Abstract
Background
Microorganisms inhabiting subterranean oil fields have recently attracted much attention. Since intact groundwater can easily be obtained from the bottom of underground oil-storage cavities without contamination by surface water, studies on such oil-storage cavities are expected to provide valuable information to understand microbial ecology of subterranean oil fields.
Results
DNA was extracted from the groundwater obtained from an oil-storage cavity situated at Kuji in Iwate, Japan, and 16S rRNA gene (16S rDNA) fragments were amplified by PCR using combinations of universal and Bacteria-specific primers. The sequence analysis of 154 clones produced 31 different bacterial sequence types (a unique clone or group of clones with sequence similarity of > 98). Major sequence types were related to Desulfotomaculum, Acetobacterium, Desulfovibrio, Desulfobacula, Zoogloea and Thiomicrospira denitrificans. The abundance in the groundwater of bacterial populations represented by these major sequence types was assessed by quantitative competitive PCR using specific primers, showing that five rDNA types except for that related to Desulfobacula shared significant proportions (more than 1%) of the total bacterial rDNA.
Conclusions
Bacteria inhabiting the oil-storage cavity were unexpectedly diverse. A phylogenetic affiliation of cloned 16S rDNA sequences suggests that bacteria exhibiting different types of energy metabolism coexist in the cavity.
Background
Underground cavities have been used for long-term storage of crude oil in several countries, and one of such facilities is situated at Kuji in Iwate, Japan. These cavities have been constructed in groundwater-rich rocky strata, where high groundwater pressure confines the stored oil in the cavities [1]. Consequently, groundwater migrates into and accumulates at the bottom of a cavity (cavity groundwater), and this cavity groundwater is discharged to maintain the oil storage capacity of the cavity (this system has been detailed in our previous study [1]). Our previous study [1] has also shown active growth of microorganisms in groundwater accumulating at the bottom of the cavities; the total count of microorganisms in the cavity groundwater was constantly more than 106 cells per ml (densities 100 times higher than those in groundwater around the cavities). This habitat can be characterized by (i) immediate contact with a large quantity of crude oil and (ii) an excess of electron donors for microbial growth (i.e., hydrocarbons) but a shortage of electron acceptors [1]. These characteristics may be similar to those of microbial habitats associated with subterranean oil reservoirs, which have recently attracted much attention in microbiology [2-6]. Since groundwater can easily be obtained from the bottom of the oil-storage cavities using standing sampling facilities without its contamination by surface water [1], studies on the oil-storage cavity are considered to provide valuable information to understand the microbial ecology of subterranean oil fields.
Our previous study applied rRNA approaches, namely cloning and sequencing of 16S rRNA gene fragments, denaturing gradient gel electrophoresis and fluorescent in situ hybridization (FISH), to analyze bacterial populations that occurred in the cavity groundwater obtained at Kuji [1]. As a result, a group of bacteria (called cluster-1 bacteria) affiliated with the Thiovulum subgroup in the ε subclass of the class Proteobacteria was consistently detected as a major population. Quantitative comparison of the results of these approaches, however, revealed a large bias associated with the cloning and sequencing approach; it was thus considered that the bacterial biodiversity has not sufficiently been assessed yet.
The present study was conducted to obtain a more reliable view on the bacterial biodiversity in the Kuji cavity groundwater. For this purpose, this study employed widely used universal primers [7] and a recently-modified Bacteria-specific primer [8] for PCR amplification of 16S rRNA gene (16S rDNA) fragments from the cavity groundwater, followed by cloning and sequencing. In addition, in order to quantitatively assess results of the cloning approach, competitive PCR (cPCR) assays were developed for some of abundantly obtained 16S rDNA types that were considered important for anaerobic petroleum degradation.
Results
FISH
In order to examine the abundance in the cavity groundwater of microorganisms affiliated with the Bacteria, FISH using probe EUB338 (Table 1) was carried out. The result was compared with that of FISH using an Archaea-specific probe, ARCH915 (Table 1). In the groundwater sample obtained in 1999, EUB338-labeled cells represented 53.5% ± 3.5% (mean ± standard deviation) of the DAPI-stained cells, while ARCH915-labeled cells represented 8.4% ± 1.8%. The FISH results for the cavity groundwater obtained in 2001 were almost the same. The result indicates that microorganisms affiliated with the Bacteria are the major constituents in the cavity groundwater.
Table 1.
Primers and probes used in this study.
| Primer and probe | Sequence | Position (5' to 3')a | Specificity | Reference |
| EUB338 | 5'-GCTGCCTCCCGTAGGAGT-3' | 355 to 338 | Bacteria | [9] |
| ARCH915 | 5'-GTGCTCCCCCGCCAATTCCT-3' | 934 to 915 | Archaea | [10] |
| I-341f | 5'-CCTACGGGIGGCIGCA-3' | 341 to 356 | Bacteria | [8] |
| U515f | 5'-GTGYCAGCMGCCGCGGTAA-3' | 515 to 533 | Universal | [7] |
| U1492r | 5'-GGTTACCTTGTTACGACTT-3' | 1510 to 1492 | Universal | [7] |
| B341f | 5'-CCTACGGGAGGCAGCAG-3' | 341 to 357 | Bacteria | [11] |
| U533r | 5'-TTACCGCGGCKGCTGRCAC-3' | 533 to 515 | Bacteria | [11] |
| B968f | 5'-AACGCGAAGAACCTTAC-3' | 968 to 984 | Bacteria | [12] |
| DV387f | 5'-CCTGACGCAGCGACG-3' | 387 to 401 | δ-Proteobacteria | [13] |
| DV701r | 5'-ACGGATTTCACTCCTACACC-3' | 701 to 682 | Desulfovibrio | [14] |
| EP710r | 5'-CAGTATCATCCCAGCAGA-3' | 710 to 693 | Cluster-1 bacteria | [1] |
| DT1041r | 5'-CACCCGAIGGTGAAAACGTAC-3' | 1041 to 1021 | Part of Desulfotomaculum | This study |
| AB1173r | 5'-GTGTTATCCACGGCAGTCTGCT-3' | 1173 to 1152 | Acetobacterium | This study |
| AZ665r | 5'-CTGCCGTACTCTAGTCATAC-3' | 665 to 646 | Parts of Azoarcus and Zoogloea | This studyb |
| DB1286r | 5'-AGGATTGGCTCCCCCTCACA-3' | 1286 to 1267 | Part of Desulfobacula | This study |
| T7W | 5'-TAATACGACTCACTATAGGGC-3' | pGEM-T vector | [1] | |
| SP6W | 5'-ATTTAGGTGACACTATAGAATACTC-3' | pGEM-T vector | [1] |
aCorresponding to the numbering in the sequence of the 16S rRNA gene of E. coli. bThis primer is a modified form of probe ZRA [38].
Analyses of cloned 16S rDNA fragments
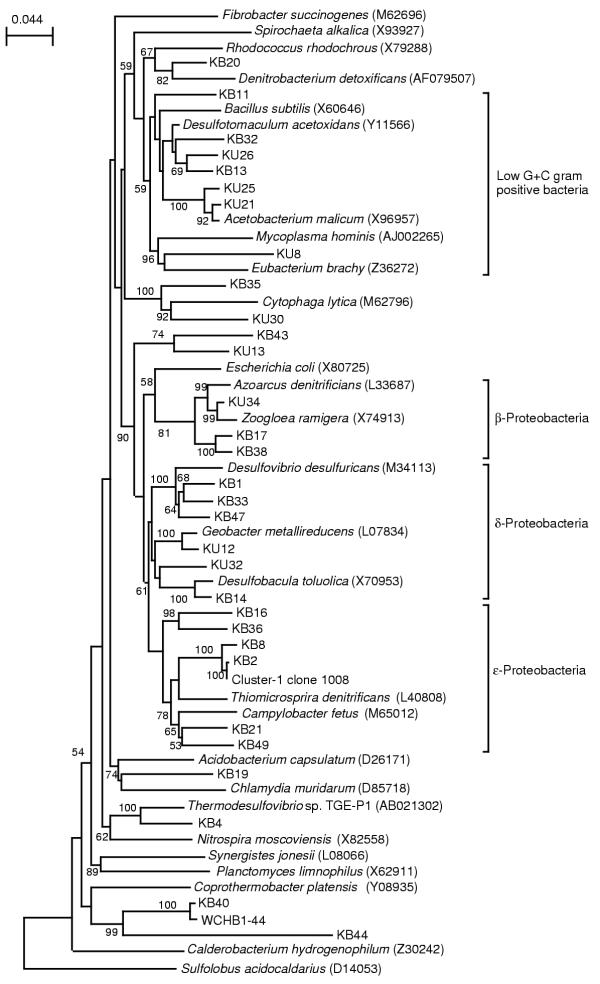
The 16S rDNA fragments amplified by PCR from groundwater obtained in 1999 were cloned into E. coli, and two libraries of clones, namely 341/1492 and 515/1492 (designated according to the primers used, see Table 1), were constructed. Nucleotide sequences of a total of 154 clones were determined, which included 81 clones from library 341/1492 and 73 clones from 515/1492 (Table 2). Results of the sequence analysis are summarized in Table 2. As shown in this table, 31 different sequence types (a unique clones or group of clones with sequence similarity of > 0.98) affiliated with the Bacteria were obtained. The database search (Table 2) and phylogenetic analysis (Fig. 1) identified the phylogenetic positions of these sequence types. The sequence types containing more than several clones were related to Desulfotomaculum, Acetobacterium, Desulfovibrio, Desulfobacula, Zoogloea and Thiomicrospira denitrificans (this sequence type was affiliated with the cluster-1 bacteria [1]). Some sequence types showed homology to 16S rDNA clones obtained from contaminated groundwater and anaerobic consortia degrading petroleum constituents (Table 2).
Table 2.
16S rDNA sequence types obtained in this study.
| Sequence type | No of clones in each librarya,b | Putative division | Database match (> 90% identity) | |
| 341/1492 | 515/1492 | |||
| KB20 | 2 (2.4) | 0 (0) | Actinobacteria | No match |
| KB11 | 2 (2.4) | 0 (0) | Low-G+C gram positive | 96% Syntrophomonas sp. MGB-C1 (AB021306) |
| KB32 | 1 (1.2) | 0 (0) | Low-G+C gram positive | 91% Hydrocarbon-contaminated groundwater clone (AF050587) |
| KU26 | 1 (1.2) | 24 (32.9) | Low-G+C gram positive | 90% Hydrocarbon-contaminated groundwater clone (AF050587) |
| 90% Desulfotomaculum thermobenzoicum DSM 6193 (AJ294430) | ||||
| KB13 | 17 (21.0) | 2 (2.7) | Low-G+C gram positive | 94% Hydrocarbon-contaminated groundwater clone (AF050587) |
| 91% Toluene-degrading methanogenic consortium clone Eub 1 (AF423181) | ||||
| 91% Desulfotomaculum thermobenzoicum DSM 6193 (AJ294430) | ||||
| KU25 | 0 (0) | 1 (1.4) | Low-G+C gram positive | 97% Acetobacterium paludosum DSM 8237 (X96958) |
| KU21 | 0 (0) | 14 (19.2) | Low-G+C gram positive | 99% Groundwater clone Aspo4 (X95232) |
| 99% Acetobacterium malicum DSM 4132 (X96957) | ||||
| KU8 | 0 (0) | 1 (1.4) | Low-G+C gram positive | No match |
| KB35 | 1 (1.2) | 0 (0) | CFB group | 97% Marine clone agg32 (L10944) |
| KU30 | 0 (0) | 1 (1.4) | CFB group | 90% Benzene mineralizing consortium clone SB-5 (AF029041) |
| KB43 | 2 (2.4) | 0 (0) | Spirochete-likec | 94% Soil clone PBS-18 (AJ390485) |
| KU13 | 0 (0) | 1 (1.4) | Spirochete-likec | No match |
| KU34 | 1 (1.2) | 9 (12.3) | β-Proteobacteria | 96% Zoogloea ramigera ATCC 19544 (X74913) |
| KB17 | 1 (1.2) | 0 (0) | β-Proteobacteria | 96% Hydrogenophaga palleronii DSM 63 (AF019073) |
| KB38 | 1 (1.2) | 0 (0) | β-Proteobacteria | 97% Acidovorax defluvii BSB411 (Y18616) |
| KB1 | 11 (13.6) | 1 (1.4) | δ-Proteobacteria | 98% Toluene-degrading methanogenic consortium clone Eub 5 (AF423185) |
| 96% Desulfovibrio aminophilus ALA-3 (AF067964) | ||||
| KB33 | 1 (1.2) | 0 (0) | δ-Proteobacteria | 99% Desulfovibrio sp. Mlhm (AF193026) |
| KB47 | 8 (9.9) | 0 (0) | δ-Proteobacteria | 91% Desulfovibrio sp. DR14 (Y17756) |
| KU12 | 0 (0) | 2 (2.7) | δ-Proteobacteria | 97% Geobacter chapelleii 172 (U41561) |
| KU32 | 0 (0) | 1 (1.4) | δ-Proteobacteria | 96% Chlorobenzene-contaminated groundwater clone GOUTB20 (AY050605) |
| KB14 | 6 (7.4) | 0 (0) | δ-Proteobacteria | 94% Desulfobacula toluolica Tol2 (X70953) |
| KB16 | 1 (1.2) | 0 (0) | ε-Proteobacteria | No match |
| KB36 | 2 (2.4) | 0 (0) | ε-Proteobacteria | 99% Kuji groundwater clone 1070 (AB030590) |
| KB8 | 1 (1.2) | 1 (1.4) | ε-Proteobacteria | 99% Kuji cluster-1 bacteria clone 1023 (AB030608) |
| KB2 | 7 (8.6) | 2 (2.7) | ε-Proteobacteria | 99% Kuji cluster-1 bacteria clone 1033 (AB030608) |
| KB21 | 1 (1.4) | 0 (0) | ε-Proteobacteria | 99% Kuji groundwater clone 1014 (AB030587) |
| 99% Geospirillium sp. KoFum (Y18254) | ||||
| KB49 | 1 (1.4) | 0 (0) | ε-Proteobacteria | 99% Kuji groundwater clone 1065 (AB030598) |
| 94% Arcobacter cryaerophilus CCUG 17801 (L14624) | ||||
| KB19 | 2 (2.4) | 0 (0) | Chlamydia group | No match |
| KB4 | 1 (1.2) | 0 (0) | Nitrospira group | 94% Chlorobenzene-contaminated groundwater clone GOUTA19 (AY050588) |
| KB40 | 2 (2.4) | 0 (0) | Green nonsulfur bacteria | 98% Hydrocarbon-contaminated groundwater clone WCHB1-44 (AF050565) |
| KB44 | 1 (1.2) | 0 (0) | Green nonsulfur bacteria | 99% Trichlorobenzene-transforming consortium clone SJA-117 (AJ009488) |
| 97% Toluene-degrading methanogenic consortium bacterium Eub 4 (AF423184) | ||||
| Archaea | 0 (0) | 5 (6.8) | ||
| Chimera | 1 (1.2) | 0 (0) | ||
| Total | 81 (100) | 73 (100) | ||
Figure 1.
Neighbor-joining tree for rDNA sequences types. Sequences corresponding to nucleotide positions 515 to 1492 of the E. coli sequence were used for calculations. Sulfolobus acidocaldarius is used as the outgroup. Accession numbers of the sequences retrieved from the databases are indicated in parentheses. The numbers at the branch nodes are bootstrap values (per 100 trials); only values greater than 50 are shown. The scale bar indicates 0.044 substitution per site.
Quantification of rDNA copies by competitive PCR
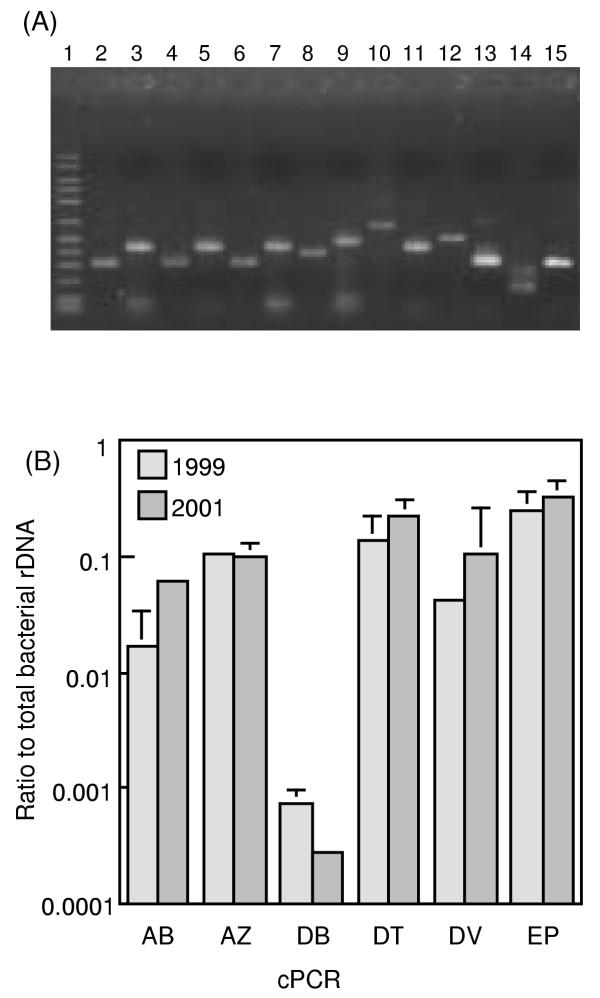
It has been suggested that the PCR-amplification and cloning procedures may cause biases towards some specific 16S rDNA types [1,15]. In order to examine the abundance of bacteria represented by the major sequence types (those related to Desulfotomaculum, Acetobacterium, Desulfovibrio, Desulfobacula, Zoogloea and Thiomicrospira denitrificans), competitive PCR (cPCR) assays using specific primers were developed (Table 3). Fig. 2a shows that the PCR assays successfully amplified the target bands from the cavity groundwater. Results of cPCR for the groundwater obtained in 1999 and 2001 are shown in Fig. 2b. In the two groundwater samples, rDNA copies of the cluster-1 bacteria were most abundant, and those related to Desulfotomaculum, Acetobacterium, Desulfovibrio and Zoogloea also shared significant proportions (more than 1%) of the total bacterial rDNA copies.
Table 3.
Summary of cPCR assay.
| cPCR type | Primer used | Annealing Tm (°C) | Length of DNA fragment (bp) | Specificity | ||
| Taget | Competitor | Sequence type | Phylogenetic group | |||
| AB | U515f and AB1173r | 60 | 658 | 431 | KU21, KU25 | Acetobacterium |
| AZ | B341f and AZ665r | 55 | 324 | 437 | KU34 | Parts of Azoarcus and Zoogloea |
| BC | B341f and U533r | 55 | 170–210 | 336 | All | Bacteria |
| DB | B968f and DB1286r | 55 | 318 | 437 | KB14 | Part of Desulfobacula |
| DT | U515f and DT1047r | 60 | 526 | 340 | KB13, KB32, KU26 | Part of Desulfotomaculum |
| DV | DV387f and DV701r | 55 | 314 | 415 | KB1, KB33, KB38 | Desulfovibrio |
| EP | B341f and EP710r | 55 | 369 | 465 | KB2, KB8 | Cluster-1 bacteria |
Figure 2.
cPCR assays for quantifying rDNA copies of major sequence types. The 6 cPCR systems used, namely AB, AZ, DB, DT, DV and EP, are explained in Table 3. (A) Target fragments amplified by PCR from the groundwater DNA (obtained in 1999) and competitor fragments. Lane 1, 50–2500 molecular size marker (FMC corp.); lanes 2, 4, 6, 8, 10, 12 and 14, target fragments; lanes 3, 5, 7, 9, 11, 13 and 15; competitor fragments. Lanes 2 and 3, DV; lanes 4 and 5, DB; lanes 6 and 7, AZ; lanes 8 and 9, EP; lanes 10 and 11, AB; lanes 12 and 13, DT; lanes 14 and 15, BC. (B) Rations of specific rDNA copies to the total bacterial rDNA copies (determined by the BC system in Table 3) in DNA extracted from the cavity groundwater. The total bacterial counts estimated by FISH were approximately 3 × 106 cells ml-1. The mean of three determinations is shown, and an error bar indicates a standard deviation.
Discussion
The present study used the two combinations of PCR primers for amplifying 16S rDNA fragments from the cavity groundwater. The sequence analysis of the amplified fragments suggests that the bacterial diversity in the cavity groundwater is much larger than the diversity found in our previous study [1]. The present study abundantly detected the sequence types related to Desulfotomaculum, Acetobacterium, Desulfovibrio, Desulfobacula, Zoogloea and Thiomicrospira denitificans, while major 16S rDNA sequences obtained in our previous study were solely affiliated with the ε-Proteobacteria. This comparison clearly indicates that methods employed in the molecular ecological approaches largely affect the results obtained. Differences in the methods employed in these two studies included primers used for PCR amplification of 16S rDNA fragments from the groundwater DNA and selection of E. coli clones harboring groundwater 16S rDNA fragments. The PCR primers used in our previous study were two Bacteria-specific primers (called 8f and 1546r [1]). We assume that organisms undetected in our previous study have 16S rDNA sequences that include mismatches to these primers, since this study did not determine sequences of the 5'- and 3'-end regions of 16S rDNA where 8f and 1546r exist. In addition to the primer selection, different from our previous study [1], the present study did not employ the β-galactosidase colony screening when selecting E. coli clones, because it has been reported that the vector containing a DNA fragment up to 2 kb in-frame produces a blue colony. These two modifications may have circumvented the bias toward the ε-Proteobacteria, which occurred in our previous study [1].
For estimating the abundance of bacterial populations represented by the major sequence types, this study employed cPCR assays. We did not use FISH, because a possible bias that causes the underestimation of slow-growing bacteria has been suggested [1]. Besides, in the present study, FISH with the Bacteria- and Archaea-specific probes detected only 60% in total of the DAPI-stained cells; in addition, a large portion of labeled cells exhibited weak signals. One possible explanation for this result would be that the probes used (e.g., EUB338) failed to detect some phylogenetic groups of microorganisms. The utility of cPCR for this purpose has been demonstrated in several studies [16,17], while attention should be paid to the variation in the number of 16S rDNA copies in a bacterial cell. For example, some Clostridium strains (affiliated with the low G+C gram positive bacteria) have been known to harbor more than 10 rDNA copies per cell [18,19], while the mean copy number in whole-genome-analyzed bacteria is 3.6 [20]. We assume that cPCR may have overestimated the Acetobacterium rDNA copies, since Acetobacterium belongs to the Clostridiaceae. In contrast, rDNA copy numbers per cell of bacteria related to the other major sequence types were not so different from the mean value for all bacteria [20]. We therefore consider that the results of cPCR reflected the abundance in the cavity groundwater of the cluster-1 bacteria and those related to Desulfotomaculum, Desulfovibrio and Zoogloea. Comparison of the cloning and cPCR results indicated that relative abundances determined by these two methods were inconsistent with each other in several cases (Table 2 and Figure 2). Furthermore, clones obtained in abundance in one library were minorities in another library in many cases (Table 2). These results suggest that the rDNA cloning approach employed in this study also included some biases.
Environmental rDNA sequencees have been used to infer some properties of the organisms that they represent [1,7,21], although this type of analysis is reliable only if some common properties can be recognized for phylogenetically related organisms. In the present study, we assume that the organism represented by sequence types KU21 and KU25 are acetogens [22], and those represented by KB1, KB14, KB33 and KB47 are likely sulfate-reducing bacteria [23]. KU34 is closely related to the genus Zoogloea that has been known to grow by denitrification under anaerobic conditions [24]. The Zoogloea belong to the Rhodocyclus group in the β subclass of the class Proteobacteria, which also includes the genera Rhodocyclus and Azoarcus[25]. Since organisms in these genera can commonly perform denitrification, organisms corresponding to clone KU34 are assumed to be capable of denitirfication. Sequence types KU26 and KB13 are related to the genus Desulfotomaculum that belongs to the Syntrophomonas/Thermoanaerobacter group in the low G+C gram-positive bacteria [25]. Typical members of this group are sulfate-reducing bacteria represented by Desulfotomaculum spp. [26], fermentative bacteria represented by Sporotomaculum hydroxybenzoicum[27] and acid-oxidizing syntrophs represented by Syntrophomonas spp. [28]. It is likely that organisms represented by KU26 and KB13 were engaged in some steps in organic compound decomposition. Similar types of bacteria, including δ-Proteobacteria, ε-Proteobacteria and low G+C gram-positive bacteria, have been detected by rDNA cloning analyses of mesophilic oil reservoirs in Canada [3], while β-Proteobacteria were not detected from there. We thus assume that understanding physiology of organisms represented by clone KU34 would provide valuable information regarding differences of ecosystem functions in these two subsurface oil fields.
We have recently isolated bacterial strains that belonged to the cluster-1 bacteria [Kodama, Y. and Watanabe, K. Unpublished results]. It has been found that these strains are capable of anaerobic chemolithotrophic growth by oxidizing reduced sulfur species (sulfide, elemental sulfur and thiosulfate) coupled to nitrate reduction. This finding suggests that this group of bacteria mainly consumed nitrate that was supplied by groundwater flowing into the oil-storage cavities.
Conclusions
The present study unveiled the large diversity of bacteria in the cavity groundwater. The phylogenetic affiliation of cloned 16S rDNA sequences suggests that bacteria exhibiting different types of energy metabolisms coexist as major proportions. This finding is considered interesting, because the chemical analyses of the groundwater have suggested that nitrate reduction is the major electron accepting process [1]. In particular, although sulfate was not significantly consumed [1] and sulfide was always undetectable (below 0.01 mg per liter [unpublished data]) in the cavity groundwater pool, the results of the present study indicate that sulfate-reducing bacteria were present at significant levels. One possible explanation for these findings would be the existence of a sulfur cycle in the cavity groundwater that involves sulfate reducers and anaerobic sulfur oxidizers that regenerated sulfate from sulfide. We assume that similar sulfur cycles may operate in subterranean oil fields, since sulfate-reducing bacteria have been isolated from subterranean oil fields whose formation water contained very low concentrations of sulfate [29,30]. In order to elucidate ecological niches and interactions among these bacteria, their isolation and physiological analyses in axenic cultures are needed. These studies will draw novel insights into the ecological interactions among microorganisms in subterranean oil fields.
Methods
Groundwater samples
The cavity groundwater was obtained in March 1999 and July 2001 from the TK101 underground crude oil storage cavity situated at Kuji in Iwate, Japan. Characteristics of the groundwater were reported previously [1].
Fluorescent in situ hybridization (FISH)
Rhodamine-labeled oligonucleotide probes, EUB338 and ARCH915 (Table 3) were used for fluorescence in situ hybridization (FISH). Microbial cells in groundwater were collected by centrifugation at 10,000 × g for 10 min, suspended in phosphate-buffered saline [31], and fixed in a 4% (wt/vol) paraformaldehyde solution for 5 h at 0°C. The cells were attached to gelatin-coated slides [31] and dehydrated by sequential washes in 50, 80, and 98% (vol/vol) ethanol (3 min each). Subsequently, 8 μl of hybridization solution (0.9 M NaCl, 20 mM Tris-HCl [pH 7.2], 0.01% [wt/vol] sodium dodecyl sulfate and formamide at a concentration [wt/vol] of 30% for EUB338 or 35% for ARCH915) containing 50 ng of probe was added to each hybridization well. It was incubated at 40°C (EUB338) or 46°C (ARCH915) for 3 h in a humid chamber. Slides were washed in the hybridization solution at 42°C (EUB338) or 48°C (ARCH915) for 20 min before all cells on the slide were stained with 4',6'-diamidino-2-phenylindole (DAPI) [1]. More than 1,000 DAPI-stained cells were counted to determine the ratio of probe-labeled cells to DAPI-stained cells. Slides were prepared in triplicate.
PCR amplification, cloning and sequencing of 16S rDNA fragments
DNA was extracted from the groundwater as described previously [1]. PCR amplification of 16S rDNA fragments was performed using either I-341f or 515f as a forward primer and 1492r as a reverse primer (Table 1). A PCR solution (50 μl) contained 1.25 U of Taq DNA polymerase (Amplitaq Gold; Applied Biosystems), 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.001% (wt/vol) gelatin, each deoxynucleoside triphosphate at a concentration of 200 μM, 50 pmol of each primer and 10 ng of DNA. The amplification conditions were as follows: 10 min of activation of the polymerase at 94°C, followed by 30 cycles consisting of 1 min at 94°C, 1 min at 50°C, and 2 min at 72°C, and finally 10 min of extension at 72°C. Amplified fragments were purified by electrophoresis, ligated into the pGEM-T vector (Promega) and cloned into Escherichia coli as described previously [1]. Vector-harboring clones were selected on Luria-Bertani plates [32] supplemented with ampicillin (50 μg ml-1). PCR-amplified 16S rDNA fragments were recovered from colonies by PCR using primers T7W and SP6W (the primers targeted the pGEM-T vector sequences flanking the insertion, see Table 1) as described previously [1]. Clones containing appropriate sizes of the insertion were selected by the electrophoresis analysis, and their nucleotide sequences were determined as described previously [1].
Sequence analysis
Database searches with 16S rDNA sequences were conducted by using the BLAST program [33] and the GenBank database. The profile alignment technique of ClustalW version 1.7 [34] was used to align the sequences, and the alignments were refined by visual inspection; secondary structures were considered for the refinement analysis [35]. A phylogenetic tree was constructed by the neighbor-joining method [36]using the njplot software in ClustalW, version 1.7. Nucleotide positions at which any sequence had an ambiguous base were not included in the phylogenetic calculations. Checks for chimeric sequences were conducted by using the chimera check in the RDP database [25].
cPCR
The primers used for cPCR (Table 1) were selected or designed by comparing the 16S rDNA sequences obtained in this study and those stored in the RDP database [25]. The specificity of the primers thus obtained was checked by using the probe match program in the RDP database [25]. Competitor fragments were produced by using a competitive DNA construction kit (Takara Shuzo). The composition of PCR solution was described above except for the competitor fragment being added at a known copy number. The PCR conditions used were as follows: 10 min of activation of the polymerase at 94°C, followed by 30 cycles consisting of 1 min at 94°C, 1 min at the annealing temperatures described Table 3, and 2 min at 72°C, and finally 10 min of extension at 72°C. Two microliters of the PCR product was electrophoresed through a 1.5% (wt/vol) agarose gel with TBE buffer, and the gel was photographed after it was stained with SYBR Gold (FMC Bioproducts). The band intensities of the target and competitor fragments were quantified by using the Multianalyst software supplied with Gel Doc 2000 (Bio-rad). A copy number of the target 16S rDNA fragment was estimated by considering the band intensity, length of the fragment and copy number of the competitor as described by Lee et al. [37].
Nucleotide sequence accession numbers
The nucleotide sequence data reported in this paper have been deposited in the GSDB, DDBJ, EMBL, and NCBI nucleotide sequence databases under accession no. AB074931 to AB074961.
Authors' contributions
KW carried out cloning and sequencing of 16S rDNA fragments and subsequent phylogenetic analyses. YK carried out sampling of groundwater and FISH analyses. NK participated in the design and coordination of this study.
Acknowledgments
Acknowledgements
We thank Yoichi Matsumura (Japan Under-ground Oil Storage Co.) for help in the groundwater sampling and Ikuko Hiramatsu for technical assistance. This work was supported in part by the New Energy and Industrial Technology Development Organization (NEDO).
Contributor Information
Kazuya Watanabe, Email: kazuya.watanabe@mbio.jp.
Yumiko Kodama, Email: yumiko.kodama@mbio.jp.
Nobuo Kaku, Email: nobuo.kaku@mbio.jp.
References
- Watanabe K, Watanabe K, Kodama Y, Syutsubo K, Harayama S. Molecular characterization of bacterial populations in petroleum-contaminated groundwater discharged from underground crude-oil-storage cavities. Appl Environ Microbiol. 2000;66:4803–4809. doi: 10.1128/AEM.66.11.4803-4809.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Haridon S, Reysenbach AL, Glenat P, Prieur D, Jeanthon C. Hot subterranean biosphere in a continental oil reservoir. Nature. 1995;377:223–224. doi: 10.1038/377223a0. [DOI] [PubMed] [Google Scholar]
- Voordouw G, Armstrong SM, Reimer MF, Fouts B, Telang AJ, Shen Y, Gevertz D. Characterization of 16S rRNA genes from oil field microbial communities indicates the presence of a variety of sulfate-reducing, fermentative, and sulfide-oxidizing bacteria. Appl Environ Microbiol. 1996;62:1623–1629. doi: 10.1128/aem.62.5.1623-1629.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen RK, Beeder J, Thorstenson T, Torsvik T. Distribution of thermophilic marine sulfate reducers in North Sea oil field waters and oil reserviors. Appl Environ Microbiol. 1996;62:1793–1798. doi: 10.1128/aem.62.5.1793-1798.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumholz LR, McKinley JP, Ulrich GA, Suflita JM. Confined subsurface microbial communities in Cretaceous rock. Nature. 1997;386:64–66. doi: 10.1038/386064a0. [DOI] [Google Scholar]
- Margot M, Ollivier B, Patel BKC. Microbiology of petroleum reservoirs. Antonie van Leeuwenhoek. 2000;77:103–116. doi: 10.1023/A:1002434330514. [DOI] [PubMed] [Google Scholar]
- Dojka MA, Hugenholtz P, Haack SK, Pace NR. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl Environ Microbiol. 1998;64:3869–3877. doi: 10.1128/aem.64.10.3869-3877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Kodama Y, Harayama S. Design and evaluation of PCR primers to amplify bacterial 16S ribosomal DNA fragments used for community fingerprinting. J Microbiol Methods. 2001;44:253–262. doi: 10.1016/S0167-7012(01)00220-2. [DOI] [PubMed] [Google Scholar]
- Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin L, Stromley JM, Rittmann BE, Stahl DA. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl Environ Microbiol. 1994;60:1232–1240. doi: 10.1128/aem.60.4.1232-1240.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nubel U, Engelen B, Felske A, Snaidr J, Wieshuber A, Amann RI, Ludwig W, Backhaus H. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel elec-trophoresis. J Bacteriol. 1996;178:5636–5643. doi: 10.1128/jb.178.19.5636-5643.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabus R, Fukui M, Wilkers H, Widdel F. Degradative capacities and 16S rRNA-targeted whole-cell hybridization of sulfate-reducing bacteria in an anaerobic enrichment culture utilizing alkylbenzenes from crude oil. Appl Environ Microbiol. 1996;62:3605–3613. doi: 10.1128/aem.62.10.3605-3613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux R, Kane MD, Winfrey J, Stahl DA. Genus- and group-specific hybridization probes for determinative and environmental studies of sulfate-reducing bacteria. Syst Appl Microbiol. 1992;15:601–609. [Google Scholar]
- Wintzingerode F, Göbel UB, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1016/S0168-6445(97)00057-0. [DOI] [PubMed] [Google Scholar]
- Lee SY, Bollinger J, Bezdicek D, Ogram A. Estimation of the abundance of an uncultured soil bacterial strain by a competitive quantitative PCR method. Appl Environ Microbiol. 1996;62:3787–3793. doi: 10.1128/aem.62.10.3787-3793.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Teramoto M, Harayama S. An outbreak of non-flocculating catabolic populations caused the breakdown of a phenol-digesting activated-sludge process. Appl Environ Microbiol. 1999;65:2813–2819. doi: 10.1128/aem.65.7.2813-2819.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson SR, Young M. Physical map of the Clostridium beijerinckii (formerly Clostridium acetobutylicum) NCIMB 8052 chromosome. J Bacteriol. 1995;177:439–448. doi: 10.1128/jb.177.2.439-448.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornillot E, Croux C, Soucaille P. Physical and genetic map of the Clostridium acetobutylicum ATCC 824 chromosome. J Bacteriol. 1997;179:7426–7434. doi: 10.1128/jb.179.23.7426-7434.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klappenbach JA, Saxman PR, Cole JR, Schmidt TM. rrndb: the Ribosomal RNA Operon Copy Number Database. Nucleic Acids Res. 2001;29:181–184. doi: 10.1093/nar/29.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugenholtz P, Goebel BM, Pace NR. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekert G. The acetogenic bacteria. In: Balows A, Trüper HG, Dworkin M, Harder W, Schleifer KH, editor. The prokaryotes. New York, Springer-Verlag; 1992. pp. 517–533. [Google Scholar]
- Widdel F, Hansen TA. The dissimilatory sulfate- and sulfur-reducing bacteria. In: Balows A, Trüper HG, Dworkin M, Harder W, Schleifer KH, editor. The prokaryotes. New York, Springer-Verlag; 1992. pp. 583–624. [Google Scholar]
- Shin YK, Hiraishi A, Sugiyama J. Molecular systematics of the genus Zoogloea and emendation of the genus. Int J Syst Bacteriol. 1993;43:826–831. doi: 10.1099/00207713-43-4-826. [DOI] [PubMed] [Google Scholar]
- Maidak BL, Cole JR, Parker CT, Jr, Garrity GM, Larsen N, Li B, Lilburn TG, McCaughey MJ, Olsen GJ, Overbeek R, Pramanik S, Schmidt TM, Tiedje JM, Woese CR. A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widdel F. The genus Desulfotomaculum. In: Balows A, Trüper HG, Dworkin M, Harder W, Schleifer KH, editor. The prokaryotes. New York, Springer-Verlag; 1992. pp. 1792–1799. [Google Scholar]
- Brauman A, Müller JA, Garcia JL, Brune A, Schink B. Fermentative degradation of 3-hydroxybenzoate in pure culture by a novel strictly anaerobic bacterium, Sporotomaculum hydroxybenzoicum gen. nov., sp. nov. Int J Syst Bacteriol. 1998;48:215–221. doi: 10.1099/00207713-48-1-215. [DOI] [PubMed] [Google Scholar]
- Mcinerney MJ. The genus Syntrophomonas and other syntrophic anaerobes. In: Balows A, Trüper HG, Dworkin M, Harder W, Schleifer KH, editor. The prokaryotes. New York, Springer-Verlag; 1992. pp. 2049–2057. [Google Scholar]
- Dickey PA, Soto C. Chemical composition of deep subsurface waters of the western Anadarko Basin. SPE-AIME Paper. 1974;SPE 5178:17. [Google Scholar]
- Hunt JM. Petroleum Geochemistry and Geology. San Francisco, WH Freeman & Co. 1979.
- Amann RI. In situ identification of microorganisms by whole cell hybridization with rRNA-targeted nucleic acid probes. In: Akkermans ADL., van Elsas JD, de Bruijn FJ, editor. Molecular microbial ecology manual. Dordrecht, Kluwer Academic Publishers; 1995. pp. 3.3.6:1–3.3.6:15. [Google Scholar]
- Sambrook J, Fitsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, Cold Spring Harbor Press. 1989.
- Karlin S, Altschul SF. Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proc Natl Acad Sci USA. 1990;87:2264–2268. doi: 10.1073/pnas.87.6.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nuceic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell RR. Collection of small subunit (16S and 16S-like) ribosomal RNA structures. Nucleic Acids Res. 1994;22:3502–3507. doi: 10.1093/nar/22.17.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Lee SY, Bollinger J, Bezdicek D, Ogram A. Estimation of the abundance of an uncultured soil bacterial strain by a competitive quantitative PCR method. Appl Environ Microbiol. 1996;62:3787–3793. doi: 10.1128/aem.62.10.3787-3793.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossello-Mora RA, Wagner M, Amann R, Schleifer KH. The abundance of Zoogloea ramigera in sewage treatment plants. Appl Environ Microbiol. 1995;61:702–707. doi: 10.1128/aem.61.2.702-707.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]