Abstract
We recently described the isolation and structural characterization of 2′-fluoropyrimidine-substituted RNA aptamers that bind to gp120 of R5 strains of human immunodeficiency virus type 1 and thereby potently neutralize the infectivity of phylogenetically diverse R5 strains. Here we investigate the physical basis of their antiviral action. We show that both N-linked oligosaccharides and the variable loops V1/V2 and V3 are not required for binding of one aptamer, B40, to gp120. Using surface plasmon resonance binding analyses, we show that the aptamer binds to the CCR5-binding site on gp120 in a relatively CD4-independent manner, providing a mechanistic explanation for its neutralizing potency.
The entry of human immunodeficiency virus type 1 (HIV-1) into target cells depends on the sequential interaction of the surface envelope glycoprotein, gp120, with the primary receptor, CD4, and the coreceptor, CCR5 (1, 5, 7, 9, 15, 29), and is an attractive target for antiviral intervention. However, gp120 is a challenging target because of the conformational flexibility and sequence variability it has evolved to counter host antibody responses (21, 37). The CD4-binding site is recessed and partially obscured from antibody access by the V1/V2 hypervariable loops and associated carbohydrate structures (37). Considerable conformational flexibility of the CD4-unbound state of gp120 further masks the conserved receptor binding site and hinders antibody access (19, 22, 27). The coreceptor binding site on gp120, which is thought to be composed of highly conserved elements of the β19 strand and parts of the V3 hypervariable loop (28, 29, 39), is also protected by the V1/V2 loops from antibody access in the CD4-unbound state (4, 34, 36). Binding of CD4 induces conformational changes in gp120 that include the displacement of the V1/V2 stem-loop structure, thereby revealing a functional coreceptor binding site (26, 31, 38). Recent studies have also shown that this CD4-induced (CD4i) epitope on gp120 is protected from antibodies by steric hindrance and that the size of the neutralizing agent directed towards this region inversely correlates with its ability to neutralize (22). Mutational variation and extensive glycosylation of gp120 further add to the ineffectiveness of the humoral immune response.
In response to these challenges, one rational antiviral approach is to generate small ligands directed to the conserved core regions of gp120 that are critical for viral entry. We hypothesized that aptamers (11, 35), by virtue of their small size compared to antibodies, could access the conserved pockets in gp120, thereby blocking infection. Recently, we isolated and structurally characterized 2′F RNA aptamers that bind gp120 of HIV-1 R5 strain Ba-L and potently neutralize a broad range of HIV-1 primary isolates (8, 18). In order to test whether, as we had hypothesized, the mechanism of neutralization by aptamers was via ligation of critical receptor-binding regions, we mapped the binding site of one such aptamer (B40) using competition with gp120's natural ligands by deletion analysis and by deglycosylation.
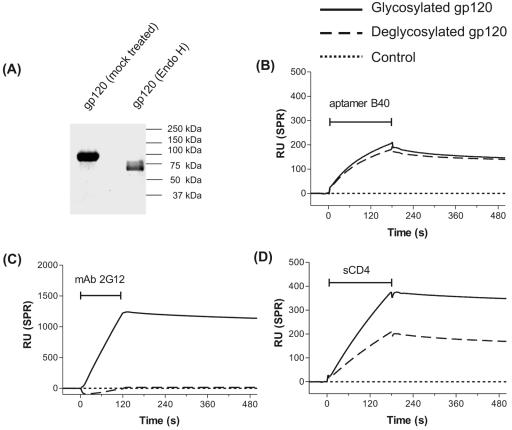
The binding of aptamer to gp120 is independent of N-linked glycosylation.
Recombinant monomeric Ba-L gp120 was expressed in insect cells using a baculovirus expression system, as previously described (18, 24), and deglycosylated using endo-β-N-acetylglucosaminidase H (endo H) under native conditions as previously described (20). When analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, endo H treatment of gp120 was found to have resulted in the complete disappearance of the fully glycosylated material (apparent mass ∼ 92 kDa) with the majority now migrating with an apparent mass of ∼63 kDa (Fig. 1A). Equimolar amounts of both the glycosylated and deglycosylated gp120 were then analyzed for ability to bind aptamer by BIAcore surface plasmon resonance analysis as previously described (8). The aptamer bound to both glycosylated and deglycosylated gp120 in a similar manner (Fig. 1B), indicating that the aptamer binding site on gp120 is not dependent on the N-linked glycans on the glycoprotein.
FIG. 1.
The binding of aptamer to gp120 is independent of N-linked glycosylation. (A) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of the endo H-treated deglycosylated gp120. The molecular mass (∼63 kDa) of the endo H-treated gp120, which is reduced compared to that of the mock-treated gp120 (∼92 kDa) indicates near-complete deglycosylation of the native glycoprotein. (B-D) Overlay of BIAcore sensorgrams to show the binding of the indicated analytes to glycosylated (solid line) and deglycosylated (dashed line) Ba-L monomeric gp120 immobilized on a CM5 biosensor chip. The signal from a control cell for each analyte (dotted line) has been subtracted from all sensorgrams in this and subsequent figures. The horizontal I-shaped bars show the injection period of the analytes. RU, response units; SPR, surface plasmon resonance.
In contrast, monoclonal antibody (MAb) 2G12, which binds to a glycan-dependent epitope (30, 33), bound to the glycosylated but not the deglycosylated gp120, as expected (Fig. 1C). Soluble CD4 (sCD4) bound better to the glycosylated gp120 than to the deglycosylated gp120 (Fig. 1D), consistent with previous reports (16, 23, 25).
Role of V1/V2 and V3 loops in aptamer binding.
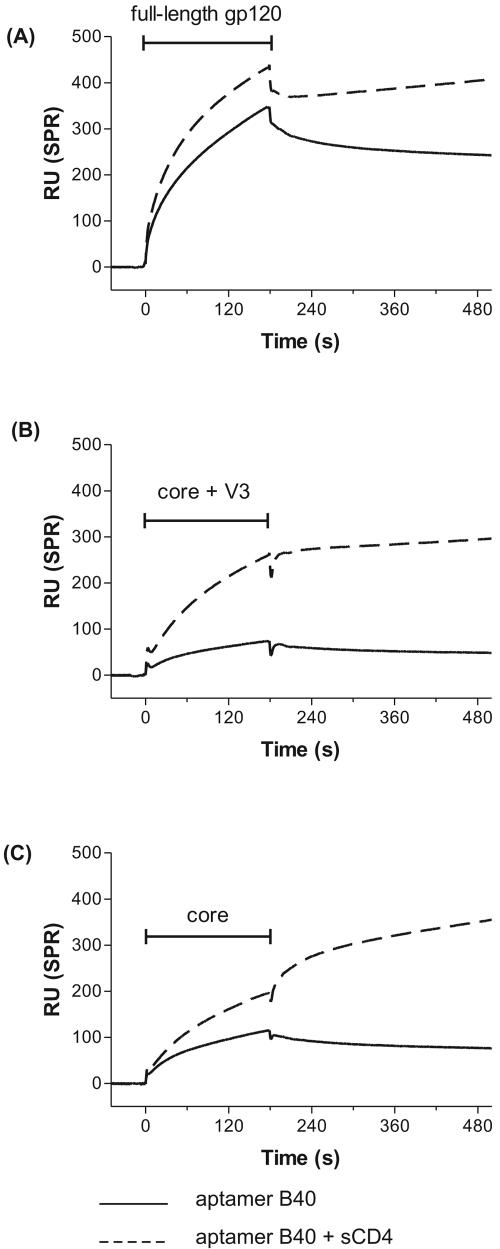
Our results, above, show that one can remove the majority of N-linked sugars from gp120 without affecting the binding site for aptamer B40 but do not exclude the possibility that a specific oligosaccharide, refractory to deglycosylation, plays an important role. The most likely candidates for such an oligosaccharide lie within the hypervariable loops V1/V2 and V3, as they would be close to sites already known to be important for interaction of gp120 with receptor and coreceptor molecules (21, 28, 29, 37). In order to simultaneously investigate this possibility of a residual oligosaccharide and the potential that aptamers bound directly to these loop sequences, we used protein constructs, derived from the R5 strain Yu2, in which one or both loops are deleted and which lack complex sugars (17, 38). The results of a BIAcore analysis (Fig. 2C) show that aptamer B40 binds to the loop-deleted core of gp120, and therefore its binding site does not lie within the V1/V2 or V3 loops nor the oligosaccharides associated with them. It appears that the aptamer binding site is somewhat obscured in the core as well as the core plus V3 constructs of gp120 and becomes fully exposed only following the binding of CD4 (Fig. 2B and 2C). This is in marked contrast to the ability of aptamer B40 to bind full-length gp120 in a largely CD4-independent manner (Fig. 2A and Table 1). We tentatively conclude from this that the aptamer binding site, though located in core regions of gp120, is maintained in its aptamer-competent state by multiple interloop interactions and further that, when deletion of the loop causes the partial collapse of the aptamer binding pocket, the binding of CD4 allosterically reopens the pocket.
FIG. 2.
Effect of hypervariable loop deletion on binding of aptamer to gp120. BIAcore sensorgrams showing binding of aptamer B40 to different gp120 constructs, with (dashed lines) and without (solid lines) prior binding of CD4 to gp120. (A) Full-length monomeric Ba-L gp120. (B) Monomeric Yu2 core plus V3 gp120 (i.e., deletion of V1/V2). (C) Monomeric Yu2 core gp120 (i.e., deletions of V1/V2 and V3). The horizontal I-shaped bars show the injection period of the analytes. RU, response units; SPR, surface plasmon resonance.
TABLE 1.
Summary of aptamer B40 binding data
| Construct bound | Maximala
|
Relativeb
|
||
|---|---|---|---|---|
| −CD4 | +CD4 | In comparison to full-length gp120 | Fold increase in presence of CD4 | |
| Full-length gp120 | 330 | 412 | 1 | 1.2 |
| Core+ V3 | 55 | 235 | 0.2 | 4.3 |
| Core | 101 | 184 | 0.3 | 1.8 |
Maximal binding of the aptamer, expressed in response units, at 180 s. −CD4, in the absence of sCD4; +CD4, in the presence of sCD4.
Relative binding of the aptamer, expressed as a unitless ratio to show the comparison with binding to full-length gp120 and fold increase in aptamer binding in the presence of CD4.
The aptamer binding site overlaps with the CCR5-binding site on gp120.
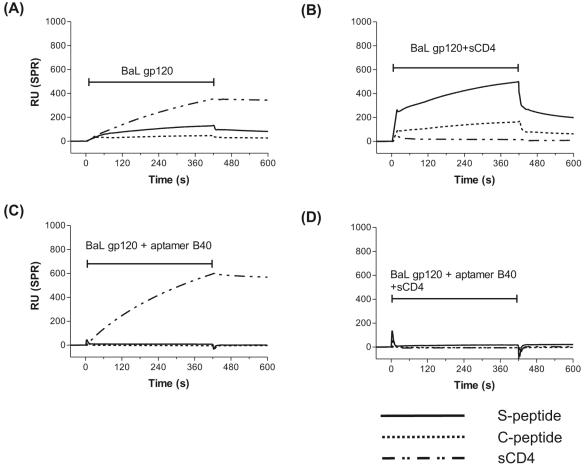
We have previously shown that aptamer B4, which shares critical structural motifs with aptamer B40, competes for binding to gp120 with the CD4i MAb 17b (8, 18). Taken together with the CD4 dependence of efficient binding of aptamer B40 to the core gp120 construct described above, this pointed strongly to the possibility that the aptamer binding site was close to that of CCR5. To test this, we carried out a BIAcore surface plasmon resonance competition binding analysis using the 22-mer tyrosine-sulfated peptide (S-peptide) corresponding to the N terminus of CCR5 that acts as an essential component of the coreceptor for HIV-1 on the host cell (2, 6, 10, 12-14), as well as an unsulfated version of the same peptide (C-peptide) and sCD4, as alternate immobilized binding partners for gp120 (Fig. 3). As controls, RANTES, a physiological ligand for both CCR3 and CCR5 (3), and the anti-CCR5 MAb 3A9, but not monocyte chemoattractant protein 1, a ligand for CCR2 and CCR4, bound to the CCR5 peptides, as expected (data not shown).
FIG. 3.
Effect of preincubation with aptamer or sCD4 on gp120 binding to immobilized receptors. BIAcore sensorgram overlays show the competition binding of (A) monomeric Ba-L gp120, (B) gp120-sCD4 complex, (C) gp120-aptamer B40 complex, and (D) gp120-sCD4-aptamer B40 complex to immobilized equimolar amounts of sulfated N-terminal peptide of CCR5 (S-peptide; solid line), desulfated N-terminal peptide of CCR5 (C-peptide; dotted line) and soluble human CD4 ectodomain (sCD4; dashed line) on a CM5 biosensor chip. The S-peptide and C-peptide are biotinylated at the C terminus and captured by use of streptavidin, while sCD4 is directly coupled via amine coupling. The horizontal I-shaped bars show the injection period of the analytes. RU, response units; SPR, surface plasmon resonance.
Monomeric Ba-L gp120 bound well to immobilized CD4 but weakly to the S-peptide and negligibly to the C-peptide (Fig. 3A). However, preincubating gp120 with CD4 increased binding to both CCR5 peptides by more than 2.5-fold (Fig. 3B), confirming the CD4-induced binding property of the peptides to gp120 (14). Moreover, the approximately threefold difference in binding of both gp120 and gp120-sCD4 complex to the S-peptide versus C-peptide confirm that the tyrosine sulfate moieties contribute substantially to the association of the gp120-sCD4 complexes, as previously reported (13). As expected, the gp120-sCD4 complex did not bind to immobilized CD4 (Fig. 3B). When the gp120-17b MAb complex was injected, it bound only to the immobilized CD4 and not to the CCR5 peptides (data not shown), also as expected. The gp120-aptamer B40 complex also bound to the immobilized CD4 but not to the CCR5 peptides (Fig. 3C). The ternary complexes, gp120-sCD4-17 MAb (data not shown) and gp120-sCD4-aptamer B40 (Fig. 3D), did not bind to either the immobilized CCR5 peptides or human CD4.
Taken together, these results show that the aptamer competes with the N-terminal ectodomain of CCR5 for binding to gp120 and hence the aptamer binding site (aptatope) must overlap with the basic, conserved, coreceptor-binding region on gp120. This is consistent with the inability of aptamer B40 to bind the gp120 of the X4 strain, HXB2 (data not shown) in contrast to aptamers raised against X4 strains (32). The ability of aptamer B40 to bind to the coreceptor-binding site without prior ligation of CD4 by gp120, in contrast to the CD4 dependence of the binding of CD4i antibodies and of CCR5 itself, may result from a combination of its small size and high charge density and confer on it remarkable antiviral potential.
Acknowledgments
This work was supported by the Wellcome Trust. A.K.D. was funded by a Felix Scholarship.
We thank Peter D. Kwong and Chih-chin Huang for helpful discussions and for supplying the Yu2 core constructs and George Gao for supplying the Ba-L gp120 expression vector.
REFERENCES
- 1.Alkhatib, G., C. Combadiere, C. C. Broder, Y. Feng, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1996. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955-1958. [DOI] [PubMed] [Google Scholar]
- 2.Atchison, R. E., J. Gosling, F. S. Monteclaro, C. Franci, L. Digilio, I. F. Charo, and M. A. Goldsmith. 1996. Multiple extracellular elements of CCR5 and HIV-1 entry: dissociation from response to chemokines. Science 274:1924-1926. [DOI] [PubMed] [Google Scholar]
- 3.Cairns, J. S., and M. P. D'Souza. 1998. Chemokines and HIV-1 second receptors: the therapeutic connection. Nat. Med. 4:563-568. [DOI] [PubMed] [Google Scholar]
- 4.Chen, B., E. M. Vogan, H. Gong, J. J. Skehel, D. C. Wiley, and S. C. Harrison. 2005. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433:834-841. [DOI] [PubMed] [Google Scholar]
- 5.Choe, H., M. Farzan, Y. Sun, N. Sullivan, B. Rollins, P. D. Ponath, L. Wu, C. R. Mackay, G. LaRosa, W. Newman, N. Gerard, C. Gerard, and J. Sodroski. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135-1148. [DOI] [PubMed] [Google Scholar]
- 6.Cormier, E. G., M. Persuh, D. A. Thompson, S. W. Lin, T. P. Sakmar, W. C. Olson, and T. Dragic. 2000. Specific interaction of CCR5 amino-terminal domain peptides containing sulfotyrosines with HIV-1 envelope glycoprotein gp120. Proc. Natl. Acad. Sci. USA 97:5762-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalgleish, A. G., P. C. Beverley, P. R. Clapham, D. H. Crawford, M. F. Greaves, and R. A. Weiss. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312:763-767. [DOI] [PubMed] [Google Scholar]
- 8.Dey, A. K., C. Griffiths, S. M. Lea, and W. James. 2005. Structural characterization of an anti-gp120 RNA aptamer that neutralizes R5 strains of HIV-1. RNA 11:873-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dragic, T., V. Litwin, G. P. Allaway, S. R. Martin, Y. Huang, K. A. Nagashima, C. Cayanan, P. J. Maddon, R. A. Koup, J. P. Moore, and W. A. Paxton. 1996. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381:667-673. [DOI] [PubMed] [Google Scholar]
- 10.Dragic, T., A. Trkola, S. W. Lin, K. A. Nagashima, F. Kajumo, L. Zhao, W. C. Olson, L. Wu, C. R. Mackay, G. P. Allaway, T. P. Sakmar, J. P. Moore, and P. J. Maddon. 1998. Amino-terminal substitutions in the CCR5 coreceptor impair gp120 binding and human immunodeficiency virus type 1 entry. J. Virol. 72:279-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellington, A. D., and J. W. Szostak. 1990. In vitro selection of RNA molecules that bind specific ligands. Nature 346:818-822. [DOI] [PubMed] [Google Scholar]
- 12.Farzan, M., H. Choe, L. Vaca, K. Martin, Y. Sun, E. Desjardins, N. Ruffing, L. Wu, R. Wyatt, N. Gerard, C. Gerard, and J. Sodroski. 1998. A tyrosine-rich region in the N terminus of CCR5 is important for human immunodeficiency virus type 1 entry and mediates an association between gp120 and CCR5. J. Virol. 72:1160-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farzan, M., T. Mirzabekov, P. Kolchinsky, R. Wyatt, M. Cayabyab, N. P. Gerard, C. Gerard, J. Sodroski, and H. Choe. 1999. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell 96:667-676. [DOI] [PubMed] [Google Scholar]
- 14.Farzan, M., N. Vasilieva, C. E. Schnitzler, S. Chung, J. Robinson, N. P. Gerard, C. Gerard, H. Choe, and J. Sodroski. 2000. A tyrosine-sulfated peptide based on the N terminus of CCR5 interacts with a CD4-enhanced epitope of the HIV-1 gp120 envelope glycoprotein and inhibits HIV-1 entry. J. Biol. Chem. 275:33516-33521. [DOI] [PubMed] [Google Scholar]
- 15.Feng, Y., C. C. Broder, P. E. Kennedy, and E. A. Berger. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872-877. [DOI] [PubMed] [Google Scholar]
- 16.Fenouillet, E., B. Clerget-Raslain, J. C. Gluckman, D. Guetard, L. Montagnier, and E. Bahraoui. 1989. Role of N-linked glycans in the interaction between the envelope glycoprotein of human immunodeficiency virus and its CD4 cellular receptor. Structural enzymatic analysis. J. Exp. Med. 169:807-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grundner, C., M. Pancera, J. M. Kang, M. Koch, J. Sodroski, and R. Wyatt. 2004. Factors limiting the immunogenicity of HIV-1 gp120 envelope glycoproteins. Virology 330:233-248. [DOI] [PubMed] [Google Scholar]
- 18.Khati, M., M. Schuman, J. Ibrahim, Q. Sattentau, S. Gordon, and W. James. 2003. Neutralization of infectivity of diverse R5 clinical isolates of human immunodeficiency virus type 1 by gp120-binding 2′F-RNA aptamers. J. Virol. 77:12692-12698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwong, P. D., M. L. Doyle, D. J. Casper, C. Cicala, S. A. Leavitt, S. Majeed, T. D. Steenbeke, M. Venturi, I. Chaiken, M. Fung, H. Katinger, P. W. Parren, J. Robinson, D. Van Ryk, L. Wang, D. R. Burton, E. Freire, R. Wyatt, J. Sodroski, W. A. Hendrickson, and J. Arthos. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678-682. [DOI] [PubMed] [Google Scholar]
- 20.Kwong, P. D., R. Wyatt, E. Desjardins, J. Robinson, J. S. Culp, B. D. Hellmig, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1999. Probability analysis of variational crystallization and its application to gp120, the exterior envelope glycoprotein of type 1 human immunodeficiency virus (HIV-1). J. Biol. Chem. 274:4115-4123. [DOI] [PubMed] [Google Scholar]
- 21.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Labrijn, A. F., P. Poignard, A. Raja, M. B. Zwick, K. Delgado, M. Franti, J. Binley, V. Vivona, C. Grundner, C. C. Huang, M. Venturi, C. J. Petropoulos, T. Wrin, D. S. Dimitrov, J. Robinson, P. D. Kwong, R. T. Wyatt, J. Sodroski, and D. R. Burton. 2003. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J. Virol. 77:10557-10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lifson, J., S. Coutre, E. Huang, and E. Engleman. 1986. Role of envelope glycoprotein carbohydrate in human immunodeficiency virus (HIV) infectivity and virus-induced cell fusion. J. Exp. Med. 164:2101-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin, C. L., A. K. Sewell, G. F. Gao, K. T. Whelan, R. E. Phillips, and J. M. Austyn. 2000. Macrophage-tropic HIV induces and exploits dendritic cell chemotaxis. J. Exp. Med. 192:587-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matthews, T. J., K. J. Weinhold, H. K. Lyerly, A. J. Langlois, H. Wigzell, and D. P. Bolognesi. 1987. Interaction between the human T-cell lymphotropic virus type IIIB envelope glycoprotein gp120 and the surface antigen CD4: role of carbohydrate in binding and cell fusion. Proc. Natl. Acad. Sci. USA 84:5424-5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore, J. P., Q. J. Sattentau, H. Yoshiyama, M. Thali, M. Charles, N. Sullivan, S.-W. Poon, M. S. Fung, F. Traincard, M. Pinkus, G. Robey, J. E. Robinson, D. D. Ho, and J. Sodroski. 1993. Probing the structure of the V2 domain of human immunodeficiency virus type 1 surface glycoprotein gp120 with a panel of eight monoclonal antibodies: human immune response to the V1 and V2 domains. J. Virol. 67:6136-6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Myszka, D. G., R. W. Sweet, P. Hensley, M. Brigham-Burke, P. D. Kwong, W. A. Hendrickson, R. Wyatt, J. Sodroski, and M. L. Doyle. 2000. Energetics of the HIV gp120-CD4 binding reaction. Proc. Natl. Acad. Sci. USA 97:9026-9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rizzuto, C., and J. Sodroski. 2000. Fine definition of a conserved CCR5-binding region on the human immunodeficiency virus type 1 glycoprotein 120. AIDS Res. Hum. Retrovir. 16:741-749. [DOI] [PubMed] [Google Scholar]
- 29.Rizzuto, C. D., R. Wyatt, N. Hernandez-Ramos, Y. Sun, P. D. Kwong, W. A. Hendrickson, and J. Sodroski. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280:1949-1953. [DOI] [PubMed] [Google Scholar]
- 30.Sanders, R. W., M. Venturi, L. Schiffner, R. Kalyanaraman, H. Katinger, K. O. Lloyd, P. D. Kwong, and J. P. Moore. 2002. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 76:7293-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sattentau, Q. J., J. P. Moore, F. Vignaux, F. Traincard, and P. Poignard. 1993. Conformational changes induced in the envelope glycoproteins of the human and simian immunodeficiency viruses by soluble receptor binding. J. Virol. 67:7383-7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sayer, N., J. Ibrahim, K. Turner, A. Tahiri-Alaoui, and W. James. 2002. Structural characterization of a 2′F-RNA aptamer that binds a HIV-1 SU glycoprotein, gp120. Biochem. Biophys. Res. Commun. 293:924-931. [DOI] [PubMed] [Google Scholar]
- 33.Scanlan, C. N., R. Pantophlet, M. R. Wormald, E. O. Saphire, R. Stanfield, I. A. Wilson, H. Katinger, R. A. Dwek, P. M. Rudd, and D. R. Burton. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of α1→2 mannose residues on the outer face of gp120. J. Virol. 76:7306-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trkola, A., T. Dragic, J. Arthos, J. M. Binley, W. C. Olson, G. P. Allaway, C. Cheng-Mayer, J. Robinson, P. J. Maddon, and J. P. Moore. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184-187. [DOI] [PubMed] [Google Scholar]
- 35.Tuerk, C., and L. Gold. 1990. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249:505-510. [DOI] [PubMed] [Google Scholar]
- 36.Wu, L., N. P. Gerard, R. Wyatt, H. Choe, C. Parolin, N. Ruffing, A. Borsetti, A. A. Cardoso, E. Desjardin, W. Newman, C. Gerard, and J. Sodroski. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179-183. [DOI] [PubMed] [Google Scholar]
- 37.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705-711. [DOI] [PubMed] [Google Scholar]
- 38.Wyatt, R., J. Moore, M. Accola, E. Desjardin, J. Robinson, and J. Sodroski. 1995. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 69:5723-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]