Abstract
Host cell range, or tropism, combined with coreceptor usage defines viral phenotypes as macrophage tropic using CCR5 (M-R5), T-cell-line tropic using CXCR4 (T-X4), or dually lymphocyte and macrophage tropic using CXCR4 alone or in combination with CCR5 (D-X4 or D-R5X4). Although envelope gp120 V3 is necessary and sufficient for M-R5 and T-X4 phenotypes, the clarity of V3 as a dominant phenotypic determinant diminishes in the case of dualtropic viruses. We evaluated D-X4 phenotype, pathogenesis, and emergence of D-X4 viruses in vivo and mapped genetic determinants in gp120 that mediate use of CXCR4 on macrophages ex vivo. Viral quasispecies with D-X4 phenotypes were associated significantly with advanced CD4+-T-cell attrition and commingled with M-R5 or T-X4 viruses in postmortem thymic tissue and peripheral blood. A D-X4 phenotype required complex discontinuous genetic determinants in gp120, including charged and uncharged amino acids in V3, the V5 hypervariable domain, and novel V1/V2 regions distinct from prototypic M-R5 or T-X4 viruses. The D-X4 phenotype was associated with efficient use of CXCR4 and CD4 for fusion and entry but unrelated to levels of virion-associated gp120, indicating that gp120 conformation contributes to cell-specific tropism. The D-X4 phenotype describes a complex and heterogeneous class of envelopes that accumulate multiple amino acid changes along an evolutionary continuum. Unique gp120 determinants required for the use of CXCR4 on macrophages, in contrast to cells of lymphocytic lineage, can provide targets for development of novel strategies to block emergence of X4 quasispecies of human immunodeficiency virus type 1.
Human immunodeficiency virus type 1 (HIV-1) is comprised of closely related but not identical viruses, or quasispecies. Changes in environmental pressures select viral variants that evolve and fluctuate throughout the course of infection within the host. Although a spectrum of postentry events can modulate virus replication, the primary determinants of host cell range for HIV-1 map to the envelope glycoproteins, which mediate entry through sequential, ordered interactions with host cell receptor complexes that include CD4, the primary receptor, and a coreceptor belonging to a family of seven-transmembrane, G-protein-coupled CC or CXC chemokine receptors (12, 15, 39). The major coreceptors used by primary isolates of HIV-1 are CCR5 and/or CXCR4, which are differentially expressed by a variety of CD4+ cells of lymphocytic and monocytic lineages (1, 2, 12, 35). All strains of HIV-1 display tropism for CD4+ T lymphocytes, both in vivo and ex vivo, but vary in their ability to infect and replicate in macrophages or T-cell lines in culture. Ex vivo tropism and coreceptor usage are characteristics that define virus phenotype (3). Viruses that use CCR5 to infect lymphocytes and macrophages display an M-R5 phenotype, while viruses that use CXCR4 to infect primary lymphocytes and T-cell lines have a T-X4 phenotype. Many primary X4 viruses are actually dually tropic and infect macrophages as well as T lymphocytic cells (1, 18, 54). Dual tropic viruses can use CXCR4 alone or in combination with CCR5 and are phenotypically either D-X4 or D-R5X4. Current models propose that CXCR4 variants of HIV-1 evolve in vivo from CCR5 viruses via intermediate phenotypes that use both CCR5 and CXCR4 (26, 55, 56).
The use of coreceptors by viruses not only determines host cell range but can also modulate the evolution of virus quasispecies, influence the type of disease sequelae, contribute to fitness, and provide a significant biomarker for HIV-1 pathogenesis (38, 59). M-R5 viruses predominate during acute, early, and asymptomatic infection, while CXCR4 strains are associated with severe immune deficiency and advanced disease in many individuals (52, 55, 63, 64). While emergence of dualtropic viruses in vivo appears to be related to disease progression in infected individuals (26), chimeric simian immunodeficiency viruses with envelopes from HIV-1DH12 or HIV-189.6, prototypic D-R5X4 viruses, are highly pathogenic in animals, indicating not only an association but a causal relationship between dually tropic viruses and advanced disease (40, 63).
Determinants of virus phenotype map to HIV-1 Env gp120, which is organized into five hypervariable domains, designated V1 through V5, that are interspersed with conserved regions (34, 47). In contrast to initial interactions between CD4 and gp120, which require a region localized to a conserved Env domain between V4 and V5 (31), a principal determinant of subsequent gp120 interactions with CCR5 or CXCR4 coreceptors is the V3 hypervariable domain (14). Exchange between a variety of T-X4 and M-R5 V3 domains in envelope constructs provides clear functional evidence that V3 is necessary and sufficient for M-R5 and T-X4 viral phenotypes (7, 53). The amino acid composition of V3 domains provides a mechanism to predict with reasonable accuracy the use of CCR5 or CXCR4 coreceptors (5, 29, 49). In general, CCR5-interactive V3 loops have net positive charges less than or equal to 4, reflecting both acidic and basic amino acid residues, while CXCR4-interactive V3 domains display net positive charges equal to or greater than 5, reflecting replacement of multiple acidic or uncharged amino acids by basic residues.
The role played by V3 as a major determinant of phenotype is less clear in the case of dually tropic viruses (45). V3 domains from dually tropic envelopes generally have high positive charges (≥5) and are both necessary and sufficient for chimeric envelopes to use CXCR4 on CD4+ T lymphocytes. Yet chimeric envelopes with high-positive-charge V3 domains derived from D-X4 gp120 are unable to use CXCR4 on macrophages, indicating that V3 amino acids other than charged residues, as well as regions of gp120 in addition to V3, may contribute to cell-type-specific coreceptor usage. For example, amino acid residue 326 in V3 provides some predictive value to models that distinguish between D-X4 and T-X4 phenotypes (5), but whether or not the residue at 326 contributes to actual coreceptor usage is unclear. In general, optimal CXCR4-mediated infection of macrophages appears to require entire gp120 domains from the prototypic dualtropic viruses HIV-189.6 or HIV-1DH12 (8, 58). Although studies implicate complex interactions between V1, V2, and V3 hypervariable loops and elements of the CD4-binding site as cell-type-specific regulators of viral entry (32), the precise domains of HIV-1 gp120 required to mediate CXCR4-dependent infection of macrophages remain to be defined.
We designed a study to evaluate the relationship between a panel of primary viruses with D-X4 phenotypes and immune suppression in a cohort of HIV-1-infected children and adolescents, to assess emergence of D-X4 viruses over time in peripheral blood and tissues from infected individuals, and to identify the genetic determinants in gp120 that mediate CXCR4-dependent entry into macrophages by a strategy that systematically interchanged continuous or discontinuous hypervariable domains between primary D-X4 and HIV-1LAI T-X4 envelopes.
MATERIALS AND METHODS
Samples.
Peripheral blood mononuclear cells (PBMC) were collected from a cross-sectional group of 50 HIV-1-infected subjects who were enrolled prospectively from the University of Florida Pediatric HIV Clinic from 1996 to 2001 or the Yale-Newborn Hospital from 1993 to 1997, according to protocols approved by the Institutional Review Boards of the University of Florida, Yale University School of Medicine, and/or Yale-New Haven Hospital, and after informed consent from the guardian and assent from children who were older than 7 years was received. Blood collection has been described previously (19, 20, 57). Tissue specimens were collected at autopsy 24 to 48 h after death, snap-frozen, and stored at −80°C. Viruses designated D01, D02, M03, T04, D05, and D07 were isolated from patients 01, 02, 03, 04, 05, and 07, who had advanced immune suppression, with <15% CD4+ T cells. All except subject 07 were not treated with protease inhibitor.
Plasma viral RNA and cell-associated viral DNA.
DNA was extracted from multiple biopsies from each tissue using the DNeasy tissue extraction kit (QIAGEN, Valencia, CA). DNA from primary PBMC, cultured PBMC, and monocyte-derived macrophages (MDM) was extracted as described previously (63, 65). A TaqMan real-time PCR assay was performed to quantify the number of infected cells (viral DNA copy number) with HIV-1 gag as the analytical parameter and human apoB as a genomic template control, as previously described (9).
Plasma viral RNA was extracted with the QIAamp viral RNA kit (QIAGEN). Reverse transcription was carried out using 1 μg total RNA mixed with 2 μg random hexamer according to the manufacturer's protocol (Gibco-BRL, Carlsbad, CA). The 1.4-kb V1-V5 hypervariable region of envelope was amplified, cloned into pGEM-T vector (Promega, Madison, WI), and sequenced as previously described (63). Nucleic acid sequences of HIV-1 Env obtained from V1-V2 and V3 regions were stored and edited using HIVbase (51). For each domain, an amino acid alignment was obtained with the Clustal algorithm (61) and edited manually using our motif-based alignment method (33), followed by adjustments of nucleic acid alignments to parallel amino acid alignments.
Phylogenetic analysis and phenotype prediction.
Phylogenetic analysis was performed by using neighbor-joining and maximum parsimony in the PHYLIP software package (17, 46). Bootstrap values were based on generation of 100 replicate trees. The integrity of genetic data was verified by using amino acid alignments and construction of phylogenetic trees, which compared new sequences with all env sequences generated in our laboratory. Phenotype (coreceptor usage and cell tropism) was predicted using a formula we developed that evaluates the number of basic or acidic amino acids, net charge, and the type of residue (isoleucine or methionine) at position 326 in the V3 loop (residues 296 to 331 in HIV-1HXB2 genome) (5). Envelopes were classified into one of three predicted phenotypes: M-R5, D-(R5)X4, or T-X4. M-R5 envelopes have V3 loops with a net positive charge of ≤4 (range, 2 to 4); D-(R5)X4 and T-X4 envelopes have a V3 net charge of ≥5 (range, 5 to 9) and an isoleucine or a methionine/leucine residue at position 326, respectively.
Cells.
All medium components for cell culture were obtained from Gibco BRL Invitrogen (Carlsbad, CA) except where noted. Human embryonic kidney (HEK) 293 cells (American Type Culture Collection, Manassas, VA) and MT-2 cells (AIDS Research and Reagent Program) were maintained according to supplier recommendations. 3T3.T4 cells, a NIH 3T3 murine cell line stably transduced with human CD4, selected for high CD4 expression, and subsequently transduced with either CCR3, CCR5, CXCR4, GPR15/BOB, or STRL33/Bonzo chemokine receptors, were obtained from the AIDS Research and Reagent Program (10, 13, 24) and maintained in complete Dulbecco's modified Eagle’s medium supplemented with 3 μg puromycin per ml (Sigma Diagnostics, St. Louis, MO). PBMC and monocytes were obtained from HIV-1-, cytomegalovirus-, and hepatitis B virus-negative commercial leukocyte preparations (Civitan Life South Blood Centers, Gainesville, FL) as previously described (46). Highly enriched human monocytes were isolated directly by negative selection using Rosette Sep human monocyte enrichment cocktail (Stem Cell Technologies, Vancouver, British Columbia, Canada) according to the manufacturer's instructions. Cultures of PBMC and monocytes were established as described (64). MDM were more than 98% CD14+ and less than 1% CD3+ by flow cytometry analysis (data not shown).
Construction of HIV-1 envelope chimeras.
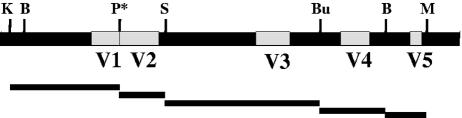
Expression plasmids containing envelope sequences derived from HIV-1LAI or HIV-1JR-FL have been described previously (63). Expression plasmids for envelopes contained gp120 sequences obtained from patient PBMC or primary isolates in combination with gp41 from the M-R5 virus HIV-1JR-FL. To construct chimeric envelopes, regions of the D-X4 gp120 gene which included no coding sequences for accessory or regulatory genes or known regulatory elements were introduced into the gp120 gene of HIV-1LAI by using the following unique restriction sites (numbers indicate coordinates in the HIV-1HXB2 genome with Gene bank accession number K03455): KpnI, 6351; BstAPI, 6393; StuI, 6834; Bsu36I, 7316; BstAPI, 7527; MfeI, 7654 (63) (Fig. 1). A PstI site was introduced by site-directed mutagenesis of nucleotides 6696 and 6697, which preserved the amino acid sequence at the junction of the V1 and V2 hypervariable regions.
FIG. 1.
Strategy to construct chimeric envelope gp120 regions. The organization of gp120 with hypervariable domains V1, V2, V3, V4, and V5 (white boxes) and conserved regions (black boxes) is shown with the restriction endonuclease sites used for construction of chimeric envelopes. Abbreviations: K, KpnI; B, BstAPI; S, StuI; Bu, Bsu36I; M, MfeI; P*, PstI site introduced by mutagenesis without changing the amino acid sequence. Bars below the gp120 diagram indicate the actual regions evaluated in the chimeric constructs: V1 includes C1; V3 includes C2 and part of C3; V4 includes part of C3 and C4; V5 includes part of C4.
Envelope-pseudotyped virus stocks.
To generate single-cycle virus tagged with luciferase (Luc), the murine heat stable antigen (HSA) CD24 gene from pNL4-3.HSA.E−R+ was replaced by the firefly luciferase (luc) gene from pNL4-3.luc.E−R− (AIDS Research and Reference Reagent program, Division of AIDS, NIAID, NIH) (10, 24) using NotI and XhoI restriction enzymes. To generate single-cycle virus tagged with enhanced green fluorescence protein (EGFP), the envelope gene from NLENG1-1RES (a gift from David Levy) (30) was replaced by the envelope gene from pNL4-3.luc.E−R− using BamHI and SalI restriction enzymes. Single-cycle Env-pseudotyped recombinant viruses were produced as described previously (63). Virus stocks were quantified using p24 antigen enzyme-linked immunosorbent assay (Coulter, Miami, FL) and gp120 antigen enzyme-linked immunosorbent assay (Advanced Biotechnologies, Inc., Columbia, MD). Standard virus inoculum contained 60 ng p24 antigen; the amount of virion-associated gp120 per infection was calculated by multiplying gp120 concentration (pg per μl) by the volume of virus inoculum that contained 60 ng p24.
To determine the relationship between virus inoculum and either Luc or EGFP, PBMC were infected for 18 h by increasing concentrations of single-cycle viruses that were tagged with either Luc or EGFP and pseudotyped with either D-X4 or T-X4 envelopes. Cell cultures were assayed for infection by measuring luciferase enzyme activity in cell lysates at 72 to 96 h postinfection, as previously described (64). To determine the number of infected cells, PBMC were fixed 4 days postinfection and assayed for EGFP-expressing cells by fluorescence-activated cell sorting analysis (FACSCalibur; BD Biosciences, San Jose, CA).
Concentrations of Luc-tagged virus inoculum that ranged between 30 ng and 120 ng p24 produced 0.1 × 106 to 1 × 106 relative light units (RLU) of luciferase enzyme activity per 150,000 PBMC (R = 0.99). The proportion of PBMC expressing EGFP following infection by 30 ng to 120 ng p24 of EGFP-tagged virus ranged from 0.2% to 0.5% (R = 0.94 and 0.99 for D-X4 and T-X4 Env-pseudotyped viruses, respectively). A standard virus inoculum of 60 ng p24 produced infection in approximately 0.3% to 0.4% of PBMC and 0.1% of MDM.
Viral phenotype.
3T3.T4 coreceptor indicator cells were seeded in 48-well plates at 104 cells per well, PBMC and MT-2 cells were seeded in 96-well plates at 105 cells per well, and MDM were seeded in 48-well plates at 106 cells per well. Sixty nanograms of p24 equivalent of virus was added to each of three replicate wells per experiment and incubated overnight. Entry was measured in the absence and presence of either AMD-3100 (a competitive antagonist of CXCR4) (4, 11) or monoclonal antibodies (MAb) specific for CD4 (RPA-T4) (BD Pharmingen, San Diego, CA), CCR5 (2D7), or CXCR4 (12G5) (AIDS Research and Reference Reagent program, Division of AIDS, NIAID, NIH) (16, 68). Actual phenotype was determined based on a combination of chemokine receptor use, CXCR4 (X4) and CCR5 (R5), and tropism for PBMC, macrophages, and/or T-cell lines and categorized as M-R5 (macrophage tropic) using CCR5, D-X4 (dually tropic) using only CXCR4, or T-X4 (T-cell-line tropic) using only CXCR4. No envelopes with D-R5X4 phenotypes were identified. The mean 50% inhibitory concentration (IC50) for AMD3100 or CD4 MAb was calculated for each chimeric envelope-pseudotyped virus using GraphPad Prism version 4 (GraphPad Software, Inc.)
Fusion assays were performed as described by Hsu et al. (25) with modifications. Briefly, effector cells expressing gp120 were prepared by cotransfection of MT-2 cells with envelope expression and psv2tat72 plasmids (AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH) (60). Target cells were prepared by transfection of MT-2 cells with pblue3/LTR-LUC (AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH) (28). Effectors and target cells were cocultured at a ratio of 3:1 for 48 h before cells were collected, lysed, and analyzed for luciferase enzyme activity. Background expression of luciferase enzyme from pblue3/LTR-LUC plasmid was determined by coculture of target cells and effector cells that were transfected with psv2tat72 plasmid alone. Optimal expression of luciferase enzyme from pblue3/LTR-LUC plasmid was determined by target cells cultivated in the absence of effector cells but cotransfected with psv2tat72 plasmid. Fusion mediated by binding of gp120 from different chimeric envelopes was expressed as a percentage of luciferase enzyme activity obtained from target cells cultured without effector cells but cotransfected with psv2tat72 plasmid (100% activity).
Construction of site-directed Env-Luc recombinants.
Site-directed mutagenesis to change methionine 326 in the HIV-1LAI and virus T04 V3 domain to isoleucine or isoleucine 326 in the HIV-1JR-FL or virus T02 V3 envelope to methionine was performed using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions with the following primers: for HIVJR-FL, N8745E04 (forward; 5′-CAGGAGAAATAATAGGAGATATGAGACAAGCACATTG-3′) and N8745E05 (reverse; 5′-GAATGTGCTTGTCTCATATCTCCTATTATTTCTCCTC-3′); for virus D02, N8745E06 (forward; 5′-GCAACAAAAACAGGAGATATGAGACAAGCATATTG-3′) and N8745E07 (reverse; 5′-CAATATGCTTGTCTCATATCTCCTGTTTTTGTTGC-3′); for virus D04, N8745E08 (forward; 5′-GACAAGTGGAAGGAGATATAAGACAAGCACATTGTTAAC-3′) and N8745E09 (reverse; 5′-GTTACAATGTGCTTGTCTTATATCTCCTTCCACTTGTC-3′); and for HIVLAI, N8745E10 (forward; 5′-CAATAGGAAAAATAGGAAATATAAGACAAGCACATTG-3′) and N8745E11 (reverse; 5′-CAATGTGCTTGTCTTATATTTCCTATTTTTCCTATTG-3′).
Statistical analysis.
Statistical analysis was performed using SigmaStat 3.0 software (Jandel Scientific Corporation, San Rafael, CA).
RESULTS
Dualtropic viruses and pathogenesis.
To assess the relationship between pathogenesis and HIV-1 phenotype, V3 envelope sequences from peripheral blood cells, plasma, and culture from a cross-sectional study of 50 HIV-infected infants, children, and adolescents, including 39 subjects who were naïve to combination antiretroviral therapy (ART), were analyzed using a formula that we developed to predict phenotype (5). Viruses predicted to use CCR5 and to have M-R5 phenotypes represented the only identifiable quasispecies in 28 of 50 (56%) subjects, while viruses predicted to use CXCR4 and have D-(R5)X4 phenotypes were identified in 22 of 50 (44%) subjects. Viruses predicted to have T-X4 phenotypes were found, but only in combination with viruses that had M-R5 or D-(R5)X4 phenotype, in two individuals. Functional analysis of multiple env quasispecies from each individual verified M-R5 and T-X4 phenotypes and demonstrated that predicted D-(R5)X4 phenotypes used CXCR4 exclusively (reference 63 and data not shown).
Virus phenotypes were related to immune suppression among 39 ART-naïve individuals. For example, among 22 subjects with D-X4 viruses, 19 (86%) had ≤15% CD4 T cells, while only 3 had ≥20% CD4 T cells at the time when dually tropic viruses were identified (Table 1). In contrast, viruses with M-R5 phenotype were found among 17 of 39 ART-naïve individuals. Fourteen of 17 (82%) had ≥20% CD4 T cells, and only 3 of 17 had ≤15% CD4 T cells. The relationship between advanced immune suppression (CD4 ≤ 15%) and dually tropic virus phenotype was significant (P < 0.001, chi-square test).
TABLE 1.
D-X4 phenotype is related to immune suppression
| % CD4+ cells | No. of subjects
|
||
|---|---|---|---|
| Total | M-R5 | D-X4 | |
| ≥20 | 17 | 14 | 3 |
| ≤15 | 22 | 3 | 19 |
| Total | 39 | 17 | 22 |
In vivo evolution of D-X4 viruses.
To evaluate an evolutionary relationship in vivo among viruses with M-R5 and D-X4 phenotypes, env quasispecies were evaluated in a longitudinal study of more than 100 V3 sequences in PBMC and postmortem tissues (thymus, lymph node, lung, spleen, and brain) from children 01 and 04, who were naïve to ART (Fig. 2A and B). Viruses in the earliest peripheral blood samples obtained from subject 01 at 1 year of age were M-R5, while X4 variants appeared 6 years later in peripheral blood and in some, but not all, postmortem tissues (Fig. 2A). D-X4 variants represented the predominant phenotype in peripheral blood at the time of terminal illness but commingled in time and space with M-R5 viruses in thymus and lymphoid tissues. Mixtures of quasispecies were found in subject 04 when D-X4 appeared in combination with M-R5 viruses in peripheral blood over the course of 2 years or in combination with T-X4 viruses in thymus (Fig. 2B).
FIG. 2.
Evolution of virus phenotype in tissues and PBMC. (A and B) Neighbor-joining trees of representative V3 nucleotide sequences from PBMC and tissues from subjects (A) 01 and (B) 04; (C) PBMC from subject 07 prior to and following ART; (D) model of evolution based on V3 net charge and viral phenotype (R5 and X4 and CCR5 and CXCR4, respectively; M, macrophage tropism; D, dual tropism; T, T-cell-line tropism). Survival times for subjects 01 and 04 were 7 and 2 years, respectively. Symbols: open, V3 loop net charge ≤ 4; closed, V3 net charge ≥ 5; *, boot strap > 90; #, boot strap > 75; PB 1, 2, or 3, PBMC at different time points; TH, thymus; SP, spleen; LN, lymph nodes; LU, lung; BR, brain; PTx1 or PTx2, PBMC at 24 weeks or 48 weeks after initiation of combination therapy with nelfinavir and reverse transcriptase inhibitors.
In a third individual, child 07, M-R5 viruses predominated in peripheral blood during the first 3 to 4 years of infection, when the proportion of CD4+ cells was >15%, but were subsequently displaced by D-X4 quasispecies close to initiation of ART when the proportion of CD4+ T cells dropped below 10% (Fig. 2C). D-X4 viruses persisted as the predominant phenotype in PBMC following 24 or 48 weeks of combination therapy with protease inhibitor, even though the proportion of CD4+ T cells increased to 20% and plasma virus levels were reduced.
In general, evolution of V3 sequences followed two major lineages with M-R5 V3 sequences (net charge ≤ 4) localized to branches that were distinct from X4-using V3 domains (net charge ≥ 5) (Fig. 2D). Evolution of V3 net charge by X4-using envelopes was unrelated to macrophage tropism. For example, envelopes with D-X4 or T-X4 phenotypes from subject 04 had V3 loops with a similar +6 net positive charge, while all V3 loops from subject 07 displayed similar D-X4 phenotypes even though the net charge ranged from +6 to +8 (data not shown). Characteristics of V3 amino acids, in addition to charged residues, and gp120 hypervariable domains, in addition to V3, could contribute to D-X4 phenotypes.
Amino acid residue 326 in V3 is a critical determinant of dual or T tropism.
The amino acid residue at position 326 in V3 displays a relationship to viral phenotype (6). Virtually all D-X4 viruses have an isoleucine at residue 326, while T-X4 viruses contain a methionine or leucine. To test the hypothesis that residue 326 contributed to viral phenotype, M326 was mutated to isoleucine (M326I) in V3 domains from HIV-1LAI and a T-X4 primary isolate, T04, which had net charges of +8 or +5, respectively (Fig. 3A). Single-cycle luciferase-tagged viruses, which were pseudotyped by M-R5 HIV-1JR-FL, T-X4 wild-type, or M326I mutant envelopes, were assayed for CCR5- and/or CXCR4-dependent entry into PBMC and MDM (Fig. 3B). As expected, HIV-1JR-FL Env-pseudotyped viruses infected PBMC (lane 1) and macrophages (lane 2). The CCR5 MAb 2D7 neutralized macrophage infection by more than 99% (lane 3), while the CXCR4 MAb 12G5 failed to diminish macrophage infection (lane 4). Viruses pseudotyped by wild-type envelopes from T-X4 HIV-1LAI or T04 and from D-X4 D02 infected PBMC with equal efficiency (1 × 104 to 2 × 104 RLU per 60 ng p24) (Fig. 3B, lanes 1), although only viruses pseudotyped by wild-type D02 envelopes infected primary macrophages (Fig. 3B, lanes 2). Viruses with variant HIV-1LAI or T04 envelopes with the M326I mutation maintained an ability to infect PBMC at levels that were indistinguishable from those of viruses with parental wild-type envelopes (Fig. 3B) and dependent exclusively on CXCR4 (data not shown). In addition, M326I variants of HIV-1LAI or T04 envelopes mediated virus entry into macrophages (>103 RLU) to levels that were about 10- to 20-fold greater than those of M326 wild-type envelopes. Macrophage infection by the M326I variants was resistant to neutralization by the CCR5 MAb but sensitive to treatment with the CXCR4 MAb, which diminished infection by more than 90% (Fig. 3B). Results demonstrated that a single change in a noncharged amino acid residue in T-X4 envelopes was sufficient to expand tropism to macrophages.
FIG. 3.
Residue 326 in V3 contributes to T-X4 or D-X4 phenotype. (A) V3 amino acid sequences from HIV-1LAI, patient-derived T-X4 (04) and D-X4 (02) envelopes, and envelopes in which residue 326 was changed by site-directed mutagenesis (indicated by asterisks) were aligned to HIV-1JR-FL. Residue 326, indicated by the box, was mutated from methionine (M) to isoleucine (I) in T-X4 or from I to M in D-X4 envelopes. Net+ indicates V3 net charge; phenotype summarizes tropism and coreceptor usage. (B) Env pseudotype luciferase-tagged single-cycle viruses used for infection of PBMC (lane 1), primary MDM (lane 2), MDM in the presence of CCR5 monoclonal antibody 2D7 (lane 3), and MDM in the presence of CXCR4 monoclonal antibody 12G5 (lane 4). Viral entry was determined by measuring luciferase enzyme activity and expressed as mean RLU, with error bars indicating standard errors of the means obtained for triplicate wells. Results are representative from experiments using macrophages from three independent donors.
In a reciprocal experiment, isoleucine was replaced by methionine (I326M) in a D-X4 envelope from subject 02. Viruses pseudotyped by either parental or variant envelopes maintained tropism for both PBMC and macrophages (Fig. 3B, virus D02, lanes 1 and 2). Infection by viruses containing the I326M variant of the D-X4 envelope was mediated exclusively by CXCR4, and not CCR5, similar to infection by viruses with the parental D-X4 envelope with isoleucine at position 326 (Fig. 3B, virus D02, lanes 3 and 4). Although T-X4 envelopes gained an ability to use CXCR4 coreceptors on macrophages as a result of a single V3 amino acid alteration, a D-X4 envelope maintained macrophage tropism independent of the amino acid at position 326. Taken together, results indicate that the D-X4 phenotype requires determinants not only in V3 but elsewhere in gp120 as well.
D-X4 phenotype maps to V1 to V5 of gp120.
To map genetic determinants in gp120 that contributed to the use of CXCR4 on macrophages, single-round viruses were pseudotyped by chimeric envelopes constructed initially with regions V1 to V5 from HIV-1JR-FL (M-R5), HIV-1LAI (T-X4), or primary D-X4 viruses from three individuals, D01, D02, and D05 (Fig. 4). Viruses pseudotyped by envelopes with D01, D02, or D03 V1 to V5 domains infected PBMC, macrophages, or MT-2 cells, consistent with D-X4 phenotypes (Fig. 4, lanes 3 to 5). D-X4 envelopes exclusively used the CXCR4 coreceptor, with no evidence for use of CCR5, on either PBMC or macrophages (data not shown).
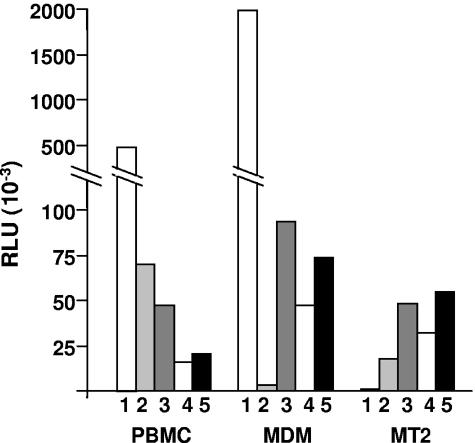
FIG. 4.
Infection of lymphocytes and macrophages by viruses pseudotyped with envelopes of different phenotypes. PBMC, macrophages, and MT-2 cells were infected by viruses pseudotyped with envelopes containing V1 to V5 domains from HIV-1JR-FL (1), HIV-1LAI (2), D01 (3), D02 (4), and D05 (5). Viral entry was determined by measuring luciferase enzyme activity, expressed in RLU. Results are representative from experiments using PBMC and macrophages from three to five independent donors.
The phenotypes of the original virus isolates, whether M-R5, D-X4, or T-X4, mapped to Env V1 to V5 (Table 2). To localize domains required for envelope phenotype, V3 in HIV-1LAI was replaced by V3 regions from HIV-1JR-FL or primary virus M03, which conferred an M-R5 phenotype, while V3 regions from HIV-1LAI or primary virus T04 were sufficient to convert HIV-1JR-FL from an M-R5 to a T-X4 phenotype (Table 2). In contrast, chimeric HIV-1LAI envelopes with V3 domains from D-X4 primary viruses failed to display a D-X4 phenotype and were invariably T-X4. A reciprocal chimera, with the V3 region from D-X4 envelope D02 in an M-R5 envelope from HIV-1JR-FL, also displayed a T-X4 phenotype (data not shown). Overall, the data support a requirement for domains other than V3 in the D-X4 phenotype. V1 to V3 from D01 provided limited tropism for macrophages, although regions V1 to V3 from D02 or D05 were insufficient to mediate any detectable macrophage infection (Table 2). Although V3 domains from D-X4 envelopes were sufficient to mediate CXCR4-dependent infection of lymphocytes, extensive regions of gp120 were required for use of CXCR4 on macrophages.
TABLE 2.
D-X4 phenotype maps to V1 to V5
| Characteristic | Result for chimeric gp120 with indicated hypervariable domain in regiona
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| V1 to V5
|
V3b
|
V1 to V3c
|
|||||||
| JR-FL or M03 | LAI or T04 | D05, D01, or D02 | JR-FL or M03 | LAI or T04 | D05, D01, or D02 | JR-FL or M03 | LAI or T04 | D05, D01, or D02 | |
| Phenotype | M-R5 | T-X4 | D-X4 | M-R5 | T-X4 | T-X4 | M-R5 | T-X4 | D- or T-X4 |
| Tropism | |||||||||
| PBMC | + | + | + | + | + | + | + | + | + |
| MT-2 | − | + | + | − | + | + | − | + | + |
| MDM | + | − | + | + | − | − | + | − | +/−d |
| Coreceptor | |||||||||
| CCR5 | + | − | − | + | − | − | + | − | − |
| CXCR4 | − | + | + | − | + | + | − | + | + |
Chimeric envelopes were constructed in JR-FL gp160 by exchanging gp120 V1 to V5, V3, or V1 to V3.
V3 in HIV-1LAI or HIV-1JR-FL gp120 was replaced by V3 regions from R5 (HIV-1JR-FL or primary virus M03), X4 (HIV-1LAI or primary virus T04), and D-X4 (D05, D01, or D02) viruses.
V1 to V3 in HIV-1LAI or HIV-1JR-FL gp120 was replaced by V1-V3 regions from R5 (HIV-1JR-FL or primary virus M03), X4 (HIV-1LAI or primary virus T04), and D-X4 (D05, D01, or D02) viruses.
V1 to V3 from D01 provided limited tropism for macrophages, while V1 to V3 domains from D02 or D05 were unable to mediate infection of macrophages.
Differential sensitivity of D-X4 and T-X4 envelopes to entry inhibition.
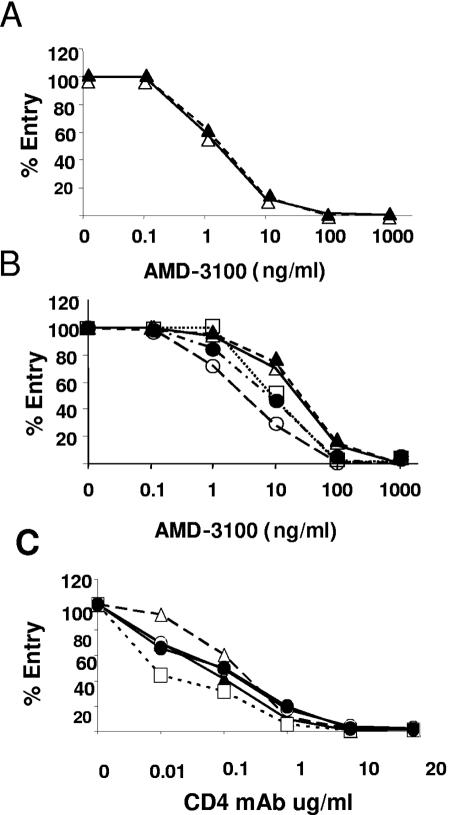
To evaluate interactions between X4-using envelopes and CXCR4 expressed on macrophages or T lymphocytic cells, sensitivity to the CXCR4 small-molecule antagonist AMD-3100 or CD4 neutralizing monoclonal antibody was measured. Macrophage infection by D-X4 V1-to-V5 envelope constructs D01 and D02 was inhibited by virtually identical concentrations of AMD3100 (IC50, 1.5 and 1.3 ng per ml, respectively) (Fig. 5A). Likewise, infection of T lymphocytic MT-2 cells mediated by either D01 or D02 V1 to V5 envelopes was inhibited by similar concentrations of AMD3100 (IC50, 27 or 23 ng per ml, respectively) (Fig. 5B). In contrast to envelopes with D-X4 phenotypes, viruses pseudotyped with envelopes by T-X4 phenotypes displayed increased sensitivity to inhibition by AMD3100 (IC50 of 3, 8.5, or 9 ng per ml for D01 or D02 V1 to V3 chimeras or HIV-1LAI, respectively). Envelopes with D-X4, rather than T-X4, phenotypes displayed more efficient utilization of CXCR4 on MT-2 cells. When infection of MT-2 cells through CD4 was evaluated, the T-X4 envelope from HIV-1LAI was more sensitive to neutralization by a CD4 monoclonal antibody than either the wild-type D01 or D02 D-X4 envelopes or the T-X4 chimeric envelopes constructed with D01 or D02 V1 to V3 domains. When results were evaluated together, efficient use of CXCR4 on MT-2 cells by D-X4 envelopes was related to an ability of D-X4 envelopes to use CXCR4 on macrophages.
FIG. 5.
AMD3100 and CD4 MAb inhibition of CXCR4- and CD4-mediated entry into macrophages and T-lymphocytic cells. (A and B) Relative sensitivity of luciferase tagged, single-cycle pseudotyped viruses to CXCR4-specific antagonist AMD3100 (0.1 to 1,000 ng per ml) macrophages and MT-2 T cells. (C) Relative sensitivity of luciferase tagged, single-cycle pseudotyped viruses to neutralization by CD4 MAb RPA-T4 (0.01 to 20 μg/ml) in MT-2 T cells. Symbols: open squares, V1 to V5 HIV-1LAI; triangles and circles, V1 to V5 and V1 to V3 from subject D02 (open symbols) or D01 (closed symbols). Results are expressed as percent luciferase enzyme production relative to infection with no inhibitors. Macrophage results are representative of triplicate determinations from three or four independent donors. Results from MT-2 cells are representative of triplicate determinations from three independent experiments.
Genetic determinants of D-X4 phenotype.
The restricted ability of chimeric envelopes with D-X4 regions V1 to V3 to mediate infection of macrophages, while retaining an ability to use CXCR4 for lymphocyte infection, provided a rationale to evaluate a systematic exchange of regions of gp120 between D-X4 envelopes D01 or D02 and a prototype T-X4 envelope from HIV-1LAI. In the first series of constructs, two or three adjacent domains from D01 or D02 envelopes were introduced into the HIV-1LAI V1 to V5 background (Fig. 6). Infection of PBMC, MT-2 cells, or macrophages mediated by virions pseudotyped with chimeric envelopes Env3 to Env8 was compared to Env2, the T-X4 parental envelope, and Env1 D-X4 parental envelopes (Fig. 6). Some chimeric envelopes, for example, Env6 or Env8, with D01 or D02 V1V2 or V4V5 domains were unable to mediate infection of any cell type. In contrast, chimeric constructs Env3, Env4, Env5, and Env7 maintained infectivity for lymphocytic cells but little, if any, infectivity for macrophages.
FIG. 6.
Entry mediated by chimeric envelopes into PBMC, MT-2 cells, or macrophages. One, two, or three domains from D-X4 envelopes were introduced into the T-X4 V1 to V5 background of HIV-1LAI using the strategy outlined in Fig. 1. Black boxes, D-X4 regions; white boxes, T-X4 regions. Entry into PBMC, MT-2 cells, or macrophages mediated by chimeric envelopes was compared to entry mediated by D-X4 V1 to V5 parental envelopes that produced levels of luciferase enzyme activity ranging from 0.3 × 105 to 5 × 105 RLU (100%). Luciferase enzyme levels produced by mock-pseudotyped viruses were ≤2 × 102 RLU.
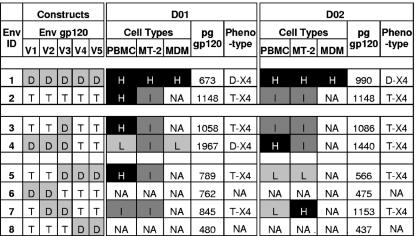
To rule out donor variability as a factor in PBMC or macrophage infection, the panel of pseudotyped viruses was evaluated in PBMC and macrophages from eight independent donors and in triplicate experiments in MT-2 cells (Fig. 7). Chimeric envelopes Env3, Env5, and Env7 with D01 or D02 V1 to V3, V3 to V5, or V2 and V3, respectively, displayed T-X4 phenotypes. Even though D01 Env4 retained limited tropism for macrophages, infection was always less than 20% of levels mediated by parental D01 Env1.
FIG. 7.
Phenotype of chimeric envelopes with one, two, or three D-X4 hypervariable domains. Infection of PBMC or macrophages from eight independent donors or MT-2 T lymphocytic cells mediated by virions pseudotyped with chimeric envelopes developed from subjects D01 and D02 was compared to D-X4 V1 to V5. Mean percent luciferase activity for each chimeric envelope relative to D-X4 parental envelope in each cell type was calculated and classified as follows: no activity (NA), ≤10%; low (L), >10% to 30%; intermediate (I), >30% to 60%; or high (H), >60% to 100%. The amount of virus inoculum (μl) equivalent to 60 ng p24 antigen was multiplied by gp120 concentration (pg/μl) to calculate the amount of virus-associated gp120 per infection. Phenotype is summarized for each chimeric construct based on the pattern of entry into three cell types.
To rule out the possibility that infectivity merely reflected levels of virion-associated envelope glycoproteins, gp120 levels in virus stocks were measured (Fig. 7). All pseudotyped viruses contained gp120 at levels that ranged between 437 and almost 2,000 pg per 60 ng p24 antigen. Some inactive chimeric envelopes (for example, Env6 or Env8) displayed the lowest levels of virion-associated gp120, suggesting a requirement for a threshold amount of gp120 for entry. To determine if Env6 or Env8 gp120 might be sufficient to initiate interaction with CD4/CXCR4, the fusion function of D02 Env6 and Env8 envelope glycoproteins was measured relative to T-X4 Env2 and D-X5 D02 Env1 in a luciferase enzyme expression-based assay. Fusion mediated by D02 Env1 produced 12,000 RLU above background values of about 3,000 RLU, while T-X4 Env2 fusion produced 3,000 RLU above background. In contrast, D02 Env6 or Env8 envelopes failed to mediate fusion. In general, the levels of gp120 appeared unrelated to D-X4 virus tropism for CXCR4-expressing cells, indicating that the use of CXCR4 on macrophages was dependent on factors in addition to the quantity of virion-associated gp120.
Discontinuous determinants of CXCR4 usage on macrophages.
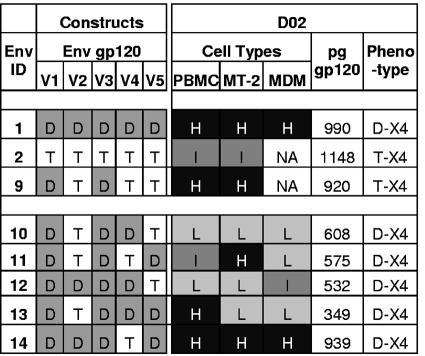
To test if CXCR4-dependent entry into macrophages involved discontinuous determinants in gp120, an extended panel of constructs (Env9 to Env14) with two, three, or four domains from D-X4 envelope D02 was developed (Fig. 8). Env9 with D02 V1 and V3 domains maintained levels of entry into PBMC and MT-2 T cells that were equivalent to levels of entry by Env1 with D02 V1 to V5 but failed to mediate infection of macrophages (T-X4 phenotype). In contrast, chimeric envelope constructs with either three or four D02 domains displayed D-X4 phenotypes (Fig. 8).
FIG. 8.
Mapping determinants in V1 to V5 required for X4-mediated infection of macrophages. Infection of PBMC or macrophages from three independent donors and MT-2 cells mediated by six chimeric envelopes constructed with two, three, or four discontinuous hypervariable domains from subject 02 was compared to infection mediated by D-X4 V1 to V5 envelope from subject 02. Mean percent luciferase activity for each chimeric envelope relative to D-X4 parental envelope in each cell type was calculated and classified as in Fig. 7.
Even though chimeric envelopes had D-X4 phenotypes, levels of entry into various cell types differed from parental D02 Env1. For example, Env11 (V1/V3/V5) maintained high-level entry into T-lymphocytic cells that was comparable to entry by Env1, while Env13 (V1/V3/V4/V5) displayed Env1-level entry into primary lymphocytes. Env14 with V1-V3/V5 was a unique D-X4 chimera that mediated entry into all cell types at levels equivalent to the parental Env1 envelope (Fig. 8).
Taken together, results indicated that V1 and V3 comprise a core region of Env gp120, which mediates efficient use of CXCR4 on CD4 T lymphocytes (Env9). Efficient entry into macrophages required a D-X4 V1 and V3 core in combination with V2 and V5 (Env14). An unexpected finding was that individual hypervariable regions could suppress, as well as enhance, use of CXCR4 usage. For example, D02 V2 domain displayed a dramatic positive effect for CXCR4 usage on primary cells of lymphocyte or macrophage lineages (Env11 versus Env14), while the D02 V5 domain (Env10) enhanced macrophage infection by a chimeric envelope Env9 with a D-X4 V1 and V3 core. In contrast, a D02 V4 hypervariable domain in chimera Env10 suppressed CXCR4-mediated entry into lymphocytes mediated by Env9, while high-level entry into lymphocytes and macrophages mediated by Env14 was diminished by reciprocal exchange of V4 and V5 in chimera Env12.
Complex genotypes in D-X4 V1 to V5 domains.
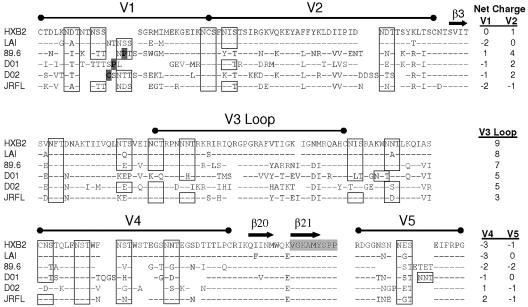
To identify genetic characteristics of V1 to V5 domains that might distinguish D-X4 from T-X4 envelopes, amino acid sequences of hypervariable domains from D-X4 D01 and D02 primary envelopes, as well as reference D-R5X4 envelope from HIV-189.6 or M-R5 envelope from HIV-1JR-FL, were aligned relative to HIV-1HXB2 (Fig. 9). Primary determinants of CD4 binding localized adjacent to the V4 hypervariable domain were highly conserved among the X4-using envelopes. In addition, N-linked glycosylation motifs were conserved in position and number, with only rare differences among the envelopes. The lengths of the hypervariable domains varied independently; for example, the length of V2, V3, and V5 varied by only two or three amino acids, while V1 and V4 varied by eight and six amino acids, respectively. Although V3 loops among X4-using envelopes displayed net positive charges that reflected an excess of basic amino acid residues, V1, V4, and V5 domains tended to have an excess of acidic amino acids. Neither glycosylation motif, length variation, nor charged residues in V1, V4, or V5 provided biomarkers that distinguished envelope phenotype.
FIG. 9.
Alignment of V1 to V5 amino acids from envelopes with D-X4 phenotypes. Amino acid sequences from V1V2 (amino acids 131 to 196 in HIV-1HXB2), V3 (274 to 347), V4 (385 to 418), or V5 (456 to 471) regions from primary D-X4 viruses from subjects 01 and 02 and reference strains of HIV-1 [LAI, 89.6, and JR-FL) were aligned with that from HIV-1HXB2. N-linked-glycosylation motifs are indicated by boxes. Novel cysteine and proline residues in V1 are highlighted. The net positive charge of each domain is indicated. CD4 binding site is highlighted in gray. β3, β20, and β21 located in conserved sites are shown.
The most distinctive features of primary D-X4 envelopes were found in V2 and V1 domains. D-X4 V2 domains were characterized by net positive charges that ranged from 2 to 4, while V1 domains included unusual proline or cysteine residues localized in the region of length polymorphism (Fig. 9). The features of V1 and V2 from D-X4 envelopes differed from those of V1 and V2 in either M-R5 or T-X4 envelopes but were shared with V1 and V2 domains in a prototype D-R5X4 envelope from HIV-189.6.
DISCUSSION
HIV-1 preference for CCR5 or CXCR4 coreceptors determines host cell range, modulates evolution of virus quasispecies, influences disease sequelae, contributes to viral fitness, and provides a significant biomarker for HIV-1 pathogenesis. Dualtropic HIV-1 variants that use CXCR4 coreceptors to infect cells of myeloid, as well as lymphoid, lineages were associated significantly with severe immune suppression and pathogenesis among children and adolescents in our cohort, indicating that an ability to use CCR5 is a nonessential characteristic of viral fitness, at least in late-stage disease. A small number of individuals in our study had D-X4 viruses prior to severe CD4 T-cell depletion, suggesting that early emergence of D-X4 viruses in peripheral blood provides a prognostic indicator of disease progression, a possibility we are investigating in prospective studies.
Viral phenotype based on coreceptor use and cell tropism describes a continuum of heterogeneous envelope characteristics. For example, the prototypic dualtropic viruses HIV-1DH12 and HIV-189.6 and some primary isolates can use CCR5, in addition to CXCR4, to infect indicator cells or macrophages, but are restricted to the CXCR4 pathway for infection of primary lymphocytes (70). In contrast, the primary parental or chimeric X4 envelopes that we evaluated used the CXCR4 coreceptor exclusively and were unable to use the CCR5 pathway to infect either primary lymphocytes or macrophages. D-X4 viruses emerged over periods of years in our study and predominated in lymphoid tissues including thymus, as well as in peripheral blood, by the time of advanced disease or terminal illness. The relationship between R5 and X4 viruses within individuals in our cohort supports a model of X4 viruses evolving from R5 progenitor viruses (56). Although envelope variants that used both CCR5 and CXCR4 were not detected, intermediates in the emergence of D-X4 viruses in vivo may be transient, present at low levels in peripheral blood, or sequestered in tissues.
The principal determinant in gp120 that is both necessary and sufficient to mediate CXCR4-dependent infection of T lymphocytes is the V3 hypervariable domain. In contrast, the use of CXCR4 to mediate infection of primary macrophages requires V3, in combination with complex determinants in gp120 (37, 58). While a dominant factor for the use of CXCR4 on macrophages is an excess of basic amino acid residues in the V3 loop, leading to a high net positive charge, uncharged amino acid residues in V3 also contribute to tropism without changing coreceptor use. For example, differences among the capacity of D-X4 envelopes to mediate CXCR4-dependent infection of human thymus mapped to a single amino acid substitution at residue 316 in V3 (41). In addition, our experiments provide direct evidence that another amino acid, at position 326 in V3, is a functional determinant of cell-type-specific CXCR4 coreceptor use that is sufficient to convert a T-X4 envelope to a D-X4 phenotype.
Envelope determinants other than V3 domains also contribute to the use of CXCR4 to mediate macrophage infection. Chimeric envelopes that combined regions of HIV-1DH12 or HIV-189.6 with envelopes from R5 HIV-1AD8 or T-X4 HIV-1HXB2 implicated the V1/V2 hypervariable region either directly or indirectly as a contributor to infection of macrophages by the CXCR4 pathway (32, 55, 58, 69). In our studies, chimeric envelopes constructed between primary D-X4 and prototype T-X4 HIV-1LAI envelopes defined V1 to V3 plus V5 as the minimal functional unit of gp120 that mediated CXCR4-dependent infection of macrophages at levels equivalent to those seen with parental D-X4 envelopes. Several novel aspects of our study design, such as independent exchange of V1 and V2 or V4 and V5 hypervariable domains, as well as screening chimeric envelopes for the ability to infect both lymphocytes and macrophages, were essential for defining the regions of gp120 required for interactions with CXCR4 on macrophages.
Viral entry into a cell involves CD4 and coreceptor engagement, conformational changes in gp120, and gp41-mediated membrane fusion. Although modulation of gp41 fusion function by different gp120 regions could be a potential variable in assessment of phenotype, the gp41 region from HIVJR-FL in our recombinant envelopes provided a constant genetic background that was permissive for infection mediated by either CCR5 or CXCR4. The amount of chimeric gp120 incorporated into virions might provide a quantitative explanation for differences among viruses in CXCR4-dependent entry into lymphocytes or macrophages (41). In our studies, the ability of chimeric envelopes to mediate CXCR4-dependent infection of different cell types was unrelated to the amounts of virion-associated gp120, at least above some threshold level, which argues against quantity of gp120 as the sole modulator of CXCR4 use on different cell types.
The overall amino acid composition of envelope glycoproteins determines conformation, which optimizes interactions between gp120, CD4, and the CXCR4 coreceptor expressed on different cell types. Our studies identified unique characteristics of amino acid sequences in D-X4 envelopes that included cysteine or proline in V1 and charged residues in V2, which support a model of gp120 conformation as a factor in CXCR4-mediated infection of macrophages. If gp120 conformation determines cell-type-specific use of CXCR4, then CXCR4, alone or in complex with CD4 or other molecules, could differ quantitatively and/or qualitatively between macrophages and lymphocytes (42, 50, 71). The neutralization effect of the CXCR4-specific monoclonal antibody 12G5 is both cell type and virus strain dependent, suggesting that CXCR4 is processed or presented in a cell-type-specific way that would facilitate neutralization escape by some HIV-1 quasispecies (40). Complexities of trimeric gp120/gp41 associations on virions and absence of a crystal structure for complete gp120 presents challenges for predicting precise structural requirements for gp120 interactions with the CD4/CXCR4 receptor complex. Alternative strategies to probe gp120 conformation by panels of monoclonal antibodies that relate immunotypes of D-X4 envelopes to CXCR4-dependent macrophage tropism could provide insights into characteristics of gp120 domains essential for CXCR4-mediated infection of different cell types (21, 22, 44, 48).
Multiple genetic changes distinguish D-X4 viruses from X4 viruses that fail to infect macrophages and indicate that the selective advantage of CXCR4 variants of HIV-1 during the course of infection is multifactorial. Reduced numbers of CD4 CCR5 cells, down regulation of CD4 expression, or increased CXCR4 expression could provide advantages for viruses that use CD4 and/or CXCR4 with highest efficiency (6, 43, 62). Certainly, an increased repertoire of CD4-expressing host cells is available to viruses that can use CXCR4. For example, only a minority of CD4 T lymphocytes expresses CCR5, while CXCR4 is found on the majority of CD4 T cells in vivo (65). CCR5 or CXCR4 subsets of macrophages might provide different evolutionary niches that could quantitatively or qualitatively alter production and subsequent transmission of viruses to cells of lymphocytic lineages. Signal transduction effected by viral interactions with CD4 and one chemokine receptor or the other on different host cells could directly modulate the intracellular milieu to diminish fitness of R5 viruses, favor replication by X4 viruses, and contribute to progressive and global immune dysfunction (9, 23, 27, 36, 66, 67). Determining the sequence of envelope changes that contribute to evolution of a D-X4 phenotype in longitudinal studies is essential to identify biomarkers for evolving D-X4 envelopes and to develop novel strategies that can block the emergence of pathogenic quasispecies of HIV-1.
Acknowledgments
The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: bicyclam JM-2987 (hydrobromide salt of AMD-3100) from AnorMED, Inc.; 3T3.CD4 CXCR4, 3T3.CD4 CCR5, 3T3.CD4 CCR3, 3T3.CD4 BOB, and 3T3.CD4 Bonzo (STRL33) from Dan R. Littman; pNL4-3.HAS.E−R+ and pNL4-3.luc.E−R− from Nathaniel Landau; CCR5 monoclonal antibody (2D7) from Millennium Pharmaceuticals, Inc., and PharMingen; CXCR4 monoclonal antibody (12G5) from James Hoxie.
Research was supported in part by PHS awards R01 HD032259, AI047723, and AI065265; T32 AI007110; T32 CA009126 from the University of Florida Shands Cancer Center; the Pediatric Clinical Research Center of All Children's Hospital, the University of South Florida, and the Maternal and Child Health Bureau, R60 MC 00003-01, Department of Health and Human Services, Health Resources and Services Administration; the General Clinical Research Center of the University of Florida MO1-RR00082; the University of Florida Children's Miracle Network and the Center for Research in Pediatric Immune Deficiency; Elizabeth Glaser Pediatric AIDS Foundation PG51154, the McClamma Research Fellowship, the Robert A. Good Professorship (J.W.S.), and the Stephany W. Holloway Professorship for AIDS Research (M.M.G.).
REFERENCES
- 1.Aquino-De Jesus, M. J., C. Anders, G. Miller, J. W. Sleasman, M. M. Goodenow, and W. A. Andiman. 2000. Genetically and epidemiologically related “non-syncytium-inducing” isolates of HIV-1 display heterogeneous growth patterns in macrophages. J. Med. Virol. 61:171-180. [DOI] [PubMed] [Google Scholar]
- 2.Bazan, H. A., G. Alkhatib, C. C. Broder, and E. A. Berger. 1998. Patterns of CCR5, CXCR4, and CCR3 usage by envelope glycoproteins from human immunodeficiency virus type 1 primary isolates. J. Virol. 72:4485-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger, E. A., R. W. Doms, E. M. Fenyo, B. T. Korber, D. R. Littman, J. P. Moore, Q. J. Sattentau, H. Schuitemaker, J. Sodroski, and R. A. Weiss. 1998. A new classification for HIV-1. Nature 391:240. [DOI] [PubMed] [Google Scholar]
- 4.Bridger, G. J., R. T. Skerlj, D. Thornton, S. Padmanabhan, S. A. Martellucci, G. W. Henson, M. J. Abrams, N. Yamamoto, K. De Vreese, R. Pauwels, and. 1995. Synthesis and structure-activity relationships of phenylenebis(methylene)-linked bis-tetraazamacrocycles that inhibit HIV replication. Effects of macrocyclic ring size and substituents on the aromatic linker. J. Med. Chem. 38:366-378. [DOI] [PubMed] [Google Scholar]
- 5.Briggs, D. R., D. L. Tuttle, J. W. Sleasman, and M. M. Goodenow. 2000. Envelope V3 amino acid sequence predicts HIV-1 phenotype (co-receptor usage and tropism for macrophages). AIDS 14:2937-2939. [DOI] [PubMed] [Google Scholar]
- 6.Chen, S., D. Tuttle, J. T. Oshier, S. J. Wolfgang, M. Goodenow, and J. K. Harrison. 2005. Transforming growth factor-beta increase CXCR4 expression, stromal-derived factor-1 a-stimulated signalling and human immunodeficiency virus-1 entry in human monocyte-derived macrophages. Immunology 114:565-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chesebro, B., K. Wehrly, J. Nishio, and S. Perryman. 1996. Mapping of independent V3 envelope determinants of human immunodeficiency virus type 1 macrophage tropism and syncytium formation in lymphocytes. J. Virol. 70:9055-9059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho, M. W., M. K. Lee, M. C. Carney, J. F. Berson, R. W. Doms, and M. A. Martin. 1998. Identification of determinants on a dualtropic human immunodeficiency virus type 1 envelope glycoprotein that confer usage of CXCR4. J. Virol. 72:2509-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coberley, C. R., J. J. Kohler, J. N. Brown, J. T. Oshier, H. V. Baker, M. P. Popp, J. W. Sleasman, and M. M. Goodenow. 2004. Impact on genetic networks in human macrophages by a CCR5 strain of human immunodeficiency virus type 1. J. Virol. 78:11477-11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 11.De Clercq, E., N. Yamamoto, R. Pauwels, J. Balzarini, M. Witvrouw, K. De Vreese, Z. Debyser, B. Rosenwirth, P. Peichl, R. Datema, and. 1994. Highly potent and selective inhibition of human immunodeficiency virus by the bicyclam derivative JM3100. Antimicrob. Agents Chemother. 38:668-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng, H., R. Liu, W. Ellmeier, S. Choe, D. Unutmaz, M. Burkhart, P. Di Marzio, S. Marmon, R. E. Sutton, C. M. Hill, C. B. Davis, S. C. Peiper, T. J. Schall, D. R. Littman, and N. R. Landau. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661-666. [DOI] [PubMed] [Google Scholar]
- 13.Deng, H. K., D. Unutmaz, V. N. Kewalramani, and D. R. Littman. 1997. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature 388:296-300. [DOI] [PubMed] [Google Scholar]
- 14.Doms, R. W., and D. Trono. 2000. The plasma membrane as a combat zone in the HIV battlefield. Genes Dev. 14:2677-2688. [DOI] [PubMed] [Google Scholar]
- 15.Dragic, T., V. Litwin, G. P. Allaway, S. R. Martin, Y. Huang, K. A. Nagashima, C. Cayanan, P. J. Maddon, R. A. Koup, J. P. Moore, and W. A. Paxton. 1996. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381:667-673. [DOI] [PubMed] [Google Scholar]
- 16.Endres, M. J., P. R. Clapham, M. Marsh, M. Ahuja, J. D. Turner, A. McKnight, J. F. Thomas, B. Stoebenau-Haggarty, S. Choe, P. J. Vance, T. N. Wells, C. A. Power, S. S. Sutterwala, R. W. Doms, N. R. Landau, and J. A. Hoxie. 1996. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell 87:745-756. [DOI] [PubMed] [Google Scholar]
- 17.Felsenstein, J. 1989. PHYLIP phylogeny package. Cladistics 5:164-166. [Google Scholar]
- 18.Foda, M., S. Harada, and Y. Maeda. 2001. Role of V3 independent domains on a dualtropic human immunodeficiency virus type 1 (HIV-1) envelope gp120 in CCR5 coreceptor utilization and viral infectivity. Microbiol. Immunol. 45:521-530. [DOI] [PubMed] [Google Scholar]
- 19.Ghaffari, G., D. J. Passalacqua, B. S. Bender, D. J. Briggs, M. M. Goodenow, and J. W. Sleasman. 2001. Human lymphocyte proliferation responses following primary immunization with rabies vaccine as neoantigen. Clin. Diagn. Lab. Immunol. 8:880-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghaffari, G., D. J. Passalacqua, J. L. Caicedo, M. M. Goodenow, and J. W. Sleasman. 2004. Two-year clinical and immune outcomes in human immunodeficiency virus-infected children who reconstitute CD4 T cells without control of viral replication after combination antiretroviral therapy. Pediatrics 114:e604-e611. [DOI] [PubMed] [Google Scholar]
- 21.Gorny, M. K., K. Revesz, C. Williams, B. Volsky, M. K. Louder, C. A. Anyangwe, C. Krachmarov, S. C. Kayman, A. Pinter, A. Nadas, P. N. Nyambi, J. R. Mascola, and S. Zolla-Pazner. 2004. The v3 loop is accessible on the surface of most human immunodeficiency virus type 1 primary isolates and serves as a neutralization epitope. J. Virol. 78:2394-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorny, M. K., C. Williams, B. Volsky, K. Revesz, S. Cohen, V. R. Polonis, W. J. Honnen, S. C. Kayman, C. Krachmarov, A. Pinter, and S. Zolla-Pazner. 2002. Human monoclonal antibodies specific for conformation-sensitive epitopes of V3 neutralize human immunodeficiency virus type 1 primary isolates from various clades. J. Virol. 76:9035-9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta, S. K., K. Pillarisetti, and N. Aiyar. 2001. CXCR4 undergoes complex lineage and inducing agent-dependent dissociation of expression and functional responsiveness to SDF-1alpha during myeloid differentiation. J. Leukoc. Biol. 70:431-438. [PubMed] [Google Scholar]
- 24.He, J., S. Choe, R. Walker, P. Di Marzio, D. O. Morgan, and N. R. Landau. 1995. Hum. immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J. Virol. 69:6705-6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsu, M., J. M. Harouse, A. Gettie, C. Buckner, J. Blanchard, and C. Cheng-Mayer. 2003. Increased mucosal transmission but not enhanced pathogenicity of the CCR5-tropic, simian AIDS-inducing simian/human immunodeficiency virus SHIVSF162P3 maps to envelope gp120. J. Virol. 77:989-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu, Q. X., A. P. Barry, Z. X. Wang, S. M. Connolly, S. C. Peiper, and M. L. Greenberg. 2000. Evolution of the human immunodeficiency virus type 1 envelope during infection reveals molecular corollaries of specificity for coreceptor utilization and AIDS pathogenesis. J. Virol. 74:11858-11872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito, Y., J. C. Grivel, S. Chen, Y. Kiselyeva, P. Reichelderfer, and L. Margolis. 2004. CXCR4-tropic HIV-1 suppresses replication of CCR5-tropic HIV-1 in human lymphoid tissue by selective induction of CC-chemokines. J. Infect. Dis. 189:506-514. [DOI] [PubMed] [Google Scholar]
- 28.Jeeninga, R. E., M. Hoogenkamp, M. Armand-Ugon, M. de Baar, K. Verhoef, and B. Berkhout. 2000. Functional differences between the long terminal repeat transcriptional promoters of human immunodeficiency virus type 1 subtypes A through G. J. Virol. 74:3740-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jensen, M. A., F.-S. Li, A. B. van't Wout, D. C. Nickle, D. Shriner, H.-X. He, S. McLaughlin, R. Shankarappa, J. B. Margolick, and J. I. Mullins. 2003. Improved coreceptor usage prediction and genotypic monitoring of R5-to-X4 transition by motif analysis of human immunodeficiency virus type 1 env V3 loop sequences. J. Virol. 77:13376-13388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kutsch, O., E. N. Benveniste, G. M. Shaw, and D. N. Levy. 2002. Direct and quantitative single-cell analysis of human immunodeficiency virus type 1 reactivation from latency. J. Virol. 76:8776-8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Labrosse, B., C. Treboute, A. Brelot, and M. Alizon. 2001. Cooperation of the V1/V2 and V3 domains of human immunodeficiency virus type 1 gp120 for interaction with the CXCR4 receptor. J. Virol. 75:5457-5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lamers, S. L., J. W. Sleasman, and M. M. Goodenow. 1996. A model for alignment of Env V1 and V2 hypervariable domains from human and simian immunodeficiency viruses. AIDS Res. Hum. Retrovir. 12:1169-1178. [DOI] [PubMed] [Google Scholar]
- 34.Lamers, S. L., J. W. Sleasman, J. X. She, K. A. Barrie, S. M. Pomeroy, D. J. Barrett, and M. M. Goodenow. 1994. Persistence of multiple maternal genotypes of human immunodeficiency virus type I in infants infected by vertical transmission. J. Clin. Investig. 93:380-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lathey, J. L., D. Brambilla, M. M. Goodenow, M. Nokta, S. Rasheed, E. B. Siwak, J. W. Bremer, D. D. Huang, Y. Yi, P. S. Reichelderfer, and R. G. Collman. 2000. Co-receptor usage was more predictive than NSI/SI phenotype for HIV replication in macrophages: is NSI/SI phenotyping sufficient? J. Leukoc. Biol. 68:324-330. [PubMed] [Google Scholar]
- 36.Lee, C., Q. H. Liu, B. Tomkowicz, Y. Yi, B. D. Freedman, and R. G. Collman. 2003. Macrophage activation through CCR5- and CXCR4-mediated gp120-elicited signaling pathways. J. Leukoc. Biol. 74:676-682. [DOI] [PubMed] [Google Scholar]
- 37.Lee, M. K., J. Heaton, and M. W. Cho. 1999. Identification of determinants of interaction between CXCR4 and gp120 of a dual-tropic HIV-1DH12 isolate. Virology 257:290-296. [DOI] [PubMed] [Google Scholar]
- 38.Li, S., J. Juarez, M. Alali, D. Dwyer, R. Collman, A. Cunningham, and H. M. Naif. 1999. Persistent CCR5 utilization and enhanced macrophage tropism by primary blood human immunodeficiency virus type 1 isolates from advanced stages of disease and comparison to tissue-derived isolates. J. Virol. 73:9741-9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maddon, P. J., A. G. Dalgleish, J. S. McDougal, P. R. Clapham, R. A. Weiss, and R. Axel. 1986. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell 47:333-348. [DOI] [PubMed] [Google Scholar]
- 40.McKnight, A., D. Wilkinson, G. Simmons, S. Talbot, L. Picard, M. Ahuja, M. Marsh, J. A. Hoxie, and P. R. Clapham. 1997. Inhibition of human immunodeficiency virus fusion by a monoclonal antibody to a coreceptor (CXCR4) is both cell type and virus strain dependent. J. Virol. 71:1692-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller, E. D., K. M. Duus, M. Townsend, Y. Yi, R. Collman, M. Reitz, and L. Su. 2001. Human immunodeficiency virus type 1 IIIB selected for replication in vivo exhibits increased envelope glycoproteins in virions without alteration in coreceptor usage: separation of in vivo replication from macrophage tropism. J. Virol. 75:8498-8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murdoch, C. 2000. CXCR4: chemokine receptor extraordinaire. Immunol. Rev. 177:175-184. [DOI] [PubMed] [Google Scholar]
- 43.Naif, H. M., A. L. Cunningham, M. Alali, S. Li, N. Nasr, M. M. Buhler, D. Schols, E. de Clercq, and G. Stewart. 2002. A human immunodeficiency virus type 1 isolate from an infected person homozygous for CCR5Δ32 exhibits dual tropism by infecting macrophages and MT2 cells via CXCR4. J. Virol. 76:3114-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nyambi, P. N., M. K. Gorny, L. Bastiani, G. van der Groen, C. Williams, and S. Zolla-Pazner. 1998. Mapping of epitopes exposed on intact human immunodeficiency virus type 1 (HIV-1) virions: a new strategy for studying the immunologic relatedness of HIV-1. J. Virol. 72:9384-9391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ogert, R. A., M. K. Lee, W. Ross, A. Buckler-White, M. A. Martin, and M. W. Cho. 2001. N-linked glycosylation sites adjacent to and within the V1/V2 and the V3 loops of dualtropic human immunodeficiency virus type 1 isolate DH12 gp120 affect coreceptor usage and cellular tropism. J. Virol. 75:5998-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perez, E. E., S. L. Rose, B. Peyser, S. L. Lamers, B. Burkhardt, B. M. Dunn, A. D. Hutson, J. W. Sleasman, and M. M. Goodenow. 2001. Human immunodeficiency virus type 1 protease genotype predicts immune and viral responses to combination therapy with protease inhibitors (PIs) in PI-naive patients. J. Infect. Dis. 183:579-588. [DOI] [PubMed] [Google Scholar]
- 47.Ping, L. H., J. A. Nelson, I. F. Hoffman, J. Schock, S. L. Lamers, M. Goodman, P. Vernazza, P. Kazembe, M. Maida, D. Zimba, M. M. Goodenow, J. J. Eron, Jr., S. A. Fiscus, M. S. Cohen, and R. Swanstrom. 1999. Characterization of V3 sequence heterogeneity in subtype C human immunodeficiency virus type 1 isolates from Malawi: underrepresentation of X4 variants. J. Virol. 73:6271-6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pinter, A., W. J. Honnen, Y. He, M. K. Gorny, S. Zolla-Pazner, and S. C. Kayman. 2004. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J. Virol. 78:5205-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Resch, W., N. Hoffman, and R. Swanstrom. 2001. Improved success of phenotype prediction of the human immunodeficiency virus type 1 from envelope variable loop 3 sequence using neural networks. Virology 288:51-62. [DOI] [PubMed] [Google Scholar]
- 50.Rossi, D., and A. Zlotnik. 2000. The biology of chemokines and their receptors. Annu. Rev. Immunol. 18:217-242. [DOI] [PubMed] [Google Scholar]
- 51.Salemi, M., K. Strimmer, W. W. Hall, M. Duffy, E. Delaporte, S. Mboup, M. Peeters, and A. M. Vandamme. 2001. Dating the common ancestor of SIVcpz and HIV-1 group M and the origin of HIV-1 subtypes using a new method to uncover clock-like molecular evolution. FASEB J. 15:276-278. [DOI] [PubMed] [Google Scholar]
- 52.Schuitemaker, H., M. Koot, N. A. Kootstra, M. W. Dercksen, R. E. de Goede, R. P. van Steenwijk, J. M. Lange, J. K. Schattenkerk, F. Miedema, and M. Tersmette. 1992. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J. Virol. 66:1354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shioda, T., J. A. Levy, and C. Cheng-Mayer. 1991. Macrophage and T cell-line tropisms of HIV-1 are determined by specific regions of the envelope gp120 gene. Nature 349:167-169. [DOI] [PubMed] [Google Scholar]
- 54.Simmons, G., J. D. Reeves, A. McKnight, N. Dejucq, S. Hibbitts, C. A. Power, E. Aarons, D. Schols, E. De Clercq, A. E. Proudfoot, and P. R. Clapham. 1998. CXCR4 as a functional coreceptor for human immunodeficiency virus type 1 infection of primary macrophages. J. Virol. 72:8453-8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simmons, G., D. Wilkinson, J. D. Reeves, M. T. Dittmar, S. Beddows, J. Weber, G. Carnegie, U. Desselberger, P. W. Gray, R. A. Weiss, and P. R. Clapham. 1996. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic and most can use either Lestr or CCR5 as coreceptors for virus entry. J. Virol. 70:8355-8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singh, A., and R. G. Collman. 2000. Heterogeneous spectrum of coreceptor usage among variants within a dualtropic human immunodeficiency virus type 1 primary-isolate quasispecies. J. Virol. 74:10229-10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sleasman, J. W., B. H. Leon, L. F. Aleixo, M. Rojas, and M. M. Goodenow. 1997. Immunomagnetic selection of purified monocyte and lymphocyte populations from peripheral blood mononuclear cells following cryopreservation. Clin. Diagn. Lab. Immunol. 4:653-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smyth, R. J., Y. Yi, A. Singh, and R. G. Collman. 1998. Determinants of entry cofactor utilization and tropism in a dualtropic human immunodeficiency virus type 1 primary isolate J. Virol. 72:4478-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spencer, L. T., M. T. Ogino, W. M. Dankner, and S. A. Spector. 1994. Clinical significance of human immunodeficiency virus type 1 phenotypes in infected children. J. Infect. Dis. 169:491-495. [DOI] [PubMed] [Google Scholar]
- 60.Subramani, S., R. Mulligan, and P. Berg. 1981. Expression of the mouse dihydrofolate reductase complementary deoxyribonucleic acid in simian virus 40 vectors. Mol. Cell. Biol. 1:854-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. Improved sensitivity of profile searches through the use of sequence weights and gap excision. Comput. Appl. Biosci. 10:19-29. [DOI] [PubMed] [Google Scholar]
- 62.Tokunaga, K., M. L. Greenberg, M. A. Morse, R. I. Cumming, H. K. Lyerly, and B. R. Cullen. 2001. Molecular basis for cell tropism of CXCR4-dependent human immunodeficiency virus type 1 isolates. J. Virol. 75:6776-6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tuttle, D. L., C. B. Anders, M. J. Aquino-De Jesus, P. P. Poole, S. L. Lamers, D. R. Briggs, S. M. Pomeroy, L. Alexander, K. W. Peden, W. A. Andiman, J. W. Sleasman, and M. M. Goodenow. 2002. Increased replication of non-syncytium-inducing HIV type 1 isolates in monocyte-derived macrophages is linked to advanced disease in infected children. AIDS Res. Hum. Retrovir. 18:353-362. [DOI] [PubMed] [Google Scholar]
- 64.Tuttle, D. L., C. R. Coberley, X. Xie, Z. C. Kou, J. W. Sleasman, and M. M. Goodenow. 2004. Effects of human immunodeficiency virus type 1 infection on CCR5 and CXCR4 coreceptor expression on CD4 T lymphocyte subsets in infants and adolescents. AIDS Res. Hum. Retrovir. 20:305-313. [DOI] [PubMed] [Google Scholar]
- 65.Tuttle, D. L., J. K. Harrison, C. Anders, J. W. Sleasman, and M. M. Goodenow. 1998. Expression of CCR5 increases during monocyte differentiation and directly mediates macrophage susceptibility to infection by human immunodeficiency virus type 1. J. Virol. 72:4962-4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vlahakis, S. R., A. Algeciras-Schimnich, G. Bou, C. J. Heppelmann, A. Villasis-Keever, R. C. Collman, and C. V. Paya. 2001. Chemokine-receptor activation by env determines the mechanism of death in HIV-infected and uninfected T lymphocytes. J. Clin. Investig. 107:207-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vlahakis, S. R., A. Villasis-Keever, T. Gomez, M. Vanegas, N. Vlahakis, and C. V. Paya. 2002. G protein-coupled chemokine receptors induce both survival and apoptotic signaling pathways. J. Immunol. 169:5546-5554. [DOI] [PubMed] [Google Scholar]
- 68.Wu, L., W. A. Paxton, N. Kassam, N. Ruffing, J. B. Rottman, N. Sullivan, H. Choe, J. Sodroski, W. Newman, R. A. Koup, and C. R. Mackay. 1997. CCR5 levels and expression pattern correlate with infectability by macrophage-tropic HIV-1, in vitro. J. Exp. Med. 185:1681-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yi, Y., S. N. Isaacs, D. A. Williams, I. Frank, D. Schols, E. De Clercq, D. L. Kolson, and R. G. Collman. 1999. Role of CXCR4 in cell-cell fusion and infection of monocyte-derived macrophages by primary human immunodeficiency virus type 1 (HIV-1) strains: two distinct mechanisms of HIV-1 dual tropism. J. Virol. 73:7117-7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yi, Y., F. Shaheen, and R. G. Collman. 2005. Preferential use of CXCR4 by R5X4 human immunodeficiency virus type 1 isolates for infection of primary lymphocytes. J. Virol. 79:1480-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zou, Y. R., A. H. Kottmann, M. Kuroda, I. Taniuchi, and D. R. Littman. 1998. Function of the chemokine receptor CXCR4 in haematopoiesis and in cerebellar development. Nature 393:595-599. [DOI] [PubMed] [Google Scholar]