Abstract
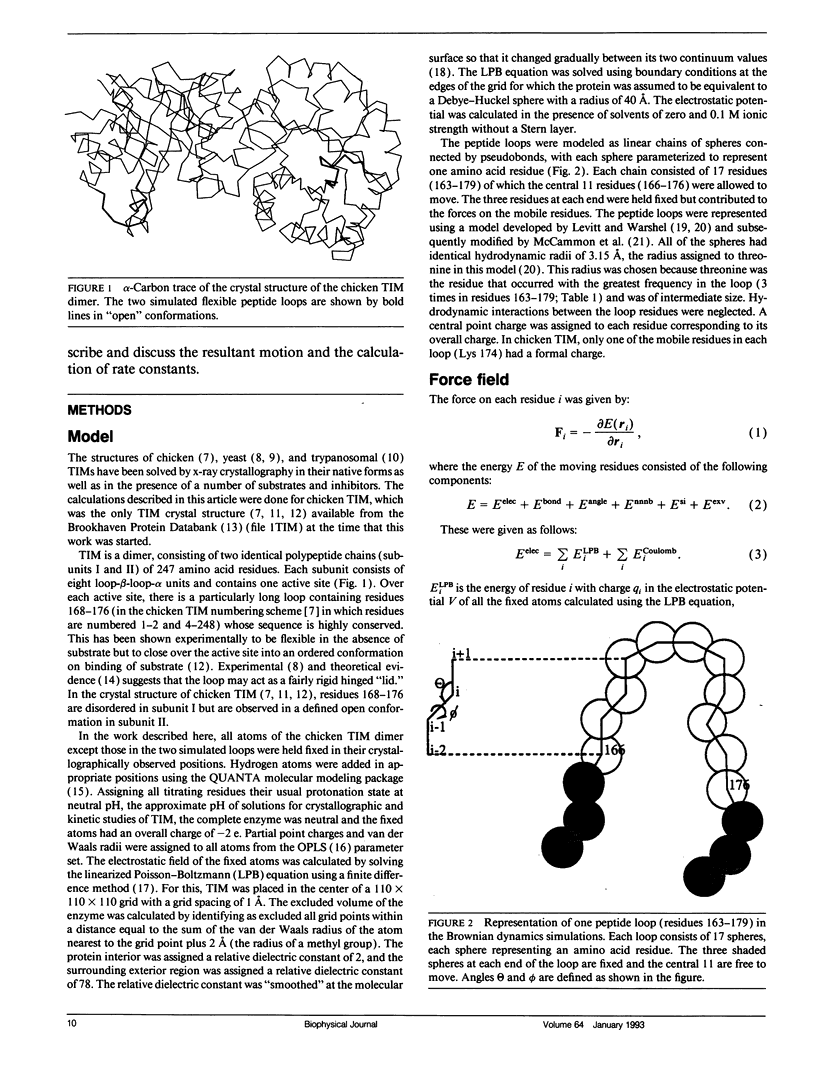
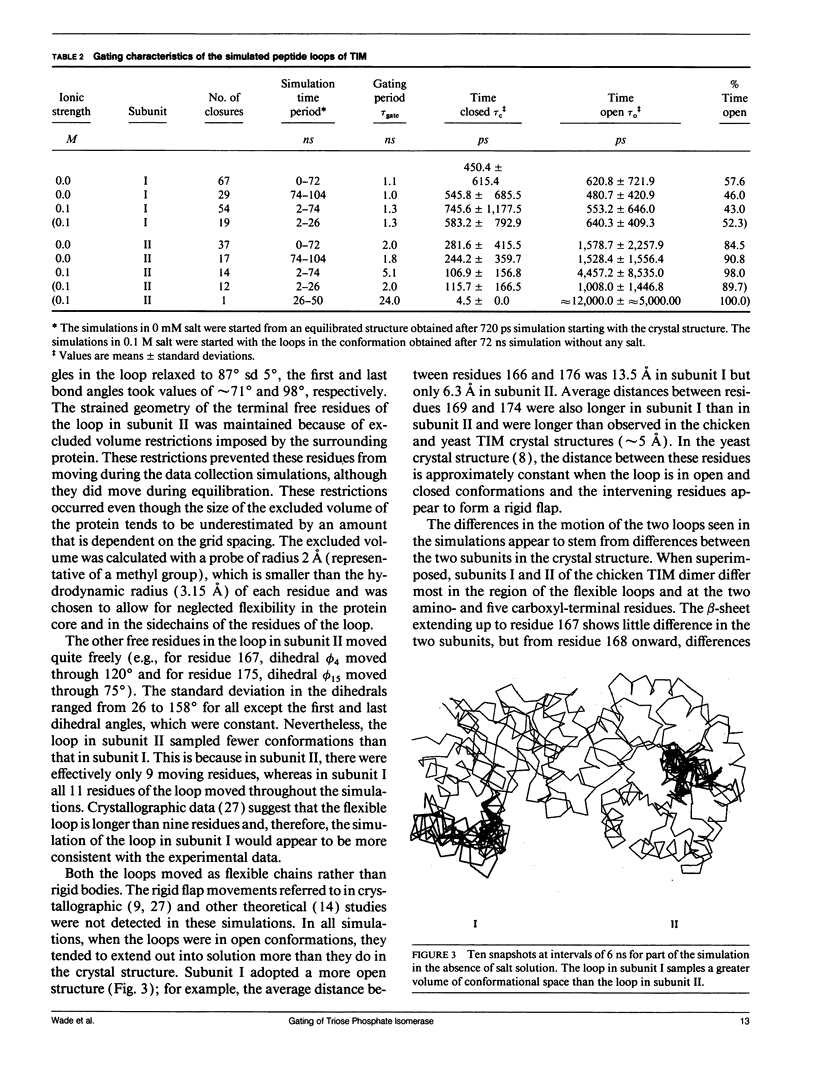
The enzyme triose phosphate isomerase has flexible peptide loops at its active sites. The loops close over these sites upon substrate binding, suggesting that the dynamics of the loops could be of mechanistic and kinetic importance. To investigate these issues, the loop motions in the dimeric enzyme were simulated by Brownian dynamics. The two loops, one on each monomer, were represented by linear chains of appropriately parameterized spheres, each sphere corresponding to an amino acid residue. The loops moved in the electrostatic field of the rest of the enzyme, which was held rigid in its crystallographically observed conformation. In the absence of substrate, the loops exhibited gating of the active site with a period of about 1 ns and occupied "closed" conformations for about half of the time. As the period of gating is much shorter than the enzyme-substrate relaxation time, the motion of the loops does not reduce the rate constant for the approach of substrate from its simple diffusion-controlled value. This suggests that the flexible loops may have evolved to create the appropriate environment for catalysis while, at the same time, minimizing the kinetic penalty for gating the active site.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alber T., Banner D. W., Bloomer A. C., Petsko G. A., Phillips D., Rivers P. S., Wilson I. A. On the three-dimensional structure and catalytic mechanism of triose phosphate isomerase. Philos Trans R Soc Lond B Biol Sci. 1981 Jun 26;293(1063):159–171. doi: 10.1098/rstb.1981.0069. [DOI] [PubMed] [Google Scholar]
- Alber T., Gilbert W. A., Ponzi D. R., Petsko G. A. The role of mobility in the substrate binding and catalytic machinery of enzymes. Ciba Found Symp. 1983;93:4–24. doi: 10.1002/9780470720752.ch2. [DOI] [PubMed] [Google Scholar]
- Albery W. J., Knowles J. R. Free-energy profile of the reaction catalyzed by triosephosphate isomerase. Biochemistry. 1976 Dec 14;15(25):5627–5631. doi: 10.1021/bi00670a031. [DOI] [PubMed] [Google Scholar]
- Banner D. W., Bloomer A. C., Petsko G. A., Phillips D. C., Pogson C. I., Wilson I. A., Corran P. H., Furth A. J., Milman J. D., Offord R. E. Structure of chicken muscle triose phosphate isomerase determined crystallographically at 2.5 angstrom resolution using amino acid sequence data. Nature. 1975 Jun 19;255(5510):609–614. doi: 10.1038/255609a0. [DOI] [PubMed] [Google Scholar]
- Banner D. W., Bloomer A. c., Petsko G. A., Phillips D. C., Wilson I. A. Atomic coordinates for triose phosphate isomerase from chicken muscle. Biochem Biophys Res Commun. 1976 Sep 7;72(1):146–155. doi: 10.1016/0006-291x(76)90972-4. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Blacklow S. C., Raines R. T., Lim W. A., Zamore P. D., Knowles J. R. Triosephosphate isomerase catalysis is diffusion controlled. Appendix: Analysis of triose phosphate equilibria in aqueous solution by 31P NMR. Biochemistry. 1988 Feb 23;27(4):1158–1167. doi: 10.1021/bi00404a013. [DOI] [PubMed] [Google Scholar]
- Davis M. E., Madura J. D., Sines J., Luty B. A., Allison S. A., McCammon J. A. Diffusion-controlled enzymatic reactions. Methods Enzymol. 1991;202:473–497. doi: 10.1016/0076-6879(91)02024-4. [DOI] [PubMed] [Google Scholar]
- Joseph D., Petsko G. A., Karplus M. Anatomy of a conformational change: hinged "lid" motion of the triosephosphate isomerase loop. Science. 1990 Sep 21;249(4975):1425–1428. doi: 10.1126/science.2402636. [DOI] [PubMed] [Google Scholar]
- Knowles J. R. Enzyme catalysis: not different, just better. Nature. 1991 Mar 14;350(6314):121–124. doi: 10.1038/350121a0. [DOI] [PubMed] [Google Scholar]
- Levitt M. A simplified representation of protein conformations for rapid simulation of protein folding. J Mol Biol. 1976 Jun 14;104(1):59–107. doi: 10.1016/0022-2836(76)90004-8. [DOI] [PubMed] [Google Scholar]
- Levitt M., Warshel A. Computer simulation of protein folding. Nature. 1975 Feb 27;253(5494):694–698. doi: 10.1038/253694a0. [DOI] [PubMed] [Google Scholar]
- Lolis E., Alber T., Davenport R. C., Rose D., Hartman F. C., Petsko G. A. Structure of yeast triosephosphate isomerase at 1.9-A resolution. Biochemistry. 1990 Jul 17;29(28):6609–6618. doi: 10.1021/bi00480a009. [DOI] [PubMed] [Google Scholar]
- Lolis E., Petsko G. A. Crystallographic analysis of the complex between triosephosphate isomerase and 2-phosphoglycolate at 2.5-A resolution: implications for catalysis. Biochemistry. 1990 Jul 17;29(28):6619–6625. doi: 10.1021/bi00480a010. [DOI] [PubMed] [Google Scholar]
- McCammon J. A., Northrup S. H. Gated binding of ligands to proteins. Nature. 1981 Sep 24;293(5830):316–317. doi: 10.1038/293316a0. [DOI] [PubMed] [Google Scholar]
- Noble M. E., Wierenga R. K., Lambeir A. M., Opperdoes F. R., Thunnissen A. M., Kalk K. H., Groendijk H., Hol W. G. The adaptability of the active site of trypanosomal triosephosphate isomerase as observed in the crystal structures of three different complexes. Proteins. 1991;10(1):50–69. doi: 10.1002/prot.340100106. [DOI] [PubMed] [Google Scholar]
- Pompliano D. L., Peyman A., Knowles J. R. Stabilization of a reaction intermediate as a catalytic device: definition of the functional role of the flexible loop in triosephosphate isomerase. Biochemistry. 1990 Apr 3;29(13):3186–3194. doi: 10.1021/bi00465a005. [DOI] [PubMed] [Google Scholar]
- Wierenga R. K., Noble M. E., Postma J. P., Groendijk H., Kalk K. H., Hol W. G., Opperdoes F. R. The crystal structure of the "open" and the "closed" conformation of the flexible loop of trypanosomal triosephosphate isomerase. Proteins. 1991;10(1):33–49. doi: 10.1002/prot.340100105. [DOI] [PubMed] [Google Scholar]
- Wierenga R. K., Noble M. E., Vriend G., Nauche S., Hol W. G. Refined 1.83 A structure of trypanosomal triosephosphate isomerase crystallized in the presence of 2.4 M-ammonium sulphate. A comparison with the structure of the trypanosomal triosephosphate isomerase-glycerol-3-phosphate complex. J Mol Biol. 1991 Aug 20;220(4):995–1015. doi: 10.1016/0022-2836(91)90368-g. [DOI] [PubMed] [Google Scholar]


