Abstract
Double-stranded RNA (dsRNA) is a common by-product of viral infections and a potent inducer of innate antiviral immune responses in vertebrates. In the marine shrimp Litopenaeus vannamei, innate antiviral immunity is also induced by dsRNA in a sequence-independent manner. In this study, the hypothesis that dsRNA can evoke not only innate antiviral immunity but also a sequence-specific antiviral response in shrimp was tested. It was found that viral sequence-specific dsRNA affords potent antiviral immunity in vivo, implying the involvement of RNA interference (RNAi)-like mechanisms in the antiviral response of the shrimp. Consistent with the activation of RNAi by virus-specific dsRNA, endogenous shrimp genes could be silenced in a systemic fashion by the administration of cognate long dsRNA. While innate antiviral immunity, sequence-dependent antiviral protection, and gene silencing could all be induced by injection of long dsRNA molecules, injection of short interfering RNAs failed to induce similar responses, suggesting a size requirement for extracellular dsRNA to engage antiviral mechanisms and gene silencing. We propose a model of antiviral immunity in shrimp by which viral dsRNA engages not only innate immune pathways but also an RNAi-like mechanism to induce potent antiviral responses in vivo.
Double-stranded RNA (dsRNA) is a hallmark of viral infections, and thus, it is not surprising that the immune system has evolved the capacity to recognize dsRNA and respond to it by mounting antiviral responses. In vertebrates, these innate antiviral responses rely in part on the recognition of dsRNA by Toll-like receptor 3 and by RNA-dependent protein kinase (32, 47). The consequences of dsRNA recognition include activation of the interferon system, initiation of apoptosis, and inhibition of cellular protein synthesis. From an evolutionary perspective, innate immune activation by dsRNA has long been thought to be exclusive to vertebrates. This view has been encouraged by the fact that genes encoding homologues of interferons, their receptors, and most of the prominent interferon-regulated genes are absent in fully sequenced invertebrate genomes (1, 7, 10, 11). Nevertheless, it is a reasonable expectation that invertebrates should have an innate immune system capable of recognizing dsRNA as a signature of viral infection. A previous study suggested such a capability by demonstrating that exposure of a marine shrimp to dsRNA induced innate antiviral immunity in a sequence-independent manner (36). The mechanisms underlying this phenomenon as well as its occurrence in otherinvertebrate taxa remain unknown, but it is clear that the recognition of dsRNA by another pathway, RNA interference (RNAi), is widely distributed among invertebrates and likely an important component of the invertebrate antiviral response.
RNAi comprises a set of related cellular processes by which dsRNA molecules direct the suppression of gene expression based on sequence homology between the dsRNA trigger and the target gene. The specific mechanisms used by dsRNA to exert its silencing effects are diverse and include target RNA degradation, chromatin modification, and translational repression. The biological consequences of this set of phenomena are significant: RNAi has been implicated in the control of gene expression during development (6, 9, 17), heterochromatin establishment and maintenance (13, 18, 30, 46), suppression of transposon activity (25, 33, 38, 41, 49), control of mRNA turnover in plants (14), and antiviral immunity (24, 28). The unifying mechanistic feature of all RNAi-related pathways seems to be the processing of dsRNAs by members of the Dicer family, RNase III type endonucleases that generate 21-to 23-bp duplexes with 5′ phosphates and 3′ dinucleotide overhangs (3). These short dsRNA duplexes are recruited by protein complexes, which then recognize cellular (or viral) RNA targets by virtue of their homology to the complex-bound short RNA. Regardless of the specific mechanism used (translational repression, mRNA degradation, or chromatin modification), these events result in sequence-specific gene silencing.
Several lines of evidence support the notion that the RNAi pathway plays a role in antiviral immunity in metazoa. The requirement for the RNAi machinery for transposon silencing in Caenorhabditis elegans (41) and Drosophila melanogaster (38) implies evolution of RNAi as a defense mechanism against foreign nucleic acids. More significantly, suppressors of RNAi, whose activity is essential for viral replication in cell culture, have been found in insect and vertebrate viruses (22, 28, 29). Furthermore, infection of animal cells with RNA viruses can lead to the accumulation of virus-specific short RNA species (ca. 20 bp) of both sense and antisense polarities, suggesting processing of viral dsRNA intermediates into short interfering RNAs (siRNAs) (28). The in vivo role of the RNAi pathway in insect antiviral immunity has been recently demonstrated by experiments where silencing of Argonaute protein family members involved in RNAi rendered mosquitoes more permissive to replication of an RNA virus (24). Finally, the feasibility of using exogenous dsRNAs and siRNAs targeting viral sequences to effectively control viral infections in vivo suggests that the antiviral functions of the RNAi pathway can be exploited for therapeutic purposes (4, 15, 43-45).
While RNAi has been clearly shown to function as an antiviral mechanism in several insects, its role in antiviral immunity in other invertebrates has remained unexplored. Moreover, until recently, the role of dsRNA as a sequence-independent inducer of innate immunity in invertebrates was unrecognized. It has previously been shown that dsRNA induces broad-spectrum antiviral immunity in a sequence-independent manner in a marine invertebrate, the shrimp Litopenaeus vannamei (36). This observation, together with the striking evolutionary conservation of the RNAi pathway, prompted us to consider the potential for the convergence of innate immunity and RNAi as two distinct antiviral mechanisms induced by a common virus-associated molecular pattern, dsRNA. Here, we show that dsRNA can engage both innate immune pathways and an RNAi-like mechanism to stimulate potent antiviral immunity in a marine shrimp.
MATERIALS AND METHODS
Shrimp, viruses, and challenge systems.
L. vannamei shrimp (1 to 2 g) were kept in artificial seawater (Marine Environment) and challenged using either individual or collective challenge systems. In the individual system, single animals were kept in 260-ml tissue culture flasks with ca. 100 ml of artificial seawater and 100% daily water exchange (34). Infection in this individual system allows good resolution of subtle differences between experimental treatments, mainly because the viral dose is not amplified via cannibalism. Alternatively, a collective infection system was used in which shrimp were kept in groups of 10 to 12 in 10-liter tanks connected to a water recirculation system with mechanical and biological filtration as well as UV sterilization. Collective challenge allows reinfection events and dose amplification to occur due to cannibalism during the course of the experiment. While less control over the total viral dose per shrimp is achieved in the collective challenge system, initial exposure is the same for all treatments. Most importantly, this system allows assessment of the effects of antiviral stimulation under conditions that mimic high population density (like those encountered in aquaculture systems and under certain environmental conditions). The white spot syndrome virus (WSSV)-containing extract used to challenge shrimp by intramuscular injection has been described elsewhere previously (34). This viral extract was used at a 4 × 10−8 dilution (weight of infected tissue:volume of saline) to typically yield mortalities between 60 and 80% or at higher concentrations to induce 90 to 100% mortality in unstimulated shrimp. Volumes of injected infective material were 20 μl per animal for all experiments, unless otherwise stated. For per os infection experiments, WSSV or Taura syndrome virus (TSV)-containing tissues were generated by infecting specific-pathogen-free L. vannamei shrimp (3 to 4 g) and collecting moribund and dead animals over a period of 7 to 10 days. Infected tissue was fed to experimental shrimp at a rate of 10% biomass per day for 3 consecutive days. Shrimp were fed artificial dry food throughout the experiments, except on days of viral infection. Statistical statements (one-tailed P values, unless otherwise indicated) were derived from pairwise comparisons of the fractions of dead and alive animals in experimental and control groups using Fisher's exact test. To silence endogenous genes, shrimp were injected with 5 μg of dsRNA, a dose determined to be effective for mRNA knockdown over a large number of experiments (data not shown). For sequence-independent antiviral immunity assays, 5 μg of dsRNA was also used, based on previous work showing that doses of 1 μg or more are consistently effective for inducing innate antiviral protection (reference 36 and data not shown). The doses of dsRNA required for sequence-specific antiviral protection were empirically determined as part of the present study and were found to be gene target specific.
dsRNA.
dsRNAs were generated by in vitro transcription as described previously (36), using recombinant pGEM-TE (Promega), pCR4.0 (Invitrogen), or pBluescript (Stratagene) as a template. Sequences representing portions of the ribonucleotide reductase small subunit (RR2), DNA polymerase (DP), and open reading frame (ORF) WSV252 from WSSV were obtained from the L. vannamei expressed sequence tag (EST) collection at http://www.marinegenomics.org. These WSSV-encoded sequences were generated from subtractive hybridization libraries from infected shrimp, and as such, they represent fragments of WSSV-expressed genes and not full-length cDNAs. When the ESTs overlapped with more than one of the computationally predicted ORFs from the WSSV genome, the annotations given corresponded to those ORFs representing the most significant match based on BLASTx (2). DP and RR2 inserts were amplified by PCR from the appropriate EST clone and subcloned into pGEM-TE for dsRNA preparation using T7 and SP6 RNA polymerases. WSV252 was similarly amplified and cloned into pCR4.0 for dsRNA synthesis using T3 and T7 RNA polymerases. NCBI accession numbers of the ESTs used as templates and insert sizes are as follows: WSV186/ribonucleotide reductase small subunit (755 bp), CK572927; WSV514/DNA polymerase (311 bp), CK572497; and WSV252 (446 bp), CX769772. vp19 DNA (366 bp) was amplified by PCR from a homogenate of WSSV-infected shrimp and cloned into pCR4.0 for dsRNA synthesis using T3 and T7 RNA polymerases. Primers for vp19 were CGAAGCTTG GCCACCACGACTAACACTC (forward) and CGGAGCTCCTGCCTCCTC TTGGGGTAAGAC(reverse). vp28 DNA (615 bp) was amplified similarly to the vp19 sequence, except that it was cloned into pCR2.1 and then subcloned into pBluescript KS for dsRNA synthesis using T3 and T7 RNA polymerases. Primers for vp28 were CGGGATCCATTGAAGGCCGCGCCATGGATC TTTCTTTCACTCT(forward) and CGGAGCTCTTACTCGGTCTCAGTG CCAGA (reverse). The TSV genomic fragment (542 bp) encoding a portion of the predicted protease gene (31) was cloned by reverse transcription-PCR from total RNA of TSV-infected shrimp using primers CGTATGGACGAAGCTTGTTGATGCC(forward) and CTGGGGGACCACTCTTGCTGTTC (reverse). The control dsRNA for duck immunoglobulin υ (Igυ) has been reported previously as an inducer of innate antiviral protection in shrimp (36). The dsRNA targeting hemocyanin was prepared from pBluescript KS hosting a fragment of the hemocyanin cDNA (NCBI accession number CK572713). The dsRNA targeting CDP (CUB domain protein) was prepared from pCR4.0 hosting a fragment of the CDP cDNA that had been amplified with primers CGCGTC ACTCGTTGAAGCTCA (forward) and CAGCTGCAGTCTGTGTTGTTC (reverse) from the full-length CDP cDNA (NCBI accession number AY907539). siRNAs with UU 3′ overhangs (sequences shown in Table 1) were purchased from Dharmacon. Poly(C-G) was from Sigma-Aldrich. Diced dsRNA was prepared by incubating dsRNA with human recombinant Dicer followed by purification by size exclusion according to the manufacturer's instructions (Ambion).
TABLE 1.
siRNAs used in this study
| Target gene | Target sequencea |
|---|---|
| vp19 | AGAAGGACAGCGAUUCUGA |
| vp19 | CGAUGAUGAGGACAAAUAU |
| vp19 | UUACACCAUGGAAGAUCUU |
| Hemocyanin | CACACCAUAUCGAGCGUAA |
| Hemocyanin | GUUGAUGUUAGCAAUAACA |
| Hemocyanin | AUACAUGGAUAACAUCUUC |
| Control siRNA | ACCAUCGAGCAUGAACAUC |
siRNAs contained the shown sequence as a 19-bp RNA duplex, with additional 3′ UU overhangs.
RNA blots.
Total RNA was resolved in denaturing agarose gels containing formaldehyde, transferred to nylon membranes (S&S), and probed using 32P-labeled randomly primed DNA (37). Hybridizations were performed overnight at 65°C in 1 mM EDTA-7% lauryl sulfate-0.5 M sodium phosphate dibasic, pH 7.2, followed by washing in decreasing concentrations of buffer SSC (150 mM sodium chloride-15 mM sodium citrate) in the presence of 0.1% lauryl sulfate. Signal on membranes was recorded with a Typhoon 9410 phosphorimager (Amersham).
RESULTS
Potent and specific antiviral immunity induced by dsRNA in vivo.
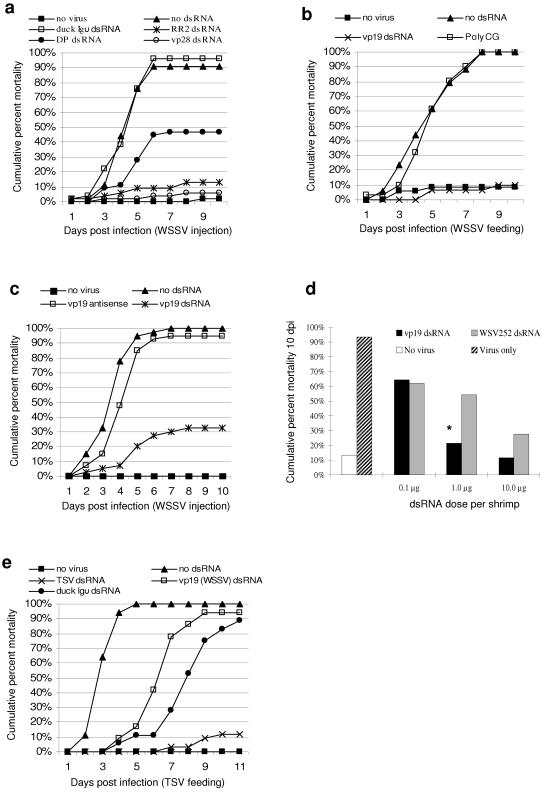
It has previously been shown that dsRNA induces innate antiviral immunity in shrimp in a sequence-independent manner (36). To test whether dsRNA also induces sequence-specific antiviral immunity, we took advantage of the fact that the innate antiviral protection afforded by dsRNA in shrimp is overcome when the virus is delivered shortly after dsRNA injection and when the dose of virus used is high (36). Thus, in an initial series of experiments, shrimp were injected with long dsRNA of either arbitrary sequence or WSSV-specific sequence and challenged by injection with a high dose of WSSV. As shown in Fig. 1a, a nonspecific dsRNA (for duck Igυ) failed to protect shrimp against challenge with this dose of virus, but treatment with dsRNAs derived from any one of three WSSV genes was significantly protective. The magnitude of antiviral protection (relative to no-dsRNA controls) afforded by the different WSSV-specific dsRNAs was varied: 78% for RR2 dsRNA, 85% for vp28 dsRNA, and only 44% for the DNA polymerase dsRNA. While this experiment (Fig. 1a) indicated viral sequence-specific dsRNA-mediated protection from a WSSV challenge administered by injection, the normal route of infection of shrimp with WSSV is the oral route, associated with the cannibalism of moribund and dead cohorts. Thus, the effect of injected dsRNA on the response of shrimp to oral WSSV infection was tested in a collective challenge system where shrimp were infected per os and allowed to further prey on infected sick and dead animals throughout the experiment. The results shown in Fig. 1b indicate that the sequence-independent innate immune response [induced in this case by poly(C-G)] was insufficient to protect against this high-dose oral challenge but that essentially complete protection was achieved by the administration of the same dose of dsRNA specific for vp19, a protein of the WSSV envelope. The ability of virus sequence-specific dsRNA to induce potent antiviral immunity is likely a dsRNA-mediated phenomenon, since treatment of shrimp with RNA antisense to vp19 was not sufficient to recapitulate the strong protective effects of vp19 dsRNA (Fig. 1c).
FIG. 1.
Potent and specific antiviral immunity induced by dsRNA in vivo. (a) Shrimp (1 to 2 g) were injected intramuscularly with either saline (no-virus and no-dsRNA controls), 12 μg of dsRNA for duck Igυ, or 12 μg of dsRNA for the WSSV gene RR2, DP, or vp28. dsRNAs were mixed with 20 μl of diluted WSSV-containing homogenate (6.6 × 10−7, wt/vol). An individual flask challenge system was used (n = 46 to 47). Every WSSV-specific dsRNA afforded significant antiviral immunity 10 dpi, compared to duck Igυ dsRNA (P < 0.0001 in every case). (b) Controls were as described above for a. One microgram of the indicated dsRNAs was injected per shrimp, followed by feeding of WSSV-containing tissue at 10% biomass daily for 3 consecutive days, starting 4 h after dsRNA injection. A collective challenge system was used (n = 30 to 35). vp19 dsRNA afforded significant antiviral immunity 10 dpi compared to poly(C-G) (P < 0.0001). (c) Twenty microliters of a mixture containing 3 μg of ssRNA or dsRNA and WSSV-positive homogenate (1 × 10−6, wt/vol) was injected into each of 40 shrimp. Controls and challenge system were as described above for a. vp19 dsRNA afforded significant antiviral protection 10 dpi compared to the no-dsRNA control (P < 0.001) (n = 40). (d) Injections and viral infections were as described above for b. The indicated doses of dsRNA for the WSSV genes vp19 and WSV252 were injected into individual shrimp. Bars represent cumulative percent mortality 10 days after initial viral exposure. A collective challenge system was used (n = 26 to 30). * indicates a significant difference (two-tailed; P < 0.05) between vp19 and WSV252 dsRNAs used at the same dose. Every dsRNA treatment afforded antiviral protection relative to the virus-only control (P < 0.01 in every case). (e) Shrimp (1 to 2 g) were injected with saline (no-virus and no-dsRNA controls) or with 5 μg of the indicated dsRNA and were then fed TSV-containing tissues at 10% biomass daily for 3 consecutive days, starting 4 h after dsRNA injection. The TSV dsRNA represents a portion of a putative protease encoded in the TSV genome. A collective challenge system was used (n = 34 to 36). Protection from viral infection by nonspecific dsRNAs was observed until 7 dpi (P < 0.01 in both cases), but by 11 dpi, only TSV-specific dsRNA was protective (P < 0.0001).
The dose dependence of the immunity induced by WSSV-specific dsRNA was tested in the oral challenge system using the vp19 sequence and WSV252, a viral ORF of unknown function. The results, shown in Fig. 1d, indicate that the induced protection was indeed dose dependent. vp19 dsRNA was highly protective at doses of 10 μg (81% protection) and 1 μg (72% protection), while at 0.1 μg, the level of protection was only 29%. When WSV252 dsRNA was used, a dose- dependent response was also observed, but the overall efficacy of this dsRNA was inferior to that of vp19. These data further demonstrate that dsRNA with viral sequence specificity induces potent antiviral immunity in a dose-dependent manner.
While the data shown in Fig. 1a and b are suggestive of sequence specificity in dsRNA-induced antiviral immunity, one alternative explanation for the dramatically different antiviral properties of dsRNAs of viral sequence and dsRNAs of arbitrary sequence is that the former induce stronger innate (nonspecific) immune responses than the latter due to some feature of dsRNAs derived from viruses that is not found in nonviral dsRNAs (e.g., biased base content or short virus-specific motifs). To address this possibility, the sequence specificity of antiviral protection was tested across two unrelated viruses. Shrimp were challenged with a high dose of TSV by the per os route after injection with duck Igυ dsRNA, vp19 dsRNA (from WSSV), or dsRNA for a portion of the putative protease encoded in the TSV genome (31). It was observed in these experiments that dsRNA of TSV-specific sequence provided effective protection from TSV infection (Fig. 1e), while vp19 dsRNA treatment resulted in only transient anti-TSV protection, comparable to that afforded by innate stimulation by duck Igυ dsRNA. Both non-TSV dsRNAs caused only a delay in the onset of mortality, which by 11 days postinfection (dpi) reached 89% and 94% for duck Igυ and vp19 dsRNA treatments, respectively. The ability of anti-TSV dsRNA to induce immunity against WSSV was also tested, and similar results were obtained (i.e., no cross-protection between TSV and WSSV) (data not shown). These data further demonstrate that strong dsRNA-mediated antiviral immunity is dependent upon sequence homology between the dsRNA trigger and viral targets. Collectively, the data shown in Fig. 1 demonstrate that (i) activation of a dsRNA-induced, sequence-dependent immune response provides strong and specific antiviral protection and (ii) the degree of protection is influenced by both the identity and dose of the dsRNA administered.
dsRNA-induced gene silencing in shrimp.
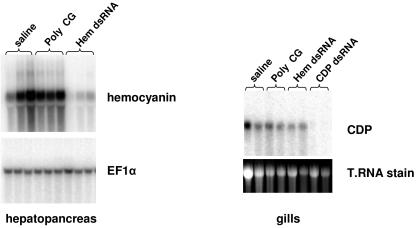
The data shown in Fig. 1 suggest engagement of an RNAi-like mechanism in antiviral protection induced by dsRNA against high doses of virus. Since functional RNAi in shrimp has not been previously reported, experiments were conducted to test the existence of RNAi-like responses in L. vannamei. Thus, long dsRNA specific for each of two endogenous genes was injected intramuscularly into the abdomen of shrimp and was followed 48 h later by analysis of the expression of the targeted mRNAs. One of the targeted genes encodes an isoform of hemocyanin (NCBI accession number AJ250830), the oxygen carrier protein in shrimp whose expression is restricted to the hepatopancreas. The other target is a novel gene of unknown function which we have termed CDP (NCBI accession number AY907539), for CUB (complement subcomponents C1r-C1s/sea urchin protein Uegf/bone morphogenetic protein 1) domain protein. The expression patterns and tissue distribution of CDP are not fully known, but EST analysis suggests that its mRNA is present in at least two tissues, hemocytes and gills (http://www.marinegenomics.org). The results shown in Fig. 2 indicate that injection of gene-specific dsRNA leads to substantial and specific depletion of the cognate mRNAs in hepatopancreas and gills. Injection of the dsRNA analogue poly(C-G) had no apparent effect on hemocyanin or CDP expression relative to injection of saline only. Likewise, injection of hemocyanin dsRNA had no effect on CDP expression. These results demonstrate that a dsRNA-induced gene-silencing pathway exists in shrimp and that it can be triggered systemically by injection of dsRNA into the abdominal muscle. While these results do not provide molecular mechanistic evidence of RNAi in shrimp, it is clear that dsRNA induces specific down-regulation of gene expression in these animals. These observations, taken together with the data shown in Fig. 1, suggest that a gene-silencing phenomenon induced by dsRNA can be mobilized in shrimp to provide strong antiviral immunity. Thus, dsRNA induces three distinct phenomena in shrimp: (i) an innate general immunity to viral infection (36) (Fig. 3c) that can be overwhelmed by high-dose viral challenge (Fig. 1a, b, and e), (ii) a much stronger sequence-specific antiviral response (Fig. 1), and (iii) specific down-regulation of gene expression (Fig. 2). All of these phenomena were induced by long dsRNA, raising the question of whether or not they can also be induced by siRNAs, the intermediates of the RNAi pathway downstream of initiating long dsRNA. This issue was addressed in the series of experiments described below.
FIG. 2.
dsRNA-induced gene silencing in shrimp. Individual shrimp were injected with saline or 5 μg of dsRNA as indicated. Forty-eight hours after injections, hemocyanin (Hem) expression and CDP expression were analyzed by Northern blot in hepatopancreas and gills, respectively. Expression of elongation factor 1α (EF1α) or total RNA (T.RNA) stained with ethidium bromide is shown as a reference. Each lane represents RNA from a single shrimp.
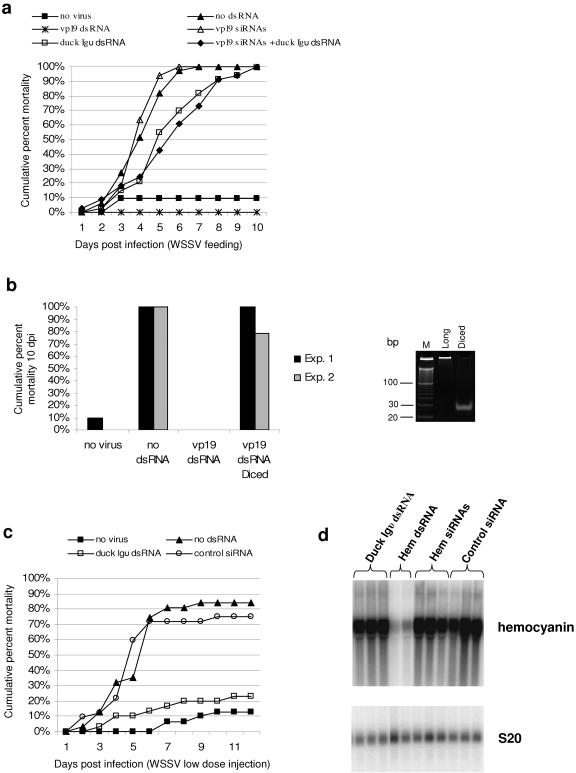
Injection of ectopic naked siRNAs does not induce antiviral immunity or gene silencing. (a) Shrimp (1 to 2 g) were injected with either saline (no-virus and no-dsRNA controls) or 2 μg of dsRNA as indicated. vp19 siRNAs are an equimolar cocktail of three synthetic siRNAs targeting vp19 (Table 1). WSSV-containing tissue was fed at 10% biomass daily for 3 consecutive days, starting 4 h after dsRNA injection. A collective challenge system was used (n = 32 to 33). vp19 long dsRNA afforded significant antiviral immunity 10 dpi compared to duck Igυ dsRNA, vp19 siRNA, vp19 siRNA plus duck Igυ dsRNA, and no-dsRNA treatments (P < 0.0001 in every case). (b) All controls were as described above for a. In experiment 1 (Exp. 1), shrimp were injected with 2 μg of dsRNA and challenged per os in a collective challenge system (n = 32 to 33). In experiment 2, shrimp were injected with 2.5 μg of dsRNA and challenged per os in the individual challenge system (n = 28 to 30). The inset on the right shows long vp19 dsRNA and diced vp19 dsRNA resolved by polyacrylamide gel electrophoresis and stained with ethidium bromide. M, 10-bp dsDNA ladder. (c) Shrimp (1 to 2 g) were injected with either saline (no-virus and no-dsRNA controls) or 5 μg of the indicated dsRNAs 72 h prior to infection by intramuscular injection of WSSV-homogenate (diluted 4 × 10−8, wt/vol). The control siRNA is of arbitrary sequence (Table 1). An individual challenge system was used (n = 30 to 32). Duck Igυ dsRNA afforded significant antiviral immunity 12 dpi compared to siRNAs and the virus-only control (P < 0.0001 in both cases). (d) Individual shrimp were injected with 5 μg of dsRNA as indicated, and hemocyanin (Hem) expression was analyzed by Northern blot in the hepatopancreas 48 h after injection. Expression of ribosomal protein S20 (NCBI accession number CK591746) is shown as a reference. Hemocyanin siRNAs are an equimolar cocktail of three siRNAs targeting the hemocyanin mRNA (Table 1); the same siRNA of arbitrary sequence used in panel c was used as a control. Each lane represents RNA from a single shrimp.
siRNAs fail to induce antiviral immunity when injected into shrimp.
The ability of siRNAs to induce a strong antiviral response was compared to that of long dsRNA. A control dsRNA for duck Igυ (309 bp), dsRNA for vp19 (366 bp), and mixtures of siRNAs specific for vp19 (Table 1) were compared in a high-dose WSSV challenge (Fig. 3a). As predicted, the long dsRNA for vp19 gave complete protection against viral challenge, while the nonspecific dsRNA (duck Igυ), although it reduced the rate at which animals died, ultimately showed 100% mortality. Surprisingly, the cocktail of three anti-vp19 synthetic siRNAs failed to induce measurable antiviral protection. Because, in vertebrates, siRNAs seem less capable of inducing some of the innate immune responses that long dsRNA molecules activate (and this seems to be analogous to observations of L. vannamei) (Fig. 3c), the hypothesis that a lack of engagement of the innate immune response by mixtures of virus-specific siRNAs could account for their lack of antiviral activity was tested. The data in Fig. 3a indicate that inclusion of duck Igυ dsRNA into a cocktail of anti-vp19 siRNAs does not complement their poor antiviral activity and instead results in significant mortality, similar in magnitude and kinetics to that observed when duck Igυ dsRNA was used alone. Thus, these data suggest that siRNAs are poor inducers of sequence-specific antiviral immunity and are incapable of mediating antiviral protection even when the innate immune response is activated concomitantly by long dsRNA molecules. For these experiments, siRNA targets in vp19 were selected using an algorithm that incorporates criteria derived empirically from silencing experiments in model organisms (http://www.dharmacon.com/sidesign/) (35). While rational design of siRNAs can significantly improve the chances of selecting effective targets (12, 21, 35), it remains possible that regions of vp19 mRNA that are most susceptible to RNAi were not targeted by the pool of rationally designed siRNAs used in the present study. To address this issue, experiments were performed in which pools of siRNAs spanning the entire vp19 coding region were generated in vitro by digesting long vp19 dsRNA with recombinant mammalian Dicer, followed by purification of small RNA species. Figure 3b summarizes the results of two independent experiments in which the antiviral activity of long vp19 dsRNA was compared to that of siRNA pools with equivalent sequence content. These data demonstrate that siRNAs injected into L. vannamei are poor inducers of antiviral immunity compared to long sequence-specific dsRNA. It seems likely that the failure of siRNAs to mediate antiviral silencing in this system is related to inadequate delivery into cells or limited half-life in the extracellular environment of the shrimp, since all known mechanisms of dsRNA-mediated homologous gene silencing utilize small RNA duplexes as intermediates.
In a second series of experiments, the ability of siRNA analogues to induce the innate (sequence-independent) antiviral immune response in shrimp was tested. In these experiments (Fig. 3c), the injected dose of WSSV was titrated to kill only 60 to 80% of control shrimp. As previously reported (36), a dsRNA representing the duck Igυ was capable of stimulating effective anti-WSSV protection under these conditions (64% relative protection) (Fig. 3c). However, an siRNA analogue of arbitrary sequence (control siRNA) (Table 1) was completely inactive in this antiviral assay. In mammals, the potential of siRNAs to induce innate antiviral responses is varied, somewhat sequence dependent, and influenced by dose and by the nature of the 5′ ends (5, 20, 26, 39). The experiments reported here do not comprehensively explore these possibilities in the shrimp system, and thus, it remains possible that siRNAs prepared in different ways and representing different sequences or sequence contents are capable of stimulating nonspecific antiviral immunity in the shrimp. The results shown in Fig. 3, however, suggest that the innate (sequence-independent) antiviral immunity as well as the sequence-specific antiviral immunity induced by long dsRNA are not effectively recapitulated by siRNAs. These results also raise the question of whether or not siRNAs can induce genetic interference in vivo in shrimp, an issue that was addressed in the set of experiments described below.
siRNAs fail to induce genetic interference when injected into shrimp.
Hemocyanin was selected as the target to test the ability of siRNAs to down-regulate gene expression. The silencing activity of long hemocyanin dsRNA (633 bp) was compared with that of a cocktail of three anti-hemocyanin synthetic siRNAs (Table 1). The results, shown in Fig. 3d, indicate that siRNAs are poor inducers of specific mRNA degradation when delivered by injection in vivo. While the long hemocyanin dsRNA induced strong down-regulation of the hemocyanin mRNA, there was no down-regulation observed with the cocktail of hemocyanin-specific synthetic siRNAs. As controls, shrimp were injected with long dsRNA of arbitrary sequence (duck Igυ) or with a control siRNA (Table 1). The anti-hemocyanin siRNAs were designed based on the same criteria used to select anti-vp19 siRNAs (http://www.dharmacon.com/sidesign/) (35).
Nonspecific long dsRNA does not enhance endogenous gene silencing or antiviral immunity induced by sequence-specific dsRNA.
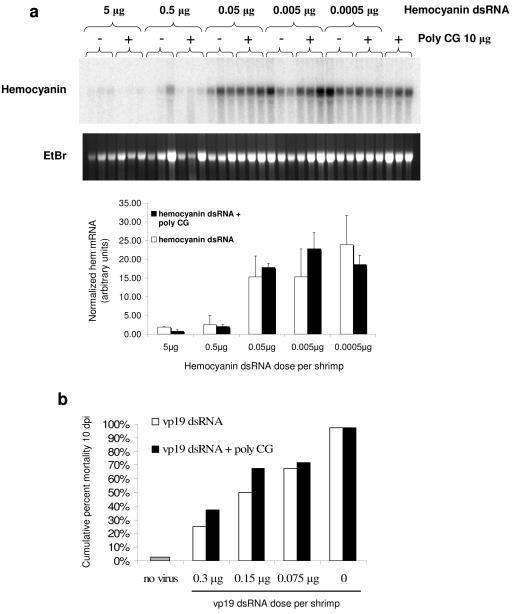
Together with the present study, previous work suggested the existence of two antiviral pathways induced by dsRNA in shrimp: an RNAi-based pathway and a broad-spectrum innate antiviral pathway. However, the fact that both kinds of responses are triggered by the same viral inducer, dsRNA, raises the question of whether nonviral dsRNA exerts its antiviral effects by simply capacitating a natural RNAi-based antiviral immune system. This could potentially explain why some (relatively low) level of antiviral immunity is observed in infected shrimp that have been treated with dsRNA of arbitrary sequence (Fig. 3c). To address this issue, the effects of dsRNA of arbitrary sequence on the RNAi response in shrimp were analyzed by assaying hemocyanin mRNA down-regulation after injection of different amounts of cognate hemocyanin dsRNA in the presence or absence of the dsRNA analogue poly(C-G) (an inducer of innate antiviral immunity) (36). The results indicate that there is no discernible effect of poly(C-G) treatment on the efficacy of hemocyanin down-regulation by hemocyanin dsRNA (Fig. 4a). These data suggest that stimulation of innate immunity by dsRNA does not further enhance the RNAi response directed against endogenous genes. To address more directly whether dsRNA of arbitrary sequence can capacitate a natural RNAi-mediated antiviral response, shrimp were treated simultaneously with both WSSV-specific and nonspecific dsRNAs, followed by infection with WSSV. As shown in Fig. 4b, poly(C-G) does not enhance the immunity to WSSV induced by vp19 dsRNA. Taken together, the data shown in Fig. 4 support the hypothesis that two distinct pathways of antiviral immunity are induced by dsRNA in shrimp: an innate immunity-based, sequence-independent response and an RNAi-based antiviral silencing pathway.
FIG. 4.
Nonspecific dsRNA does not enhance endogenous gene silencing or antiviral immunity induced by sequence-specific dsRNA. (a) Individual shrimp (1 to 2 g) were injected with the indicated amounts of hemocyanin dsRNA either alone or together with 10 μg of poly(C-G). Hemocyanin mRNA levels were analyzed by Northern blot 48 h after dsRNA injection. rRNA stained with ethidium bromide (EtBr) is shown as a reference. Each lane contains RNA from a single shrimp. The bottom panel shows densitometric analysis of hemocyanin mRNA levels normalized to rRNA (ethidium bromide stain) in each of the experimental groups. The mean ratio and standard deviations for each group of three shrimp are shown. (b) Shrimp (1 to 2 g) were coinjected with the indicated amounts of vp19 dsRNA and 10 μl of WSSV-infective extract (diluted 2 × 10−6, wt/vol) in the presence or absence of 10 μg of poly(C-G). An individual challenge system was used (n = 38 to 40). Bars indicate the cumulative percent mortality 10 dpi. No significant differences were observed 10 dpi between treatments with vp19 dsRNA alone and vp19 dsRNA combined with poly(C-G).
DISCUSSION
These experiments systematically demonstrate that the in vivo administration of long dsRNA down-regulates the expression of endogenous genes in a marine shrimp in a sequence-specific manner. In addition, the administration of dsRNA specific for viral genes induces a potent and virus-specific antiviral response that results in highly effective control of viral disease. Using WSSV as a model, it was shown that the degree of protection afforded by specific dsRNAs varies between different viral genes targeted. Although the reason for these differences was not addressed in the present study, it seems likely that different mRNAs are differentially susceptible to sequence-dependent targeting. Alternatively, the viral gene products encoded by the targetedRNAs may be differentially required for WSSV replication and/or pathogenesis. We have observed an even stronger gene-specific effect in anti-TSV immunity induced by TSV-specific dsRNAs (data not shown), supporting the idea that target selection is a crucial parameter for successful suppression of viral disease by dsRNA. While the present paper was under review, another study reported the use of virus-specific dsRNA to suppress yellow head virus replication in cultured cells of the shrimp Penaeus monodon (42). The work of Tirasophon and colleagues is in agreement with our studies in several ways: (i) dsRNAs derived from viral genomic sequence were observed to inhibit replication of a cognate virus, (ii) viral gene target selection was shown to significantly influence the potency of dsRNA-induced antiviral protection, and (iii) dsRNA of arbitrary sequence was shown to evoke a significant antiviral response, albeit much lower in potency than that induced by virus-specific dsRNA.
The present study also compared the biological activities of long dsRNAs and siRNAs delivered by injection into shrimp. It was found that endogenous gene silencing and virus-specific immunity are not induced by siRNAs injected intramuscularly. Infections with low doses of WSSV indicated that siRNAs lack the significant sequence-independent innate antiviral properties of long dsRNA molecules. Taken together, these data strongly suggest that siRNAs delivered naked into shrimp have poor biological activity, not only in terms of gene silencing but also as inducers of both sequence-specific and sequence-independent antiviral responses. This lack of activity could be the result of instability or poor intracellular delivery of naked siRNAs injected in the extracellular environment of the shrimp. For instance, it seems likely that shrimp cells, like those of other animals, are susceptible to gene silencing when siRNAs are delivered intracellularly (e.g., by transfection). If internalization into cells accounts for the functional differences we have observed between long dsRNA and siRNAs, the data presented here may indicate the existence of mechanisms for uptake of long dsRNA that do not act on very short (i.e., ca. 20 bp) substrates. The lack of activity of the siRNAs tested in these studies in terms of gene silencing and antiviral immunity should be interpreted with caution, since in mammalian systems, a great deal of dependency on dose, sequence, and chemical structure has been documented (5, 20, 26, 39), and similar issues may apply to shrimp in vivo.
One of the more interesting hypotheses derived from our observations of activation of RNAi and innate immunity by dsRNA is that these two pathways interact functionally to mount immunity to a viral pathogen. Such a possibility was previously unrecognized among invertebrates, as L. vannamei is the only invertebrate in which dsRNA has been shown to induce both innate immune reactions and RNAi-like antiviral immunity. The data suggest, however, that dsRNA analogues that are capable of inducing innate nonspecific immunity do not significantly influence the silencing activity of cognate dsRNAs. These same dsRNA analogues were also not capable of enhancing the antiviral properties of virus-specific long dsRNA or of complementing the lack of antiviral activity of siRNAs delivered by injection. This argues against a mechanism by which induction of broad-spectrum antiviral immunity by dsRNA of arbitrary sequence is due to some level of nonspecific stimulation of the RNAi pathway. However, it remains unknown whether the strong antiviral protection afforded by virus-specific long dsRNAs is the result of RNAi mechanisms alone or the combination of innate immune activation and RNAi. This issue is currently difficult to address because it has not been possible to experimentally induce RNAi in shrimp with molecules other than long dsRNA (which induces both specific mRNA degradation and innate antiviral immunity). Thus, formal testing of a model by which induction of the two antiviral pathways is responsible for the potent antiviral immunity mounted in response to viral dsRNA awaits a better understanding of the molecular bases for innate immunity and RNAi in shrimp.
The demonstration of sequence-specific dsRNA-mediated antiviral immunity in a marine shrimp further supports the recently established role of RNAi as an antiviral system in invertebrates. In mosquitoes, components of the RNAi machinery have been shown to be required for immunity against an RNA virus (24), and small RNAs that resemble siRNAs derived from viral genomes have been found in virally infected cells in culture (28). Viral suppression of RNAi has also been demonstrated for insect viruses (22, 28), further implicating RNAi as a natural antiviral mechanism in invertebrates. Similarly, an RNAi-based antiviral mechanism in shrimp predicts that suppressors of RNAi will be found in viruses affecting crustacea. This also suggests the possibility of links between viral pathogenesis and viral manipulation of RNAi (as has been shown in plants [23]), given the essential role that this pathway plays in regulating gene expression in metazoa. The view of RNAi as an immune system also challenges the idea that invertebrates rely exclusively on the more broad-spectrum and less specific mechanisms of innate immunity. In fact, in plant biology, the term “adaptive” has been suggested to better describe the type of immunity that RNAi represents (16).
In contrast to the adaptive antiviral RNAi system, nothing is known about molecular mechanisms of innate antiviral immunity in invertebrates. It is expected that just as antimicrobial peptides under the control of known signal transduction pathways are part of the antibacterial and antifungal defense response (19), analogous systems with viral specificity will also operate in invertebrates. Phenomena suggesting that such responses exist in crustacea, such as induction of antiviral immunity by microbial cell wall components and by dsRNA, have been reported (8, 36, 40). Immune phenomena reminiscent of vertebrate adaptive immunity have also been documented in crustacea from observations of memory of pathogen-specific features (27, 48). Elucidation of the molecular mechanisms behind these immune phenomena promises to uncover novel immune pathways in invertebrates.
The practical implications of the results presented in this study are threefold: (i) it should be possible to use naked long dsRNA (but not naked siRNAs) to induce RNAi in shrimp, allowing for the first time to test gene function in vivo in this model; (ii) such experimental use of RNAi should take into consideration the possible effects of unintended stimulation of innate immune responses; and (iii) joint manipulation of RNAi and innate immune pathways is a promising approach to the development of antiviral therapeutics for the control of shrimp disease. From the perspective of understanding the evolution of immunity, this study suggests, for the first time, the possibility of dual stimulation of innate immunity and antiviral silencing by dsRNA in an invertebrate.
Acknowledgments
This work was partially supported by Shrimp Improvement Systems LLC, the National Science Foundation (grant MCB0315393 to G.W.W.), the National Marine Fisheries Service (grant NA03NMF4720362 to R.W.C., C.L.B., P.S.G., and G.W.W.), the South Carolina Sea Grant Consortium (grant R/MT-6 to P.S.G.), the United States Marine Shrimp Farming Consortium, the United States Department of Agriculture (USDA NRICGP-CSREES/AREA grant number 2002-35201-11620 to P.S.G.), the South Carolina Department of Natural Resources, Escuela Superior Politécnica del Litoral, and the Fundación para la Ciencia y Tecnología (Ecuador).
Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the supporting bodies mentioned herein.
We thank Caroline Payne, Laxminath Tumburu, Adrienne Metz, Melinda Stilley, Jeremy Landers, Justin Yost, Michelle Pate, James Powell, and Selena Kirby for help with rearing and challenging shrimp.
This is contribution number 566 from the Marine Resources Research Institute (SC-DNR) and contribution number 21 from the Marine Biomedicine and Environmental Sciences Center (MUSC).
REFERENCES
- 1.Adams, M. D., S. E. Celniker, R. A. Holt, C. A. Evans, J. D. Gocayne, P. G. Amanatides, S. E. Scherer, P. W. Li, R. A. Hoskins, R. F. Galle, R. A. George, S. E. Lewis, S. Richards, M. Ashburner, S. N. Henderson, G. G. Sutton, J. R. Wortman, M. D. Yandell, Q. Zhang, L. X. Chen, R. C. Brandon, Y. H. Rogers, R. G. Blazej, M. Champe, B. D. Pfeiffer, K. H. Wan, C. Doyle, E. G. Baxter, G. Helt, C. R. Nelson, G. L. Gabor, J. F. Abril, A. Agbayani, H. J. An, C. Andrews-Pfannkoch, D. Baldwin, R. M. Ballew, A. Basu, J. Baxendale, L. Bayraktaroglu, E. M. Beasley, K. Y. Beeson, P. V. Benos, B. P. Berman, D. Bhandari, S. Bolshakov, D. Borkova, M. R. Botchan, J. Bouck, P. Brokstein, P. Brottier, K. C. Burtis, D. A. Busam, H. Butler, E. Cadieu, A. Center, I. Chandra, J. M. Cherry, S. Cawley, C. Dahlke, L. B. Davenport, P. Davies, B. de Pablos, A. Delcher, Z. Deng, A. D. Mays, I. Dew, S. M. Dietz, K. Dodson, L. E. Doup, M. Downes, S. Dugan-Rocha, B. C. Dunkov, P. Dunn, K. J. Durbin, C. C. Evangelista, C. Ferraz, S. Ferriera, W. Fleischmann, C. Fosler, A. E. Gabrielian, N. S. Garg, W. M. Gelbart, K. Glasser, A. Glodek, F. Gong, J. H. Gorrell, Z. Gu, P. Guan, M. Harris, N. L. Harris, D. Harvey, T. J. Heiman, J. R. Hernandez, J. Houck, D. Hostin, K. A. Houston, T. J. Howland, M. H. Wei, C. Ibegwam, et al. 2000. The genome sequence of Drosophila melanogaster. Science 287:2185-2195. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein, E., A. A. Caudy, S. M. Hammond, and G. J. Hannon. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363-366. [DOI] [PubMed] [Google Scholar]
- 4.Bitko, V., A. Musiyenko, O. Shulyayeva, and S. Barik. 2005. Inhibition of respiratory viruses by nasally administered siRNA. Nat. Med. 11:50-55. [DOI] [PubMed] [Google Scholar]
- 5.Bridge, A. J., S. Pebernard, A. Ducraux, A. L. Nicoulaz, and R. Iggo. 2003. Induction of an interferon response by RNAi vectors in mammalian cells. Nat. Genet. 34:263-264. [DOI] [PubMed] [Google Scholar]
- 6.Carrington, J. C., and V. Ambros. 2003. Role of microRNAs in plant and animal development. Science 301:336-338. [DOI] [PubMed] [Google Scholar]
- 7.C. elegans Sequence Consortium. 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282:2012-2018. [DOI] [PubMed] [Google Scholar]
- 8.Chang, C. F., M. S. Su, H. Y. Chen, and I. C. Liao. 2003. Dietary beta-1,3-glucan effectively improves immunity and survival of Penaeus monodon challenged with white spot syndrome virus. Fish Shellfish Immunol. 15:297-310. [DOI] [PubMed] [Google Scholar]
- 9.Chen, C. Z., L. Li, H. F. Lodish, and D. P. Bartel. 2004. MicroRNAs modulate hematopoietic lineage differentiation. Science 303:83-86. [DOI] [PubMed] [Google Scholar]
- 10.Christophides, G. K., E. Zdobnov, C. Barillas-Mury, E. Birney, S. Blandin, C. Blass, P. T. Brey, F. H. Collins, A. Danielli, G. Dimopoulos, C. Hetru, N. T. Hoa, J. A. Hoffmann, S. M. Kanzok, I. Letunic, E. A. Levashina, T. G. Loukeris, G. Lycett, S. Meister, K. Michel, L. F. Moita, H. M. Muller, M. A. Osta, S. M. Paskewitz, J. M. Reichhart, A. Rzhetsky, L. Troxler, K. D. Vernick, D. Vlachou, J. Volz, C. von Mering, J. Xu, L. Zheng, P. Bork, and F. C. Kafatos. 2002. Immunity-related genes and gene families in Anopheles gambiae. Science 298:159-165. [DOI] [PubMed] [Google Scholar]
- 11.Dehal, P., Y. Satou, R. K. Campbell, J. Chapman, B. Degnan, A. De Tomaso, B. Davidson, A. Di Gregorio, M. Gelpke, D. M. Goodstein, N. Harafuji, K. E. Hastings, I. Ho, K. Hotta, W. Huang, T. Kawashima, P. Lemaire, D. Martinez, I. A. Meinertzhagen, S. Necula, M. Nonaka, N. Putnam, S. Rash, H. Saiga, M. Satake, A. Terry, L. Yamada, H. G. Wang, S. Awazu, K. Azumi, J. Boore, M. Branno, S. Chin-Bow, R. DeSantis, S. Doyle, P. Francino, D. N. Keys, S. Haga, H. Hayashi, K. Hino, K. S. Imai, K. Inaba, S. Kano, K. Kobayashi, M. Kobayashi, B. I. Lee, K. W. Makabe, C. Manohar, G. Matassi, M. Medina, Y. Mochizuki, S. Mount, T. Morishita, S. Miura, A. Nakayama, S. Nishizaka, H. Nomoto, F. Ohta, K. Oishi, I. Rigoutsos, M. Sano, A. Sasaki, Y. Sasakura, E. Shoguchi, T. Shin-i, A. Spagnuolo, D. Stainier, M. M. Suzuki, O. Tassy, N. Takatori, M. Tokuoka, K. Yagi, F. Yoshizaki, S. Wada, C. Zhang, P. D. Hyatt, F. Larimer, C. Detter, N. Doggett, T. Glavina, T. Hawkins, P. Richardson, S. Lucas, Y. Kohara, M. Levine, N. Satoh, and D. S. Rokhsar. 2002. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science 298: 2157-2167. [DOI] [PubMed] [Google Scholar]
- 12.Elbashir, S. M., J. Martinez, A. Patkaniowska, W. Lendeckel, and T. Tuschl. 2001. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 20:6877-6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukagawa, T., M. Nogami, M. Yoshikawa, M. Ikeno, T. Okazaki, Y. Takami, T. Nakayama, and M. Oshimura. 2004. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 6:784-791. [DOI] [PubMed] [Google Scholar]
- 14.Gazzani, S., T. Lawrenson, C. Woodward, D. Headon, and R. Sablowski. 2004. A link between mRNA turnover and RNA interference in Arabidopsis. Science 306:1046-1048. [DOI] [PubMed] [Google Scholar]
- 15.Ge, Q., L. Filip, A. Bai, T. Nguyen, H. N. Eisen, and J. Chen. 2004. Inhibition of influenza virus production in virus-infected mice by RNA interference. Proc. Natl. Acad. Sci. USA 101:8676-8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldbach, R., E. Bucher, and M. Prins. 2003. Resistance mechanisms to plant viruses: an overview. Virus Res. 92:207-212. [DOI] [PubMed] [Google Scholar]
- 17.Grishok, A., A. E. Pasquinelli, D. Conte, N. Li, S. Parrish, I. Ha, D. L. Baillie, A. Fire, G. Ruvkun, and C. C. Mello. 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106:23-34. [DOI] [PubMed] [Google Scholar]
- 18.Hall, I. M., G. D. Shankaranarayana, K. Noma, N. Ayoub, A. Cohen, and S. I. Grewal. 2002. Establishment and maintenance of a heterochromatin domain. Science 297:2232-2237. [DOI] [PubMed] [Google Scholar]
- 19.Hetru, C., L. Troxler, and J. A. Hoffmann. 2003. Drosophila melanogaster antimicrobial defense. J. Infect. Dis. 187(Suppl. 2):S327-S334. [DOI] [PubMed] [Google Scholar]
- 20.Hornung, V., M. Guenthner-Biller, C. Bourquin, A. Ablasser, M. Schlee, S. Uematsu, A. Noronha, M. Manoharan, S. Akira, A. de Fougerolles, S. Endres, and G. Hartmann. 2005. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat. Med. 11:263-270. [DOI] [PubMed] [Google Scholar]
- 21.Jagla, B., N. Aulner, P. D. Kelly, D. Song, A. Volchuk, A. Zatorski, D. Shum, T. Mayer, D. A. De Angelis, O. Ouerfelli, U. Rutishauser, and J. E. Rothman. 2005. Sequence characteristics of functional siRNAs. RNA 11:864-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson, K. L., B. D. Price, L. D. Eckerle, and L. A. Ball. 2004. Nodamura virus nonstructural protein B2 can enhance viral RNA accumulation in both mammalian and insect cells. J. Virol. 78:6698-6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kasschau, K. D., Z. Xie, E. Allen, C. Llave, E. J. Chapman, K. A. Krizan, and J. C. Carrington. 2003. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA unction. Dev. Cell 4:205-217. [DOI] [PubMed] [Google Scholar]
- 24.Keene, K. M., B. D. Foy, I. Sanchez-Vargas, B. J. Beaty, C. D. Blair, and K. E. Olson. 2004. RNA interference acts as a natural antiviral response to O'nyong-nyong virus (Alphavirus; Togaviridae) infection of Anopheles gambiae. Proc. Natl. Acad. Sci. USA 101:17240-17245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ketting, R. F., T. H. Haverkamp, H. G. van Luenen, and R. H. Plasterk. 1999. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell 99:133-141. [DOI] [PubMed] [Google Scholar]
- 26.Kim, D. H., M. Longo, Y. Han, P. Lundberg, E. Cantin, and J. J. Rossi. 2004. Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nat. Biotechnol. 22:321-325. [DOI] [PubMed] [Google Scholar]
- 27.Kurtz, J., and K. Franz. 2003. Innate defence: evidence for memory in invertebrate immunity. Nature 425:37-38. [DOI] [PubMed] [Google Scholar]
- 28.Li, H., W. X. Li, and S. W. Ding. 2002. Induction and suppression of RNA silencing by an animal virus. Science 296:1319-1321. [DOI] [PubMed] [Google Scholar]
- 29.Li, W. X., H. Li, R. Lu, F. Li, M. Dus, P. Atkinson, E. W. Brydon, K. L. Johnson, A. Garcia-Sastre, L. A. Ball, P. Palese, and S. W. Ding. 2004. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc. Natl. Acad. Sci. USA 101:1350-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lippman, Z., and R. Martienssen. 2004. The role of RNA interference in heterochromatic silencing. Nature 431:364-370. [DOI] [PubMed] [Google Scholar]
- 31.Mari, J., B. T. Poulos, D. V. Lightner, and J. R. Bonami. 2002. Shrimp Taura syndrome virus: genomic characterization and similarity with members of the genus Cricket paralysis-like viruses. J. Gen. Virol. 83:915-926. [DOI] [PubMed] [Google Scholar]
- 32.Matsumoto, M., K. Funami, H. Oshiumi, and T. Seya. 2004. Toll-like receptor 3: a link between Toll-like receptor, interferon and viruses. Microbiol. Immunol. 48:147-154. [DOI] [PubMed] [Google Scholar]
- 33.Pal-Bhadra, M., B. A. Leibovitch, S. G. Gandhi, M. Rao, U. Bhadra, J. A. Birchler, and S. C. Elgin. 2004. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303:669-672. [DOI] [PubMed] [Google Scholar]
- 34.Prior, S., C. L. Browdy, E. F. Shepard, R. Laramore, and P. G. Parnell. 2003. Controlled bioassay systems for determination of lethal infective doses of tissue homogenates containing Taura syndrome or white spot syndrome virus. Dis. Aquat. Organ. 54:89-96. [DOI] [PubMed] [Google Scholar]
- 35.Reynolds, A., D. Leake, Q. Boese, S. Scaringe, W. S. Marshall, and A. Khvorova. 2004. Rational siRNA design for RNA interference. Nat. Biotechnol. 22:326-330. [DOI] [PubMed] [Google Scholar]
- 36.Robalino, J., C. L. Browdy, S. Prior, A. Metz, P. Parnell, P. Gross, and G. Warr. 2004. Induction of antiviral immunity by double-stranded RNA in a marine invertebrate. J. Virol. 78:10442-10448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Sarot, E., G. Payen-Groschene, A. Bucheton, and A. Pelisson. 2004. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics 166:1313-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sledz, C. A., M. Holko, M. J. de Veer, R. H. Silverman, and B. R. Williams. 2003. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 5:834-839. [DOI] [PubMed] [Google Scholar]
- 40.Song, Y. L., J. J. Liu, L. C. Chan, and H. H. Sung. 1997. Glucan-induced disease resistance in tiger shrimp (Penaeus monodon). Dev. Biol. Stand. 90:413-421. [PubMed] [Google Scholar]
- 41.Tabara, H., M. Sarkissian, W. G. Kelly, J. Fleenor, A. Grishok, L. Timmons, A. Fire, and C. C. Mello. 1999. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99:123-132. [DOI] [PubMed] [Google Scholar]
- 42.Tirasophon, W., Y. Roshorm, and S. Panyim. 2005. Silencing of yellow head virus replication in penaeid shrimp cells by dsRNA. Biochem. Biophys. Res. Commun. 334:102-107. [DOI] [PubMed] [Google Scholar]
- 43.Tompkins, S. M., C. Y. Lo, T. M. Tumpey, and S. L. Epstein. 2004. Protection against lethal influenza virus challenge by RNA interference in vivo. Proc. Natl. Acad. Sci. USA 101:8682-8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uprichard, S. L., B. Boyd, A. Althage, and F. V. Chisari. 2005. Clearance of hepatitis B virus from the liver of transgenic mice by short hairpin RNAs. Proc. Natl. Acad. Sci. USA 102:773-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valdes, V. J., A. Sampieri, J. Sepulveda, and L. Vaca. 2003. Using double-stranded RNA to prevent in vitro and in vivo viral infections by recombinant baculovirus. J. Biol. Chem. 278:19317-19324. [DOI] [PubMed] [Google Scholar]
- 46.Volpe, T. A., C. Kidner, I. M. Hall, G. Teng, S. I. Grewal, and R. A. Martienssen. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297:1833-1837. [DOI] [PubMed] [Google Scholar]
- 47.Williams, B. R. 2001. Signal integration via PKR. Sci. STKE 2001:RE2. [DOI] [PubMed] [Google Scholar]
- 48.Witteveldt, J., C. C. Cifuentes, J. M. Vlak, and M. C. van Hulten. 2004. Protection of Penaeus monodon against white spot syndrome virus by oral vaccination. J. Virol. 78:2057-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yao, M. C., P. Fuller, and X. Xi. 2003. Programmed DNA deletion as an RNA-guided system of genome defense. Science 300:1581-1584. [DOI] [PubMed] [Google Scholar]