Abstract
To identify nucleotides in or near the active site, we have used a circularly permuted version of the VS ribozyme capable of cleavage and ligation to incorporate a single photoactive nucleotide analog, 4-thio- uridine, immediately downstream of the scissile bond. Exposure to UV light produced two cross-linked RNAs, in which the 4-thio-uridine was cross-linked to A756 in the 730 loop of helix VI. The cross-links formed only under conditions that support catalytic activity, suggesting that they reflect functionally relevant conformations of the RNA. One of the cross-linked RNAs contains a lariat, indicative of intramolecular cross-linking in the ligated RNA; the other is a branched molecule in which the scissile phosphodiester bond is cleaved, but occupies the same site in the ribozyme–substrate complex. These are the two forms of the RNA expected to be the ground state structures on either side of the transition state. This localization of the active site is consistent with previous mutational, biochemical and biophysical data, and provides direct evidence that the cleavage site in helix I interacts with the 730 loop in helix VI.
Keywords: active site/cross-linking/RNA structure/4-thio-uridine/VS RNA
Introduction
The discovery that RNA molecules can catalyze chemical reactions revolutionized traditional thinking about the nature of enzymes and the biological role of RNA. Several naturally occurring ribozymes that cleave and/or ligate phosphodiester bonds during RNA splicing or processing reactions have been discovered (Gesteland and Atkins, 1993). Under some conditions, spliceosomal snRNAs participate directly in chemical reactions (Valadkhan and Manley, 2001), and circumstantial evidence supports the idea that the eukaryotic pre-mRNA splicing may be catalyzed by the snRNAs (for a recent review, see Villa et al., 2002). The crystal structure of the large ribosomal subunit shows that the peptidyl transferase center contains only RNA, implying that peptide bond formation is also RNA catalyzed (Ban et al., 2001). A large number of small, non-coding RNAs, apparently involved in a variety of cellular functions, continue to be discovered at a rapid pace (Eddy, 2002; Kiss, 2002; Wassarman, 2002). Little is yet known about the molecular mechanisms by which many of these RNAs function; however, it is becoming increasingly clear that a complete understanding of cell biology will require an understanding of RNA function. Studies of the small and large ribozymes over the last two decades have provided substantial information about RNA structure. The nature of RNA active sites, however, remains poorly understood (Simons and Grunberg-Manago, 1998).
We have made use of the Varkud Satellite (VS) ribozyme, identified in the mitochondria of certain natural isolates of Neurospora crassa (Saville and Collins, 1990, 1991), as a model system with which to investigate the relationship between RNA structure and function. Like other small ribozymes, VS RNA performs a site-specific cleavage and ligation reaction involving 2′3′ cyclic phosphate and 5′ OH termini (Saville and Collins, 1990). However, VS RNA has a secondary and presumably tertiary structure, different from the other characterized ribozymes (Beattie et al., 1995). One unanswered question about VS RNA is the physical location of the ribozyme active site. Based on substitution and deletion mutagenesis and low resolution modeling, we proposed that the 730 loop in helix VI contributed to the active site (Sood et al., 1998). Subsequent mutagenesis (Lafontaine et al., 2001b; Sood and Collins, 2002), hydroxyl radical protection (Hiley and Collins, 2001) and fluorescence resonance energy transfer (FRET) experiments (Lafontaine et al., 2001a) are consistent with this proposal; however, no physical evidence placing the cleavage site (between nucleotides 620 and 621 in helix I) in close proximity to the 730 loop in helix VI has been presented.
Site-specific photo-cross-linking has been used to identify tertiary interactions in other RNAs. Photoactive nucleotide analogs, such as 4-thio-uridine (sU), have been used extensively as probes of ribozyme tertiary structure because they generate covalent cross-links between RNA functional groups that are close in the tertiary structure of the folded RNA, but distant in the primary and secondary structure (Favre, 1990; Favre and Fourrey, 1995). Mapping the nucleotides involved in the cross-links reveals which region(s) of the RNA interact closely with the site of incorporation of the analog; the small size of the sU moiety and the nature of cross-link formed limits the tertiary interactions it detects to ∼4 Å (Bravo et al., 1999 and references therein). This technique has been used to map many RNA–RNA and RNA–protein interactions (Favre, 1990; Favre and Fourrey, 1995; Bravo et al., 1996; Pinard et al., 1999).
We discovered a circularly permuted version of VS RNA that displays a unique Mg2+-dependent cleavage– ligation equilibrium. This permitted us to generate a cis-cleaving ribozyme with an sU substitution at position 621, immediately 3′ to the scissile bond in the cleavage and ligation reactions. We discovered two cross-linked RNAs that form only under conditions that support catalytic activity, and identified the nucleotides involved in both cross-links to be sU621 and A756. The cross-linked RNAs were shown to be the cleaved and ligated forms of VS RNA, having a branched and lariat structure, respectively. These data provide strong evidence that nucleotides near the cleavage site in stem–loop I interact with the 730 loop in helix VI to form the active site of the VS ribozyme.
Results and discussion
Changing the cleavage–ligation equilibrium of the VS ribozyme
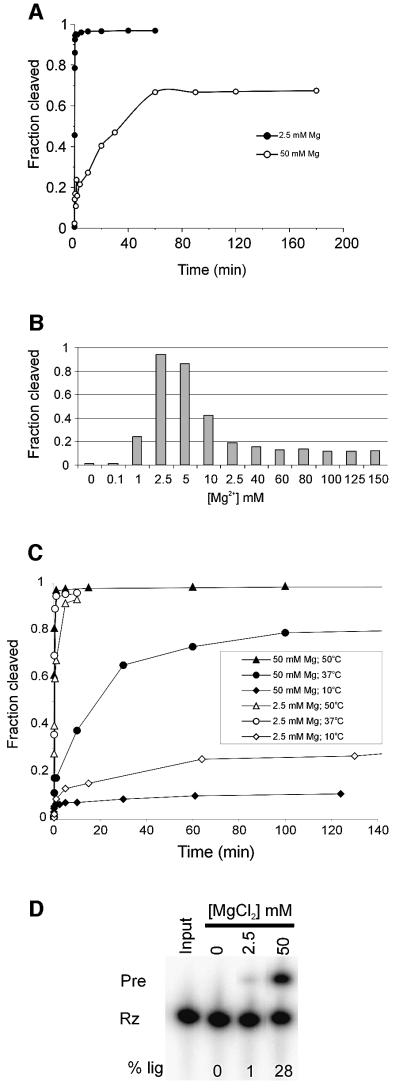
Previously, a circular permutation of the VS ribozyme, clone RS19, was designed that was capable of more efficient ligation than the well-characterized G11 construct (Andersen and Collins, 2000). We have now found that a variant of RS19, called Jen1 (Figure 1), displays unusual kinetics in that the relative preference for cleavage compared with ligation can be substantially altered by changing the concentration of Mg2+ in the reaction solution and/or changing the temperature. In the presence of 2.5 mM Mg2+, Jen1 precursor (Jen1Pre) cleaves rapidly to a final extent of >90%, with a first-order rate constant of ∼4 min–1 (Figure 2A, filled circles). In the presence of 50 mM Mg2+, Jen1Pre cleaves more slowly and the reaction does not fit to a simple kinetic scheme: there is an initial fast phase (to ∼20% cleavage), followed by a slower phase, then the reaction reaches a plateau at <70% cleaved (Figure 2A, open circles). We investigated the Mg2+-dependence of self-cleavage by performing cleavage assays over a range of Mg2+ concentrations (Figure 2B). The apparent self-cleavage rate reached a maximum at 2.5 mM Mg2+ and decreased sharply above 5 mM Mg2+. These data indicate that Jen1Pre has a low Mg2+ optimum for self-cleavage and that the observed cleavage is inhibited at higher concentrations.
Fig. 1. Secondary structure of Jen1 precursor (Pre). Pre undergoes a reversible cleavage–ligation reaction involving the substrate (for the ligation reaction; gray box) and the ribozyme (Rz; the remainder of the sequence shown). Nucleotides involved in the kissing interaction (Rastogi et al., 1996) between loops I and V are outlined with circles and joined by the dashed line, and the 24 nucleotide linker connecting the 3′ end of helix II and the 5′ end of helix Ib is shown by the solid line. The site of self-cleavage and ligation is indicated by the arrowhead. The 730 loop is enclosed by a rectangle. Nucleotides are numbered as in Beattie et al. (1995), and the position of 621 is highlighted by a solid triangle.
Fig. 2. Magnesium-dependent cleavage–ligation equilibrium of Jen1. (A) Time courses of Jen1Pre cleavage in the presence of 2.5 or 50 mM Mg2+. Internally labeled Jen1Pre RNA was incubated at 37°C in the presence of 2.5 or 50 mM Mg2+. (B) Magnesium titration of self- cleavage reaction. Internally labeled Jen1Pre was incubated at 37°C in the presence of 0–150 mM Mg2+ for 1 min. (C) Time courses of self-cleavage at various temperatures. Jen1Pre was incubated in the presence of 2.5 or 50 mM Mg2+ and allowed to cleave at 10, 37 or 50°C. (D) Ligation of ribozyme and substrate. Internally labeled Rz (2 µM) and unlabeled substrate (2 µM) were incubated for 1 h at 25°C in the presence of 0, 2.5 or 50 mM Mg2+. The reaction was quenched by the addition of 2 vol of formamide loading dye and the products were electrophoresed on a denaturing polyacrylamide gel. The percentage of total counts in ligated Pre is indicated below each lane (% lig).
Decreasing the reaction temperature at a given concentration of Mg2+ also affected the final extent of cleavage, suggesting either that an increasing fraction of the RNA is inactive at lower temperatures or that lower temperatures favor ligation (Figure 2C). To test directly the ability of Jen1 to undergo ligation, we incubated internally labeled ribozyme (Rz; Figure 1) with unlabeled substrate in the presence of 0, 2.5 or 50 mM Mg2+ at an intermediate temperature (25°C) for 1 h (Figure 2D). In the presence of 50 mM Mg2+, 28% of the Rz ligated to form Jen1Pre, a small amount of ligation was detectable at 2.5 mM Mg2+ and none occurred in the absence of Mg2+. Ligation at 4°C in 50 mM Mg2+ proceeded to a similar extent as at 25°C (data not shown), indicating that the low extent of cleavage observed at the low temperatures (Figure 2C) reflects an equilibrium in favor of ligation, rather than a large fraction of non-functional RNA. Taken together, these data indicate that conditions that would be expected to favor the stability of the cleaved ribozyme–substrate complex (high Mg2+, low temperature) favor ligation.
A cross-linking strategy to identify the active site of the VS ribozyme
We hypothesized that we could take advantage of the efficient ligation reaction of Jen1 to incorporate a short-range photo-cross-linking agent at a single location adjacent to the scissile bond in a fully functional precursor RNA. Performing cross-linking under conditions where some of the RNA exists in equilibrium between ligated precursor and cleaved (or not yet ligated) ribozyme– substrate complex should maximize the chance of obtaining functionally relevant cross-links. Also, this approach avoids the use of non-cleavable substrate analogs that might form a slightly different structure than the native RNA. Mapping the ends of cross-links that form only under conditions that support catalytic activity should identify nucleotides in or very near the active site.
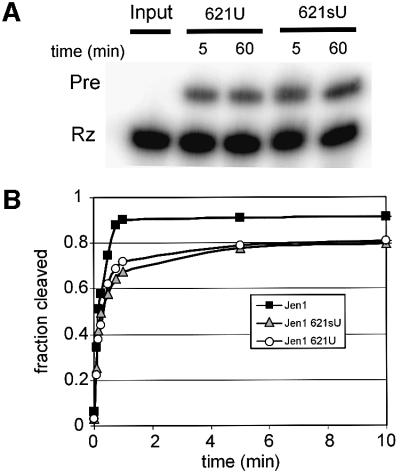
Previous in vitro selector and mutagenesis of other VS RNA constructs showed that position 621, immediately downstream of the scissile bond, is tolerant to some base substitutions (Andersen and Collins, 2000) (Figure 1). In the current work, we ligated in vitro-transcribed ribozyme and chemically synthesized substrates that contained either uridine or the photoactive base analog sU at position 621 to obtain U621Pre and sU621Pre RNAs (Figure 3A). Approximately 20% of the total Rz was ligated to form Pre in each case, similar to that observed with the wild type, which contains an A at position 621 (Figure 2D). Figure 3B shows that RNAs containing a U or sU at position 621 self-cleaved with indistinguishable rates, only slightly slower than the rate of wild-type RNA. Together, the cleavage and ligation data of the Jen1 variants show that sU incorporation at position 621 has little effect on catalytic activity and that sU621-substituted RNA should serve as an accurate probe of VS RNA structure in cross-linking analyses.
Fig. 3. Characterization of U and sU substitution at nucleotide 621. (A) Ligation of U621 and sU621 substrates and ribozyme. Unlabeled substrates containing U or sU at position 621 were incubated with internally labeled ribozyme (2 µM each ribozyme and substrate) in the presence of 50 mM Mg2+ at 25°C. Aliquots were removed and the reaction quenched by the addition of 2 vol of formamide loading dye at the indicated time points, and the reaction products were electrophoresed on a denaturing polyacrylamide gel. (B) Cleavage time courses of Jen1Pre and variants. Pre-RNAs generated by in vitro transcription (Jen1) or ligation of U621 or sU621 substrates to internally labeled ribozyme (U621, sU621) were gel purified and incubated at 37°C in the presence of 2.5 mM Mg2+.
Cross-linking results in covalent attachment of two RNA functional groups that are close to one another in the tertiary structure. Cross-linked RNAs therefore have more complex backbone connectivities than linear RNA molecules; this results in aberrant electrophoretic mobilities on denaturing polyacrylamide gels (Sanger et al., 1976).
Ligation reactions containing 32P-labeled ribozyme and unlabeled substrate containing sU at position 621 were allowed to reach equilibrium, and were then exposed to 365 nm light for up to 10 min and separated by gel electrophoresis. We observed three novel bands (XL-A, XL-B, and XL-C), whose formation depended upon both sU and UV exposure; these bands were not observed after 10 min of UV exposure in the U621 RNA (i.e. lacking the thio group; Figure 4A, lane 1) or in the sU lane not exposed to UV light (lane 2), but all three increased significantly with increasing UV exposure (lanes 3–5). XL-A and XL-B showed anomalous mobility compared with linear Pre and Rz on denaturing gels containing different polyacrylamide concentrations (Figure 4A and B; data not shown), as expected for non-linear RNAs.
Fig. 4. Cross-linking of 4sU substituted RNA. (A) Irradiation at 365 nm generates novel RNAs. Total ligation reactions containing internally labeled ribozyme and unlabeled substrate were incubated at 25°C for 5 min, exposed to 365 nm light for 0–10 min and electrophoresed on a 4% denaturing polyacrylamide gel (Figure 3A). Bands on the gel corresponding to ribozyme and Pre are indicated to the left of the gel. Ligation reactions with 4sU substituted substrate give rise to three bands of aberrant mobility when exposed to 365 nm light (XL-A, XL-B and XL-C; indicated by arrows to the right of the gel). (B) Magnesium dependence of crosslinking. sU621Pre, gel purified following a ligation reaction with internally labeled ribozyme and unlabeled substrate, was incubated for 5 min at 25°C in the presence of increasing amounts of Mg2+, as indicated at the top of the gel. The reactions were irradiated at 365 nm for 10 min and electrophoresed on a 6% denaturing polyacrylamide gel. The input RNA, which was not incubated at 25°C or irradiated, is shown in lane 1. (C) Magnesium dependence of ligation. Internally labeled ribozyme (2 µM) was incubated at 25°C with sU621 substrate (2 µM) for 1 h in the presence of increasing amounts of Mg2+, as indicated at the top of the gel. Reactions were quenched by the addition of 2 vol of formamide loading dye and separated by electrophoresis on a denaturing polyacrylamide gel. (D) Cation dependence of cross-linking. 621sU Pre was incubated in 1 mM EDTA plus 0 or 50 mM MgCl2, 0.6 mM Co(NH3)6Cl3 or 4 M LiCl for 15 min, irradiated with 365 nm UV light for 7.5 min and electrophoresed on a 6% denaturing polyacrylamide gel.
To determine which of the cross-linked RNAs are relevant to the productive folding and catalytic pathway of the RNA, we compared the cation requirements for cross-link formation with those for ligation activity. To survey cross-linking over a range of Mg2+ concentrations, sU621Pre RNA (whose formation by ligation requires high concentrations of Mg2+; Figures 2D and 4C) was prepared by gel purification after a ligation reaction. Figure 4B (lane 2) shows that XL-C forms in the absence of Mg2+, where no catalytic activity is detected, and was not characterized further. Formation of both XL-A and XL-B is dependent on Mg2+: they begin to appear faintly at ∼2.5 mM Mg2+, convincingly at 5 mM, and maximally between 25 and 50 mM Mg2+. Similarly, in the ligation reactions, sU621Pre is clearly visible at 5 mM Mg2+ and is maximal between 25 and 50 mM Mg2+ (Figure 4C). The similarity in the Mg2+ dependence of ligation and the formation of XL-A and XL-B is consistent with these cross-links identifying conformations of the RNA that are stabilized under conditions that favor ligation.
Experiments using other cations also demonstrate a correlation between XL-A, XL-B and catalytic activity (Figure 4D). Cobalt hexamine does not support catalytic activity of VS RNA, but chemical modification structure probing showed that the RNA folds into a structure that closely resembles that formed in magnesium (Maguire and Collins, 2001). Although the structure formed in Co(NH3)63+ is only subtly different to the structure formed in Mg2+, neither XL-A nor XL-B form in Co(NH3)63+, and no cleavage of sU621Pre was detected (above the background of cleaved RNA in the input material; Figure 4D, lane 4). Thus, XL-A and XL-B are sensitive reporters of a conformation which forms only under conditions that support catalytic activity. In contrast, lithium chloride has been observed to support the cleavage activity of a trans-acting VS ribozyme (Murray et al., 1998). Figure 4D (lane 5) shows that sU621Pre also self-cleaves in lithium chloride, and this cation also supports formation of both XL-A and XL-B. In other experiments (data not shown), concentrations of MnCl2, CaCl2, NH4Cl, NaCl and KCl that supported cleavage and ligation also supported cross-linking, with XL-B being more abundant than XL-A. Taken together, the formation of XL-A and XL-B only in the presence of cations that support catalytic activity strongly suggests that these cross-links reflect functionally relevant conformations of the RNA.
Identification of the active site
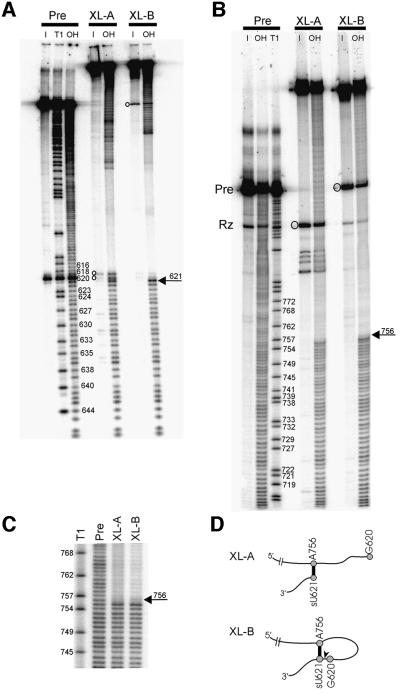
We used partial alkaline hydrolysis of 5′- or 3′-end-labeled RNAs to map the ends of the cross-linked RNAs and to identify the nucleotide(s) to which the cleavage site is cross-linked (Figure 5). Partial hydrolysis of the uncross-linked pre-RNA produces an even ladder corresponding to hydrolysis at each position in the RNA backbone; partial T1 ribonuclease digestion provides markers used to identify bands in the hydrolysis ladder. Partial hydrolysis of a cross-linked RNA produces an even ladder only between the end-label and the end of the cross-link closest to the label; hydrolysis of a phosphodiester bond past this point gives rise to a branched, full-length RNA because of the covalent attachment between the ends of the cross-link. These RNAs will migrate through the gel much more slowly. Figure 5A confirms that the 3′ ends of XL-A and XL-B are at position 621, as expected from the location of the sU substitution. Bands migrating slightly slower than 621 (indicated by open circles in Figure 5A, XL-A input lane) result from breakage of the RNA near the cross-link during gel purification (see below). Despite the different electrophoretic mobilities of XL-A and XL-B, the 5′ end of the cross-link in both RNAs maps to A756 in the 730 loop of helix VI (Figures 5B, C and 1), suggesting that XL-A and XL-B are structural isomers containing the same cross-linked nucleotides.
Fig. 5. Mapping the ends of the cross-links. (A) 3′ ends. XL-A, XL-B and uncross-linked Pre, obtained from a UV-irradiated ligation reaction containing unlabeled ribozyme and 3′-end-labeled sU621 substrate, were gel-purified (I, input) and subjected to partial alkaline hydrolysis (OH) and/or RNase T1 digestion (T1; see Materials and methods). The band at G620 in Pre corresponds to a small amount of self-cleavage during the gel purification. Circles indicate bands produced by breakage of cross-linked nucleotides during purification. (B) 5′ ends. 5′-end- labeled XL-A, XL-B and uncross-linked Pre were obtained as described in Materials and methods and treated as above. Circles indicate bands produced by breakage of cross-linked nucleotides during purification from the gel. (C) A longer electrophoretic run of the 5′-end-labeled RNAs in (B) near the point of discontinuity in the OH ladders; the last nucleotide before the break is C755, indicating that the cross-linked nucleotide is A756 in both XL-A and XL-B. (D) Schematic outlining the proposed structures of XL-A and XL-B. The RNA backbone (not to scale) is indicated as a thin black line. The positions of G620, sU621 and A756 are indicated by gray circles, and the cross-link formed between sU621 and A756 is shown as a thick black line. The scissile bond between G620 and sU621 is indicated by a black arrowhead in XL-B.
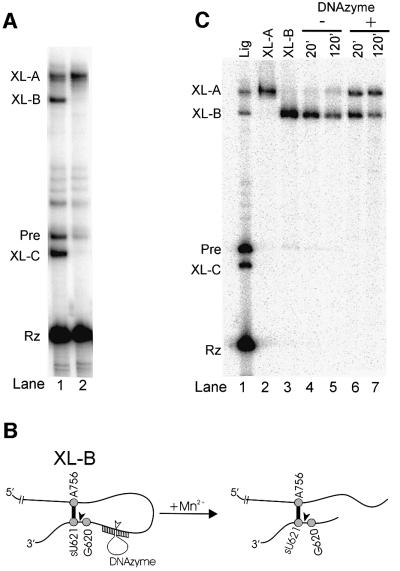
The data in Figure 3A showed that, in 50 mM Mg2+, a portion of ribozyme and sU621 substrate exists in the ligated form (Pre). Analysis of similar ribozyme–substrate combinations by non-denaturing gel electrophoresis suggests that a portion also exists as a non-covalent ribozyme–substrate complex (R.Zamel and R.A.Collins, unpublished observations). If both the Pre and the ribozyme–substrate complex form the same cross-link (sU621–A756), cross-linked Pre would contain a lariat, and cross-linked ribozyme–substrate complex would form a branched structure (Figure 5D). Indeed, when cross-linking reactions were enriched for Pre (Figure 4B, lane 8), XL-B was formed in greater abundance than XL-A; when the proportion of Pre was lower (Figure 4A, lane 5), XL-B constituted a smaller fraction of the cross-linked RNA. This suggests that XL-B is the cross-link derived from Pre that contains the lariat. When cross-linking reactions were performed using ribozyme from which the 2′3′ cyclic phosphate had been removed to prevent ligation (see Materials and methods), only XL-A was formed, suggesting that XL-A corresponds to the branched species derived from the ribozyme–substrate complex (Figure 6A, lane 2).
Fig. 6. Identity of XL-A and XL-B. (A) Selective formation of XL-A when ligation is prevented. Unlabeled 621sU substrate was incubated under ligation conditions with internally labeled ribozyme containing (lane 1) or lacking (lane 2) a 2′3′ cyclic phosphate and irradiated at 365 nm; products were separated by gel electrophoresis. (B) Diagram of cleavage within the lariat of XL-B by a 10-23 DNAzyme. The ribozyme cleavage site between G620 and sU621 is indicated by the filled arrowhead and the predicted DNAzyme cleavage site is indicated by the open arrowhead in XL-B. (C) Conversion of XL-B from lariat to branched form by site-specific DNAzyme cleavage. Lanes 4–7: gel purified, 5′-labeled XL-B was incubated with (+) or without (–) DNAzyme at 37°C in the presence of 3 mM MnCl2 for 20 or 120 min, as indicated at the top of the figure. Lanes 1–3: a cross-linked ligation reaction (Lig) and gel-purified XL-A and XL-B were included as markers on the gel.
Further evidence for these interpretations of the cross-linked RNAs comes from the nature of the small proportion of molecules in which the cross-linked nucleotides broke during gel purification (see Materials and methods). Although 4sU cross-links are not thought to be specifically photoreversible (Sontheimer, 1994), release of cross-linked species during purification has been observed in other RNA cross-links involving sU and other photoactive agents (Wower et al., 1989; Burgin and Pace, 1990; Pinard et al., 1999). As predicted by the model in Figure 5D, breakage of 5′-end-labeled XL-A released an RNA that co-migrated with Rz (indicated by a circle in Figure 5B, XL-A input lane). 3′-end-labeled XL-A produced two bands that migrated slightly slower than free substrate (indicated by circles in Figure 5A, XL-A input lane), suggesting that the cross-link does not simply reverse, but that some nearby bond(s) breaks, resulting in release of the substrate with a nucleotide fragment(s) from the ribozyme attached to it. Release of the cross-link in 5′- or 3′-end-labeled XL-B produced a band that co-migrated with Pre (Figure 5A and B, XL-B input lanes), as would be expected if either of the cross-linked nucleotides broke, converting the lariat RNA into a linear RNA.
To confirm the assignments of the cross-linked RNAs, XL-B was purified and incubated with a 10-23 DNAzyme (Santoro and Joyce, 1997, 1998) designed to cleave at a specific location within the lariat to produce a branched structure similar to that of XL-A (Figure 6B). Figure 6C shows that DNAzyme cleavage of XL-B produced an RNA with the mobility of XL-A. Taken together, these data identify XL-B as cross-linked Pre, and XL-A as the cross-linked ribozyme–substrate complex in which the scissile bond is in the cleaved state.
XL-A and XL-B represent the cleaved and ligated forms of the RNA in which the cleavage site is close enough to the active site to be cross-linked, and similar enough in conformation to be cross-linked to exactly the same nucleotide. These are precisely the two forms of the RNA that would be expected to be the ground state structures on either side of the transition state, a conformation that itself is unlikely to be occupied enough to be detected by cross-linking of a population of RNA. Cross-links between nucleotides near the active site and the cleavage site in either the precursor or the cleavage product have also been identified in other ribozymes (Burgin and Pace, 1990; Bravo et al., 1996, 1999; Kufel and Kirsebom, 1996; Christian et al., 1998). Most of these cross-linked RNAs were not catalytically active, as might be expected due to interference with structures or conformational changes required for catalysis (Bravo et al., 1996); this is also the case for the VS cross-links under all conditions tested to date (data not shown). Nonetheless, subsequent X-ray crystal structures and/or models based on biochemical and mutational data have supported the functional relevance of active site cross-links in several ribozymes (Pley et al., 1994; Scott et al., 1996; Ferré-d’Amare et al., 1998; Rupert and Ferré-d’Amare, 2001). The results in the current study provide evidence for the location of the VS active site, independent of previous models. Nonetheless, data from substitution and deletion mutagenesis, chemical modification protection and interference, FRET and modeling experiments are all consistent with the 730 loop in helix VI making an essential contribution to the active site of the VS ribozyme (Beattie et al., 1995; Beattie and Collins, 1997; Rastogi and Collins, 1998; Sood et al., 1998; Hiley and Collins, 2001; Lafontaine et al., 2001a,b).
Our finding that the nucleotide adjacent to the cleavage site in helix I cross-links to the 730 loop in helix VI in both the cleaved and ligated states of VS RNA, specifically under conditions that support catalytic activity, provides direct evidence that the 730 loop forms part of the active site of the VS ribozyme. This information will be useful in the design of more detailed studies of the chemical mechanism of VS RNA cleavage, the evaluation of future high resolution structures of this ribozyme and the understanding of how the structure of the RNA confers its ability to perform catalysis. The experimental approach of enriching for functionally relevant cross-links using ribozymes designed to be in equilibrium between the ligated state and a non-covalent ribozyme–substrate complex may be applicable to other ribozymes.
Materials and methods
Cloning and characterization of Jen1
VS RNA clone Jen1 was derived from RS19, a circular permutation of VS RNA in which the 3′ end of helix II is connected to the 5′ end of helix Ia by a 24 nucleotide linker (5′-ATTTTACTAACAAGATCTCCAGCT-3′) (Andersen and Collins, 2000). PCR-mediated mutagenesis was used to remove the 5′ and 3′ primer binding sites previously used for in vitro selection experiments in RS19 and introduce substitutions in the substrate which mutationally lock helix Ib in the active, or shifted, conformation (Figure 1). RNA was transcribed from EcoRI-digested plasmid DNA according to standard procedures in 100 µl reactions containing ∼10 µg of linearized DNA, 1.5 µg of T7 RNA polymerase, 40 mM Tris–HCl pH 8.0, 25 mM NaCl, 2 mM spermidine, 1 mM each NTP, 4 mM dithiothreitol, 20 mM MgCl2 and 50 µCi of [α-32P]GTP (3000 Ci/mmol; Amersham). The reactions were incubated at 37°C for at least 45 min, and the Pre and/or Rz were purified from a 4% polyacrylamide/8.3 M urea gel, visualized by UV shadowing and excised and eluted into 2 ml of diethylpyro carbonate (DEPC) treated H2O at 65°C for 1 h. Eluted RNAs were filtered through 0.8 µM/0.2 µM acrodiscs (Gelman Sciences). Then they were ethanol precipitated and dissolved in DEPC-treated H2O. Time courses of self-cleavage were determined with 50 nM RNA pre-incubated for 2 min in 40 mM Tris–HCl pH 8.0 and 50 mM KCl at the reaction temperature, as indicated. Cleavage reactions were initiated by the addition of MgCl2 to the final concentrations indicated in the figures, and aliquots were quenched at the time points indicated by mixing with 2 vol of formamide loading dye. The products were resolved on denaturing polyacrylamide gels and exposed to a PhosphorImager screen. The results were quantified using ImageQuant 3.3 (Molecular Dynamics).
Ligation reactions and characterization of (s)U621 substitutions
Ligation reactions contained internally labeled ribozyme, prepared and purified as described above, except that the concentration of MgCl2 in the transcription reaction was 8 mM to maximize the yield of cleaved RNA. Ribozyme (2 µM) was pre-incubated in 50 mM Tris–HCl pH 8.0, 50 mM KCl and 50 mM MgCl2 unless otherwise indicated. Chemically synthesized substrate RNA (2 µM; Dharmacon Reseach Inc.) containing A, U or sU at position 621 in H2O was added to start the reactions, which were incubated at 25°C for 5–60 min. Reactions were quenched in loading dye and the products analyzed on denaturing gels as described above. Self-cleavage assays of purified Pre containing a (s)U621 substitution were performed and analyzed as described above for in vitro-transcribed Jen1.
Photo-cross-linking analysis
Total ligation reactions (described above) were placed as aliquots onto Parafilm in 10 µl drops and exposed, on ice, to 365 nm light (Blak-ray B100 AP; UVP, Upland, CA) at a distance of ∼4 cm for 7.5 min unless indicated otherwise. Cross-linking was performed on ice to minimize evaporation caused by heating of the sample by the UV light; cross-linking at room temperature or 25°C did not change the identity or abundance of the cross-links. When purified Pre was cross-linked, ∼100 nM RNA was incubated in 50 mM Tris–HCl, 50 mM KCl and MgCl2, Co(NH3)6Cl3 or LiCl as indicated for 5 min at 25°C, and irradiated as described for the ligation reactions. Cross-linked products were diluted in 1.5–3 vol of formamide loading dye and resolved on either 4% (Figure 3A) or 6% (all other) polyacrylamide/8.3 M urea gels and visualized by exposure to a PhosphorImager screen.
Mapping the ends of the cross-links using partial alkaline hydrolysis
RNAs for mapping the 3′ ends of the cross-links were prepared by ligation (see above) of unlabeled ribozyme and sU621 substrate that had been labeled at the 3′ end using RNA ligase and 5′ [32P]pCp. The ligation reaction was irradiated as described above. To map the 5′ end of the cross-links, a ligation reaction containing unlabeled ribozyme and substrate was irradiated as described above. Following irradiation, the reaction was precipitated with ethanol, redissolved and treated with calf intestinal alkaline phosphatase (New England Biolabs, Beverly, MA) according to the manufacturer’s instructions. The reaction products were labeled at the 5′ end using polynucleotide kinase and [γ-32P]ATP (6000 Ci/mmol); they were then gel purified and dissolved in DEPC-treated H2O. This procedure avoids the loss of the 2′3′ cyclic phosphate on the ribozyme (which would prevent ligation with the substrate) during 5′ end-labeling of free ribozyme using polynucleotide kinase.
We noticed that purified 5′-labeled XL-A contained a small amount of a product in a band that comigrated with free Rz (Figure 5B, XL-A input lane). This was not the result of contamination in the gel, because these bands were widely separated and neither XL-B nor Pre, which run between XL-A and Rz, were present in gel-purified XL-A. Similarly, XL-B contained a small amount of a product in a band that comigrated with Pre, even though these RNAs were well separated on the preparative gel. The amounts of these uncross-linked products were affected by temperature; elution at 65°C yielded more Pre and Rz from XL-B and XL-A, respectively, than did elution at 4°C (see the amount of Pre in XL-B lanes in Figure 5A compared with B; and data not shown), and the short 3′-labeled fragment released from XL-A increased in abundance during the incubation at 95°C, which was used to 3′-map the end of the cross-link (Figure 5A, compare XL-A input and OH lanes). These observations suggest that the cross-linked nucleotides are more susceptible to chemical breakage that uncross-linked nucleotides.
Uncross-linked Pre, XL-A and XL-B were partially hydrolyzed by incubation in 0.2 M NH4OH for 3 min at 95°C and lyophilized. RNAs were resolved by electrophoresis on an 8% denaturing polyacrylamide gel and visualized by exposure to a PhosphorImager screen. Partial RNase T1 digestion of the uncross-linked Pre was used to identify bands in the alkaline hydrolysis ladders: the cross-linked nucleotide is to the nucleotide 3′ to the discontinuity in the hydrolysis ladder. Note that, because the guanosine at a T1 cleavage site of a 3′-end-labeled RNA is actually attached to the unlabeled cleavage product, for clarity we have numbered the corresponding visible bands produced from 3′-labeled RNAs. The ligation reaction was cross-linked and the RNAs were purified, hydrolyzed and resolved as described for the 5′ mapping experiment, except that during gel purification the RNA was eluted overnight at 4°C instead of for 1 h at 65°C.
Removal of the 2′3′ cyclic phosphate from the ribozyme
Internally labeled ribozyme was treated with T4 polynucleotide kinase (New England Biolabs) according to the manufacturer’s instructions, with the exception that no ATP was added to the reaction. The 2′3′ cyclic phosphodiesterase and phosphatase activities of polynucleotide kinase removes the 2′3′ cyclic phosphate on the ribozyme.
10-23 DNAzyme cleavage of XL-B
A version of the 10-23 deoxyribozyme (5′-CTGGAGAGGCTAGCTACAACGACTTGTTA) (Santoro and Joyce, 1997, 1998) was designed to anneal to, and specifically cleave between, linker nucleotides 15 and 16 in Jen1. Gel-purified, 5′-labeled XL-B was incubated with a large excess of DNAzyme in 50 mM Tris–HCl pH 8.0, 150 mM NaCl, 0.01% SDS and 3 mM MnCl2 for 2 h at 37°C. Reactions were quenched by the addition of 2 vol of formamide loading dye and the products were resolved by electrophoresis.
Acknowledgments
Acknowledgements
We are grateful to Drs Paul Sadowski and Deborah Field, and members of the Collins laboratory for helpful discussions and critical reading of the manuscript. This work was supported by a grant from the Canadian Institutes of Health Research. S.L.H. was supported in part by a PGS B award from the National Sciences Engineering and Research Council of Canada (NSERC). R.A.C is a Canada Research Chair and a fellow of the Canadian Institute for Advanced Research Program in Evolutionary Biology.
References
- Andersen A.A. and Collins,R.A. (2000) Rearrangement of a stable RNA secondary structure during VS ribozyme catalysis. Mol. Cell, 5, 469–478. [DOI] [PubMed] [Google Scholar]
- Ban N., Nissen,P., Hansen,J., Moore,P.B. and Steitz,T.A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science, 289, 905–920. [DOI] [PubMed] [Google Scholar]
- Beattie T.L. and Collins,R.A. (1997) Identification of functional domains in the self-cleaving Neurospora VS ribozyme using damage selection. J. Mol. Biol., 267, 830–840. [DOI] [PubMed] [Google Scholar]
- Beattie T.L., Olive,J.E. and Collins,R.A. (1995) A secondary-structure model for the self-cleaving region of Neurospora VS RNA. Proc. Natl Acad. Sci. USA, 92, 4686–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo C., Lescure,F., Laugaa,P., Fourrey,J.L. and Favre,A. (1996) Folding of the HDV antigenomic ribozyme pseudoknot structure deduced from long-range photocrosslinks. Nucleic Acids Res., 24, 1351–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo C., Woisard,A., Fourrey,J.L., Laugaa,P. and Favre,A. (1999) A Y form of hammerhead ribozyme trapped by photo-cross-links retains full cleavage activity. Biochimie, 81, 201–212. [DOI] [PubMed] [Google Scholar]
- Burgin A.B. and Pace,N.R. (1990) Mapping the active site of ribonuclease P RNA using a substrate containing a photoaffinity agent. EMBO J., 9, 4111–4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian E.L., McPheeters,D.S. and Harris,M.E. (1998) Identification of individual nucleotides in the bacterial ribonuclease P ribozyme adjacent to the pre-tRNA cleavage site by short-range photo-cross-linking. Biochemistry, 37, 17618–17628. [DOI] [PubMed] [Google Scholar]
- Eddy S. (2002) Computational genomics of noncoding RNA genes. Cell, 109, 137–140. [DOI] [PubMed] [Google Scholar]
- Favre A. (1990) 4-thiouridine as an intrinsic photoaffinity probe of nucleic acid structure and interactions. In Morrison,H. (ed.), Bioorganic Photochemistry: Photochemistry and the Nucleic Acids. Wiley, New York, NY.
- Favre A. and Fourrey,J.L. (1995) Structural probing of small endonucleolytic ribozymes in solution using thio-substituted nucleobases as intrinsic photolabels. Acc. Chem. Res., 28, 375–382. [Google Scholar]
- Ferré-d’Amare A.R., Zhou,K. and Doudna,J.A. (1998) Crystal structure of a hepatitis delta virus ribozyme. Nature, 395, 567–574. [DOI] [PubMed] [Google Scholar]
- Gesteland R.F. and Atkins,J.F. (1993) The RNA World. Cold Spring Harbor Laboratory Press, Plainview, NY.
- Hiley S.L. and Collins,R.A. (2001) Rapid formation of a solvent-inaccessible core in the Neurospora Varkud satellite ribozyme. EMBO J., 20, 5461–5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T. (2002) Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell, 109, 145–148. [DOI] [PubMed] [Google Scholar]
- Kufel J. and Kirsebom,L.A. (1996) Different cleavage sites are aligned differently in the active site of M1 RNA, the catalytic subunit of Escherichia coli RNase P. Proc. Natl Acad. Sci. USA, 93, 6085–6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine D.A., Norman,D.G. and Lilley,D.M. (2001a) Structure, folding and activity of the VS ribozyme: importance of the 2-3-6 helical junction. EMBO J., 20, 1415–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine D.A., Wilson,T.J., Norman,D.G. and Lilley,D.M. (2001b) The 730 loop is an important component of the active site of the VS ribozyme. J. Mol. Biol., 312, 663–674. [DOI] [PubMed] [Google Scholar]
- Maguire J.L. and Collins,R.A. (2001) Effects of cobalt hexammine on folding and self-cleavage of the Neurospora VS ribozyme. J. Mol. Biol., 309, 45–56. [DOI] [PubMed] [Google Scholar]
- Murray J.B., Seyhan,A.A., Walter,N.G., Burke,J.M. and Scott,W.G. (1998) The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem. Biol., 5, 587–595. [DOI] [PubMed] [Google Scholar]
- Pinard R., Heckman,J.E. and Burke,J.M. (1999) Alignment of the two domains of the hairpin ribozyme–substrate complex defined by interdomain photoaffinity crosslinking. J. Mol. Biol., 287, 239–251. [DOI] [PubMed] [Google Scholar]
- Pley H.W., Flaherty,K.M. and McKay,D.B. (1994) Three-dimensional structure of a hammerhead ribozyme. Nature, 372, 68–74. [DOI] [PubMed] [Google Scholar]
- Rastogi T. and Collins,R.A. (1998) Smaller, faster ribozymes reveal the catalytic core of Neurospora VS RNA. J. Mol. Biol., 277, 215–224. [DOI] [PubMed] [Google Scholar]
- Rastogi T., Beattie,T.L., Olive,J.E. and Collins,R.A. (1996) A long-range pseudoknot is required for activity of the Neurospora VS ribozyme. EMBO J., 15, 2820–2825. [PMC free article] [PubMed] [Google Scholar]
- Rupert P.B. and Ferré-d’Amare,A.R. (2001) Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature, 410, 780–786. [DOI] [PubMed] [Google Scholar]
- Sanger H.L., Klotz,G., Riesner,D., Gross,H.J. and Kleinschmidt,A.K. (1976) Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl Acad. Sci. USA, 73, 3852–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro S.W. and Joyce,G.F. (1997) A general purpose RNA-cleaving DNA enzyme. Proc. Natl Acad. Sci. USA, 94, 4262–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro S.W. and Joyce,G.F. (1998) Mechanism and utility of an RNA-cleaving DNA enzyme. Biochemistry, 37, 13330–13342. [DOI] [PubMed] [Google Scholar]
- Saville B.J. and Collins,R.A. (1990) A site-specific self-cleavage reaction performed by a novel RNA in Neurospora mitochondria. Cell, 61, 685–696. [DOI] [PubMed] [Google Scholar]
- Saville B.J. and Collins,R.A. (1991) RNA-mediated ligation of self-cleavage products of a Neurospora mitochondrial plasmid transcript. Proc. Natl Acad. Sci. USA, 88, 8826–8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott W.G., Murray,J.B., Arnold,J.R., Stoddard,B.L. and Klug,A. (1996) Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme. Science, 274, 2065–2069. [DOI] [PubMed] [Google Scholar]
- Simons R.W. and Grunberg-Manago,M. (1998) RNA Structure and Function. Cold Spring Harbor Laboratory Press, Plainview, NY.
- Sontheimer E.J. (1994) Site-specific RNA crosslinking with 4-thiouridine. Mol. Biol. Rep., 20, 35–44. [DOI] [PubMed] [Google Scholar]
- Sood V.D., and Collins,R.A. (2002) Identification of the catalytic subdomain of the VS ribozyme and evidence for remarkable sequence tolerance in the active site loop. J. Mol. Biol., 320, 443–454. [DOI] [PubMed] [Google Scholar]
- Sood V.D., Beattie,T.L. and Collins,R.A. (1998) Identification of phosphate groups involved in metal binding and tertiary interactions in the core of the Neurospora VS ribozyme. J. Mol. Biol., 282, 741–750. [DOI] [PubMed] [Google Scholar]
- Valadkhan S. and Manley,J.L. (2001) Splicing-related catalysis by protein-free snRNAs. Nature, 413, 701–707. [DOI] [PubMed] [Google Scholar]
- Villa T., Pleiss,J. and Guthrie,C. (2002) Spliceosomal snRNAs: Mg2+-dependent chemistry at the catalytic core? Cell, 109, 149–152. [DOI] [PubMed] [Google Scholar]
- Wassarman K.M. (2002) Small RNAs in bacteria: diverse regulators of gene expression in response to environmental changes. Cell, 109, 141–144. [DOI] [PubMed] [Google Scholar]
- Wower J., Hixson,S.S. and Zimmermann,R.A. (1989) Labeling the peptidyltransferase center of the Escherichia coli ribosome with photoreactive tRNA(Phe) derivatives containing azidoadenosine at the 3′ end of the acceptor arm: a model of the tRNA–ribosome complex. Proc. Natl Acad. Sci. USA, 86, 5232–5236. [DOI] [PMC free article] [PubMed] [Google Scholar]