Abstract
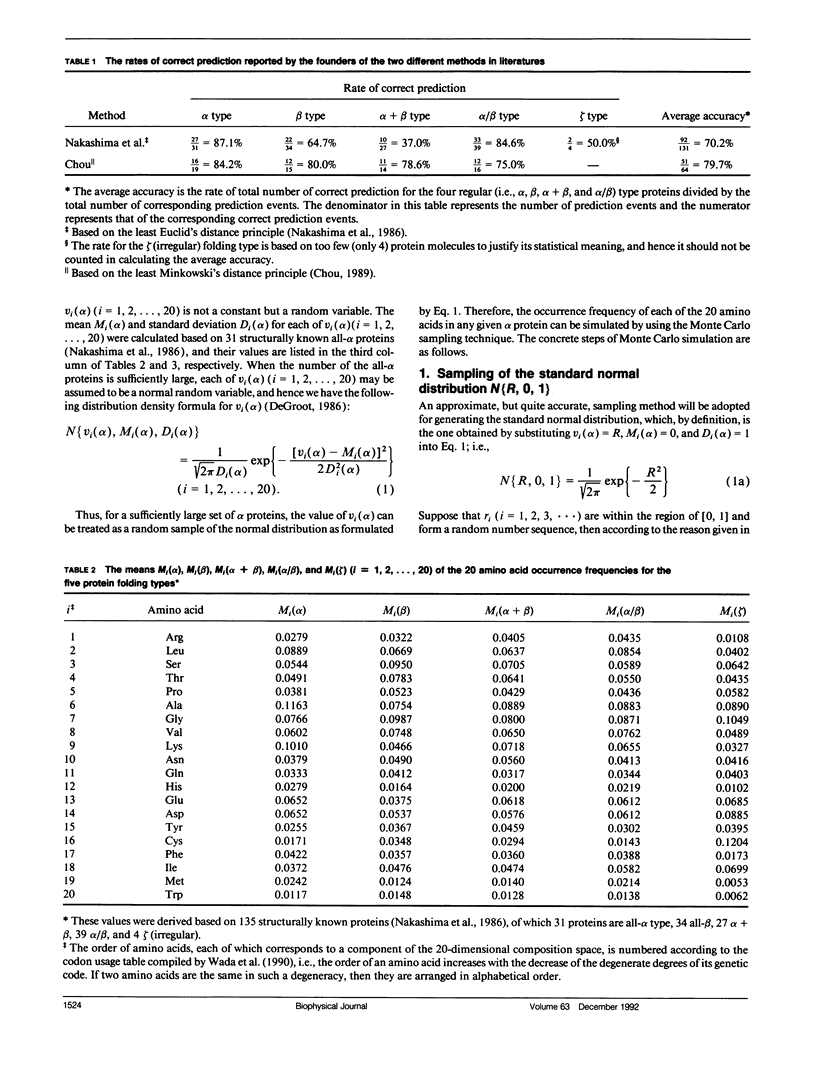
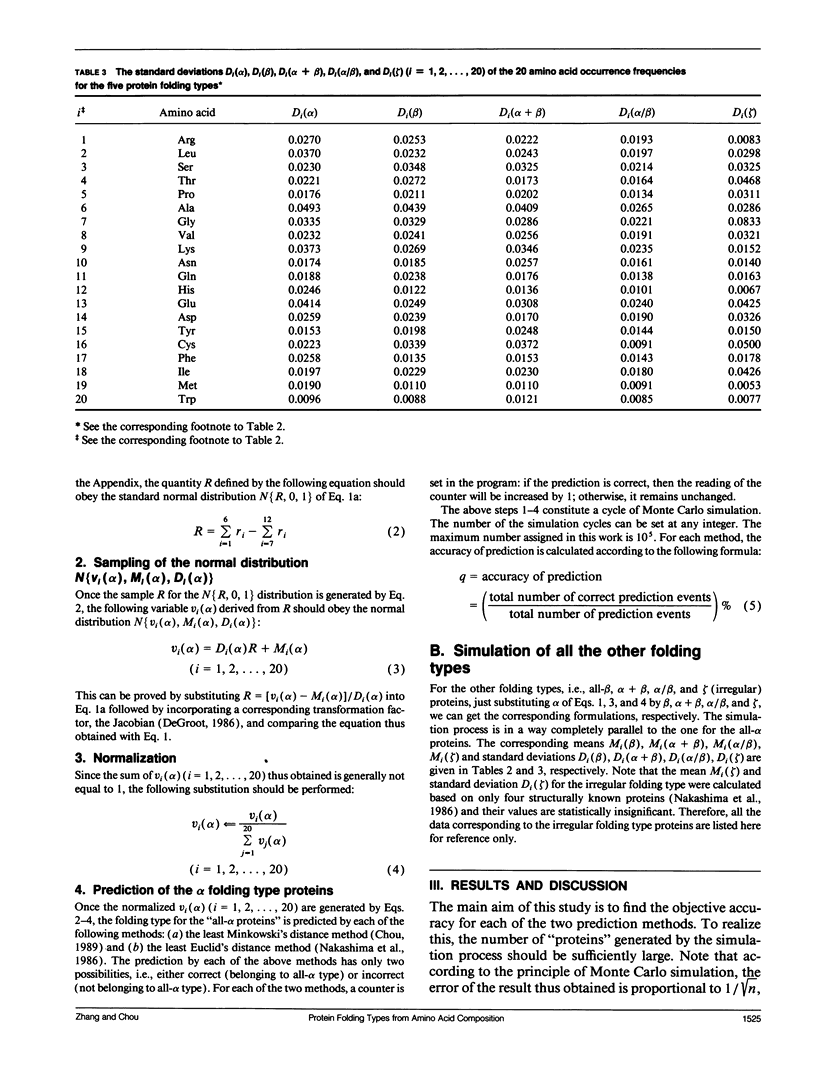
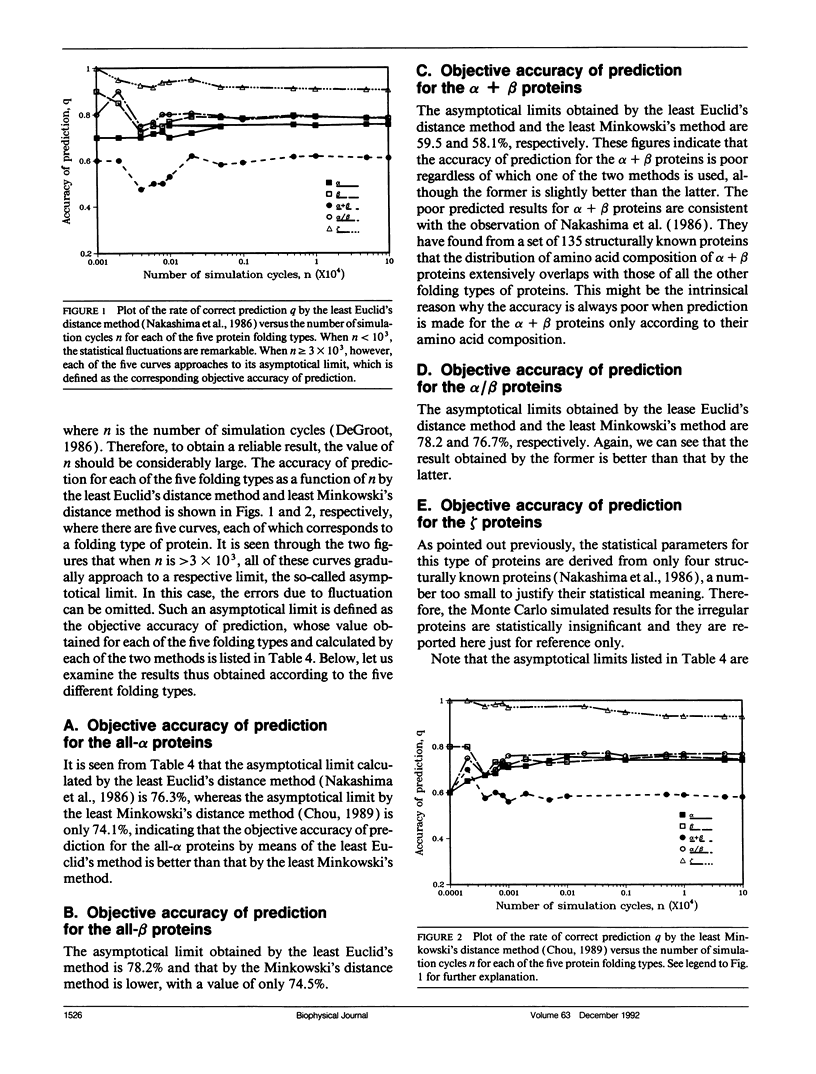
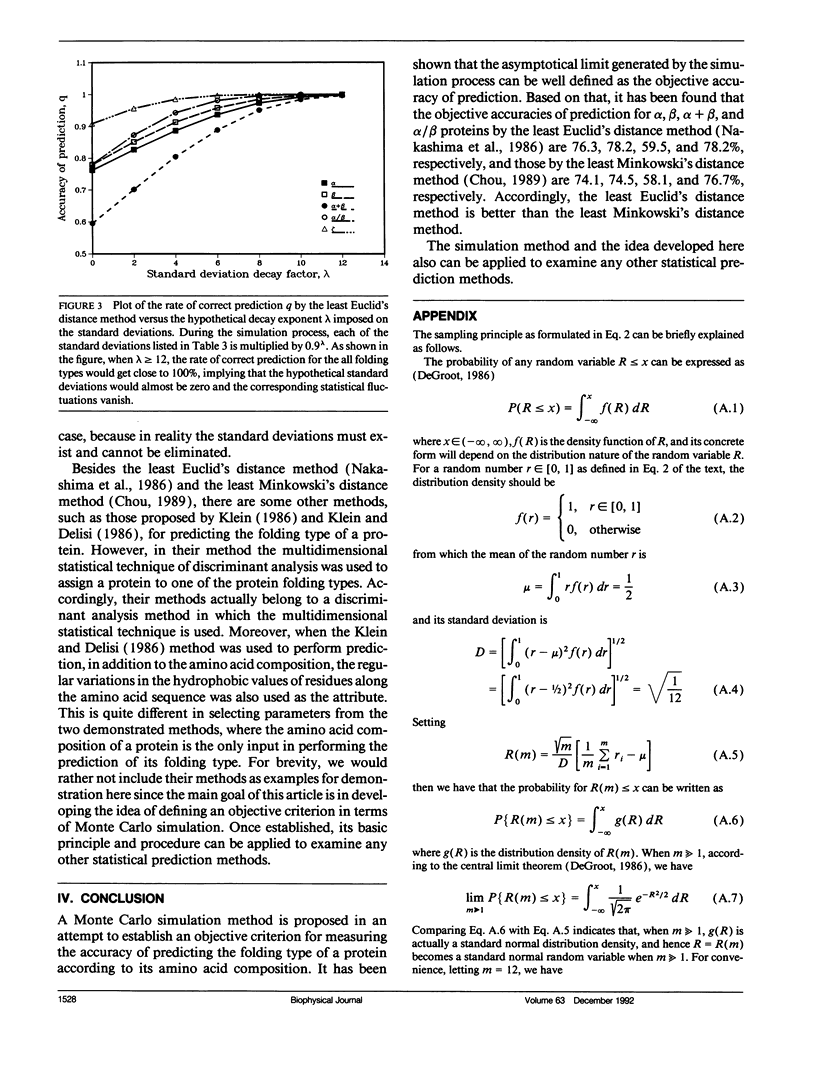
In the methodology development for statistical prediction of protein structures, the founders of different methods usually selected different sets of proteins to test their predicted results. Therefore, it is hard to make a fair comparison according to the results they reported. Even if the predictions by different methods are performed for the same set of proteins, there is still such a problem: a method better that the other for one set of proteins would not necessarily remain so when applied to another set of proteins. To tackle this problem, a Monte Carlo simulation method is proposed to establish an objective criterion to measure the accuracy of prediction for the protein folding type. Such an objective accuracy is actually corresponding to the asymptotical limit genereated during the Monte Carlo simulation process. Based on that, it has been found that the average objective accuracy for predicting the all-alpha, all-beta, alpha + beta, and alpha/beta proteins by the least Euclid's distance method (Nakashima, H., K. Nishikawa, and T. Ooi. 1986. J. Biochem. 99:152-162) is 73.0% and that by the least Minkowski's distance method (Chou, P.Y. 1989. Prediction in Protein Structure and the Principles of Protein Conformation. Plenum Press. New York. 549-586) is 70.9%, indicating that the former is better than the latter. However, according to the original reports, the latter claimed a rate of correct prediction with 79.7% but the former with only 70.2%, leading to a completely opposite conclusion. This indicates the necessity of establishing an objective criterion, and a comparison is meaningful only when it is based on the objective criterion. The simulation method and the idea developed here also can be applied to examine any other statistical prediction methods.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Klein P., Delisi C. Prediction of protein structural class from the amino acid sequence. Biopolymers. 1986 Sep;25(9):1659–1672. doi: 10.1002/bip.360250909. [DOI] [PubMed] [Google Scholar]
- Klein P. Prediction of protein structural class by discriminant analysis. Biochim Biophys Acta. 1986 Nov 21;874(2):205–215. doi: 10.1016/0167-4838(86)90119-6. [DOI] [PubMed] [Google Scholar]
- Levitt M., Chothia C. Structural patterns in globular proteins. Nature. 1976 Jun 17;261(5561):552–558. doi: 10.1038/261552a0. [DOI] [PubMed] [Google Scholar]
- Nakashima H., Nishikawa K., Ooi T. The folding type of a protein is relevant to the amino acid composition. J Biochem. 1986 Jan;99(1):153–162. doi: 10.1093/oxfordjournals.jbchem.a135454. [DOI] [PubMed] [Google Scholar]
- Wada K., Aota S., Tsuchiya R., Ishibashi F., Gojobori T., Ikemura T. Codon usage tabulated from the GenBank genetic sequence data. Nucleic Acids Res. 1990 Apr 25;18 (Suppl):2367–2411. doi: 10.1093/nar/18.suppl.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]


