is required to facilitate the access of transcription factors to their target sites. However, recent studies revealed that both SWI/SNF- and ISWI/SNF2-containing complexes are involved in transcriptional repression in vivo (Goldmark et al., 2000; Fazzio et al., 2001). Drosophila ISWI and RNA polymerase II do not co-localize on polytene chromosomes, indicating that ISWI is not associated with active genes in vivo (Deuring et al., 2000). Moreover, ACF, CHRAC and WICH are targeted to heterochromatin, suggesting that they play a role in gene silencing (Varga-Weisz, 2001).
Gene silencing and heterochromatin are often associated with repetitive DNA sequences and may be involved in stabilizing such sequences. In eukaryotic cells, two distinct chromatin structures have been observed at the rDNA locus. A fraction of transcriptionally active genes are in an open configuration and accessible to the cross-linking agent psoralen, whereas inactive rDNA repeats exhibit a more compact chromatin structure (Conconi et al., 1989). Apparently, epigenetic control mechanisms maintain ‘active’ transcription units in a euchromatic conformation and ‘inactive’ genes in a heterochromatic conformation. The results of this study suggest that NoRC plays a central role in establishing the inactive state of rRNA genes. NoRC has been found to target HDAC activity to the rDNA promoter. Histones are subject to a diverse array of post-translational modifications at their N-termini that dictate transitions between transcriptionally active and silent chromatin states. Histone acetylation, which is thought to loosen chromatin structure, is generally correlated with increased transcriptional activity (Mizzen and Allis, 1998; Kingston and Narlikar, 1999), whereas hypoacetylation is associated with gene silencing (Grunstein, 1997; Struhl, 1998). Deacetylation of nucleosomes, in turn, serves as a mark for the recruitment of proteins that methylate histone tails. Consistent with a functional link between histone deacetylation and methylation, recent ChIP experiments have revealed that nucleosomes at the promoter of silent ribosomal genes are methylated at lysine 9 of histone H3 (Santoro et al., 2002). Moreover, silent rDNA genes have been shown to be methylated at CpG residues (Santoro and Grummt, 2001). These observations are not only consistent with the ‘histone code’ hypothesis, which implies a complex interplay of different histone modifications (Strahl and Allis, 2000), but also suggest a link between DNA methylation and covalent modifications of the core histone tails. Apparently, there is a functional cross-talk between transcription factors, ATP-dependent chromatin remodelers, histone modifiers and DNA methyltransferases to either maintain an inactive chromatin state or convert an accessible chromatin conformation into an inaccessible structure.
Previous studies suggested that some of the rDNA repeats were associated with acetylated histones and that these repeats did not exhibit a nucleosomal structure (Mutskov et al., 1996). Moreover, Hirschler-Laszkiewicz et al. (2001) observed stimulation of rDNA transcription after treating NIH 3T3 cells with TSA, suggesting that acetylation of either histones or UBF may activate rRNA synthesis. Although we did not observe activation of Pol I transcription by TSA treatment (Muth et al., 2001; this study), our data support the view that the acetylation state of histones affects rDNA transcription. NoRC-mediated targeting of the SIN3 corepressor complex results in hypoacetylation of histones and transcriptional repression. SIN3 is a complex of HDAC1/2, RbAp46, RbAp48, Sin3A, SAP30, SAP18 and several other polypeptides, including the methyl CpG-binding protein MeCP2, the Rb-binding protein RBP1 and the corepressors NCoR and SMRT (Knoepfler and Eisenman, 1999). After fractionation of nuclear extracts on DEAE–Sepharose and glycerol gradient centrifugation, we observed association of NoRC with SIN3. However, we did not detect SIN3 subunits in immunopurified NoRC. Immunopurified NoRC contained TIP5, SNF2h and two additional proteins, p80 and p50 (Strohner et al., 2001). Subsequent identification of p80 and p50 by mass spectrometry revealed that these two proteins were neither genuine subunits of NoRC nor NoRC-associated proteins. Thus, like other members of ISWI/SNF2 chromatin remodeling factors, NoRC consists of two subunits, TIP5 and SNF2h. The absence of SIN3 in purified NoRC indicates that the association of SIN3 with NoRC is not stable enough to tolerate stringent purification.
Examples of gene silencing by targeted recruitment of chromatin remodelers and HDAC complexes exist both in vivo and in vitro, from yeast to man. In Saccharomyces cerevisiae, a member of the ISWI class of ATP-dependent chromatin remodeling factors, the Isw2 complex, negatively regulates transcription of early meiotic genes during mitotic growth. The repressor function of the Isw2 complex depends on Ume6p, which recruits the Sin3– Rpd3 HDAC complex to specific promoters. As a consequence, histones in the vicinity of the Ume6p binding site are deacetylated, a nuclease-inaccessible chromatin structure is established and transcription is repressed (Goldmark et al., 2000; Fazzio et al., 2001). Another example demonstrating the generality of transcription repressor-mediated recruitment is the targeting of the NuRD complex to regions of heterochromatin. Upon T-cell activation, the DNA-binding protein Ikaros recruits NuRD, a complex containing the ATP-dependent chromatin remodeler Mi-2 and HDAC activity (Kim et al., 1999; Kingston and Narlikar, 1999). This recruitment either maintains an inactive chromatin state or converts an accessible chromatin conformation into an inaccessible structure. The current view is that DNA-binding regulatory factors directly target ATP-dependent remodelers and chromatin-modifying complexes, such as HAT and HDAC complexes, to specific locations. Thus, the ‘division of labor’ of different chromatin remodeling and modifying complexes causes the establishment of the active or silent state of a given gene. Consistent with this, silencing of the rDNA locus requires NoRC, the nucleolus-specific remodeling machine, to recruit the SIN3 corepressor complex to the rDNA promoter. This finding indicates that the principles of epigenetic control are not restricted to genes transcribed by RNA polymerase II but are also used by other classes of RNA polymerases.
We postulate the following model for the formation of ‘inactive’ ribosomal chromatin. First, TTF-I bound to the promoter-proximal terminator T0 recruits NoRC to rDNA (Längst et al., 1997; Strohner et al., 2001). Secondly, NoRC induces repositioning of nucleosomes at the rDNA promoter and targets the SIN3 corepressor complex to ribosomal genes. Thirdly, histone tails are deacetylated and a repressive chromatin conformation is established. Thus, NoRC serves at least two functions in silencing rDNA transcription: (i) as a remodeling complex that positions the nucleosome at the rDNA promoter; and (ii) as a scaffold that coordinates the activities of macromolecular complexes that modify chromatin structure. The finding that NoRC links two chromatin-modifying activities, i.e. ATP-dependent nucleosome remodeling and targeting corepressor(s) to rDNA, suggests that the accessibility and transcriptional competence of individual rDNA gene copies are regulated at an epigenetic level. An important question is whether NoRC functions in de novo establishment or maintenance of rDNA silencing. What also remains to be investigated is which remodeling complex(es) is involved in the formation of the open chromatin structure that characterizes active rRNA genes. With the purification of NoRC and HDAC complexes, as well as the development of chromatin-based in vitro transcription systems, the tools are now available to address these issues.
Materials and methods
Plasmids
The cloning of full-length murine TIP5 has been described previously (Strohner et al., 2001). The C-terminal deletion mutant TIP5ΔC1509 was generated by excision of the EcoRV–XhoI fragment. A C-terminal fragment harboring amino acids 1579–1850 was inserted into pGEX (Pharmacia) to yield pGST-TIP51579–1850. pEGFP/TTF-I was cloned by insertion of full-length mTTF-I into pEGFP-C1 (Clontech). The reporter plasmid pMr1930-BH (Budde and Grummt, 1999) contains a 5′-terminal mouse rDNA fragment (from –1930 to +292) fused to a 3′-terminal rDNA fragment including two ‘Sal box’ terminator elements. pcDNA-myc-HDAC1 and pcDNA-myc-HDAC4 (Brehm et al., 1999; Miska et al., 1999) were provided by T.Kouzarides.
Cell culture, transient transfections and RNA analysis
NIH 3T3 and 293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum. Cells were transfected with 2.5 µg of pMr1930-BH and different amounts of expression vectors encoding TIP5 (pcDNA-Flag-TIP5, pcDNA-Flag-TIP5ΔC1509) and TTF-I (pEGFP/TTF-I). Total RNA was isolated after 48 h and analyzed on northern blots as described previously (Voit et al., 1999). To normalize for differences in RNA loading, the filter was also hybridized with a riboprobe complementary to cytochrome c oxidase or actin mRNA.
GST pull-down assays
GST fusion proteins were expressed in Escherichia coli BL21(DE3) pLysS and purified on glutathione–Sepharose. Ten microliters of beads were pre-incubated at room temperature in 200 µl of buffer AM-100 (100 mM KCl, 20 mM Tris–HCl pH 7.9, 5 mM MgCl2, 0.1 mM EDTA, 10% glycerol, 0.5 mM dithioerythritol, 0.5 mM PSMF) containing 2 mg/ml bovine serum albumin (BSA). After 30 min, the beads were incubated for 60 min at room temperature with in vitro translated 35S-labeled proteins or a fractionated nuclear extract (DEAE-280 fraction) and washed with buffer AM-300, and bound proteins were analyzed by SDS–PAGE followed by autoradiography or western blotting.
Immunoprecipitation assays
Anti-HDAC1 (Upstate), anti-TIP5 (Strohner et al., 2001) and anti-mSin3A (Santa Cruz) antibodies were bound to protein G–agarose beads, blocked with 2 mg/ml BSA and incubated for 4 h with a murine DEAE-280 fraction at 4°C. After washing with buffer AM-300, bound proteins were analyzed on western blots. To monitor the interaction of TIP5 and HDAC in vivo, 293T cells were transfected with expression vectors encoding the respective proteins (i.e. pcDNA-Flag-TIP5, pcDNA-Flag-TIP5ΔC1509, pcDNA-myc-HDAC1 and pcDNA-myc-HDAC4). After 48 h, the cells were lysed in 500 µl of CoIP buffer (50 mM Tris–HCl pH 8.0, 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, 0.1 mM PMSF) at 4°C for 30 min. The cleared lysate was subjected to immunoprecipitation for 4 h at 4°C using immobilized anti-Flag antibodies (M2 beads; Sigma). Precipitates were washed four times with CoIP buffer, separated on 8% SDS–polyacrylamide gels and analyzed on western blots.
Glycerol gradient centrifugation
A total of 250 µl of a fractionated mouse nuclear extract (DEAE-280 fraction) were layered on top of a 3.75 ml of 12.5–30% glycerol gradient in buffer AM-100. Samples were centrifuged at 4°C for 16 h at 45 000 r.p.m. in a SW60 rotor. Fractions of 150 µl were collected, precipitated with trichloroacetic acid and analyzed on immunoblots.
ChIP experiments
ChIP experiments were performed as described previously (Santoro and Grummt, 2001). Briefly, 4 × 105 NIH 3T3 cells were co-transfected with 1 µg of Pol I reporter (pMr1930-CBH) and 4 µg of pcDNA-Flag-TIP5. After 48 h, cells were fixed for 15 min with 1% formaldehyde, suspended in 200 µl of 1% SDS, 10 mM EDTA, 50 mM Tris–HCl pH 8.1, and sonicated to yield 0.5–1 kb DNA fragments. Chromatin was diluted 10-fold with IP buffer (16.7 mM Tris–HCl pH 8.1, 167 mM NaCl, 1.2 mM EDTA, 0.01% SDS, 1.1% Triton X-100), pre-cleared for 1 h at 4°C on protein G–agarose in the presence of 20 mg/ml sonicated salmon sperm DNA and immunoprecipitated overnight with antibodies against α-AcH4 (Upstate). After elution from protein G–agarose and reversion of cross-links by heating for 6 h at 65°C, 1 and 3% of purified DNA were amplified by 30 cycles (30 s at 95°C, 40 s at 55°C, 40 s at 72°C) in the presence of 1.5 mM MgCl2, 200 µM dNTPs and 10 pmol of the following primers: rDNA promoter forward primer 5′-GACCAGTTGTTCCTTTGAGG-3′ (–165/–145); rDNA reverse primer 5′-TCTGGTTATAGGTACAATTGAGC-3′ (+301/+318); reporter reverse primer 5′-AAAGGCCGGATAAAACTTGTGC-3′; 28S RNA forward primer 5′-GCGACCTCAGATCAGACGTGG-3′ (+8124/+8145); and 28S RNA reverse primer 5′-CTTAACGGTTTCACGCCCTC-3′ (+8529/+8549). PCR products were visualized on ethidium bromide-stained agarose gels. To quantitate the amount of DNA more accurately, the immunoprecipitated DNA was also subjected to real-time PCR using a LightCycler (Roche) and the SYBR Green detection system. In all cases, the amount of DNA in the immunoprecipitates of control or specific antibodies differed by two orders of magnitude, usually showing a 300- to 600-fold enrichment of rDNA with the specific antibodies.
HDAC assay
Immobilized GST–TIP51579–1850 was incubated with 200 µl of nuclear extract at 4°C for 4 h. After washing with buffer AM-300, beads were assayed for HDAC activity as described previously (Taunton et al., 1996; Brehm et al., 1999).
Antibodies
Polyclonal antibodies recognizing residues 1–18 of TIP5 (α-mTIP5N1– 18; Strohner et al., 2001) were affinity purified using the synthetic peptide cross-linked to UltraLink iodoacetyl column (Pierce). Anti-HDAC1 and α-AcH4 antibodies were obtained from Upstate. Anti-mSin3A antibodies (amino acids 2–19) were from Santa Cruz, and anti-myc antibodies were from Clontech. Antibodies against RbAp46 and Mi-2 (Zhang et al., 1998) were provided by D.Reinberg.
Acknowledgments
Acknowledgements
We thank Tony Kouzarides for providing HDAC clones and Danny Reinberg for antibodies against RbAp46 and Mi-2. We are grateful to all members of the laboratory for sharing reagents and advice. This work was supported by the German Cancer Research Center, the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie.
References
- Ahringer J. (2000) NuRD and SIN3 histone deacetylase complexes in development. Trends Genet., 16, 351–356. [DOI] [PubMed] [Google Scholar]
- Ayer D. (1999) Histone deacetylases: transcriptional repression with SINers and NuRDs. Trends Cell Biol., 9, 193–198. [DOI] [PubMed] [Google Scholar]
- Bozhenok L., Wade,P.A. and Varga-Weisz,P. (2002) WSTF–ISWI chromatin remodeling complex targets heterochromatic replication foci. EMBO J., 21, 2231–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm A., Nielsen,S.J., Miska,E.A., McCance,D.J., Reid,J.L., Bannister,A.J. and Kouzarides,T. (1999) The E7 oncoprotein associates with Mi2 and histone deacetylase activity to promote cell growth. EMBO J., 18, 2449–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.J. and Robinson,W.P. (2000) The causes and consequences of random and non-random X chromosome inactivation in humans. Clin. Genet., 58, 353–363. [DOI] [PubMed] [Google Scholar]
- Budde A. and Grummt,I. (1999) p53 represses ribosomal gene transcription. Oncogene, 18, 1119–1124. [DOI] [PubMed] [Google Scholar]
- Conconi A., Widmer,R.M., Koller,T. and Sogo,J.M. (1989) Two different chromatin structures coexit in ribosomal RNA genes throughout the cell cycle. Cell, 57, 753–761. [DOI] [PubMed] [Google Scholar]
- Deuring R. et al. (2000) The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol. Cell, 5, 355–365. [DOI] [PubMed] [Google Scholar]
- Fazzio T.G., Kooperberg,C., Goldmark,J.P., Neal,C., Basom,R., Delrow,J. and Tsukiyama,T. (2001) Widespread collaboration of Isw2 and SIN3–Rpd3 chromatin remodeling complexes in transcriptional repression. Mol. Cell. Biol., 21, 6450–6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmark J.P., Fazzio,T.G., Estep,P.W., Church,G.M. and Tsukiyama,T. (2000) The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell, 103, 423–433. [DOI] [PubMed] [Google Scholar]
- Grunstein M. (1997) Histone acetylation in chromatin structure and transcription. Nature, 389, 349–352. [DOI] [PubMed] [Google Scholar]
- Hirschler-Laszkiewicz I., Cavanaugh,A., Hu,Q., Catania,J., Avantaggiati,M.L. and Rothblum,L.I. (2001) The role of acetylation in rDNA transcription. Nucleic Acids Res., 29, 4114–4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Tyler,J.K., Bulger,M., Kobayashi,R. and Kadonaga,J.T. (1996) ATP-facilitated chromatin assembly with a nucleoplasmin-like protein from Drosophila melanogaster. J. Biol. Chem., 271, 25041–25048. [DOI] [PubMed] [Google Scholar]
- Jenuwein J. and Allis,C.D. (2001) Translating the histone code. Science, 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- Kim J. et al. (1999) Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity, 10, 345–355. [DOI] [PubMed] [Google Scholar]
- Kingston R.E. and Narlikar,G.J. (1999) ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev., 13, 2339–2352. [DOI] [PubMed] [Google Scholar]
- Knoepfler P.S. and Eisenman,R.N. (1999) Sin meets NuRD and other tails of repression. Cell, 99, 447–450. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A. and Reinberg,D. (2001) Role of histone deacetylase complexes in the regulation of chromatin metabolism. Curr. Top. Microbiol. Immunol., 254, 35–58. [DOI] [PubMed] [Google Scholar]
- Längst G., Blank,T.A., Becker,P.B. and Grummt,I. (1997) RNA polymerase I transcription on nucleosomal templates: the transcription termination factor TTF-I induces chromatin remodeling and relieves transcription. EMBO J., 16, 760–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Längst G., Becker,P.B. and Grummt,I. (1998) TTF-I determines the chromatin architecture of active rDNA promoter. EMBO J., 17, 3135–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDouarin B. et al. (1995) The N-terminal part of TIF1, a putative mediator of the ligand-dependent activation function (AF-2) of nuclear receptor, is fused to B-raf in the oncogenic protein. EMBO J., 14, 2020–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeRoy G., Orphanides,G., Lane,W.S. and Reinberg,D. (1998) Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science, 282, 1900–1904. [DOI] [PubMed] [Google Scholar]
- Miska E.A., Karlsson,C., Langley,E. Nielsen,J. and Kouzarides,T. (1999) HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J., 18, 5099–5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G., Tsukiyama,T., Wisniewski,J. and Wu,C. (1997) Role of nucleosome remodeling factor NURF in transcriptional activation of chromatin. Mol. Cell, 1, 141–150. [DOI] [PubMed] [Google Scholar]
- Mizzen C.A. and Allis,C.D. (1998) Linking histone acetylation to transcriptional regulation. Cell. Mol. Life Sci., 54, 6–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muth V., Nadaud,S., Grummt,I. and Voit,R. (2001) Acetylation of TAFI68, a subunit of TIF-IB/SL1, activates RNA polymerase I transcription. EMBO J., 20, 1353–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutskov V.J., Russanova,V.R., Dimitrov,S.I. and Pashev,I.G. (1996) Histones associated with nonnucleosomal rat ribosomal genes are acetylated while those bound to nucleosome-organized gene copies are not. J. Biol. Chem., 271, 11852–11857. [DOI] [PubMed] [Google Scholar]
- Ng H.H. and Bird,A. (2000) Histone deacetylases: silencers for hire. Trends Biochem. Sci., 25, 121–126. [DOI] [PubMed] [Google Scholar]
- Pazin M.J. and Kadonaga,J.T. (1997) What’s up and down with histone deacetylation and transcription? Cell, 89, 325–328. [DOI] [PubMed] [Google Scholar]
- Roth S.Y., Denu,J.M. and Allis,C.D. (2001) Histone acetyltransferases. Annu. Rev. Biochem., 70, 81–120. [DOI] [PubMed] [Google Scholar]
- Santoro R. and Grummt,I. (2001) Molecular mechanisms mediating methylation dependent silencing of ribosomal gene transcription. Mol. Cell, 8, 719–725. [DOI] [PubMed] [Google Scholar]
- Santoro R., Li,J. and Grummt,I. (2002) The nucleolar remodeling complex NoRC silences ribosomal gene transcription. Nat. Genet., in press. [DOI] [PubMed] [Google Scholar]
- Schnapp A. and Grummt,I. (1996) Purification, assay, and properties of RNA polymerase I and class I-specific transcription factors in mouse. Methods Enzymol., 273, 233–248. [DOI] [PubMed] [Google Scholar]
- Schultz D.C., Friedman.,J.R. and Rauscher,F.J.,III (2001) Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2α subunit of NuRD. Genes Dev., 15, 428–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner D.E. and Berger,S.L. (2000) Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev., 64, 435–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl B.D. and Allis,C.D. (2000) The language of covalent histone modifications. Nature, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- Strohner R., Nemeth,A., Jansa,P., Hoffmann-Rohrer,U., Santoro,R., Längst.G. and Grummt,I. (2001) NoRC—a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J., 20, 4892–4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. (1998) Histone acetylation and transcriptional regulatory mechanisms. Genes Dev., 12, 599–606. [DOI] [PubMed] [Google Scholar]
- Taunton J., Hassig,C.A. and Schreiber,S.L. (1996) A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science, 272, 408–411. [DOI] [PubMed] [Google Scholar]
- Turner B.M. (2000) Histone acetylation and an epigenetic code. BioEssays, 22, 836–845. [DOI] [PubMed] [Google Scholar]
- Tyler J.K. and Kadonaga,J.T. (1999) The ‘dark side’ of chromatin remodeling: repressive effects on transcription. Cell, 99, 443–446. [DOI] [PubMed] [Google Scholar]
- Varga-Weisz P. (2001) ATP-dependent chromatin remodeling factors: nucleosome shufflers with many missions. Oncogene, 20, 3076–3085. [DOI] [PubMed] [Google Scholar]
- Venturini L. et al. (1999) TIF1γ, a novel member of the transcriptional intermediary factor 1 family. Oncogene, 18, 1209–1217. [DOI] [PubMed] [Google Scholar]
- Voit R., Hoffmann,M. and Grummt,I. (1999) Phosphorylation by G1-specific cdk–cyclin complexes activates the nucleolar transcription factor UBF. EMBO J., 18, 1891–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., LeRoy,G., Seelig,H.P., Lane,W.S. and Reinberg,D. (1998) The dermatomyositis-specific autoantigen Mi-2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell, 95, 279–289. [DOI] [PubMed] [Google Scholar]