Abstract
The complete genomic sequence of kelp fly virus (KFV), originally isolated from the kelp fly, Chaetocoelopa sydneyensis, has been determined. Analyses of its genomic and structural organization and phylogeny show that it belongs to a hitherto undescribed group within the picorna-like virus superfamily. The single-stranded genomic RNA of KFV is 11,035 nucleotides in length and contains a single large open reading frame encoding a polypeptide of 3,436 amino acids with 5′ and 3′ untranslated regions of 384 and 343 nucleotides, respectively. The predicted amino acid sequence of the polypeptide shows that it has three regions. The N-terminal region contains sequences homologous to the baculoviral inhibitor of apoptosis repeat domain, an inhibitor of apoptosis commonly found in animals and in viruses with double-stranded DNA genomes. The second region contains at least two capsid proteins. The third region has three sequence motifs characteristic of replicase proteins of many plant and animal viruses, including a helicase, a 3C chymotrypsin-like protease, and an RNA-dependent RNA polymerase. Phylogenetic analysis of the replicase motifs shows that KFV forms a distinct and distant taxon within the picorna-like virus superfamily. Cryoelectron microscopy and image reconstruction of KFV to a resolution of 15 Å reveals an icosahedral structure, with each of its 12 fivefold vertices forming a turret from the otherwise smooth surface of the 20-Å-thick capsid. The architecture of the KFV capsid is unique among the members of the picornavirus superfamily for which structures have previously been determined.
Kelp fly virus (KFV) has small (29-nm-diameter) isometric virions and a single-stranded, positive-sense RNA genome. It was first isolated from apparently healthy adult kelp flies (Chaetocoelopa sydneyensis) collected from rocks on a headland near Wapengo Lake in the southern coastal region of New South Wales, Australia. Although isolated on several separate occasions from groups of adult flies, significant amounts of KFV virions were obtained only by infecting wax moth (Galleria mellonella) larvae (51). No other hosts, including cell lines, have been found.
The virus has been an enigma since its isolation and remains in the “unclassified” group of unenveloped, small RNA viruses of insects (12, 47) because of the unique physical characteristics of its virions. They have a high buoyant density (1.43 g/ml) in neutral CsCl and contain a single-stranded RNA genome estimated at 3.5 MDa (51). The initial study of KFV detected only two major capsid proteins with estimated molecular masses of 73 and 29 kDa, a pattern unique for a nonoccluded animal virus. When observed by negative-stain electron microscopy, the virions of KFV resemble the cores of reovirus particles in that they have small surface projections, apparently on their fivefold icosahedral axes, although the virions are much smaller in diameter than those of reoviruses.
Gene sequence comparisons usually provide an excellent view of the relationships of viruses, as they have few distinctive characteristics. Sequences from the RNA-dependent RNA polymerases (RdRps) are generally the most useful as they occur in most viral genomes and are conserved (4, 5, 35, 68). RdRp sequence comparisons place positive-strand viruses with RNA genomes into three supergroups (I, II, and III), and this grouping correlates with groupings based on other characters. Most positive-sense, single-stranded RNA viruses are classified in the alphavirus (II) or picorna-like virus (I) supergroups (35, 68). The picorna-like viruses are distinguished by their RdRp sequence alignments and the conserved helicase-protease-replicase array (36) and are divided among the Picornaviridae, Caliciviridae, Sequiviridae, Comoviridae, Potyviridae, and Dicistroviridae. These virus families comprise the “picornavirus superfamily” and are characterized by the production of an exclusive genomic RNA message, encoding one or two polyproteins which are posttranslationally cleaved to generate the structural and nonstructural viral proteins (63).
Here we characterize the genome, structural proteins, and cryoelectron microscopy (cryo-EM) capsid structure of KFV for comparison to other RNA viruses. The data show that KFV has unique genomic and capsid organization. Comparative analysis of motifs from the putative KFV viral replicase shows it to be related to, but distant from, other picornaviruses. Similarly, the KFV VP1 capsid protein is related to that of Acyrthosiphon pisum virus (APV) but distinct from all known picornavirus capsid proteins. The genome also encodes a homolog of the baculoviral inhibitor of apoptosis repeat (BIR) domain. These characteristics indicate that KFV is the only known representative of a hitherto undescribed group of viruses.
MATERIALS AND METHODS
Virus purification.
Virions of KFV were obtained by infecting midinstar Galleria mellonella larvae and purifying them as described by Scotti et al. (51) but using dichloromethane to treat the crude larval extracts and either 100 mM sodium phosphate or 100 mM Tris-HCl at pH 7.2 as the buffer. Virion concentration was estimated by UV absorption at 260- and 280-nm wavelengths, assuming an RNA content of 30%, using the equation of Layne (38) [1 mg/ml protein = (1.55 × A280) − (0.76 × A260)].
RNA preparation, synthesis of cDNA, and cloning.
Viral RNA was extracted by using the phenol-guanidinium isothiocyanate method (10). From total KFV viral RNA, cDNA was synthesized using random primers, blunt-ended with T4 DNA polymerase, and then cloned into pBluescript KS (Stratagene) as described by Hanzlik et al. (31). For an internal region of the genome that was refractory to the standard approaches, a PCR fragment was obtained by using reverse transciption-PCRwith Superscript (Gibco BRL) and primers based on the flanking sequences. The fragment was T tailed using Taq (Gibco BRL) and cloned using the TOPO-TA cloning system (Invitrogen). To obtain the terminal regions of the genome, both 5′ and 3′ rapid amplifications of cDNA ends were done using the appropriate systems bought from Gibco BRL. The PCR fragments were cloned with the TOPO-TA cloning system (Invitrogen).
Nucleotide sequencing.
The cDNA clones, a number of subclones generated by specific restriction fragment deletion, and the PCR-derived products were sequenced using dye terminator sequencing kits (ABI Prism and Beckman CEQ). Primer walking and multiple coverage of difficult regions with high AT contents were used to obtain accurate sequence data for the entire KFV genome.
Sequence analysis.
Nucleotide and amino acid sequence data were analyzed and assembled using the University of Wisconsin Genetics Computer Group (GCG) program (15), Vector NTI 5 (Informax), and CEQ Investigator (Beckman). Comparison with sequence data of other viruses utilized BioManager from Australian National Genomic Information Service (ANGIS) and NCBI with analysis using ClustalW 1.8 (58), BLAST (2), Seqboot (19), ProtDist (19), and OldDistance (GCG; Oxford Molecular Group, Inc.).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
N,N-methylenebisacrylamide-Tris gradient gels (4 to 12%) were used with the NuPAGE electrophoresis system (Invitrogen). Samples were disrupted in NuPAGE dissociation buffer, containing NuPAGE reducing agent, per the manufacturer's instructions and run at 200 V until the dye marker reached the bottom of the gel. Gels were fixed in 50% methanol-7% acetic acid for 15 min and washed with water prior to staining with a Coomassie-based reagent (GelCode Blue stain; Pierce) per the manufacturer's instructions. Molecular mass standards were made up in NuPAGE dissociation buffer (bovine serum albumin [66 kDa], carbonic anhydrase [31 kDa], and lysozyme [14.5 kDa]; Sigma).
Protein HPLC.
Reversed-phase high-performance liquid chromatography (HPLC) was done with an HP 1050 Ti-series chromatograph (Agilent) fitted with an analytical 300-Å Vydac C18 column, 25 cm by 4.6 mm internal diameter (i.d.). Samples of purified KFV in 10% acetonitrile-water were eluted with a 10 to 100% acetonitrile-in-water (vol/vol) gradient containing 0.1% trifluoroacetic acid over 60 min, recording the absorption at 218 nm. Peaks were collected, and the solvent was evaporated in a vacuum centrifuge. Samples were then dissolved in 10% aqueous acetonitrile containing 1% formic acid for molecular weight determination by mass spectrometry.
N-terminal microsequencing.
N-terminal amino acid sequences were analyzed with proteins obtained by electrophoretic transfer from SDS-PAGE gels, equilibrated in 100 mM CAPS buffer (pH 10.5) containing 10% methanol, to polyvinylidene difluoride membrane (Bio-Rad) by using an X-Cell II Blot module (Invitrogen) set at 100 V for 30 min. Protein bands were stained with Coomassie and dried, and their N termini were microsequenced at the Protein Microchemistry Unit, University of Otago, Dunedin, New Zealand.
Analysis of peptides by liquid chromatography-mass spectrometry.
Colloidal Coomassie-stained gel bands were excised from one-dimensional SDS-PAGE gels, washed three times in 1:1 acetonitrile-50 mM ammonium bicarbonate (pH 8.3), and dried in a vacuum centrifuge. Sufficient trypsin (modified sequencing grade; Roche) was added at 0.5 μg/10 μl in 25 mM ammonium bicarbonate to cover the gel plugs, and the samples were incubated at 37°C for 18 h. The resulting peptides were extracted first with water and twice with water-acetonitrile-formic acid (50/50/5), and the combined extracts were concentrated to 30 μl and subjected to nanospray mass spectrometry using an LCQ Deca ion trap mass spectrometer fitted with a nanoelectrospray interface (ThermoFinnigan, San Jose, Calif.) coupled to a Surveyor HPLC system. Twenty microliters of each digest was injected onto a reversed-phase column (Inertsil ODS-3, C18, 300 Å, 15 cm by 300 μm i.d.; LC Packings, San Francisco, Calif.). Tryptic peptides were separated at a flow rate of 4 μl/min with a linear gradient from 2 to 80% aqueous acetonitrile plus 0.1% formic acid over 50 min. The column flow rate was produced by splitting the primary flow rate of 40 μl/min from the Surveyor HPLC system via an Accurate flow splitter (LC Packings, San Francisco, Calif.). The nanospray interface was used with a 30-μm i.d. fused-silica standard coated PicoTip needle (New Objective, Woburn, Mass.), and the spray voltage was supplied directly to the coated needle tip at 2.2 kV. The mass spectrometer was operated in the positive ion mode, and the mass range acquired was an m/z of 300 to 2,000. The heated capillary temperature was set at 210°C. Data were acquired using a triple-play experiment in data-dependent mode with dynamic exclusion enabled. Before the nucleotide sequence had been determined, predicted protein sequence information was generated from peptide tandem mass spectrometry data by using the Lutefisk de novo sequencing program (57).
Tandem mass spectrometry data were analyzed using TurboSEQUEST (ThermoFinnigan), a computer program that correlates experimental data with theoretical spectra generated from known protein or virtually translated nucleotide sequences (18, 66). Spectra were searched against the KFV genome sequence, initially for the nucleotide sequence, translated virtually in all six reading frames, and then for the inferred protein sequence once the correct open reading frame (ORF) had been determined. The criteria used for a positive peptide identification for a doubly charged peptide were a correlation factor (Cn) greater than 2.0, a delta cross-correlation factor (ΔCn) greater than 0.1 (indicating a significant difference between the best match reported and the next best match) and a high preliminary scoring (Sp) (66). For triply charged peptides, the correlation factor threshold was set at 2.5. All matched peptides were confirmed by visual examination of the spectra (57).
Protein molecular weight determination by mass spectrometry.
Protein molecular weight was determined by using the LCQ Deca ion trap mass spectrometer fitted with an electrospray interface (ThermoFinnigan, San Jose, Calif.). Samples from preparative HPLC in aqueous acetonitrile acidified with formic acid were infused at 10 μl/min directly into the mass spectrometer. Electrospray mass spectrometry spectra were obtained and processed using the BIOMASSconvolution and deconvolution algorithms in the BioWorks 3.1 suite of programs (ThermoFinnigan).
Cryo-EM and image reconstruction.
KFV (3 μl) at ∼1 mg/ml were placed on a holey carbon-coated copper electron microscopy grid, blotted, and vitrified by being plunged into liquid ethane. The grid was transferred to an FEI/Phillips CM200 cryoelectron microscope operating at ∼100 K and imaged using low-dose techniques. Images were collected at a range of defocus values. Developed negatives were scanned on a UMAX PowerLook 3000 scanner with a raster size of 8.322 μm. Scanned negatives were converted to optical density values by using SPIDER(21) and interpolated by using LABEL (MRC Program Suite [14]), and individual virions were selected interactively by using XIMDISP(14). The contrast transfer function of each negative was determined as previously described (20, 25). Contrast transfer function correction and classification of images were performed with IMAGIC(60). The total data set consisted of 3,890 particles imaged at 27 different defocuses. Resolution was defined as the point at which the Fourier shell correlation fell below 0.5.
Classification of the images produced class averages which indicated that the capsid turrets were present in substoichiometric amounts. The proportion of fivefold axes that had turrets varied from 1 to 12 per virion. One reconstruction of the data set was made using the EMBL icosahedral image reconstruction programs (23). The resulting map was masked by the GAP program to remove all except one turret (29). This model was used to perform a reconstruction using SPIDER(21), and then fivefold symmetry only was applied to the map in GAP, with symmetry refinement (29). Volumes of reconstructed density to user-defined thresholds, values for the capsid and turret densities, and a fully turreted reconstruction based on the single-turret map were computed with GAP (25, 29).
The three-dimensional reconstructions and supporting electron microscopy data described in this paper have been deposited in the Macromolecular Structure Database (http://www.ebi.ac.uk/msd:1150,1153,1154).
Nucleotide sequence accession numbers.
The full genomic sequence for KFV has been deposited with GenBank under accession number DQ112227. Virus families and accession numbers of representative picorna-like viruses used in the analyses are listed in Table 1.
TABLE 1.
General classifications and GenBank accession numbers of the viruses used in this study
| Classification | Virus | GenBank no. | Reference(s) or source |
|---|---|---|---|
| Dicistroviridae | Drosophila C virus | AF014388 | 34 |
| Cricket paralysis virus | AF218039 | 56 | |
| Rhopalosiphum padi virus | AF022937 | 46 | |
| Plautia stali intestine virus | AB006531 | 50 | |
| Iflavirus | Infectious flacherie virus | AB000906 | 33 |
| Sacbrood virus | AF092924 | 24 | |
| Perina nuda picorna-like virus | AF323747 | 63 | |
| Picornaviridae | Human hepatitis A virus | MI4707 | 13 |
| Foot-and-mouth disease virus | PO3306 | 1 | |
| Sequiviridae | Rice tungro spherical virus | 9627951 | 52 |
| Comoviridae | Tobacco ringspot virus | NC005097 | 67 |
| Bean pod mottle virus | NC003496 | 41 | |
| Caliciviridae | Human calicivirus | AY081134 | 42 |
| Lordsdale virus | X86557 | 42 | |
| Norwalk virus | M87661 | 9,49 | |
| Parvoviridae | Galleria mellonella densovirus | NC004286 | 53 |
| Adeno-associated virus | VCPV3A | 65 | |
| Nodaviridae | Pariacoto virus | AF171943 | 55 |
| Unclassified | Acyrthosiphon pisum virus | AF024514 | 59 |
| Kelp fly virus | DQ112227 | This study |
RESULTS
KFV can be distinguished from other unclassified picorna-like viruses of insects by the following characteristics. First, it is only known to infect the kelp fly (Chaetocoelopa sydneyensis) and the wax moth larvae (Galleria mellonella). Second, KFV virions are 29 nm in diameter with a buoyant density of 1.43 g/ml in neutral CsCl. These contain two major capsid proteins of ∼75 kDa and 29 kDa assembled into a unique particle with projections on the fivefold icosahedral axes. Relative stoichiometric amounts of each capsid protein within the virion, determined densitometrically (51), indicate that the two largest proteins occur in a molar ratio of 1:5. The virion contains a single-stranded RNA genome of 3.5 MDa and 11,035 nucleotides (nt) in size. The genome encodes a single polypeptide with virion proteins located in the N-terminal region and other proteins located in the C-terminal region. One exception to this is the BIR motif occurring in the N-terminal region of the virus-encoded polypeptide.
KFV genome.
Total RNA was extracted from whole viral extracts, and a cDNA library was constructed using poly(A)-primed products inserted into pBlueScript SK(+). Library clones were sequenced and used to compile an ∼8-kb contiguous sequence. However, N-terminal amino acid sequence data and mass spectrometric de novo peptide sequencing analysis of KFV capsid proteins suggested that the contiguous sequence was incomplete. Reverse transcription-PCR, using primers designed to flank regions of a suspected gap in the KFV sequence, together with 5′ and 3′ rapid amplification of cDNA ends was then used to obtain the complete genomic sequence for KFV.
KFV genomic RNA has an AU content of 62.86% and carries a single large ORF from nt 344 to 10651, which accounts for 93.4% of the genome. This single ORF encodes a predicted polypeptide of 3,436 amino acids (Fig. 1), with a 343-nt 3′ untranslated region (UTR) and a 5′ noncoding region of 384 nt in length. Domains showing sequence homology to viral RNA helicase and RdRp domains are located within the C-terminal region of the complete predicted protein (Fig. 2), while the virion proteins are grouped in the distal N-terminal region (amino acids 310 to 1257 [Table 2; Fig. 2]). Unusually for an RNA virus, the N-terminal region of the polypeptide (aminoacids 159 to 230 [Fig. 2]) encodes a small domain identified by BLAST comparisons as homologous to the BIR found in inhibitor of apoptosis (IAP) proteins (43).
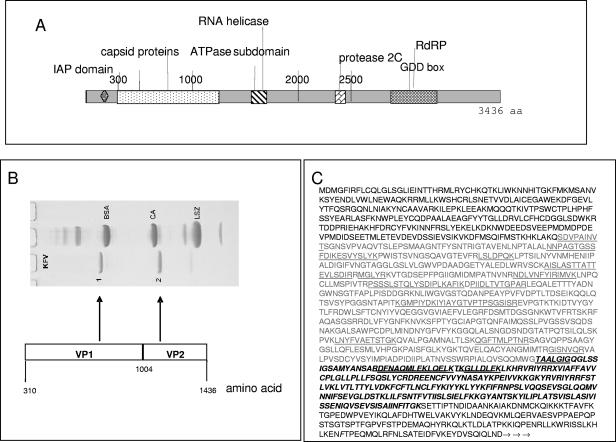
FIG. 1.
KFV genome. (A) Graphical illustration showing the peptide domains within the full-length KFV polypeptide coding sequence and UTRs. aa, amino acid. (B) Coomassie-stained SDS-PAGE illustrating the structural proteins isolated from KFV virions and the corresponding coding regions within the KFV genome. BSA, bovine serum albumin; CA, carbonic anhydrase; LSZ, lysozyme. (C) Virus capsid protein sequence. The inferred amino acid sequence begins at the start of the single KFV ORF. The light gray type indicates VP1 residues, and the bold and italic type represents the probable extent of VP2 residues. The N-terminal sequences SDVPAINVT and TAALGIG were determined as described in Materials and Methods. These and other sequences determined by mass spectrometry of tryptic digests of KFV capsid proteins are underlined.
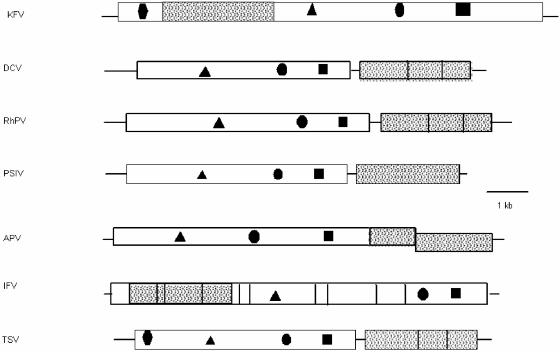
FIG. 2.
Genomic arrangement of KFV peptide domains within the full-length KFV polypeptide sequence and comparison to other picorna-like insect viruses. Lines represent UTRs, and boxes represent ORFs. Shaded areas represent capsid proteins, and objects within the boxes represent the following: RdRp (▪), protease (•), RNA helicase (▴), and IAP domain (⧫). DCV, Drosophila C virus; RhPV, Rhopalosiphum padi virus; PSIV, Plautia stali intestine virus.
TABLE 2.
Summary of virion protein informationa
| Protein | N-terminal sequence | aa Position from start of ORF | No. of aa residues | Mw from inferred, N-terminal, and/or mass spectral sequence data | Mw from SDS-PAGE |
|---|---|---|---|---|---|
| VP1 | SDVPAINVT | 310-1004 | 695 | 74.705 | 74.7 |
| VP2 | TAALGIG | 1005-1257? | 253? | 28.719 | 28.7 |
aa, amino acid. The position from the start of the ORF refers to the amino acid residue position numbered from the N terminus of the ORF. The question marks indicate that the value represents a “best guess.” See Fig. 1C for sequence data.
Both capsid and other viral proteins were identified within the deduced amino acid sequence (Fig. 2) by computer analysis prediction, de novo interpretation of mass spectral data, and N-terminal peptide analysis. The 3′ UTR following the KFV polypeptide coding sequence was not predicted to form any significant secondary structure arrangements, but a complex secondary structure was predicted for the 5′ UTR by use of the MFOLD prediction algorithm (GCG; Accelrys) (data not shown). A motif similar to the Yn-Xm-AUG motif associated with internal ribosome entry site (IRES) elements of typical picornaviruses (54) was located to the 5′ side of the first AUG in the KFV ORF (nt 299 to 346 [UCUUUUGUCUAUAUCUAUGACAGAGAAGUUGUAGCAGACUCUGUCAUG]). The context of the start codon for KFV ORF 1 (GUCAUGG) somewhat resembles the most common nonvertebrate initiation sequence, ANNAUGG, as described by Cavener and Ray (7).
The genomic arrangement of KFV therefore resembles that of the classical picornaviruses. These encode single large polypeptides with virion proteins arranged towards the N terminus of the polypeptide and other predicted enzyme proteins towards the C terminus, the latter aligned in the order helicase-protease-replicase. No subgenomic RNAs have been detected for KFV, and expression of the viral genes during its replication cycle is likely to involve exclusively monocistronic expression of the single large polypeptide, followed by posttranslational cleavage to produce the viral proteins, as is found for the picornaviruses and other members of the picorna-like virus superfamily (11). The long noncoding 5′ region of the genome, with a secondary structure probably indicative of an IRES, indicates that KFV likely employs this mechanism of initiation.
We have not investigated whether the genome is capped or uncapped, but the relationship between KFV and other picorna-like viruses suggests that KFV might employ a special viral protein (VPg) for protein priming of RNA synthesis (28).
Viral proteins not found in the virions.
Three major functional domains attributed to replication proteins of picorna-like viruses (reviewed in reference 36) were identified in the KFV genome by computer-assisted analysis of the deduced amino acid sequence, as they had the core motifs of a 2C RNA helicase, 3C-like chymotrypsin-like protease, and three-dimensional RdRp (Fig. 2).
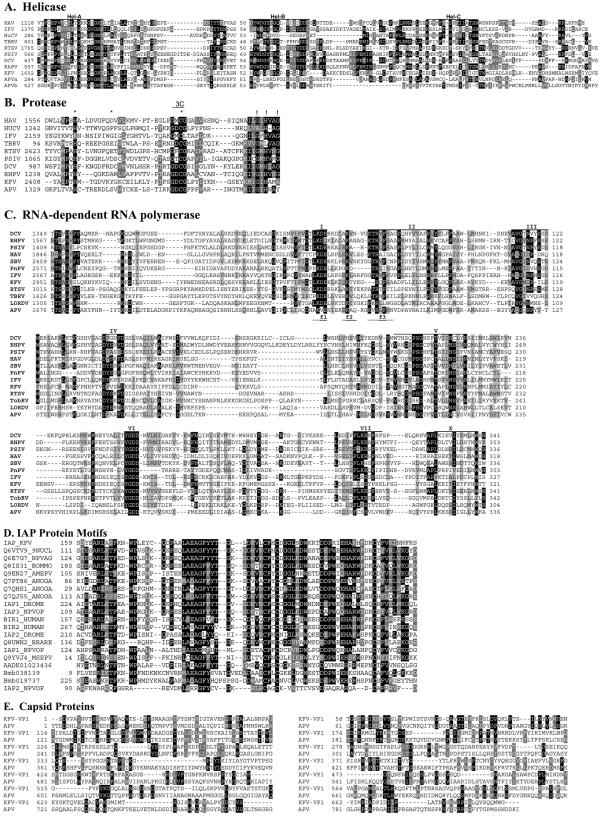
First, the core motifs of an RNA helicase from superfamily 3, as designated by Gorbalenya et al. (27), were present, including the putative nucleotide-binding domain GX4GK, with an associated ATPase chaperone functional region occurring between amino acids 1573 and 1651 (Fig. 2). This predicted helicase domain most closely resembled those of APV and the Cripavirus cricket paralysis virus (CrPV) (CrPV: 25% amino acid identity, E = 5e−16; APVa: 24% amino acid identity, E = 8e−16). Alignment of the encoded putative KFV helicase amino acid sequence with related viruses revealed the location of the three domains that are common among single-stranded positive-sense RNA viruses, as reviewed by Koonin and Dolja (36) (Fig. 3A). A pairwise distance matrix analysis based on the above alignment showed that the KFV helicase sequence is related to those of the other representative picorna-like viruses described but does not share a closer evolutionary relationship with any one particular virus or virus group (data not shown).
FIG. 3.
Comparison of the deduced amino acid sequences of the structural and nonstructural proteins of KFV and other picorna-like viruses. (A) Alignment of the conserved regions of the putative RNA helicase protein sequence from KFV with those of other picorna-like virus. The motifs recognized by Gorbalenya et al. (27) are labeled Hel-A, Hel-B, and Hel-C. (B) Alignment of KFV and picorna-like RNA virus chymotrypsin-related cysteine protease 3C-like protein sequences. The conserved residues of chymotrypsin-like proteases (26) that should form the catalytic triad are marked with asterisks, and putative substrate-binding residues are indicated by exclamation marks. The conserved catalytic cysteine motif associated with 3C proteases is indicated by the region labeled “3C.” (C) Alignment of KFV and picorna-like virus RdRp protein sequences. The amino acid sequences were aligned using the GAP program as described by Gorbalenya et al. (28). The motifs reviewed by Koonin and Dolja (36) are labeled I to VII. The f1 to f3 regions of the conserved motif F recognized by Bruenn (4) are denoted as such below the alignment sequences. (D) Comparison of the deduced amino acid sequences of representative BIR proteins of KFV, viral, dipteran, lepidopteran, and vertebrate origins. The KFV domain is labeled IAP_KFV. Other amino acid sequences are from UniProt (labeled with their entry numbers). The only exceptions are three possible novel IAPs from the Bombyx mori genome (64); these are predicted proteins Bmb038139 and Bmb019737, plus a partial sequence present in contig AADK01023436. Sequence identities of the sequences shown to the KFV sequence range from 58% identity (E = 7e−22) for the Choristoneura fumiferana defective nucleopolyhedrovirus (Q6VTV9_9NUCL) at the top down to ca. 30% for the IAP2 from the Orgyia pseudotsugata multicapsid polyhedrosis virus (IAP2_NPVOP). (E) Comparison of the deduced amino acid sequences of the structural proteins of KFV (VP1) and APV. Numbers to the initial left of alignments represent the residue numbers within the GenBank sequences. Numbers thereafter refer to residue distance from that point. Black shading indicates ≥50% identity; gray shading indicates ≥50% similarity. DCV, Drosophila C virus; RhPV, Rhopalosiphum padi virus; PSIV, Plautia stali intestine virus; PnPV, Perina nuda picorna-likevirus; HAV, human hepatitis A virus; RTSV, rice tungro spherical virus; HuCV, human calicivirus; TBRV and TobRV, tobacco ringspot virus.
A putative 3C-like chymotrypsin-related protease core motif, as described by Gorbalenya et al. (26) and reviewed by Koonin and Dolja (36), was encoded by amino acids within the region 2408 to 2802 of the genome (motifs illustrated in Fig. 3B). Comparison with other 3C-like proteases revealed that the predicted KFV protease resembled the protease domain of APV more closely than those of other picorna-like viruses (Fig. 3B).
The predicted RdRp encoded by the KFV genome was located within the C-terminal region of the KFV polypeptide and illustrated all eight conserved motifs (I to VII, and X) of RdRp found in picorna-like viruses, as designated by Koonin (35) and reviewed by Koonin and Dolja (36). Components f1 to f3 of the universally conserved RdRp motif F, involved in nucleotide binding and strand separation as defined by Bruenn (4), could also be identified between amino acids 3016 and 3036 (Fig. 3C). The motif F region was first identified in the HCV RdRp (motif nc of Lai et al. [37]), and the generality of this nucleoside triphosphate-binding site was confirmed by structural analysis (6, 39).
Comparison of the RdRp amino acid sequence of KFV to those of other viruses in the picorna-like superfamily showed levels of amino acid sequence identity ranging between 22 and 27%, with approximately 40 to 47% similarity. The deduced RdRp amino acid coding sequence of KFV was compared with all protein sequences in the NCBI database by using BLASTP. KFV RdRp and Drosophila C virus RdRp share 47% amino acid similarity (26% identity), while the KFV RdRp shares 27% amino acid identity with the Plautia stali intestine virus RdRp. Overall identities with the other picorna-like virus RdRp sequences were as follows: Plautia stali intestine virus, 27%; Drosophila C virus, 26%; Kashmir bee virus, 25%; CrPV, 24%; Rhopalosiphum padi virus, 24%; rice tungro spherical virus, 22%; acute bee paralysis virus, 25%; Perina nuda picorna-like virus, 26%; Ectropis obliqua picorna-like virus, 26%; infectious flacherie virus (IFV), 23%; APV, 23%; sacbrood virus (SBV), 22%; human hepatitis A virus, 23%; tobacco ringspot virus, 22%; and Lordsdale virus (LORDV), 22%.
The conserved 2A-like protein “cleavage” motif (DxExNPGP), described for cardioviruses, aphthoviruses, and some picorna-like viruses (16, 30, 63) and attributed to protein “cleavage” by a ribosomal skip from one codon to the next (17), was not detected within the KFV polypeptide sequence.
The putative KFV BIR motif is a single repeat that lacks the associated RING zinc finger which accompanies BIR repeats in mammalian cells. BLAST searches have shown it to most closely resemble BIR domains from a number of viral and cellular IAP proteins. An alignment of the sequence of the KFV BIR against several of those it resembles most closely is shown in Fig. 3D. The KFV BIR contains the conserved core motif GX9-11CX2C8-10E/DX5HX3-6C. Identity scores against the proteins shown in the alignment range from 50% to 58%. Shrimp Taura syndrome virus (TSV), an arthropod virus tentatively assigned to the Dicistroviridae, is the only other RNA virus encoding a similar BIR motif within the genome (44). Like the KFV BIR, the TSV BIR domain contains only a single repeat and no zinc finger; its sequence shows lower homology to those of the other BIR proteins and is not included in the alignment. The function of the TSV and KFV domains has not yet been experimentally tested.
Virion proteins.
Purified KFV virions were disrupted and separated by HPLC, and the N termini of the main peaks were microsequenced by standard Edman degradation methodology (Protein Microchemistry Facility, University of Otago, New Zealand). The N-terminal sequences obtained were SDVPAINVT and TAALGIG, which corresponded to the region of inferred amino acid sequence encoded by the 5′ end of the KFV RNA (Fig. 1). These are the N termini of VP1 and VP2, whose sequences are contiguous in the uncleaved polypeptide translated from the KFV ORF. The Mw of VP1 estimated from the inferred sequence is identical to that estimated from SDS-PAGE (Table 2).
Electrospray mass spectrometry on tryptic digests of HPLC peaks confirmed the location of the coat protein region. Two high-Mw proteins corresponding to HPLC peaks were observed by mass spectrometry of the undigested, but disrupted, virions. These peaks contained common sequences of VP1 when the constituent peptides were analyzed. The smaller Mw peak was judged to be a degradation product of VP1 since it lacked the sequences QGFTMLPTNR and GISNVQR but contained the other tryptic digest sequences listed below.
The mass spectrometer “call” sequences were as follows: for VP1, NNPAGTGSSFDIK, ESVYSLYK, LSLDPQK, AISLASTTATTEVLSDIR, MGLYR, NDLVNFYIR, IMVK, PSSSLSTQLYSDIPLK, AFIK, DPIIDLTVTGPAR, KGMPIYDK, IYIAYGTVPTPSGSISR, LNYFVAETSTGK, QGFTMLPTNR, and GISNVQR; and for VP2, DFNAQMLEK, LQELK, and GLLDLEK.
SDS-PAGE analysis was performed to estimate the molecular masses of the capsid proteins. It was important to use freshly thawed KFV, as obvious degradation of VP1 and VP2 occurred over a period of several days, even at 4°C. This was likely due to a protease not being completely removed during the virus purification steps. Two major protein bands of ∼75 kDa and 29 kDa were identified. These were denoted VP1 and VP2, respectively. Two additional protein bands, albeit minor, were also present: a 16-kDa protein and a protein of slightly lower molecular mass than VP1 (Fig. 1B), which presumably derives from a degradation of VP1,corresponding to the additional HPLC peaks observed. The major bands concur with the predicted proteins described by Scotti et al. (51), but the two minor bands were not clearly apparent at that time. Two virus capsid proteins have therefore been identified by using N-terminal and peptide mass fingerprint analysis: VP1 (695 amino acids, 74.705 kDa) and VP2 (235 amino acids, 28.719 kDa). The minor 16.6-kDa protein would appear to be a second degradation product of VP1 (Table 2; Fig. 1), since mass spectrometry confirmed that it contained VP1 peptide sequences (the “call” sequences listed above for VP1 and VP2 corresponded to the region of the single polypeptide indicated in Fig. 1).
The virion proteins of KFV are unlike those of other insect picorna-like viruses in size and number, having only two in total, comprising one large Mw protein and one small Mw protein (Table 2; Fig. 1B). In contrast, four capsid proteins are found in the virions of all members of both the iflaviruses and the Dicistroviridae, comprising three proteins of approximately equal 20- to 30-kDa size (VP1 to VP3) and one smaller cleavage product, VP4 (40, 45).
VP1 showed some homology with the capsid protein region of APV protein P1 (24% identity and 43% similarity between APV amino acids 2420 and 2579, nt 7260 and 7737, and KFV amino acids 368 and 515) in a BLASTP search against the nonredundant protein database, while VP2 did not evince any significant homology with other viral structural proteins. The homology between the capsid proteins of APV and the VP1 protein of KFV, shown in Fig. 3E, is of interest and may reflect a link between these two viruses despite other differences in genomic organization and expression strategy.
Cryo-EM reconstruction of KFV.
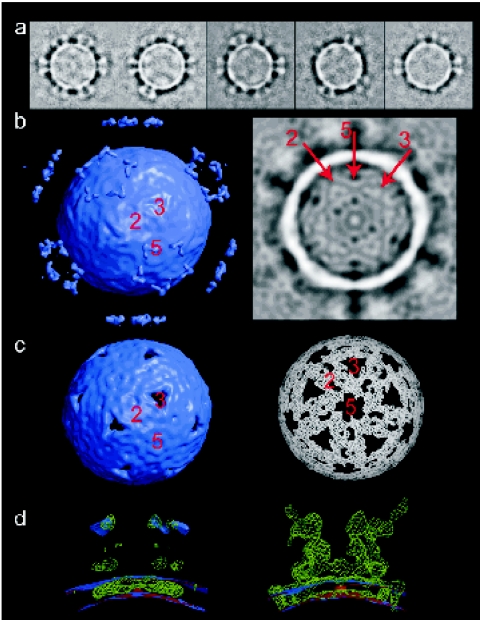
Cryo-EM and image reconstruction of KFV, initially to a resolution of 18 Å, revealed an icosahedral structure, with each of its 12 fivefold vertices projecting a turret from the otherwise smooth surface of the ∼20-Å-thick capsid. Discounting the turrets, the diameter of the virus capsid is 330 Å but extends to 450 Å including the turrets projecting 60 Å above the surface of the virion. Image classification of the particles showed that not all fivefold vertices were occupied by turrets (Fig. 4a), that loss of the turrets does not lead to a change in the capsid morphology, and that the individual turret structure is the same in fully occupied and partially occupied cases (see Materials and Methods and http://www.ebi.ac.uk/msd). This observation is reflected in the fact that the density for the turrets is much weaker than that for the capsid in the icosahedrally averaged reconstruction (Fig. 4a), where only the bulkier rim of the turret was visible at a significant density threshold above the noise level. The lower occupancy of turret density than of capsid density was also apparent in the central section through the reconstruction shown in Fig. 4b. Other features of the structure apparent from this density section included the presence of weaker density at the fivefold and threefold axes and the fact that the viral RNA (in common with that of picornaviruses such as foot-and-mouth disease virus [FMDV]) is apparently disordered within the capsid.
FIG. 4.
Three-dimensional reconstruction of KFV by cryo-EM. (a) Gallery of class averages of raw KFV images, ranked by turret occupancy. This shows that there is no change in overall capsid morphology with the loss of turrets and that the turret structure is the same when fully occupied as when partially occupied. (b) Left: surface-rendered view (contour 2σ) of an icosahedral reconstruction of KFV. Right: central section through the same map. The symmetry axes are marked in both cases. (c) Left: surface-rendered view (contour 4σ) of the map shown in panel a. Right: the same map in another view at the 5.2σ-contour level. (d) Superpositions of a reconstruction selected for low turret occupancy (red), the reconstruction shown in panels b and c (lilac), and a difference map (green), viewed across a fivefold vertex. The difference map highlights features present in the partially turreted map but absent from the map constructed from particles selected for the absence of turrets. Left: reconstructions at 2σ and difference map at 3σ. Right: these thresholds are switched.
Higher density thresholds show the underlying structure in the capsid (Fig. 4c). The threefold axes of the capsid contain a pore, plugged by weaker density which rises slightly above the viral surface (as seen in Fig. 4b). Less prominent pores also occur at the fivefold axes. At these higher threshold levels, it should have been possible to see any quasi-symmetry within the icosahedral asymmetric unit, but there is no evidence of such symmetry, suggesting that, if there is more than one protein subunit, they are not arranged in a quasi-equivalent fashion.
Figure 4d shows a superposition of the reconstruction shown in Fig. 4b and c, a reconstruction in which particles were selected for a relative absence of turrets, and a corresponding difference map. Viewed across the fivefold vertex, the reconstruction lacking turrets has thinner density than that with partial turret occupancy (Fig. 4b to d). The difference map also suggests that the shedding of turrets involves the dislodging of some protein from the viral surface, which presumably represents their foothold in it.
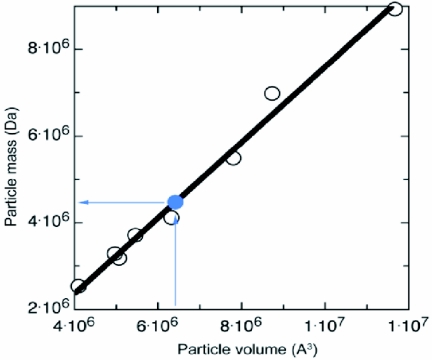
In order to address the mass and architecture of the capsid further, KFV was compared with a series of insect and other representative viruses (Fig. 5 and 6; Table 1). These viruses include insect members of the picornavirus superfamily (40), the Caliciviridae (LORDV and Norwalk virus), and other insect viruses (Pariacoto virus and Galleria mellonella densovirus), together with the noninsect viruses FMDV, bean pod mottle virus, tobacco ringspot virus, and adeno-associated virus. For each comparison, electron density was calculated from the icosahedral atomic structure and that density was used to compute the volume of the capsid shell (with GAP) (25, 29), which was compared to its known atomic mass. A linear relationship was calculated between capsid volume and atomic mass (Fig. 5), allowing the volume of the KFV capsid shell (6.41 × 106 Å3) to be converted to its mass (4.48 MDa). Thus, each icosahedral asymmetric unit of the KFV capsid shell has a mass of ∼75 kDa.
FIG. 5.
Plot of the atomic mass of virus particles (capsid shell) against the volume of low-resolution maps calculated from their atomic coordinates. The lilac plot point marks the positioning of the KFV shell in this range of masses, with the lilac arrows showing that a volume of 6.41 × 106 Å3 gives the KFV shell a mass of 4.48 MDa.
FIG. 6.
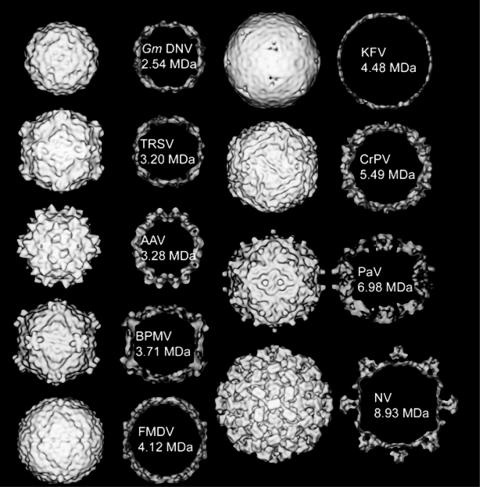
A gallery of views of virus structures, including insect members of the picornavirus superfamily, definitive picornaviruses, and other insect viruses. In each case, the rendered surface of the structure (at an 18-Å resolution) is shown, as well as a central section through the map. The masses marked are those of the atomic structures shown at lower-resolution density here, except for KFV, which is a mass value based on its capsid surface volume normalized to the mass/volume ratio of the other structures (see Fig. 5). The viruses shown are Galleria mellonella densovirus (GmDNV) (a parvovirus with a T=1 capsid; total genome size, 5 kb [53]); tobacco ringspot virus (TRSV) (a nepovirus with one coat protein making a pseudo T=3 capsid; total genome size, 15.5 kb [8]); adeno-associated virus (AAV) (with a T=1 capsid [65]); bean pod mottle virus (BPMV) (a comovirus with two coat proteins making a pseudo T=3 capsid; total genome size, 9.7 kb [41]); FMDV (a classical picornavirus with a pseudo T=3 capsid; total genome size, 8.4 kb [1]); KFV (total genome size, 11 kb); CrPV (a pseudo T=3 virus; total genome size, 9.2 kb [56]); Pariacoto virus (PaV) (a nodavirus with a T=3 capsid; total genome size, 4.3 kb [55]); and Norwalk virus (NV) (a calicivirus with a proper T=3 capsid; total genome size, 7.9 kb [9, 49]).
As previously observed, due to their low occupancy, the structure of the turrets was poorly defined in the icosahedrally averaged reconstruction. Comparison of peak density values for the turret and the (necessarily) fully occupied capsid layer indicates an average of 25% occupancy. However, the actual occupancy of any single virion varied from apparently full to zero (see deposited information at http://www.ebi.ac.uk/msd). In order to obtain a more detailed picture of the turrets, a new reconstruction was performed, aligning images to a model in which only one of the fivefold vertices projected a turret, allowing all of the virus images to be aligned such that the occupancy of the single turret was maximized. First, all but one of the turrets were removed from the reconstruction shown in Fig. 4b and c (see Materials and Methods), and then this single-turreted map was used to align the whole data set. The resulting reconstruction of a single KFV turret projecting from the capsid surface, computed to a resolution of 15 Å, illustrated that the turret structure had two major lobes of density for each of its five subunits: a core lobe and a radially projecting lobe that is less well ordered (Fig. 7a). Each turret is connected to the capsid surface through slender projections. Based on volumetric comparison with other viral turret structures, the mass of the protomeric unit of the projecting portion of the turret can be estimated as ∼70 kDa. The single-turret reconstruction was used to produce a reconstruction of the whole virus with its vertices fully occupied by turrets: an icosahedral asymmetric unit around the aligned turret was cut out, and the complete 8.70-MDa KFV capsid was generated by the application of icosahedral symmetry. The result is shown in Fig. 7b.
FIG. 7.
Resolving the KFV turret structure. (a) Views of the reconstruction of a single-turreted KFV, viewed from above the virus surface (left, surface not in view) and from the side, through the fivefold vertex (right). The map is contoured at three levels, 1.5σ, 2σ, and 4σ, to show the relative strengths of the density for different portions of the turret and the steep density gradient defining the edges of the structure. Identical features in the two views are labeled i to iii. (b) Views down fivefold, threefold, and twofold axes (left to right, respectively) of the complete KFV capsid generated using icosahedral symmetry from an asymmetric unit derived from the single-turreted reconstruction.
The architecture of the KFV capsid is unique among members of the picornavirus superfamily and insect viruses for which structures have previously been determined (Fig. 6). In particular, KFV has an unusually smooth surface which is much thinner than the viruses to which it is compared (Fig. 5 and 6). Given that the total genome size of KFV (11 kb) is the second largest of the animal viruses used for comparison (see legend for Fig. 6), the packing density will actually be slightly lower than for many picornaviruses, given the greater internal volume. The thin capsid may reflect a packaging strategy which has evolved to allow incorporation of the large RNA genome into the virus particle. The thickness of the capsid is in fact similar to that of the bacteriophage HK97 (18 Å) with its striking chain mail construction (62). From a morphological point of view, the most similar picornavirus superfamily member appears to be FMDV (pseudo T=3 with three major proteins and one minor one). Overall, the KFV reconstruction is consistent with the capsid having T=1 symmetry and with the shell being made up of 60 copies of an asymmetric unit consisting of a protein, or proteins, of ∼75 kDa, completed by the addition of up to 60 copies of the turret protein, which has a footprint in the capsid surface itself. If VP1 were the turret protein, then the observed cleavage may correlate with the lability of the turrets, and the majority of the shell would be constructed from several copies of VP2 arranged with no obvious quasi-symmetry.
Reconstruction of the whole virus with its vertices fully occupied by turrets (Fig. 7b) revealed that the turrets represent nearly half of the 8.70-MDa KFV capsid mass, consistent with a significant functional role in the virus. Turrets could be composed of receptor-binding proteins or be involved in the other known roles of turret structures in viruses, which include mRNA extrusion and genome packaging. Aside from these possible roles, the lability of the turrets and the relative disorder of the pendant turret protein domain may indicate a dynamic role in KFV function. The pendant (probably flexible) domains of the turrets are close enough to each other to interact if they are hinged at their junction with the central part of the turret.
DISCUSSION
Phylogenetic relationships.
Numerous picorna-like viruses have been isolated from insect and other arthropod hosts, and the genome sequences now available for many of these confirm them as members of the picorna-like virus superfamily. Based on genome organization, phylogeny of their replicase protein coding sequences, and structural data, two major taxonomic groups have emerged, comprising most of the sequenced viruses. The first family to achieve recognition was the Dicistroviridae family, recently described by Mayo (45), with a single recognized genus, Cripavirus, represented by cricket paralysis virus. Dicistroviruses have genomes of 9,000 to 10,000 nt in length, carrying two ORFs separated by an intragenic region which can function as an IRES to initiate translation. Dicistroviral capsid proteins (VP1, VP2, VP3, and possibly VP4) are encoded by the 3′ ORF, while nonstructural proteins are encoded by the 5′ ORF.
Several other picorna-like viruses infecting insects use the characteristic picornavirus coding strategy involving a unique large ORF with the structural proteins encoded at the 5′ region and the RdRp at the 3′ end of the genome. Although this genome organization resembles that of the mammalian picornaviruses, these insect viruses are phylogenetically distinct from the established picornaviruses. Insect viruses with this genome organization, and showing a clear phylogenetic relationship, are as follows: SBV (24), IFV (33), Perina nuda picorna-like virus (63), Ectropis obliqua picorna-like virus (61), deformed wing virus (GenBank accession no. AY292384 and AJ489744), Kakugo virus (22), and Varroa destructor virus (48). These viruses have been grouped into a “floating” genus (not a recognized family) termed Iflavirus (45).
Distinctions in the 5′ nonstructural BIR domain and capsid protein characteristics, as well as unique structural properties, separate KFV from the Dicistroviridae, iflaviruses, and other picorna-like insect viruses. The KFV virion also has only two major capsid proteins, but the large size of VP1 (75 kDa) is anomalous to the capsid protein sizes of Dicistroviridae, iflaviruses, and APV. Despite significant amino acid identity between the nonstructural proteins of KFV and some members of the Dicistroviridae family, KFV clearly differs from this family in genomic arrangement and lack of an intragenic region, as well as in capsid protein coding sequence and arrangement. Despite its superficial resemblance to the iflaviruses in genome organization, KFV shows no phylogenetic affinity with this group and therefore remains with those picorna-like insect viruses that are not able to be assigned to any specific genus or distinct group, the other currently being the aphid-infecting APV. APV resembles the Dicistroviridae in possessing a monopartite, bicistronic genome with similarly arranged structural and nonstructural proteins, but with the two ORFs overlapping and the 3′ ORF thought to be translated by a −1 frameshift (59).
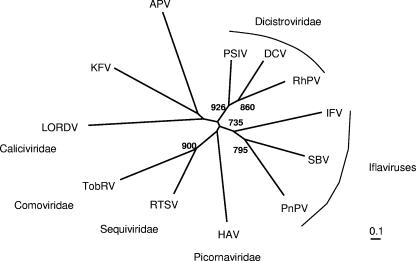
Phylogenetic analysis based on the aligned RdRp sequences in Fig. 3C failed to show any clear, close relationship between APV and KFV. Neighbor-joining analysis (Fig. 8) placed both of these viruses at the ends of adjacent and deeply rooted branches distinct from all other families in the superfamily (e.g., the dicistroviruses, the iflaviruses, the picornaviruses, the caliciviruses, and the plant viruses). Analysis using maximum likelihood showed a similar deeply branching pattern (not shown), although the calicivirus now formed a possible clade with APV over KFV. This was consistent with the low bootstrap support (316 and 531 of 1,000 trials) obtained for the branches to LORDV and APV/KFV in the neighbor-joining phenogram in Fig. 8, indicating the uncertain resolution obtainable with this limited set of distantly related sequences. In summary, the distinct phylogenetic clades derived from insect picorna-like viruses now include the Dicistroviridae, the iflaviruses, APV, and KFV.
FIG. 8.
Phylogenetic analysis of the relationship of KFV RdRp sequences to those of other virus families and viruses in the picornavirus superfamily. The phenogram is based on the alignment in Fig. 3C of polymerases from KFV and representative viruses of the Picornaviridae, Dicistroviridae, Comoviridae, Sequiviridae, and Caliciviridae families, as well as the unclassified insect virus APV. An unrooted neighbor-joining tree was inferred using the programs ClustalX 1.81 (58) and PHYLIP (19). All bifurcations with support in >700 out of 1,000 bootstraps are indicated. DCV, Drosophila C virus; RhPV, Rhopalosiphum padi virus; PSIV, Plautia stali intestine virus; PnPV, Perina nuda picorna-like virus; HAV, human hepatitis A virus; RTSV, rice tungro spherical virus; TobRV, tobacco ringspot virus.
Interestingly, the VP1 KFV capsid protein is related to the APV capsid protein, sharing 24% amino acid identity and 44% similarity over the first 108 amino acids, with overall amino acid homology of 17.2% identity and 27.5% similarity. If KFV VP1 was the turret protein, then this could correspond to a domain acting as a foothold within the capsid. This relationship suggests that KFV and APV may in fact share a distant evolutionary relationship, and their virion proteins hint at a new grouping within those of the insect picorna-like viruses.
The highly unusual presence of the BIR is a feature of the KFV genome shared only with one other arthropod RNA virus, TSV. Like KFV, TSV also encodes an unusually large 56-kDa capsid protein (VP3, Cripavirus notation) but is dissimilar to KFV in genomic arrangement and has been assigned to the dicistroviruses. IAP proteins are involved in the inhibition of apoptosis by preventing the activation of caspases and are presumed to enhance viral replication and survival in mammalian and insect cells (3, 43). Such proteins were initially discovered in baculoviruses, where their function has been well studied, but their identification on the basis of sequence homology in TSV, and now KFV, represents the first finding of the possibility of such domains in RNA viruses. Further work is needed to ask whether the KFV BIR is in fact capable of inhibiting apoptosis before it can be ascertained whether this protein domain may have any role during the KFV replication cycle. The baculoviral IAPs are thought to have been acquired from insect hosts through gene capture (32). Phylogenetic analysis of the KFV BIR domain in comparison with those from vertebrates, insects, and insect viruses was unable to clearly resolve an evolutionary origin for the KFV domain, so that its ancestry, like that of the TSV domain, remains unknown. The analysis included IAPs newly identified for the Bombyx mori genome (64) that may be ancestors of the IAPs found in baculoviruses infecting lepidoptera (listed in Fig. 3D). No close relationship to any of the IAPs previously identified in dipteran genomes was evident. Functional determination and further characterization of this protein, together with wider arthropod genome data, may eventually reveal the significance of the BIR with regard to the biology of KFV and the evolutionary origin of the domain.
Thus, KFV appears to have certain features of both the Dicistroviridae and the iflaviruses, providing some support for the theory of a common ancestor for the insect picorna-like viruses that is purported to have encoded a single coat protein within a genomic arrangement similar to that of IFV and SBV (40). However, unique KFV characteristics, such as the unusual genomic arrangement, capsid protein arrangement, and particle morphology, suggest that significant evolutionary divergence has occurred and establish KFV as representing a new group of viruses. Cryo-EM analysis of KFV particles provided an interesting basis for comparison of the viral particle structures of KFV and other insect picorna-like viruses, and the resultant model of KFV reveals further adaptations to exploit the available dimensions of viral protein:RNA space in small RNA virus particles. The origin of the BIR domains of KFV, compared and contrasted with that of TSV, may also provide additional insight into the evolution of the picorna-like virus superfamily and the links, if any, between host genomes and the small single-stranded RNA viruses.
Acknowledgments
We thank Adrian Gibbs for critical review of the manuscript and invaluable advice. D.R.G. and P.D.S. thank Janine Cooney and Dwyane Jensen (HortResearch, Ruakura) for mass spectrometer operation and the University of Otago Biochemistry Department Microsequencing Facility for N-terminal Edman sequencing. We also acknowledge Sally Dearing and Madeleine Parker for their excellent technical help.
R.J.C.G. is a Royal Society University Research Fellow, and D.I.S. is an MRC Research Professor.
REFERENCES
- 1.Acharya, R., E. E. Fry, D. I. Stuart, G. Fox, D. J. Rowlands, and F. Brown. 1989. The three-dimensional structure of foot-and-mouth disease virus at 2.9 Å resolution. Nature 337:709-716. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birnbaum, M. J., R. J. Clem, and L. K. Miller. 1994. An apoptosis-inhibiting gene from a nuclear polyhedrosis virus encoding a polypeptide with Cys/His sequence motifs. J. Virol. 68:2521-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruenn, J. A. 2003. A structural and primary sequence comparison of the viral RNA-dependent RNA polymerases. Nucleic Acids Res. 31:1821-1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruenn, J. A. 1991. Relationships among the positive strand and double-strand RNA viruses as viewed through their RNA-dependent RNA polymerases. Nucleic Acids Res. 19:217-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butcher, S. J., J. M. Grimes, E. V. Makeyev, D. H. Bamford, and D. I. Stuart. 2001. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410:235-240. [DOI] [PubMed] [Google Scholar]
- 7.Cavener, D. R., and S. C. Ray. 1991. Eukaryotic start and stop translation sites. Nucleic Acids Res. 19:3182-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandrasekar, V., and J. E. Johnson. 1997. The structure of tobacco ringspot virus: a link in the evolution of icosahedral capsids in the picornavirus superfamily. Structure 6:157-171. [DOI] [PubMed] [Google Scholar]
- 9.Chen, R., J. D. Neill, J. S. Noel, A. M. Hutson, R. I. Glass, M. K. Estes, and B. V. Venkataram Prasad. 2004. Inter- and intragenus structural variations in caliciviruses and their functional implications. J. Virol. 78:6469-6479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 11.Christian, P., E. Carstens, L. Domier, K. Johnson, N. Nakashima, P. Scotti, and F. van der Wilk. 1999. Cricket paralysis-like viruses, p. 678-683. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 12.Christian, P., and P. D. Scotti. 1998. Picornalike viruses of insects, p. 301-336. In L. K. Miller and L. A. Ball (ed.), The insect viruses. Plenum Press, New York, N.Y.
- 13.Cohen, J. I., J. R. Ticehurst, R. H. Purcell, A. Buckler-White, and B. M. Baroudy. 1987. Complete nucleotide sequence of wild-type hepatitis A virus: comparison with different strains of hepatitis A virus and other picornaviruses. J. Virol. 61:50-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crowther, R. A., R. Henderson, and J. M. Smith. 1996. MRC image processing programs. J. Struct. Biol. 116:9-16. [DOI] [PubMed] [Google Scholar]
- 15.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donnelly, M. L. L., D. Gani, M. Flint, S. Monoghan, and M. D. Ryan. 1997. The cleavage activity of aphtho- and cardiovirus 2A proteins. J. Gen. Virol. 61:51-59. [DOI] [PubMed] [Google Scholar]
- 17.Donnelly, M. L. L., G. Luke, A. Mehrotra, X. Li, L. E. Hughes, D. Gani, and M. D. Ryan. 2001. Analysis of the aphthovirus 2A/2B polyprotein “cleavage” mechanism indicates not a proteolytic reaction but a novel translational effect: a putative ribosomal “skip.” J. Gen. Virol. 82:1013-1025. [DOI] [PubMed] [Google Scholar]
- 18.Eng, J., A. L. McCormack, and J. R. Yates III. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976-989. [DOI] [PubMed] [Google Scholar]
- 19.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 20.Ferlenghi, I., B. Gowen, F. de Haas, E. J. Mancini, H. Garoff, M. Sjoberg, and S. D. Fuller. 1998. The first step: activation of Semliki Forest virus spike protein precursor causes a localized conformational change in the trimeric spike. J. Mol. Biol. 283:71-81. [DOI] [PubMed] [Google Scholar]
- 21.Frank, J., M. Radermacher, P. Penczek, J. Zhu, Y. Li, M. Ladjadj, and A. Leith. 1996. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116:190-199. [DOI] [PubMed] [Google Scholar]
- 22.Fujiyuki, T., H. Takeuchi, M. Ono, S. Ohka, T. Sasaki, A. Nomoto, and T. Kubo. 2004. Novel insect picorna-like virus identified in the brains of aggressive worker honeybees. J. Virol. 78:1093-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuller, S. D., S. J. Butcher, R. H. Cheng, and T. S. Baker. 1996. Three-dimensional reconstruction of icosahedral particles—the uncommon line. J. Struct. Biol. 116:48-55. [DOI] [PubMed] [Google Scholar]
- 24.Ghosh, R. C., B. V. Ball, M. M. Willcocks, and M. J. Carter. 1999. The nucleotide sequence of sacbrood virus of the honey bee: an insect picorna-like virus. J. Gen. Virol. 80:1541-1549. [DOI] [PubMed] [Google Scholar]
- 25.Gilbert, R. J. C., P. Fucini, S. Connell, S. Fuller, K. H. Nierhaus, C. V. Robinson, C. M. Dobson, and D. I. Stuart. 2004. Three dimensional structures of translating ribosomes by cryo-EM. Mol. Cell 14:57-66. [DOI] [PubMed] [Google Scholar]
- 26.Gorbalenya, A. E., A. P. Donchenko, V. M. Blinov, and E. V. Koonin. 1989. Cysteine proteases of positive strand RNA viruses and chymotrypsin-like serine proteases. A distinct superfamily with a common structural fold. FEBS Lett. 243:103-113. [DOI] [PubMed] [Google Scholar]
- 27.Gorbalenya, A. E., E. V. Koonin, and Y. I. Wolf. 1990. A new superfamily of putative NTP-binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett. 252:145-148. [DOI] [PubMed] [Google Scholar]
- 28.Gorbalenya, A. E., F. M. Pringle, J.-L. Zeddam, B. T. Luke, C. E. Cameron, J. Kalmakoff, T. N. Hanzlik, K. H. J. Gordon, and V. K. Ward. 2002. The palm subdomain-based active site is internally permuted in viral RNA-dependent RNA polymerases of ancient lineage. J. Mol. Biol. 324:47-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grimes, J. M., J. N. Burroughs, P. Gouet, J. M. Diprose, R. Malby, S. Zientara, P. P. Mertens, and D. I. Stuart. 1998. The atomic structure of the bluetongue virus core. Nature 395:470-478. [DOI] [PubMed] [Google Scholar]
- 30.Hahn, H., and A. C. Palmenberg. 1996. Mutational analysis of the encephalomyocarditis virus primary cleavage. J. Virol. 70:6870-6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanzlik, T. N., S. J. Dorrian, K. N. Johnson, E. M. Brooks, and K. H. J. Gordon. 1995. Sequence of RNA2 of the Helicoverpa armigera stunt virus (Tetraviridae) and bacterial expression of its genes. J. Gen. Virol. 76:799-811. [DOI] [PubMed] [Google Scholar]
- 32.Hughes, A. L. 2002. Evolution of inhibitor of apoptosis in baculoviruses and their insect hosts. Infect. Genet. Evol. 2:3-10. [DOI] [PubMed] [Google Scholar]
- 33.Isawa, H., S. Asano, K. Sahara, T. Iizuka, and H. Bando. 1998. Analysis of genetic information of an insect picorna-like virus, infectious flacherie virus of silkworm: evidence for evolutionary relationships among insect, mammalian and plant picorna(-like) viruses. Arch. Virol. 143:127-143. [DOI] [PubMed] [Google Scholar]
- 34.Johnson, K. N., and P. D. Christian. 1998. The novel genome organization of the insect picorna-like virus Drosophila C virus suggests this virus belongs to a previously undescribed virus family. J. Gen. Virol. 79:191-203. [DOI] [PubMed] [Google Scholar]
- 35.Koonin, E. V. 1991. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J. Gen. Virol. 72:2197-2206. [DOI] [PubMed] [Google Scholar]
- 36.Koonin, E. V., and V. V. Dolja. 1993. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 28:375-430. [DOI] [PubMed] [Google Scholar]
- 37.Lai, V. C. H., C. C. Kao, E. Ferrari, J. Park, A. S. Uss, J. Wright-Minogue, Z. Hong, and J. Y. N. Lau. 1999. Mutational analysis of bovine viral diarrhea virus RNA-dependent RNA polymerase. J. Virol. 73:10129-10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Layne, E. 1957. Spectrophotometric and turbidometric methods for measuring proteins. Methods Enzymol. 3:447-455. [Google Scholar]
- 39.Lesburg, C. A., M. B. Cable, E. Ferrari, Z. Hong, A. F. Mannarino, and P. C. Weber. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6:937-943. [DOI] [PubMed] [Google Scholar]
- 40.Liljas, L., J. Tate, P. Christian, and J. E. Johnson. 2002. Evolutionary and taxonomic implications of conserved structural motifs between picornaviruses and insect picorna-like viruses. Arch. Virol. 147:59-84. [DOI] [PubMed] [Google Scholar]
- 41.Lin, T., J. Cavarelli, and J. E. Johnson. 2003. Evidence for assembly-dependent folding of protein and RNA in an icosahedral virus. Virology 314:26-33. [DOI] [PubMed] [Google Scholar]
- 42.Liu, B. L., I. N. Clarke, E. O. Caul, and P. R. Lambden. 1995. Human enteric caliciviruses have a unique genome structure and are distinct from the Norwalk-like viruses. Arch. Virol. 1140:1345-1356. [DOI] [PubMed] [Google Scholar]
- 43.Maguire, T., P. Harrison, O. Hyink, J. Kalmakoff, and V. K. Ward. 2000. The inhibitors of apoptosis of Epiphyas postvittana nucleopolyhedrosis virus. J. Gen. Virol. 81:2803-2811. [DOI] [PubMed] [Google Scholar]
- 44.Mari, J., B. T. Poulos, D. V. Lightner, and J. R. Bonami. 2002. Shrimp Taura syndrome virus: genomic characterization and similarity with members of the genus Cricket paralysis-like viruses. J. Gen. Virol. 83:915-926. [DOI] [PubMed] [Google Scholar]
- 45.Mayo, A. M. 2002. Virus taxonomy—Houston 2002. Arch. Virol. 147:1071-1076. [DOI] [PubMed] [Google Scholar]
- 46.Moon, J. S., L. L. Domier, N. K. McCoppin, C. J. D'Arcy, and H. Jin. 1998. Nucleotide sequence analysis shows that Rhopalosiphum padi virus is a member of a novel group of insect-infecting RNA viruses. Virology 243:54-65. [DOI] [PubMed] [Google Scholar]
- 47.Moore, N. F., and T. W. Tinsley. 1982. The small RNA viruses of insects. Arch.Virol. 72:229-245. [DOI] [PubMed] [Google Scholar]
- 48.Ongus, J. R., D. Peters, J.-M. Bonmatin, E. Bengsch, J. M. Vlak, and M. M. van Oers. 2004. Complete sequence of a picorna-like virus of the genus Iflavirus replicating in the mite Varroa destructor. J. Gen. Virol. 85:3747-3755. [DOI] [PubMed] [Google Scholar]
- 49.Prasad, B. V. V., M. E. Hardy, T. Dokland, J. Bella, M. G. Rossmann, and M. K. Estes. 1999. X-ray crystallographic structure of the Norwalk virus capsid. Science 286:287-290. [DOI] [PubMed] [Google Scholar]
- 50.Sasaki, J., N. Nakashima, H. Saito, and H. Noda. 1998. An insect picorna-like virus, Plautia stali intestine virus, has genes of capsid proteins in the 3′ part of the genome. Virology 244:50-58. [DOI] [PubMed] [Google Scholar]
- 51.Scotti, P. D., A. J. Gibbs, and N. G. Wrigley. 1976. Kelp fly virus. J. Gen. Virol. 30:1-9. [Google Scholar]
- 52.Shen, P., M. Kanieswka, C. Smith, and R. N. Beachy. 1993. Nucleotide sequence and genomic organization of rice tungro spherical virus. Virology 193:621-630. [DOI] [PubMed] [Google Scholar]
- 53.Simpson, A. A., P. R. Chipman, T. S. Baker, P. Tijssen, and M. G. Rossmann. 1998. The structure of an insect parvovirus (Galleria mellonella densovirus) at 3.7 Å resolution. Structure 6:1355-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stewart, S. R., and B. L. Semler. 1997. RNA determinants of picornavirus cap-independent translation initiation. Semin. Virol. 8:242-255. [Google Scholar]
- 55.Tang, L., K. N. Johnson, L. A. Ball, T. Lin, M. Yeager, and J. E. Johnson. 2001. The structure of Pariacoto virus reveals a dodecahedral cage of duplex RNA. Nat. Struct. Biol. 8:77-83. [DOI] [PubMed] [Google Scholar]
- 56.Tate, J., L. Liljas, P. Scotti, P. Christian, T. Lin, and J. E. Johnson. 1999. The crystal structure of cricket paralysis virus: the first view of a new virus family. Nat. Struct. Biol. 6:765-774. [DOI] [PubMed] [Google Scholar]
- 57.Taylor, J. A., and R. S. Johnson. 2001. Implementation and uses of automated de novo peptide sequencing by tandem mass spectrometry. Anal. Chem. 73:2594-2604. [DOI] [PubMed] [Google Scholar]
- 58.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van der Wilk, F., A. M. Dullemans, M. Verbeek, and J. F. Van den Heuvel. 1997. Nucleotide sequence and genomic organization of Acyrthosiphon pisum virus. Virology 238:353-362. [DOI] [PubMed] [Google Scholar]
- 60.van Heel, M., G. Harauz, E. V. Orlova, R. Schmidt, and M. Schatz. 1996. A new generation of the IMAGIC image processing system. J. Struct. Biol. 116:17-24. [DOI] [PubMed] [Google Scholar]
- 61.Wang, X. C., C. Y. Wu, and C. F. Lo. 2004. Sequence analysis and genomic organization of a new insect picorna-like virus, Ectropis obliqua picorna-like virus, isolated from Ectropis obliqua. J. Gen. Virol. 85:1145-1151. [DOI] [PubMed] [Google Scholar]
- 62.Wikoff, W. R., L. Liljas, R. L. Duda, H. Tsuruta, R. W. Hendrix, and J. E. Johnson. 2000. Topologically linked protein rings in the bacteriophage HK97 capsid. Science 289:2129-2133. [DOI] [PubMed] [Google Scholar]
- 63.Wu, C. Y., C. F. Lo, C. J. Huang, H. T. Yu, and C. H. Wang. 2002. The complete genome sequence of Perina nuda picorna-like virus, an insect-infecting virus with a genome organization similar to that of the mammalian picornaviruses. Virology 294:312-323. [DOI] [PubMed] [Google Scholar]
- 64.Xia, Q., Z. Zhou, C. Lu, et al. 2004. A draft sequence for the genome of the domesticated silkworm (Bombyx mori). Science 306:1937-1940. [DOI] [PubMed] [Google Scholar]
- 65.Xie, Q., W. Bu, S. Bhatia, J. Hare, T. Somasundaram, A. Azzi, and M. S. Chapman. 2002. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc. Natl. Acad. Sci. USA 99:10405-10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yates, J. R., III, J. K. Eng, A. L. McCormack, and D. Schieltz. 1995. A method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal. Chem. 67:1426-1436. [DOI] [PubMed] [Google Scholar]
- 67.Zalloua, P. A., J. M. Buzayan, and G. Bruening. 1996. Chemical cleavage of 5′-linked protein from tobacco ringspot virus genomic RNAs and characterization of the protein-RNA linkage. Virology 219:1-8. [DOI] [PubMed] [Google Scholar]
- 68.Zanotto, P. M. D. A., M. J. Gibbs, E. A. Gould, and E. C. Holmes. 1996. A reevaluation of the higher taxonomy of viruses based on RNA polymerases. J. Virol. 70:6083-6096. [DOI] [PMC free article] [PubMed] [Google Scholar]