Abstract
Background
DNA microarray technology is a powerful technique that was recently developed in order to analyze thousands of genes in a short time. Presently, microarrays, or chips, of the cDNA type and oligonucleotide type are available from several sources. The number of publications in this area is increasing exponentially.
Results
In this study, microarray data obtained from two different commercially available systems were critically evaluated. Our analysis revealed several inconsistencies in the data obtained from the two different microarrays. Problems encountered included inconsistent sequence fidelity of the spotted microarrays, variability of differential expression, low specificity of cDNA microarray probes, discrepancy in fold-change calculation and lack of probe specificity for different isoforms of a gene.
Conclusions
In view of these pitfalls, data from microarray analysis need to be interpreted cautiously.
Background
Traditionally, techniques for the study of gene expression were significantly limited in both breadth and efficiency since these studies typically allowed investigators to study only one or a few genes at a time. However, the recently developed DNA microarray technique is a powerful method that provides researchers with the opportunity to analyze the expression patterns of tens of thousands of genes in a short time [1]. Presently, several vendors offer these microarray systems, also known as chips, with a variety of technologies available. Currently, DNA microarrays are manufactured using either cDNA or oligonucleotides as gene probes. cDNA microarrays are created by spotting amplified cDNA fragments in a high density pattern onto a solid substrate such as a glass slide [1,2]. Oligonucleotide arrays are either spotted or constructed by chemically synthesizing approximately 25-mer oligonucleotide probes directly onto a glass or silicon surface using photolithographic technology [3].
Due to the powerful nature of microarrays, the number of relevant publications in this burgeoning field is increasing exponentially. During the years 1995–1997, the number of reports featuring microarray data was less than ten. However, in 2001 alone approximately 800 publications featured data generated by microarray studies (according to a PubMed search).
Microarray technology certainly has the potential to greatly enhance our knowledge about gene expression, but there are drawbacks that need to be considered. As Knight [4] cautioned, it is possible that errors could be incorporated during the manufacture of the chips. Consequently, the fidelity of the DNA fragments immobilized to the microarray surface may be compromised. However, there are few studies where the majority of the gene sequences spotted on the microarrays were verified [5]. Kuo et al (2002) compared the data from two high-throughput DNA microarray technologies, cDNA microarray (Stanford type) and oligonucleotide microarray (from Affymetrix) and found very little correlation between these two platforms [6]. Unfortunately, many investigators are reporting microarray data without confirming their results by other traditional gene expression techniques such as PCR, Northern blot analysis and RNase protection assay. Raw microarray data obtained from questionable nucleotide sequences are then often manipulated using cluster and statistical analysis software and subsequently reported in scientific journals. In addition the quality of the probe sequences and the location of the probes selected for incorporation into the array are also very important. For example, if probes are selected only from the 3' end of a given gene, then there is a strong possibility that different splice variants of that gene will not be identified if the alternative splicing occurs at the 5' region of the gene.
The development of a single chip containing the complete gene set for a given tissue or for a complex organism (30,000 to 60,000 genes) is likely in the near future, so it is paramount that chip manufacturers avoid these problems [7]. In this report, we demonstrate that microarray technology continues to be a dynamic and developing process and highlight potential pitfalls that must be addressed when interpreting data.
Results
Inconsistent sequence fidelity of spotted cDNA microarrays
cDNA microarray analysis was performed using the UniGEM-V chip (IncyteGenomics, Palo Alto, CA) with mRNA isolated from peripheral blood mononuclear cells (PBMC) of a large granular lymphocyte leukemia patient and a healthy control. In this microarray, 7075 immobilized cDNA fragments (4107 from known genes and 2968 ESTs) were immobilized onto a glass slide. After careful examination of the microarray probes, it was determined that the majority of the spotted cDNA fragments were from the 3' end of the genes. Approximately 80 up-regulated and 12 down-regulated genes were identified in leukemic LGL. We then purchased seventeen clones from IncyteGenomics containing cDNA fragments that represent fourteen of the up-regulated and three of the down-regulated genes. Plasmid DNA was isolated from the clones and the sequences were verified. Unfortunately, we found several problems with the insert DNA sequences in these clones. Four of the seventeen c DNA fragments spotted on the microarray contained incorrect sequences (23.5%) (Table 1).
Table 1.
Verification of genes spotted on cDNA microarray
| Gene Name | Size in kb | Balanced Differential Expression | Sequence correct / incorrect | Northern Blots Positive / negative |
|---|---|---|---|---|
| EST | 0. 250 | 17.7 | correct | negative |
| EST | 11.8 | incorrect | negative | |
| Granzyme H | 1.2 | 6.3 | correct | positive |
| NKG2C | 1.5 | 5.5 | correct | positive |
| Lymphopain | 1.2 | 5.4 | correct | positive |
| Interferon RF | 0.8 | 5.0 | incorrect | ------- |
| SCFM | ----- | 4.6 | correct | positive |
| Pac-1 | 1.2 | 4.2 | correct | positive |
| Perforin | 0.6 | 3.8 | correct | positive |
| A-20 | 0.6 | 3.2 | correct | positive |
| CHK | 3.2 | incorrect | ------ | |
| GCSF | 2.4 | 3.1 | incorrect | ------ |
| EST | 0.250 | 3.1 | correct | positive |
| Com.comp. C2 | 1.8 | 2.8 | correct | negative |
| IHH | 0.9 | -18.6 | correct | negative |
| H transport | 1.5 | -10.4 | correct | negative |
| Ribo.26 | 0.4 | -6.2 | correct | negative |
Variable reliability of differential expression data
The cDNA fragments corresponding to differentially expressed genes spotted on the microarrays were excised from the plasmid DNA and used as probes in Northern blots. Out of the seventeen only eight provided positive results as indicated by microarray (47%). Although all the sequences for the down-regulated genes were correct, Northern blot analysis with these probes did not show any differential expression of the genes. This is in contrast to the microarray data that suggested they were down regulated (Table 1).
Low specificity of cDNA microarray probes
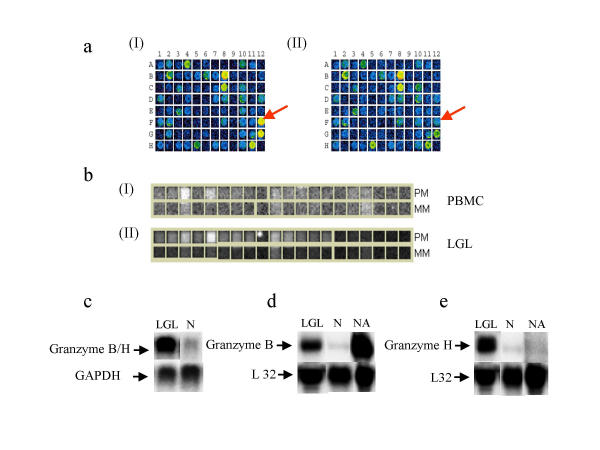
By microarray analysis, it is very difficult to distinguish between two genes that share a high degree of sequence similarity. Low specificity of probes is also a frequently encountered problem in oligonucleotide arrays. This problem is especially prevalent in instances where DNA sequences are nearly identical between two genes and the oligonucleotide probes are generated from the 3' end of the genes. For example, the 1.2 kb fragment (GB Accession No. M 57888) spotted on the cDNA microarray as granzyme B was not able to distinguish between granzyme B and H (Fig. 1a). The balanced differential expression of 6.3 was calculated. A probe set was generated by Affymetrix using the similar sequence information (GB Accession No.M28879) and according to oligonucleotide array, granzyme B was shown to be up-regulated (fold change 21.5: Fig. 1b). Northern blot analysis (using the same fragment as probe) did not discriminate between the genes for granzyme B versus granzyme H (Fig. 1c). However, by using gene-specific probes in an RNase protection assay, we were able to demonstrate the over-expression of granzyme B and granzyme H separately in leukemic LGL cells (Fig. 1d and 1e).
Figure 1.
a). A portion of a scanned cDNA microarray showing the differential expression of granzyme H. The cDNA fragment spotted on position F12 corresponds to granzyme H (indicated by arrow). (I). Hybridization profile for LGL leukemia cells. (II). Hybridization profile for control. Balanced differential expression, 6.3. b). Shows the differential expression of Granzyme B in oligonucleotide array. Calculated fold change is 20.5 (I). Hybridization pattern for the granzyme B probe set (with RNA isolated from normal PBMC), PM = Perfect match, MM = Mismatch. (II). Hybridization pattern for the granzyme B probe set (with RNA isolated from leukemic LGL), PM = Perfect match, MM = Mismatch c). Northern blot showing expression of granzyme B/H. The probe used was the same as the probes spotted on the microarray. Lane LGL = total RNA samples isolated from LGL leukemia patient Lane N = total RNA isolated from normal healthy control. NA = Total RNA isolated from PBMC of a normal healthy individual activated with IL2 and PHA. d). RNase protection assay for granzyme B. Probe set hAPO-4 obtained from PharMingen contains a specific probe for granzyme B. Note: Leukemic LGL cells over-expressed granzyme B (Lane LGL), whereas very low levels of granzyme B were observed in PBMC from normal healthy control (Lane N). Normal activated PBMC showed strong expression of granzyme B (Lane NA). e). RNase protection assay for granzyme H. Probe set hAPO-4 obtained from PharMingen contains a specific probe for granzyme H. Leukemic LGL cells showed over-expression (Lane LGL). PBMC obtained from a normal healthy individual (Lane N) and activated PBMC showed trace expression.
Discrepancy in fold change calculation for a given gene
It is very difficult to compare the exact fold change between two microarray techniques, and no standard value system is currently in place to compare the changes found in one microarray to the next. This fact was clearly demonstrated by Kuo et al (2002) in their recent publication [6]. In this paper we compared the fold change (Affymetrix) and balanced differential expression (cDNA) with Northern blot expression. For example, our IncyteGenomics cDNA microarray data demonstrated only a 3.8 differential expression in the expression of perforin (Fig. 2a), a pore-forming protein produced by cytolytic lymphocytes [8] in leukemic LGL cells, whereas the oligonucleotide microarray indicated a 103 fold increase (Fig. 2b). Using a probe identical to the one spotted on the cDNA microarray, we performed a Northern blot analysis. The blot demonstrated the up-regulation of the perforin transcript in leukemia LGL cells (Fig. 2c), but the fold increase was neither 103 as indicated by oligonucleotide array nor 3.8 as determined by the cDNA microarray data. Instead, the actual value was determined to fall between these two extreme values. These observations strongly suggest that results for significantly altered genes should be confirmed with other traditional techniques such as Northern blots or RNase protection assays prior to reporting the fold increase.
Figure 2.
a). A portion of a scanned cDNA microarray showing the differential expression of perforin in the cDNA microarray. Arrow indicates the position of spots corresponding to perforin (D2). Differential expression is only 3.8 b). Shows the differential expression of perforin in oligonucleotide array. Calculated fold change is 103. (I). Hybridization pattern for the perforin probe set (with RNA isolated from normal PBMC), PM = Perfect match, MM = Mismatch. (II). Hybridization pattern for the perforin probe set (with RNA isolated from leukemic LGL), PM = Perfect match, MM = Mismatch Note: Even though the gene is present in trace amounts in PBMC, it is calculated as absent because of the high background caused by MM hybridization. c). Northern blot showing the expression of perforin. Lane LGL = total RNA obtained from leukemic LGL. Lane N = total RNA isolated from normal healthy individuals. Sample from LGL leukemia patients showed over-expression of perforin.
Lack of probe specificity for gene isoforms
One of the genes spotted on the cDNA microarray that we are interested in is (Phosphatase in Activated Cells) PAC-1[9]. The differential expression of PAC-1 by both cDNA microarray (differential expression 4.2) and oligonucleotide arrays (fold change 1.6) is shown in Figures 3a and 3b. Using a cDNA fragment identical to the PAC-1 probe on the cDNA microarray, we performed a Northern blot analysis and confirmed the over-expression of two transcripts in leukemic LGL cells (Fig. 3c). RT-PCR was performed using total RNA from leukemic LGL and specific probes designed to amplify full-length PAC-1. We did not see amplification of any product. In addition, we found no PAC-1 expression using two different monoclonal anti-PAC-1 antibodies in Western blot analysis (data not shown). The monoclonal antibodies obtained from Santa Cruz were based on the amino acid sequence information obtained from the N-terminus and C-terminus of the PAC-1. The results of all the experiments did not confirm the over-expression of PAC-1. Therefore, to obtain more information about the structure of the PAC-1 related genes in leukemic LGL, we screened an LGL leukemia cDNA library using a 1.2 kb PAC-1 cDNA fragment and identified similar genes which are different forms of PAC-1 (GenBank Accession #AF331843, the other sequence is not deposited). Similarly an anti-apoptotic gene A20 is also over-expressed in leukemic LGL, but protein expression was absent when Western blots were performed with monoclonal antibodies raised against the amino acid sequence derived from A20 (data not shown).
Figure 3.
a). A portion of a scanned cDNA microarray showing the differential expression of PAC-1. The cDNA fragment spotted on position E6 corresponds to PAC-1. (I) Hybridization pattern for LGL patient. (II). Hybridization pattern for control. b). Shows the differential expression of PAC-1 in oligonucleotide array. Calculated fold change is 1.6 (I) Hybridization pattern for the PAC-1 probe set (RNA isolated from normal PBMC. PM = Perfect Match, MM = Mismatch. (II) Hybridization pattern for the PAC-1 probe set (RNA isolated from leukemic LGL cells. PM = Perfect Match, MM = Mismatch. c). Northern blot showing the expression of PAC-1 like genes. The probe used was the same as the one spotted on the microarray. Lane LGL = total RNA isolated from LGL leukemia patients (LGL). Lane N = total RNA isolated from normal healthy controls.
Likewise, another gene of interest, NKG2 C, showed a balanced differential expression of 5.5 (Fig. 4a). By using a probe derived from an NKG2 C clone, we identified a number of transcripts by Northern blot analysis (Fig. 4b). In order to ascertain more structural information, we again screened the LGL leukemia library and identified the presence of several members of the NKG2 gene family including NKG2 A, NKG2 D, NKG2 E and NKG2 F (GB Accession Nos. AF461812, AF461811, AF461157) [10]. Therefore, if genes similar to NKG2 family members are spotted on a microarray, it may be difficult to confirm which form of the gene is differentially expressed in a given sample.
Figure 4.
a). A portion of a scanned cDNA microarray showing the differential expression of NKG2 C. The cDNA fragment spotted on position H11 correspond to NKG2 C. (I) Hybridization pattern for LGL patient. (II). Hybridization pattern for control. b). Northern Blot showing the expression of NKG2 family members. The probe used was the same as the one spotted on the microarray. Lane LGL = total RNA isolated from LGL leukemia patients (LGL). Lanes N = total RNA isolated from normal healthy controls.
Mismatch probe sets mask the perfect match signals in oligonucleotide array (Affymetrix)
In order to accomplish the highest sensitivity and specificity in the presence of a complex background, Affymetrix introduced a system that entails the use of a series of specific and non-specific gene probe sets that are intended to result in a more accurate discrimination between true signal and random hybridization. Each probe set consists of a pair of 25-mer probes, one that represents a perfect match (PM)to the mRNA of interest, and a second probe differing by only one nucleotide, the mismatch (MM). The mismatch in the middle position theoretically provides maximal disruption of hybridization. Unfortunately, the use of the mismatch probe information can interfere with fold change calculations of gene expression. For example, perforin transcripts showed strong hybridization to both PM and MM probe sets. As a consequence, the strong MM signal masked the PM signal resulting in a low expression readout, even though the gene was present in normal PBMC (Fig. 2b). Therefore, the subsequently calculated fold increase from the test sample was extraordinarily high and deemed unreliable. Similarly, the fold change calculation was underestimated for PAC-1 due to the strong signal displayed for MM probe set (Fig. 3b). Genes such as human auto-antigen (GenBank Accession #L26339) and carboxyl ester lipase-like protein (GenBank Accession #L14813), are additional examples where these genes are present in LGL sample, but because of the strong signals associated with some of the MM probes, they are considered absent in the samples (Fig. 5a and 5b).
Figure 5.
a). Hybridization pattern for the human autoantigen probe set (with RNA isolated from normal PBMC), PM = Perfect match, MM = Mismatch. b). Hybridization pattern for probe set human carboxy ester lipase-like protein (with RNA isolated from leukemic LGL. PM = Perfect match, MM = Mismatch Note: Even though the gene is present in trace amounts, it is calculated as absent because of the high background caused by MM hybridization.
Discussion
In order to identify the differentially expressed genes in large granular lymphocytic (LGL) leukemia, we performed microarray analysis using the UniGEM-V microarray from IncyteGenomics and the HU6800 oligonucleotide array from Affymetrix. In the course of our analysis, we discovered several problems that we feel could occur in other studies that might lead to false conclusions.
Approximately 80 up-regulated genes and 12 down-regulated genes were identified by cDNA microarray analysis in leukemic LGL cells. Since microarray technology was a new tool at that time, we decided to verify the sequences of all the genes that were differentially expressed. To that end, we purchased approximately 20 clones representing the differentially expressed genes and verified the sequences. We found that only approximately 70% of the genes spotted on the microarray matched the correct sequence of the clones. Other groups reported similar observations. For example, IMAGE mouse cDNA clones (approximately 1200) were purchased from Research Genetics (Huntsville, Alabama) and sequences were verified by Halgren et al [11]. This group found that only 62% were definitely identified as a pure sample of the correct clones. In another study, PCR amplification products (previously sequence-verified cDNA clones) were re-sequenced and only 79% of the clones matched the original database [12]. In a different study, it was estimated that only 80% of the genes in a set of microarray experiments were correctly identified [5]. Therefore, we advise that when preparing cDNA microarrays (commercial or homemade), it is necessary to sequence verify each clone at the final stage before printing the microarray. If mistakes are made at this stage, it is not possible to correct them later by using the most sophisticated analytical tools.
We used cDNA microarray analysis to compare the gene expression profile of leukemic LGL cells obtained from a patient versus the expression profile of PBMC obtained from a normal healthy individual as a control. We decided to verify the microarray results using samples from more patients by employing the use of other methods such as PCR, Northern blot and RNase protection assay. To our surprise, none of the three down-regulated genes studied exhibited differential expression in Northern blots when the cDNA fragments of these genes were used as probes. In the up-regulated genes, only 47 % proved to support the results from the microarray data. The rest either displayed no signal, were not detectable in any sample or failed to reveal any differential expression whatsoever. Although some genes such as PAC-1 and A20 showed differential expression in LGL leukemia patients, no product amplification was obtained using RT-PCR with gene-specific primers.
By microarray analysis, it is very difficult to distinguish between two similar genes. The best example in our case is when granzyme B and granzyme H are compared. These two genes share approximately 80% similarity at the DNA level but have different enzymatic activities [13,14]. Using either one of the genes as a probe, both cDNA microarray and northern blot analysis indicated over-expression of both genes indiscriminately (Fig. 1). However, using gene-specific probes in an RNase protection assay, we were able to distinctly identify the over-expression of both granzyme B and H in leukemic LGL cells (Fig. 1d and 1e). In normal PBMC only trace amounts of both genes were identified, but after activation by PHA and IL2 only granzyme B was up-regulated. It is very difficult to get this information by microarray analysis alone. Therefore, caution in presenting microarray data without verification and confirmation is advised.
When the results from two different microarray technologies (cDNA and oligonucleotide arrays) were compared, the differential expression in some of the genes appeared to agree in both cases but a large variation in expression profiles between the two microarrays was clearly evident. Previously, such systematic differences in the two technologies were reported [6]. For example, perforin showed a 103-fold change in the Affymetrix array, whereas the cDNA microarray showed only a balanced differential expression of 3.8-fold. Northern blot results indicate that the genes were over-expressed, but the actual value is in between the values from the two microarrays. This problem may be due to an inaccurate fold change calculation due to the inclusion of mismatch values in the formula. We observed that many over-expressed genes were not properly identified at times. This may be the result of the introduction of mismatch values in the Affymetrix system. For example, genes for human autoantigen and human carboxyl ester lipase-like protein would be considered up-regulated in the microarray (according to PM match hybridization) if the MM hybridization values were ignored in the fold change calculation.
DNA microarray anlysis can be a powerful technique to identify differentially expressed genes but differentiating between splice variants can be problematic. For example, although the differential expression of the several genes such as PAC-1 and A20 were confirmed by northern blot analysis, we were unable to see any expression of protein corresponding to these genes by Western blot analysis. We were also unable to amplify those genes using gene-specific primers by RT-PCR. After screening the LGL library, we obtained several full-length genes that were different from both the 5' and 3' ends of PAC1. Similarly, we screened an LGL leukemia library and obtained several 1.5 kb cDNA fragments using the A20 cDNA as a probe. The deduced amino acid sequences of these genes revealed different proteins.
We found an up-regulation of NKG2C with a balanced differential expression of 5.8 in cDNA microarray (Fig. 4a). When Northern Blot analysis was performed using NKG2 C cDNA as a probe, we identified multiple transcripts. Screening the LGL leukemia library resulted in the identification of several other members of the NKG2 family such as NKG2 A, D, E, and F[10]. Therefore, it can be very difficult to distinguish different forms of genes if they are similar in certain sequence regions.
Conclusions
At the time of writing this report there were approximately 1150 articles published describing microarray results (PubMed). There is no doubt that these results will provide an overall idea of gene expression and contribute to understanding the molecular mechanisms involved in various processes. However, as demonstrated by our findings, the development of a standardized microarray system is needed to obtain more meaningful data from these experiments. The introduction of more uniform systems combined with the consideration of the above described pitfalls and alternatives will allow better utilization of this powerful technique in an expanding collection of scientific endeavors. It will be very helpful for the scientific community if the verified data is deposited in a public data base.
Methods
Isolation of PBMC and RNA
PBMC were isolated from whole blood using Ficoll-Hypaque density gradient centrifugation. These cells were suspended in Trizol reagent (GIBCO-BRL, Rockville, MD) and total RNA was isolated immediately according to the manufacturer's instructions. Poly A+ RNA was isolated from total RNA by using Oligo-Tex mini mRNA kit (Qiagen, Valencia, CA) according to the manufacturer's recommendations.
Activation of PBMC
Normal PBMC were cultured in vitro and activated by PHA, (Sigma Chemical Co. St. Louis, MO) (1 μg/ml, 2 days) and Interleukin-2 (IL-2) (100 U/ml, 10 days), then total RNA was isolated.
cDNA microarray analysis
Microarray probing and analysis was performed by IncyteGenomics. Briefly, one μg of Poly (A) + RNA isolated from PBMC of an LGL leukemia patient and healthy individual was reverse transcribed to generate Cy3 and Cy5 fluorescent labeled cDNA probes. cDNA probes were competitively hybridized to a human UniGEM-V cDNA microarray containing approximately 7075 immobilized cDNA fragments (4107 for known genes and 2968 for ESTs). Microarrays were scanned in both Cy3 and Cy5 channels with an Axon GenePix scanner (Foster City, CA) with a 10 μm resolution. P1 and P2 signals are the intensity reading obtained by the scanner for Cy3 and Cy5 channels. The balanced differential expression was calculated using the ratio between the P1 signal (intensity reading for probe 1) and the balanced P2 signal (intensity reading for probe 2 adjusted using the balanced coefficient)
Incyte GEMtools software (Incyte Pharmaceuticals, Inc., Palo Alto, CA) was used for image analysis. A gridding and region detection algorithm determined the elements. The area surrounding each element image was used to calculate a local background and was subtracted from the total element signal. Background subtracted element signals were used to calculate Cy3:Cy5 ratio. The average of the resulting total Cy3 and Cy5 signal gave a ratio that was used to balance or normalize the signals.
Oligonucleotide microarray analysis
The HU 6800 microarray was obtained from Affymetrix (Santa Clara, CA). Briefly, total RNA isolated from normal PBMC and leukemic LGL were DNase-treated and purified with a Qiagen kit (Valencia, CA). Approximately 10 μg of purified RNA was used to prepare double-stranded cDNA (Supercript GIBCO/BRL, Rockville, MD) using a T7 (dT)24 primer containing a T7 RNA polymerase promoter binding site. Biotinylated complementary RNA was prepared from 10 μg of cDNA and then fragmented to approximately 50 to 100 nucleotides. In vitro transcribed transcripts were hybridized to the HU 6800 microarray for 16 h at 45°C with constant rotation at 60 rpm. Chips were washed and stained by using the Affymetrix fluidics station. Fluorescence intensity was measured for each chip and normalized to the fluorescence intensity for the entire chip.
Verification of the clones
GEM cDNA clones (supplied as a bacterial stab) were purchased from IncyteGenomics and streaked on LB agar plates containing the appropriate antibiotic. Individual colonies were picked and grown in LB medium. Plasmid DNA was isolated and sequenced in order to verify the sequence identity.
Northern blot analysis
Northern Blotting was performed as described. Briefly 10 μg of total RNA from each sample was denatured at 65°C in RNA loading buffer, electrophoresed in a 1% agarose gel containing 2.2 M formaldehyde, then blotted onto a Nytran membrane (Schleicher & Schuell, Inc, Keene, N.H). The RNA was fixed to the membrane by UV cross-linking. cDNA was labeled with [32P] and purified using Nick columns (Amersham Pharmacia Biotech AB, Piscataway, NJ). Hybridization and washing of the blots were performed as described by Engler-Blum et al [15].
RNase protection assay (RPA)
RPAs were performed using the RNA isolated from leukemic LGL, normal PBMC and normal PBMC activated by IL-2 and PHA. Five μg of total RNA was hybridized to the in vitro transcribed hAPO-4 probe set (PharMingen, SanDiego, CA), and the RPA assay was performed according to the manufacturer's protocol. After the assay, the samples were resolved on a 5% polyacrylamide gel. The gel was dried and exposed to X-ray film. After developing the film, the bands were quantitated by using the ImageQuant program and normalized with the housekeeping gene, L32.
Western immunoblot analysis
Cells were lysed in a buffer containing 50 mM Tris-HCl (pH 7.6), 5 mM EDTA, 150 mM NaCl, 0.5 % NP-40, and 0.5% Triton X-100 containing 1 μg/ml leupeptin, aprotinin and antipain; 1 mM sodiumorthovanadate; and 0.5 mM PMSF (all reagents were obtained from Sigma Chemical Co.). Twenty-five μg of total protein from each sample was subjected to 10% SDS-PAGE. Then the proteins were transferred to a membrane and Western blotting was performed using the monoclonal antibody for PAC-1 and A20, followed by the ECL technique as recommended by the manufacturer (Amersham Biosciences, Piscataway, NJ).
Authors' contributions
RK conceived of the study along with TPL, isolated, purified RNA from the samples for microarray and performed all the experiments to validate the microarray data and analysed the data and drafted the manuscript. SJY verified the microarray data and participated in validation of the microarray. SM performed microarray analysis and analyzed the data and TPL conceived of the study, and participated in its design and coordination.
Contributor Information
Ravi Kothapalli, Email: kothapar@moffitt.usf.edu.
Sean J Yoder, Email: yodersj@moffitt.usf.edu.
Shrikant Mane, Email: Shrikantmane@yale.edu.
Thomas P Loughran, Jr, Email: Loughrat@moffitt.usf.edu.
Acknowledgements
This investigation was supported by grants from the Veterans Administration Merit Review, National Cancer Institute, Hisamitsu Pharmaceutical Co, Inc., (CA83947, CA90633, G60203). We thank Susan Nyland and Steven Enkemann for critical reading of the manuscript and helpful suggestions.
References
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Hedge P, Qi R, Abernathy K, Gay C, Dharap S, Gaspard R, Hughes JE, Snesrud E, Lee N, Quackenbush J. A concise guide to cDNA microarray analysis. Biotechniques. 2000;29:548–562. doi: 10.2144/00293bi01. [DOI] [PubMed] [Google Scholar]
- Lipshutz RJ, Morris D, Chee M, Hubbell E, Kozal MJ, Shah N, Shen N, Yang R, Fodor SPA. Using oligonucleotide probe arrays to access genetic diversity. Biotechniques. 1995;19:442–447. [PubMed] [Google Scholar]
- Knight J. When the chips are down. Nature. 2001;410:860–861. doi: 10.1038/35073680. [DOI] [PubMed] [Google Scholar]
- Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, Iyer V, Jeffrey SS, Van de Rijn M, Waltham M. Systematic variation in gene expression patterns in human cancer cell lines. Nature Genetics. 2000;24:227–234. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- Kuo WP, Jenssen T, Butte AJ, Ohno-Machado L, Kohane IS. Analysis of matched mRNA measurements from two different microarray technologies. Bioinformatics. 2002;18:405–412. doi: 10.1093/bioinformatics/18.3.405. [DOI] [PubMed] [Google Scholar]
- Jain KK. Biochips for gene spotting. Science. 2001;294:621–623. doi: 10.1126/science.294.5542.621. [DOI] [PubMed] [Google Scholar]
- Liu CC, Walsh CM, Young JD-E. Perforin: structure and function. Immunol Today. 1995;16:194–201. doi: 10.1016/0167-5699(95)80121-9. [DOI] [PubMed] [Google Scholar]
- Rohan PJ, Davis P, Moskaluk CA, Kearns M, Krutzsch H, Siebenlist U, Kelly K. PAC-1 A mitogen-induced nuclear protein tyrosine phosphatase. Science. 1993;259:1763–1766. doi: 10.1126/science.7681221. [DOI] [PubMed] [Google Scholar]
- Glienke J, Sobanov Y, Brostjan C, Steffens C, Nguyen C, Lehrach HE, Hofer E, Francies F. The genomic organization of NKG2C, E, F, and D receptor genes in the human natural killer gene complex. Immunogenetics. 1998;48:163–173. doi: 10.1007/s002510050420. [DOI] [PubMed] [Google Scholar]
- Halgren RG, Fielden MR, Fong CJ, Zacharewski TR. Assessment of clone identity and sequence fidelity for 1189 IMAGE cDNA clones. Nucleic Acids Res. 2001;29:582–588. doi: 10.1093/nar/29.2.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor E, Cogdell D, Coombes K, Hu L, Ramdas L, Tabor A, Hamilton S, Zhang W. Sequence verification as quality-control step for production of cDNA microarrays. Biotechniques. 2001;31:62–65. doi: 10.2144/01311st01. [DOI] [PubMed] [Google Scholar]
- Poe M, Blake JT, Boulton DA, Gammon M, Sigal NH, Wu JK, Zweerink HJ. Human cytotoxic lymphocyte granzyme B. Its purification from granules and the characterization of substrate and inhibitor specificity. J Biol Chem. 1991;266:98–103. [PubMed] [Google Scholar]
- Edwards EM, Kam CM, Powers JC, Trapani JA. The human cytotoxic T cell granule serine protease granzyme H has chymotrypsin-like (Chymase) activity and is taken up into cytoplasmic vesicles reminiscent of granzyme B-containing endosomes. J Biol Chem. 1999;274:30468–30473. doi: 10.1074/jbc.274.43.30468. [DOI] [PubMed] [Google Scholar]
- Engler-Blum G, Meier M, Frank J, Muller GA. Reduction of background problems in non-radioactive Northern and Southern blot analyses enables higher sensitivity than 32P-based hybridizations. Anal Biochem. 1993;210:235–244. doi: 10.1006/abio.1993.1189. [DOI] [PubMed] [Google Scholar]