Abstract
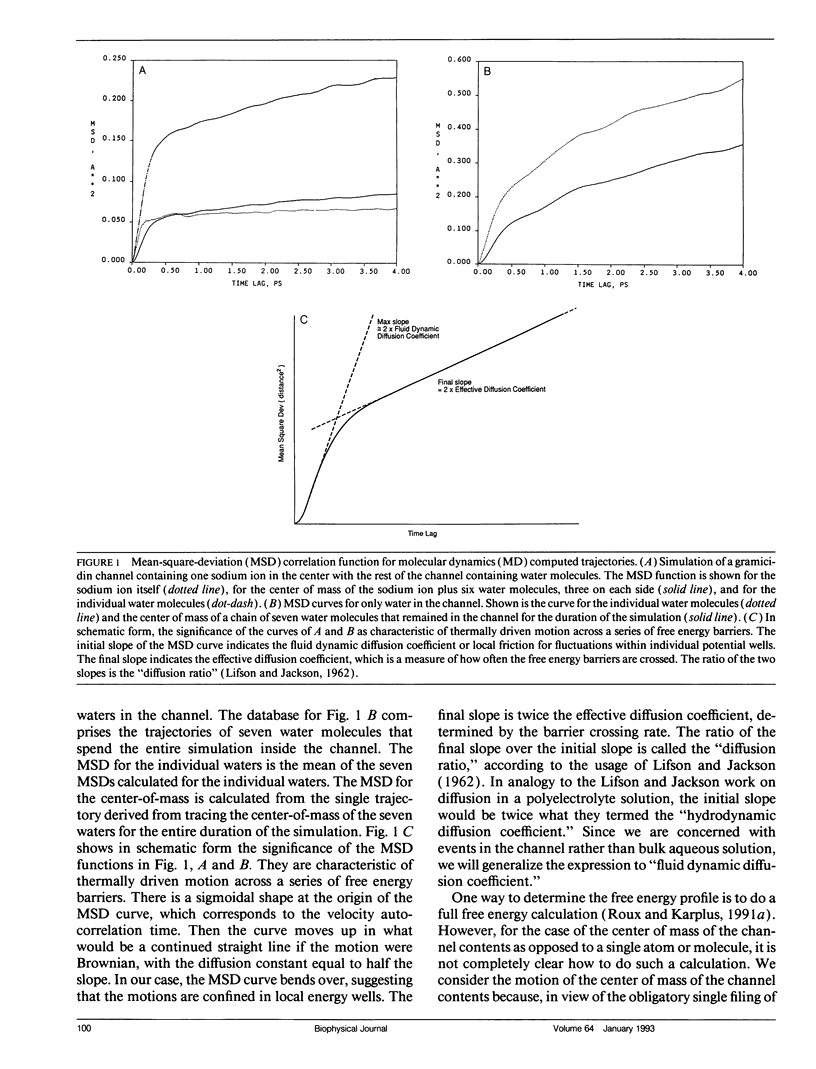
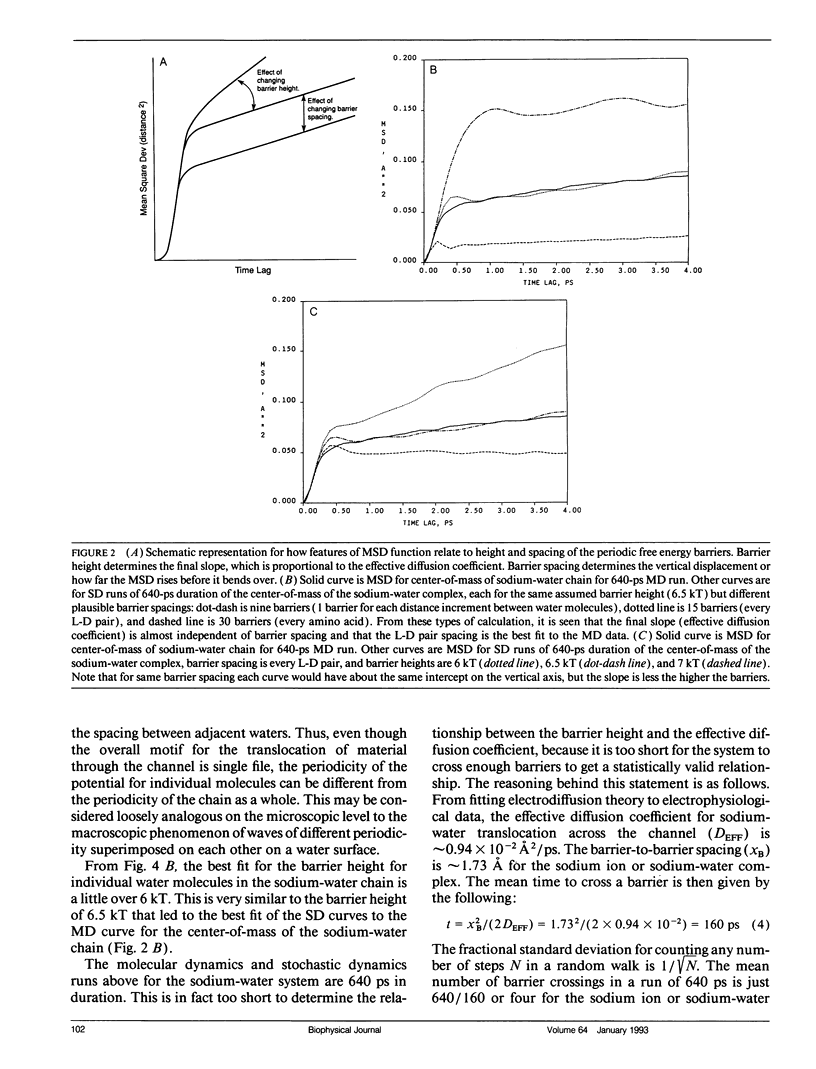
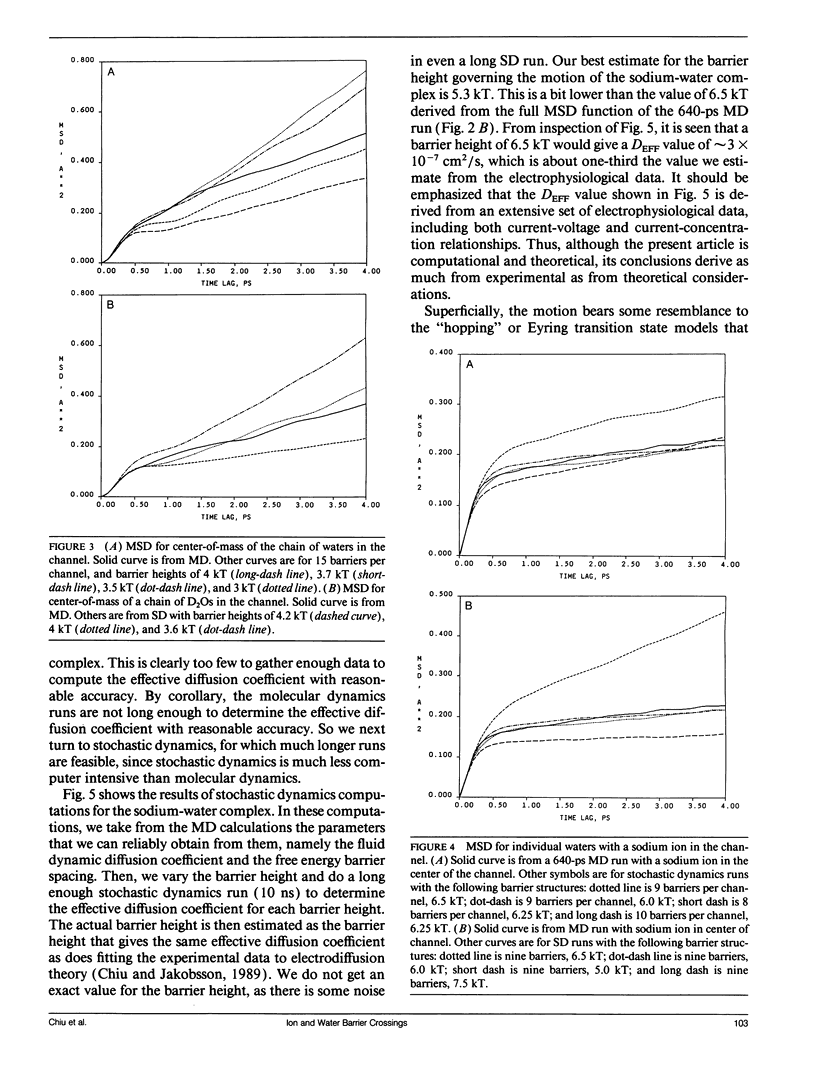
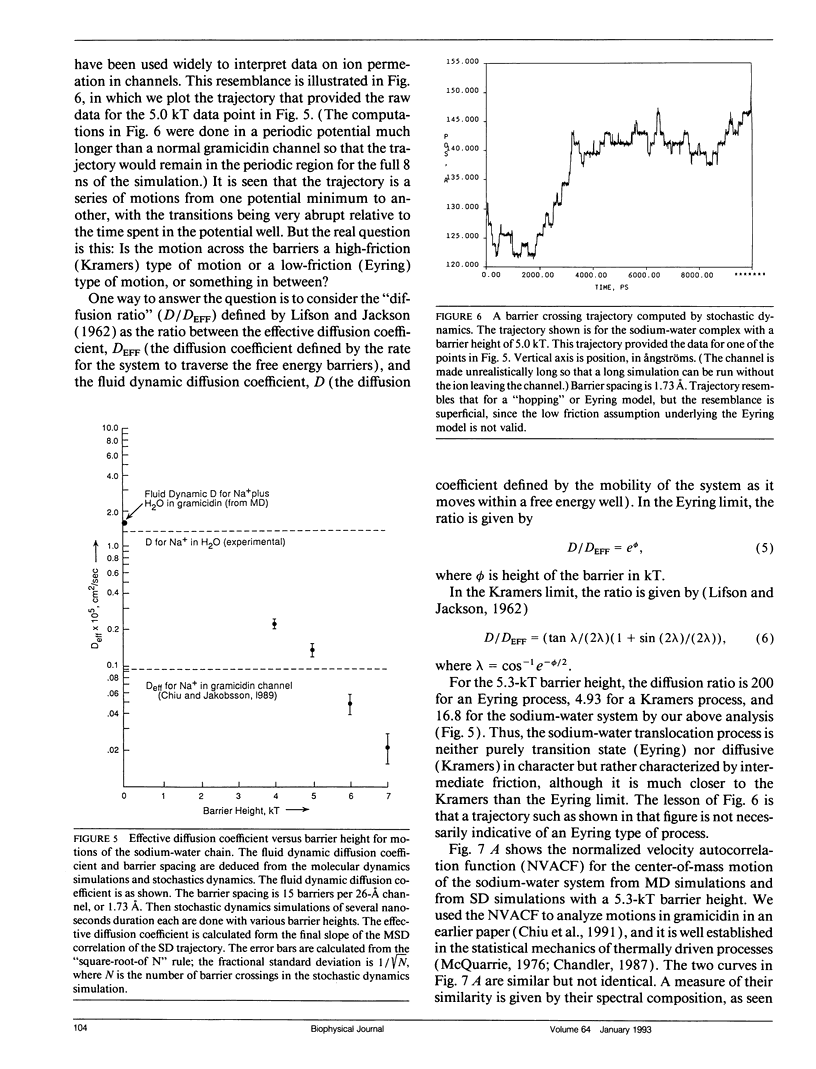
Using a combination of techniques, including molecular dynamics, time-correlation analysis, stochastic dynamics, and fitting of continuum diffusion theory to electrophysiological data, a characterization is made of thermally driven sodium, water, and D2O motion within the gramicidin A channel. Since the channel contents are constrained to move in a single-file fashion, the motion that corresponds to experimentally measurable rates of permeation of the membrane is the motion of the center of mass of the channel contents. We therefore emphasize channel contents center-of-mass motion in our analysis of molecular dynamics computations. The usual free energy calculation techniques would be of questionable validity when applied to such motion. As an alternative to those techniques, we postulate a periodic sinusoidal free energy profile (related to the periodic structure of the helical channel) and deduce the fluid dynamic diffusion coefficient and the height and spacing of the free energy barriers from the form of the mean-square-deviation function, using stochastic computations. The fluid dynamic friction in each case appears similar to that for aqueous solution. However, the diffusive motions are modulated by a spatially periodic free energy profile with a periodicity characteristic of an L-D pair of amino acids in the gramicidin helix, approximately 1.7 A in the model we use. The barrier height depends on which substance is moving in the channel, but in each case is several times thermal energy. For barriers of this width and height, the motion is intermediate between the low-friction (transition-state) and high-friction (Brownian) limits. Thus, neither of these formalisms that have been used commonly to describe membrane permeation gives an accurate picture of the underlying physical process (although the Brownian description seems closer to correct). The non-Markovian Langevin equation must be solved to describe properly the statistics of the process. The "channel state of matter" characteristic of the channel contents appears to have some properties typical of the solid and some typical of the liquid state. The magnitude of the local friction and nature of the ion solvation are similar to the liquid state, but the periodicities of structure, free energy, and dynamics are somewhat solid-like. The alignment of water dipoles in the channel bears some resemblance to the orientational ordering of a nematic liquid crystal, but unlike a nematic liquid crystal, the waters have a degree of translational order as well. Thus, the "channel state" is not adequately described by analogy to either the solid or liquid states or to liquid crystals but must be dealt with as its own characteristic type of condensed matter.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aqvist J., Warshel A. Energetics of ion permeation through membrane channels. Solvation of Na+ by gramicidin A. Biophys J. 1989 Jul;56(1):171–182. doi: 10.1016/S0006-3495(89)82662-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu S. W., Jakobsson E. Stochastic theory of singly occupied ion channels. II. Effects of access resistance and potential gradients extending into the bath. Biophys J. 1989 Jan;55(1):147–157. doi: 10.1016/S0006-3495(89)82786-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu S. W., Jakobsson E., Subramaniam S., McCammon J. A. Time-correlation analysis of simulated water motion in flexible and rigid gramicidin channels. Biophys J. 1991 Jul;60(1):273–285. doi: 10.1016/S0006-3495(91)82049-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu S. W., Subramaniam S., Jakobsson E., McCammon J. A. Water and polypeptide conformations in the gramicidin channel. A molecular dynamics study. Biophys J. 1989 Aug;56(2):253–261. doi: 10.1016/S0006-3495(89)82671-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper K. E., Gates P. Y., Eisenberg R. S. Diffusion theory and discrete rate constants in ion permeation. J Membr Biol. 1988 Dec;106(2):95–105. doi: 10.1007/BF01871391. [DOI] [PubMed] [Google Scholar]
- Cooper K., Jakobsson E., Wolynes P. The theory of ion transport through membrane channels. Prog Biophys Mol Biol. 1985;46(1):51–96. doi: 10.1016/0079-6107(85)90012-4. [DOI] [PubMed] [Google Scholar]
- Frauenfelder H., Wolynes P. G. Rate theories and puzzles of hemeprotein kinetics. Science. 1985 Jul 26;229(4711):337–345. doi: 10.1126/science.4012322. [DOI] [PubMed] [Google Scholar]
- Granick S. Motions and relaxations of confined liquids. Science. 1991 Sep 20;253(5026):1374–1379. doi: 10.1126/science.253.5026.1374. [DOI] [PubMed] [Google Scholar]
- Jakobsson E., Chiu S. W. Application of Brownian motion theory to the analysis of membrane channel ionic trajectories calculated by molecular dynamics. Biophys J. 1988 Oct;54(4):751–756. doi: 10.1016/S0006-3495(88)83012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson E., Chiu S. W. Stochastic theory of ion movement in channels with single-ion occupancy. Application to sodium permeation of gramicidin channels. Biophys J. 1987 Jul;52(1):33–45. doi: 10.1016/S0006-3495(87)83186-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P. C. Electrostatic modeling of ion pores. Energy barriers and electric field profiles. Biophys J. 1982 Aug;39(2):157–164. doi: 10.1016/S0006-3495(82)84503-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P. C. Ion-water and ion-polypeptide correlations in a gramicidin-like channel. A molecular dynamics study. Biophys J. 1990 Nov;58(5):1133–1156. doi: 10.1016/S0006-3495(90)82456-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt D. G. Electrostatic calculations for an ion channel. I. Energy and potential profiles and interactions between ions. Biophys J. 1978 May;22(2):209–219. doi: 10.1016/S0006-3495(78)85485-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay D. H., Berens P. H., Wilson K. R., Hagler A. T. Structure and dynamics of ion transport through gramicidin A. Biophys J. 1984 Aug;46(2):229–248. doi: 10.1016/S0006-3495(84)84016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux B., Karplus M. Ion transport in a model gramicidin channel. Structure and thermodynamics. Biophys J. 1991 May;59(5):961–981. doi: 10.1016/S0006-3495(91)82311-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell E. W., Weiss L. B., Navetta F. I., Koeppe R. E., 2nd, Andersen O. S. Single-channel studies on linear gramicidins with altered amino acid side chains. Effects of altering the polarity of the side chain at position 1 in gramicidin A. Biophys J. 1986 Mar;49(3):673–686. doi: 10.1016/S0006-3495(86)83694-3. [DOI] [PMC free article] [PubMed] [Google Scholar]


