Abstract
Viruses frequently use insect vectors to effect rapid spread through host populations. In plant viruses, vector transmission is the major mode of transmission, used by nearly 80% of species described to date. Despite the importance of this phenomenon in epidemiology, the specificity of the virus-vector relationship is poorly understood at both the molecular and the evolutionary level, and very limited data are available on the precise viral protein motifs that control specificity. Here, using the aphid-transmitted Cauliflower mosaic virus (CaMV) as a biological model, we confirm that the “noncirculative” mode of transmission dominant in plant viruses (designated “mechanical vector transmission” in animal viruses) involves extremely specific virus-vector recognition, and we identify an amino acid position in the “helper component” (HC) protein of CaMV involved in such recognition. Site-directed mutagenesis revealed that changing the residue at this position can differentially affect transmission rates obtained with various aphid species, thus modifying the spectrum of vector species for CaMV. Most interestingly, in a virus line transmitted by a single vector species, we observed the rapid appearance of a spontaneous mutant specifically losing its transmissibility by another aphid species. Hence, in addition to the first identification of an HC motif directly involved in specific vector recognition, we demonstrate that change of a virus to a different vector species requires only a single mutation and can occur rapidly and spontaneously.
Most plant viruses require the assistance of another organism—the vector—to spread from one host to the next (34). Insects, the main virus vectors, are frequently responsible for causing severe disease epidemics worldwide. Specialized plant feeders with piercing-sucking mouthparts from the Homoptera, particularly aphids, are responsible for transmitting around half of the >1,000 plant virus species described so far (18). The specificity of the relationship between aphids and viruses can vary widely depending on the type of virus-vector interaction, but it is generally low for the most frequent noncirculative interactions. The term noncirculative applies to cases of transmission where the virus is simply retained within (and later released from) the stylets of the vector while feeding on plants and never replicates or even circulates within the vector body (15). In fact, most aphid species tested are capable of transmitting any noncirculative virus with various degrees of efficiency, and there are only very few examples reported in the literature of a given aphid species being unable to act as the vector for a specific noncirculative virus [e.g., Lypaphis erysimi (Kaltenbach) and Tobacco etch virus (39), Nasonovia ribisnigri Mosley and Lettuce mosaic virus (26), or Brachycaudus helichrysi L. and Cauliflower mosaic virus (19)]. Whether this apparent lack of specificity is a viral adaptation to increase the chances of transmission by several vector species remains an open question.
The molecular basis of the specificity (or lack of specificity) between noncirculative viruses and their vectors is poorly documented. One reported example is that of cucumoviruses, where minor amino acid changes in the coat protein of Cucumber mosaic virus differentially modify transmission by its two main vectors, Aphis gossypii (Glover) and Myzus persicae (Sulzer) (30). In potyviruses, the molecular mechanisms of the virus-vector interaction have been studied extensively, and some level of specificity has been reported (36). Although a highly conserved KITC amino acid motif in the N-terminal domain of the helper component (HC)-Pro protein is mandatory for all potyvirus/aphid interactions, other protein sequences that affect the efficiency of transmission in a specific virus/vector couple remain completely unknown (reviewed in references 32 and 35).
Similarly, for Cauliflower mosaic virus (CaMV, genus Caulimovirus), various aphid species transmit the disease with different efficiencies or even fail to transmit it at all (7, 19, 25). CaMV is certainly the plant virus for which the molecular mechanisms of virus/vector interactions have been most thoroughly documented. However, although the biological and biochemical properties of the various viral proteins involved in aphid transmission are well characterized (for a review, see references 4 and 18), the exact domains or motifs involved, directly or indirectly, in specific recognition between the virus and one or more aphid species are still unidentified and thus remain totally uncharacterized. Among the six viral genes expressed upon CaMV infection, three (open reading frames [ORFs] II, III, and IV) are involved in vector transmission. The coat protein (P4), the product of ORF IV, has long been known to be incapable of direct interaction with the aphid mouthparts. Instead, nonstructural proteins, such as those encoded by ORFs II and III (P2 and P3), create a molecular “bridge” between virus and vector, thus linking the coat protein to attachment sites within the aphid mouthparts. P3 has been demonstrated to form a complex with the virus particle (20, 33), but it cannot bind putative receptors in aphids (9). P3, in the form of P3-virion complexes, attaches to P2 (the helper component of CaMV), which in turn directly recognizes the putative receptor sites within the aphid stylet (9). Consistently, P2 is the only viral product that is retained in the stylets when acquired alone by aphids, and its acquisition prior to that of P3-virion complexes is mandatory for successful transmission (9). Biochemical and biological characterization of P2 has revealed a number of remarkable properties, but no information on motifs or domains that could be directly involved in binding to the aphid stylet has been reported. While the C-terminal α-helical domain of P2 (from amino acids [AA] 100 to 159) was shown to be responsible both for P2-P3 binding (21) and for P2 self-association and polymerization (16), a large N-terminal region (from AA 1 to 100) remains unexplored regarding structure, biochemical properties, and biological function. It is thus tempting to hypothesize that the motif that attaches to the aphid vector resides on this end of the molecule. However, the lack of naturally nontransmissible variants of CaMV deficient in P2-aphid binding (2), together with the intrinsic instability of the N-terminal domain when isolated from the rest of the molecule (S. Blanc, unpublished results), has so far precluded further exploration of this possibility.
In this report, we present a series of converging observations indicating that a domain of P2 recognizing the attachment sites within the aphid stylets is located at the N terminus of the protein. More precisely, we identified a single amino acid position that can either abolish transmission or differentially affect transmission efficiency by various aphid species and thereby change the spectrum of vector species for CaMV. Interestingly, we also obtained evidence that changes at this amino acid position can occur spontaneously when a particular aphid species is used as a vector after a series of successive passages in host plants, indicating potential for very rapid adaptation to new vector species.
MATERIALS AND METHODS
Virus, host plants, and vectors.
The aphid-transmissible isolate Cabb-S of CaMV (12) was used as the standard isolate and is hereafter referred to as the wild type. CaMV Cabb-S and the mutant derivatives described below were propagated by serial mechanical inoculation in turnip plants (Brassica rapa L. cv. Just Right).
The aphid species used were previously characterized as being good vectors (Brevicoryne brassicae L. and Myzus persicae Sulzer), poor vectors (Macrosiphum euphorbiae Thomas and Nasonovia ribisnigri Mosley), or nonvectors (Brachycaudus helichrysi L.) with respect to transmission of CaMV (7, 19, 25). Laboratory colonies of all aphid species were initiated from a single viviparous aptera collected on cauliflower (B. brassicae), pepper (M. persicae), lettuce (N. ribisnigri and M. euphorbiae), or Senecio vulgaris L. (B. helichrysi) plants in central Spain. All aphid colonies, except N. ribisnigri, were reared in environmental growth chambers at temperatures of 23°C (day) and 18°C (night) and a photoperiod of 14 h of light and 10 h of dark. The clone of N. ribisnigri was reared at a constant temperature of 12°C and a photoperiod of 14 h of light and 10 h of dark. B. brassicae and M. persicae colonies were reared on Brassica rapa cv. Just Right, while M. euphorbiae and N. ribisnigri were cultured on lettuce (cv. Cazorla) and B. helichrysi was cultured on chrysanthemum (Chrysanthemum coronarium L.).
Plasmid construction and mutagenesis.
Clone pCa37 is the reference clone for the CaMV isolate Cabb-S (12); clone ΔII-S, where the entire coding sequence of gene II is replaced by the unique restriction site SpeI, was described elsewhere (13).
CaMV mutants with a substitution at amino acid position 6 of P2 were created by PCR-directed mutagenesis. Gene II was PCR amplified on the template pCa37 with reverse and forward primers containing an SpeI restriction site at their 5′ extremities. The PCR products were later digested by SpeI and directly cloned at the corresponding site in plasmid ΔII-S. Eight different forward primers were used, each containing a mutation inducing an amino acid change at position 6 of the P2 protein sequence. The primers were designed to substitute either glycine (G; codon, GGA), lysine (K; codon, AAA), glutamic acid (E; codon, GAA), asparagine (N; codon, AAT), methionine (M; codon, ATG), threonine (T; codon, ACA), tyrosine (Y; codon, TAT), or histidine (H; codon, CAT) for the original glutamine (Q; wild-type codon, CAA) and, after cloning, yielded mutant CaMV clones designated Q6G, Q6K, Q6E, Q6N, Q6M, Q6T, Q6Y, and Q6H, respectively.
Plasmid Top-S, containing the full genome-length CaMV Cabb-S sequence with an engineered early stop codon at amino acid position 6 of P2, was described previously (13). This plasmid is not infectious when inoculated into turnip plants unless the stop codon reverts to a coding nucleotide triplet; that infectious revertants appear spontaneously upon Top-S inoculation has been reported previously (13). Several infectious revertants were characterized and are described in Results. To ensure that these revertants were not a mixed population, viral genomes were extracted from an infected plant as described previously (13) and cloned in pUC19 using the unique SalI restriction site in the CaMV sequence. The nature of the reversion of the stop codon at amino acid position 6 of P2 was determined by sequencing the various clones produced. The clones were then inoculated back into plants, and the properties determined for each revertant were verified on individual clones.
To express P2Rev5 using a baculovirus/insect cell expression system, a DNA fragment of 621 bp released upon digestion of pTop-S-Rev5 (see Results) with BamHI and BglII was cloned between the BglII sites in the transfer plasmid p119 (17) to produce p119-P2Rev5. To create a fusion between P2Rev5 and green fluorescent protein (GFP), the GFP gene was extracted from plasmid pEGFP-C1 (8) by Eco47III-PstI double digestion and inserted into pUC19 using XbaI and PstI sites to yield pUC-GFP. The P2Rev5 coding sequence was PCR amplified from the plasmid pTop-S-Rev5 with forward and reverse primers containing BamHI and BglII sites at their 5′ extremities, respectively, with the reverse primer omitting the stop codon of P2Rev5, and the PCR product was inserted into the BamHI site of the plasmid pUC-GFP, yielding pUC-P2Rev5-GFP. The sequence encoding a P2Rev5-GFP fusion was then extracted from pUC-P2Rev5-GFP by a double BamHI-BglII digestion and inserted into p119 at the BglII cloning site to generate p119-P2Rev5-GFP.
Insect cell culture maintenance and infection with p119-P2Rev5 and p119-P2Rev5-GFP baculovirus recombinants, as well as production and purification of recombinant proteins, were as previously described (16). Plasmids containing the full-length CaMV genome or mutant derivatives were mechanically inoculated onto host plants as described previously (13).
Protein analysis and microscopy.
Accumulation of P2 in turnip plants infected with Top-S-Rev5 or the CaMV Q6x mutant series was verified by total protein extraction and 13.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by P2- or P4-specific immunodetection as previously described (13). Far-Western experiments to detect interaction between P2, P3, and P3-virion complexes were done precisely as previously described (21). Briefly, membranes from Western blots of P2 and mutant derivative proteins were incubated either with P3 alone or with a mixture of P3 and virions. The interaction of P3 and P3-virion complexes with various forms of P2 on the membrane is revealed using antibodies against P3 and antibodies against virions, respectively. Electron-lucent inclusion bodies in Top-S-Rev5-infected plants were observed using electron microscopy, as described previously (9). The paracrystals of P2Rev5 produced in the baculovirus/insect cell expression system were observed by negative staining electron microscopy as described previously (5). The association between P2Rev5-GFP and the microtubular network in insect cells was observed using an epifluorescence microscope. In this case, insect cells were infected with the corresponding baculovirus recombinant for 48 h and observed live, directly in the culture medium, without further processing.
Transmission tests.
Infected turnip plants, used as virus source plants, were selected for consistency between batches and uniformity of symptom appearance. Transmission tests were performed essentially as described previously (11). Groups of 25 to 30 young-adult aphid apterae were placed inside plastic cages for 1 h for preacquisition starving. Aphids were then released on the upper side of an infected leaf for virus acquisition. In all cases, the aphids were placed on the last expanded leaf showing vein-clearing symptoms. After a 5-min acquisition access period, groups of five aphids were transferred onto 15-day-old seedlings of Brassica rapa cv. Just Right, used as test plants, for a 3-hour inoculation period. Turnip test plants were finally sprayed with imidacloprid (Confidor, Bayer) and transferred to an aphid-free growth chamber at 26°C (day) and 20°C (night), with a photoperiod of 16 h of light and 8 h of dark, where they were checked regularly for symptom appearance for 3 to 5 weeks.
Transmission tests were conducted using all possible combinations of the selected five aphid species and the eight CaMV variants described above, as well as wild-type Cabb-S. Eleven replicates of six plants each were used for each virus variant-aphid species combination.
Statistical analysis.
The ratio corresponding to the number of infected plants divided by the total number of test plants for each of the treatments used in the study (all combinations of virus variant-vector species) was subjected to a pairwise comparison using a chi-square test. Furthermore, the transmission rate obtained in each of the individual tests was transformed by arc sine √x (where x is the observed transmission rate) to reduce heterocedasticity. The transformed data were subjected to an analysis of variance as a factorial design, where the two factors under study were the type of virus variant and the species of aphid. Multiple mean comparisons were made between treatments using the Tamhane T2 test, which allows reliable pairwise comparisons based on a t test, in cases where the variance between treatments was not the same. All statistical analyses were made using the statistical package SPSS (version 12.0) for personal computer.
RESULTS
Identification of a residue in P2 involved in interaction with the aphid vector.
We recently described the CaMV clone Top-S, which has an engineered stop codon at amino acid position 6 of P2 and which is not infectious unless this codon reverts to an amino acid coding triplet (13). Because the amino acid at position 6 lies in the uncharacterized N-terminal region of P2, which is possibly involved in mediating the P2-aphid interaction (see the Introduction), we decided to inoculate host plants with clone Top-S and systematically test spontaneous revertants for P2 accumulation and aphid transmissibility. The fifth revertant analyzed, designated Top-S-Rev5, failed to be transmitted by aphids despite significant P2 accumulation (data not shown, but see also CaMV mutant Q6Y in Fig. 2 and 3), and cloning and sequencing of the viral DNA revealed a single nucleotide change, resulting in introduction of Y at amino acid position 6 of P2 in place of the stop codon.
FIG. 2.
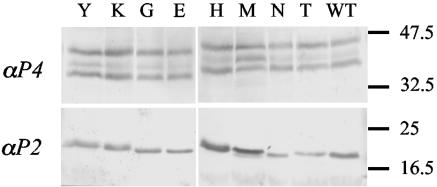
Detection of the coat protein (P4) and P2 in plants infected with various mutant derivatives of CaMV. The identity of the amino acid substituted for Q at position 6 of P2 is indicated at the top. The upper panel shows immunostaining of the coat protein (P4), whereas the lower panel is P2 specific. The molecular weight scale (in thousands) is indicated on the right.
FIG. 3.
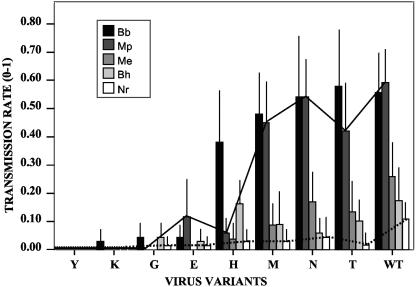
Transmission rates of different CaMV mutants by five aphid species. Means are presented together with standard errors. Bb, Brevicoryne brassicae; Mp, Myzus persicae; Me, Macrosiphum euphorbiae; Bh, Brachycaudus helichrysi; and Nr, Nasonovia ribisnigri. Note that N. ribisnigri (dotted line) transmitted all variants at similar rates, while transmission by M. persicae (plain line) varied drastically depending on the type of virus variant tested. For statistical significances of the observed differences in transmission rates, see Table 1.
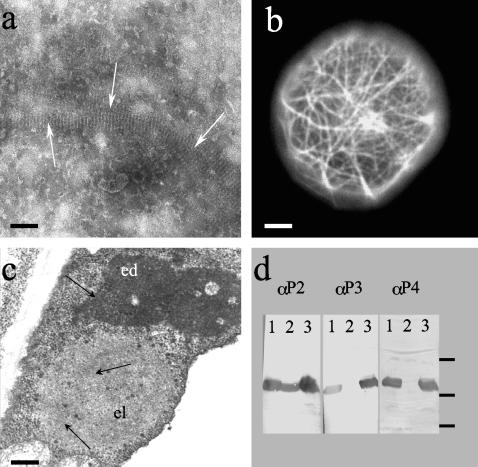
To understand the reason for the lack of aphid transmissibility of Top-S-Rev5, and hence likely explain why P2Rev5 is biologically inactive, we examined all of the well-characterized biological or biochemical features of wild-type P2 for this particular mutant. P2Rev5 was expressed in the baculovirus/insect cell expression system, and its lack of biological activity was confirmed by aphid transmission testing (not shown) as previously described (3). Large amounts of P2Rev5 accumulated as paracrystals (Fig. 1a), similar to those obtained previously with wild-type P2 (5), thus indicating that the overall structure and the polymerization of the molecule is not affected. In live Sf9 insect cells, paracrystals of P2Rev5 were clearly associated with microtubules (not shown), as described earlier for wild-type P2 (6), and a P2Rev5-GFP fusion expressed in the same system confirmed that the affinity of wild-type P2 for the microtubular network of the host cell was not abolished in P2Rev5 (Fig. 1b). Figure 1c shows that, in infected plant cells, P2Rev5 forms electron-lucent inclusion bodies similar to those formed by wild-type P2 (9, 10). Finally, we demonstrated that the capacity of P2 to bind P3 and P3-virion complexes (21) is also unaltered in P2Rev5 (Fig. 1d). Overall, the characterization of mutant Top-S-Rev5 presented in Fig. 1 indicates that none of the previously described properties of P2 are significantly affected by the mutation “Rev5,” and thus the loss of aphid transmission is most likely due to the loss of an uncharacterized function of P2, such as its capacity to recognize and bind aphid stylets.
FIG. 1.
Characterization of biochemical and biological properties of mutant P2Rev5. (a) Crude extracts of Sf9 insect cells infected with a baculovirus recombinant expressing P2Rev5, observed by negative staining and electron microscopy. White arrows indicate P2Rev5 paracrystal bundles. (b) Live Sf9 insect cell infected with a baculovirus recombinant expressing a P2Rev5-GFP fusion observed by epifluorescence microscopy. (c) Plant cell infected with CaMV Top-S-Rev5. The cell contains both electron-dense (ed) and electron-lucent (el) inclusion bodies; virions are indicated by black arrows. (d) Far-Western experiments revealing P2-P3 interaction. Ten micrograms of wild-type P2, P2157m (a negative control that can no longer bind P3 [21, 37]), and P2Rev5 were loaded in lanes 1, 2, and 3, respectively. The proteins are specifically revealed with an anti-P2 serum (3) in the left panel and tested for P3 and P3-virion binding capacity in the middle and right panels, respectively (see Materials and Methods). Molecular mass marker positions 6.5, 16.5, and 25 kDa are shown on the right. Bars represent 100, 1,000, and 500 μm in panels a, b, and c, respectively.
An independent observation later supported this hypothesis. Indeed, by maintaining the wild-type Cabb-S isolate through 10 serial aphid transmissions using B. brassicae as a vector, we obtained a variant that was very poorly transmitted by M. persicae, a species known to transmit wild-type CaMV Cabb-S with high efficiency (see Fig. 3). The transmission efficiencies were tested as described in Materials and Methods, using 28 test plants for each vector species, and were 31.7% and 5.5% for B. brassicae and M. persicae, respectively. We purified the viral DNA from the final series of infected plants, and DNA sequencing revealed a single mutation leading to a change from Q to H at amino acid position 6 of P2. This CaMV variant confirmed that the amino acid residue at position 6 of P2 is indeed involved in aphid recognition and, together with the results obtained with Top-S-Rev5, supports the view that its substitution by other residues could either totally abolish transmission or cause some alteration in vector specificity.
The amino acid residue at position 6 of P2 determines the spectrum of vector species of CaMV.
To further characterize this phenomenon and test the above hypothesis, we created a series of eight mutant clones of CaMV, all with an amino acid change at position 6 of P2. Plants inoculated with these eight different CaMV variants showed symptoms of infection 2 to 3 weeks after inoculation, and all accumulated virions and P2 (Fig. 2). Variations in the detected amounts of P2 and virions in Fig. 2 are similar to variations we routinely observed with a single CaMV clone. The fact that they do not correlate with transmission efficiencies reported in Table 1 and Fig. 3 confirm that they are not significant.
TABLE 1.
Pairwise comparisons of the transmission rates between the different variants of CaMV by each of the aphid species used in the studya
| Mutant | No. of plants infected/total no. of plants tested with:
|
||||
|---|---|---|---|---|---|
| M. persicae | B. brassicae | M. euphorbiae | N. ribisnigri | B. helychrisi | |
| Q6Y | 0/66 a | 0/65 a | 0/66 a | 0/66 a | 0/66 a |
| Q6K | 0/62 a (a) | 2/62 a (a) | 0/62 a (a) | 0/62 a (a) | 0/62 a (a) |
| Q6E | 8/66 bd (ac) | 3/64 a (a) | 0/66 a (a) | 1/66 a (a) | 2/66 a (ab) |
| Q6G | 0/65 a (a) | 3/64 a (a) | 0/66 a (a) | 1/66 a (a) | 3/66 a (ab) |
| Q6H | 4/62 ad (a) | 23/62 c (b) | 2/62 a (a) | 2/62 a (a) | 10/62 b (b) |
| Q6M | 30/66 c (bc) | 31/64 bc (b) | 6/66 b (ab) | 2/66 a (a) | 6/65 ab (ab) |
| Q6N | 36/66 c (bc) | 36/66 b (b) | 11/64 b (ab) | 3/66 a (a) | 4/66 ab (ab) |
| Q6T | 28/66 c (cd) | 38/65 b (b) | 9/66 b (ab) | 1/65 a (a) | 5/66 ab (ab) |
| WT | 38/64 c (bd) | 37/64 b (b) | 17/63 b (b) | 7/64 a (a) | 11/64 b (ab) |
Proportions (number of plants infected/total number of plants tested) followed by the same letter within each column indicate no significant differences (P < 0.05) according to pairwise comparisons using a chi-square test or a Fisher's exact test when expected values were lower than 5. The same letters in parentheses within each column indicate no significant differences (P < 0.05) on the basis of a multiple-pairwise-comparison Tamhane T2 test. WT, wild type.
Aphid transmission tests were then performed with all possible combinations between wild-type or mutant CaMV variants and the various aphid species used in this study, and the results are summarized in Table 1 and Fig. 3. The first striking observations were that none of the mutations had a positive effect on transmission efficiency and that different amino acids at this position had various impacts on the spectrum of aphid species that could successfully transmit the virus. Variant Q6Y was never transmitted, whatever the aphid species, confirming the results obtained with Top-S-Rev5. All of the other variants were transmitted by at least one aphid species and fell into three distinct categories: (i) variants that had no effect or only a minor effect on transmission and behaved as wild-type CaMV (Q6M, Q6N, and Q6T); (ii) variants for which the transmission rate by all vector species was dramatically reduced (Q6G, Q6K, and Q6E); and (iii) variants with transmission rates that were affected differentially depending on the vector species (best exemplified by Q6H). Most strikingly for the variant Q6H, the transmission rate was dramatically and specifically reduced with M. persicae, and to a lesser extent M. euphorbiae, whereas it was barely modified, if at all, for other species. For all CaMV mutants, virus DNA was PCR amplified from aphid-inoculated plants, and ORF II was sequenced, confirming the absence of both back mutation to wild type or other sequences at amino acid position 6 of P2 and second-site reversions.
On examination of the data obtained with different vector species in Fig. 3 and Table 1, it appears that the transmission rate observed with a poor vector species is not greatly sensitive to changes at amino acid position 6 of P2. For example, N. ribisnigri (dotted line in Fig. 3) transmitted all mutants except Q6Y and Q6K at a rather constant rate, as indicated by the lack of statistically significant differences between mutants in Table 1. In contrast, transmission by very efficient vectors, such as B. brassicae and, particularly, M. persicae (plain line in Fig. 3), is greatly affected by some of the mutations (statistical significances in Table 1).
DISCUSSION
The amino acid at position 6 of P2 could be directly involved in aphid binding.
Numerous studies on the noncirculative transmission of plant viruses have repeatedly shown that HC (for instance, HC-Pro in potyvirus or P2 in caulimovirus) can be acquired alone, and prior to virus particles, by the vector (3, 9, 14, 24, 38). This simple fact definitively demonstrates that HCs are viral proteins directly recognizing attachment sites in the vector mouthparts. In potyviruses, a conserved KITC motif located in the N-terminal domain of HC-Pro has been shown to be involved in aphid binding (1). However, whether this domain directly recognizes the putative receptor of the vector or indirectly affects the binding capacity of HC-Pro remains unclear. Here, in the caulimovirus CaMV, we provide the first identification of a key amino acid of P2 (Q at amino acid position 6) that is specifically involved in aphid recognition. A number of arguments support the hypothesis that we have characterized an element directly involved in the recognition of, and binding to, the putative receptor(s) in the insect. (i) The CaMV variant Top-S-Rev5 produces a mutation (equivalent to P2Q6Y) that is not active in transmission when expressed either in infected plants or in the baculovirus/insect cell system. Interestingly, P2Rev5 (Q6Y) has biochemical properties otherwise similar to wild-type P2. If the effect of the Q6Y mutation on the biological activity of P2 were a structural and/or indirect effect, one or more of the other features tested in Fig. 1 would likely also be affected. (ii) Figure 3 and Table 1 show that mutant Q6H is poorly transmitted by Myzus persicae, whereas the effect of this mutation on transmission by other vector species is less, with some species not being affected at all (i.e., B. brassicae). If the Q6H mutation induced a change in a distinct domain of P2 governing the interaction with the aphid, this change should nonspecifically and similarly affect the interaction with all aphid species. (iii) Based on the properties of amino acids (22, 23), we could not identify any structural or biochemical trends predicting the impact of the various residues tested on the biological activity of P2 (transmission rate in Fig. 3), either in size (cf. Q, N, and T versus E, which is isometric of Q), polarity (cf. Q/N/T and M versus H), charge (cf. K and E), or the presence of an aromatic ring (cf. Y and H). Again, if AA 6 of P2 were acting indirectly, residues with related biochemical properties would be expected to have comparable effects. Although direct evidence is still lacking, we believe that the involvement of AA 6 of P2 in recognizing receptor sites within the aphid stylets is more specific and is most likely direct. In this case, direct perturbation of the interface would likely be due to charge repulsion (as exemplified with E and K) or steric clashes (Y versus H), with only G potentially interfering through a distinct mechanism, such as a local destabilization of the protein structure.
The only other available example of a change of a single or a few amino acids apparently impacting the specificity of a noncirculative virus-aphid relationship comes from Cucumber mosaic virus (CMV) (30). However, it was later demonstrated that this effect is due to a change in the stability of CMV virions rather than to a differential interaction with putative receptors in the two aphid species tested (27, 28). It is important to note that CMV is transmitted according to the “capsid strategy” (for a review, see reference 31), where the coat protein is able to interact directly with putative receptors in the stylets, whereas CaMV has adopted the more frequently used “helper strategy,” in which an additional viral product, the HC, links the virus particles to these receptors. The mutations we have engineered in P2 are thus independent of the virions and cannot alter their stability.
Another important outcome of our results is that they provide invaluable tools for future attempts to isolate the putative receptor(s) of noncirculative plant viruses in insect vectors. We again stress the fact that a large number of plant virus genera are transmitted in a noncirculative manner, most often according to the helper strategy (18), and that it is very well possible that many virus species use the same or similar receptors. Unfortunately, even the chemical nature of these putative receptors remains a mystery. So far, CaMV P2 is the only viral molecule that can recognize them to be efficiently overproduced, easily purified, and biologically active in a heterologous expression system (16). The mutants described in the present study will be very useful as specific affinity targets in the search for receptors of noncirculative viruses.
Specificity of CaMV aphid transmission.
The aphid species selected for our study covered the main vectors of CaMV that occur in the field (M. persicae and B. brassicae) plus two species (M. euphorbiae and N. ribisnigri) that are commonly found landing on Brassica fields in several growing regions of Spain (26) and the United Kingdom (7). The latter two species have a high potential for spreading the virus, even if they are unable to reproduce and colonize the crop, because CaMV can be transmitted after brief superficial probes (29). For the purposes of comparison, we also included a species that has been reported to be a nonvector of CaMV: B. helichrysi (19). Surprisingly, the results of our study showed that B. helichrysi was able to transmit CaMV, although with low efficiency (Table 1). These apparently divergent results could be due to the low number of replicates used by previous authors or to differences in the transmission abilities of the different aphid clones and virus isolates used in the two studies.
The fact that the transmission rates obtained with poor vector species are not very sensitive to changes at amino acid position 6 of P2, while those obtained with good vector species are markedly affected, is difficult to explain. One possibility could be that the interaction between P2 and the putative receptors is complex and consists of a nonspecific weak binding, strengthened by more specific and precisely tuned adaptation to particular vector species. In this hypothesis, the poor vectors would transmit with low efficiency due solely to the nonspecific interaction with P2, whereas the good vectors would engage additional specific residues of P2 (possibly including that at position 6), leading to more stable binding.
One striking observation was that, when compared to wild-type CaMV Cabb-S, none of the mutations tested had a positive effect on transmission efficiency, whatever the aphid species. Although the panel of amino acids tested covers a wide range of biochemical properties, the transmission rates of all variants were either unchanged or reduced. This might indicate that the Q at amino acid position 6 of P2 is optimal for the interaction of CaMV with its vectors and that the virus has evolved to maximize its transmission by a wide range of vector species. Consistently, all of the CaMV isolates that have been collected from the field and sequenced to date (available in the GenBank database) have a glutamine at position 6 of P2. This speculation infers frequent contact in nature between CaMV populations and several alternating vector species. Indeed, under our experimental conditions, when only one vector species (B. brassicae) was involved in transmission through several serial passages, we rapidly produced a spontaneous variant, Q6H, that was no longer “optimized” and had almost lost its transmissibility by one of its best vectors (M. persicae).
Whether the above interpretation is correct or not, both the fact that we induced important changes in the transmission performance of a spectrum of vector species by mutating a single amino acid of the CaMV HC and the spontaneous appearance of a CaMV mutant at the very same position when using a single vector species certainly demonstrate that adaptation of a plant virus to fluctuations in vector populations is most likely rapid and likely to occur under field conditions.
Acknowledgments
We thank D. Gargani and Marc Ravallec for technical assistance in electron microcopy, P. Travo for fluorescence microscopy, and M. Duque for assistance in aphid rearing and transmission testing. We are very grateful to Takii Ltd. for generously providing seeds of Brassica rapa cv. Just Right.
This work was supported by the Plan Nacional de I+D+I from the Ministerio de Educación y Ciencia (AGL-2000-2006) and by the bilateral INRA-CSIC grant HF2003-0318.
REFERENCES
- 1.Blanc, S., E. D. Ammar, S. Garcia-Lampasona, V. V. Dolja, C. Llave, J. Baker, and T. P. Pirone. 1998. Mutations in the potyvirus helper component protein: effects on interactions with virions and aphid stylets. J. Gen. Virol. 79:3119-3122. [DOI] [PubMed] [Google Scholar]
- 2.Blanc, S., M. Cerutti, H. Chaabihi, C. Louis, G. Devauchelle, and R. Hull. 1993. Gene II product of an aphid-nontransmissible isolate of cauliflower mosaic virus expressed in a baculovirus system possesses aphid transmission factor activity. Virology 192:651-654. [DOI] [PubMed] [Google Scholar]
- 3.Blanc, S., M. Cerutti, M. Usmany, J. M. Vlak, and R. Hull. 1993. Biological activity of cauliflower mosaic virus aphid transmission factor expressed in a heterologous system. Virology 192:643-650. [DOI] [PubMed] [Google Scholar]
- 4.Blanc, S., E. Hébrard, M. Drucker, and R. Froissart. 2001. Molecular basis of vector transmission: caulimoviruses, p. 143-166. In K. Harris, O. P. Smith, and J. E. Duffus (ed.), Virus-insect-plant interactions. Academic Press, San Diego, Calif.
- 5.Blanc, S., I. Schmidt, G. Kuhl, P. Esperandieu, G. Lebeurier, R. Hull, M. Cerutti, and C. Louis. 1993. Paracrystalline structure of cauliflower mosaic virus aphid transmission factor produced both in plants and in a heterologous system and relationship with a solubilized active form. Virology 197:283-292. [DOI] [PubMed] [Google Scholar]
- 6.Blanc, S., I. Schmidt, M. Vantard, H. B. Scholthof, G. Khul, P. Esperandieu, M. Cerutti, and C. Louis. 1996. The aphid transmission factor of cauliflower mosaic virus forms a stable complex with microtubules in both insect and plant cells. Proc. Natl. Acad. Sci. USA 93:15158-15163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broadbent, L. 1957. Investigation of virus diseases of Brassica crops. Agric. Res. Counc. Rep. 14:94. [Google Scholar]
- 8.Cormack, B., R. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 9.Drucker, M., R. Froissart, E. Hebrard, M. Uzest, M. Ravallec, P. Esperandieu, J. C. Mani, M. Pugniere, F. Roquet, A. Fereres, and S. Blanc. 2002. Intracellular distribution of viral gene products regulates a complex mechanism of cauliflower mosaic virus acquisition by its aphid vector. Proc. Natl. Acad. Sci. USA 99:2422-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Espinoza, A. M., V. Medina, R. Hull, and P. G. Markham. 1991. Cauliflower mosaic virus gene II product forms distinct inclusion bodies in infected plant cells. Virology 185:337-344. [DOI] [PubMed] [Google Scholar]
- 11.Fereres, A., P. Perez, C. Gemeno, and F. Ponz. 1993. Transmission of Spanish Pepper-PVY isolates by aphid vectors: epidemiological implications. Environ. Entomol. 22:1260-1265. [Google Scholar]
- 12.Franck, A., H. Guilley, J. Jonard, K. Richards, and L. Hirth. 1980. Nucleotide sequence of cauliflower mosaic virus DNA. Cell 21:285-294. [DOI] [PubMed] [Google Scholar]
- 13.Froissart, R., M. Uzest, V. Ruiz-Ferrer, M. Drucker, E. Hebrard, T. Hohn, and S. Blanc. 2004. Splicing of Cauliflower mosaic virus 35S RNA serves to downregulate a toxic gene product. J. Gen. Virol. 85:2719-2726. [DOI] [PubMed] [Google Scholar]
- 14.Govier, D. A., and B. Kassanis. 1974. A virus induced component of plant sap needed when aphids acquire potato virus Y from purified preparations. Virology 61:420-426. [DOI] [PubMed] [Google Scholar]
- 15.Gray, S. M., and N. Banerjee. 1999. Mechanisms of arthropod transmission of plant and animal viruses. Microbiol. Mol. Biol. Rev. 63:128-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hébrard, E., M. Drucker, D. Leclerc, T. Hohn, M. Uzest, R. Froissart, J.-M. Strub, S. Sanglier, A. van Dorsselaer, A. Padilla, G. Labesse, and S. Blanc. 2001. Biochemical characterization of the helper component of Cauliflower mosaic virus. J. Virol. 75:8538-8546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hericourt, F., S. Blanc, V. Redeker, and I. Jupin. 2000. Evidence for phosphorylation and ubiquitinylation of the turnip yellow mosaic virus RNA-dependent RNA polymerase domain expressed in a baculovirus-insect cell system. Biochem. J. 349:417-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hull, R. 2001. Matthews' plant virology, 4th ed., vol. 1. Academic Press, San Diego, Calif.
- 19.Kennedy, J. S., M. F. Day, and V. F. Eastop. 1962. A conspectus of aphids as vectors of plant viruses. CAB, London, United Kingdom.
- 20.Leh, V., E. Jacquot, A. Geldreich, M. Haas, S. Blanc, M. Keller, and P. Yot. 2001. Interaction between cauliflower mosaic virus ORFIII product and the coat protein is required for transmission of the virus by aphids. J. Virol. 75:100-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leh, V., E. Jacquot, A. Geldreich, T. Hermann, D. Leclerc, M. Cerrutti, P. Yot, M. Keller, and S. Blanc. 1999. Aphid transmission of cauliflower mosaic virus requires the viral PIII protein. EMBO J. 18:7077-7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livingstone, C. D., and G. J. Barton. 1996. Identification of functional residues and secondary structure from protein multiple sequence alignment. Methods Enzymol. 266:497-512. [DOI] [PubMed] [Google Scholar]
- 23.Livingstone, C. D., and G. J. Barton. 1993. Protein sequence alignments: a strategy for the hierarchical analysis of residue conservation. Comput. Appl. Biosci. 9:745-756. [DOI] [PubMed] [Google Scholar]
- 24.Lung, M. C. Y., and T. P. Pirone. 1974. Acquisition factor required for aphid transmission of purified cauliflower mosaic virus. Virology 60:260-264. [DOI] [PubMed] [Google Scholar]
- 25.Markham, P. G., M. S. Pinner, B. Raccah, and R. Hull. 1987. The acquisition of a caulimovirus by different aphid species: comparison with a potyvirus. Ann. Appl. Biol. 111:571-587. [Google Scholar]
- 26.Nebreda, M., A. Moreno, N. Perez, I. Palacios, V. Seco-Fernandez, and A. Fereres. 2004. Activity of aphids associated with lettuce and broccoli in Spain and their efficiency as vectors of Lettuce mosaic virus. Virus Res. 100:83-88. [DOI] [PubMed] [Google Scholar]
- 27.Ng, J. C., C. Josefsson, A. J. Clark, A. W. Franz, and K. L. Perry. 2005. Virion stability and aphid vector transmissibility of Cucumber mosaic virus mutants. Virology 332:397-405. [DOI] [PubMed] [Google Scholar]
- 28.Ng, J. C., S. Liu, and K. L. Perry. 2000. Cucumber mosaic virus mutants with altered physical properties and defective in aphid vector transmission. Virology 276:395-403. [DOI] [PubMed] [Google Scholar]
- 29.Palacios, I., M. Drucker, S. Blanc, S. Leite, A. Moreno, and A. Fereres. 2002. Cauliflower mosaic virus is preferentially acquired from the phloem by its aphid vectors. J. Gen. Virol. 83:3163-3171. [DOI] [PubMed] [Google Scholar]
- 30.Perry, K. L., L. Zhang, and P. Palukaitis. 1998. Amino acid changes in the coat protein of cucumber mosaic virus differentially affect transmission by the aphids Myzus persicae and Aphis gossypii. Virology 242:204-210. [DOI] [PubMed] [Google Scholar]
- 31.Pirone, T. P., and S. Blanc. 1996. Helper-dependent vector transmission of plant viruses. Annu. Rev. Phytopathol. 34:227-247. [DOI] [PubMed] [Google Scholar]
- 32.Pirone, T. P., and K. L. Perry. 2002. Aphids—non-persistent transmission, p. 1-19. In R. T. Plumb (ed.), Advances in botanical research, vol. 36. Academic Press, San Diego, Calif. [Google Scholar]
- 33.Plisson, C., M. Uzest, M. Drucker, R. Froissart, C. Dumas, J. Conway, D. Thomas, S. Blanc, and P. Bron. 2005. Structure of the mature P3-virus particle complex of cauliflower mosaic virus revealed by cryo-electron microscopy. J. Mol. Biol. 346:267-277. [DOI] [PubMed] [Google Scholar]
- 34.Plumb, R. T. 2002. Plant virus vector interactions. Academic Press, San Diego, Calif.
- 35.Raccah, B., H. Huet, and S. Blanc. 2001. Potyviruses, p. 181-206. In K. Harris, J. E. Duffus, and O. P. Smith (ed.), Virus-insect-plant interactions. Academic Press, San Diego, Calif.
- 36.Sako, N., K. Yoshioka, and K. Eguchi. 1984. Mediation of helper component in aphid transmission of some potyviruses. Ann. Phytopathol. Soc. Jpn. 50:515-521. [Google Scholar]
- 37.Schmidt, I., S. Blanc, P. Esperandieu, G. Kuhl, G. Devauchelle, C. Louis, and M. Cerutti. 1994. Interaction between the aphid transmission factor and virus particles is a part of the molecular mechanism of cauliflower mosaic virus aphid transmission. Proc. Natl. Acad. Sci. USA 91:8885-8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thornbury, D. W., G. M. Hellman, R. E. Rhoads, and T. P. Pirone. 1985. Purification and characterization of potyvirus helper component. Virology 144:260-267. [DOI] [PubMed] [Google Scholar]
- 39.Wang, R. Y., G. Powell, J. Hardie, and T. P. Pirone. 1998. Role of the helper component in vector-specific transmission of potyviruses. J. Gen. Virol. 79:1519-1524. [DOI] [PubMed] [Google Scholar]