Abstract
The hepatitis C virus (HCV) encodes a large polyprotein; therefore, all viral proteins are produced in equimolar amounts regardless of their function. The aim of our study was to determine the ratio of nonstructural proteins to RNA that is required for HCV RNA replication. We analyzed Huh-7 cells harboring full-length HCV genomes or subgenomic replicons and found in all cases a >1,000-fold excess of HCV proteins over positive- and negative-strand RNA. To examine whether all nonstructural protein copies are involved in RNA synthesis, we isolated active HCV replication complexes from replicon cells and examined them for their content of viral RNA and proteins before and after treatment with protease and/or nuclease. In vitro replicase activity, as well as almost the entire negative- and positive-strand RNA, was resistant to nuclease treatment, whereas <5% of the nonstructural proteins were protected from protease digest but accounted for the full in vitro replicase activity. In consequence, only a minor fraction of the HCV nonstructural proteins was actively involved in RNA synthesis at a given time point but, due to the high amounts present in replicon cells, still representing a huge excess compared to the viral RNA. Based on the comparison of nuclease-resistant viral RNA to protease-resistant viral proteins, we estimate that an active HCV replicase complex consists of one negative-strand RNA, two to ten positive-strand RNAs, and several hundred nonstructural protein copies, which might be required as structural components of the vesicular compartments that are the site of HCV replication.
Hepatitis C virus (HCV) is an enveloped positive-strand RNA virus belonging to the genus Hepacivirus in the family Flaviviridae. The genome of HCV encompasses a single ∼9600-nucleotide (nt) RNA molecule carrying one large open reading frame (ORF) that is flanked by nontranslated regions (NTRs). In addition to the polyprotein, the expression of a novel HCV protein with a yet-unknown function has recently been described, the so-called F-protein, which is generated by ribosomal frameshifting (78, 79). The 5′ NTR contains an internal ribosome entry site (IRES) that directs translation of the ORF (74). In addition, the 5′ NTR is required for RNA replication, as is the case with the 3′ NTR (25, 27, 42, 80). HCV proteins generated from the polyprotein precursor are cleaved by cellular and viral proteases into at least 10 different products (for reviews, see references 8 and 66). The structural proteins core, E1 and E2, are located in the amino terminus of the polyprotein (37), followed by p7, a hydrophobic peptide that is supposed to be a viroporin, forming an ion channel with a yet-unknown function (33, 63), and the nonstructural proteins (NS) NS2, NS3, NS4A, NS4B, NS5A, and NS5B. NS2 and the amino terminus of NS3 comprise the NS2-3 protease responsible for cleavage between NS2 and NS3 (31, 38). NS3 is a multifunctional protein, consisting of an amino-terminal protease domain required for processing of the NS3 to 5B region (6, 32) and a carboxy-terminal helicase/nucleoside triphosphatase domain (39, 71). NS4A is a cofactor that activates the NS3 protease function by forming a heterodimer (9, 21, 48, 72). The hydrophobic protein NS4B induces the formation of a cytoplasmic vesicular structure, designated the membranous web, that appears to contain the replication complex of HCV (18, 30). NS5A is a phosphoprotein that seems to play an important role in viral replication since most of the cell culture-adaptive mutations described thus far are located within the central region of NS5A (12, 34, 43, 50). NS5B is the RNA-dependent RNA polymerase of HCV (11, 52).
Since the establishment of HCV replicons (53), the understanding of the mechanisms underlying HCV RNA replication has increased tremendously (for a review, see reference 7). It is clear that nonstructural proteins NS3 to 5B are necessary and sufficient for HCV RNA replication. They build up a multiprotein complex that in analogy to other positive-strand RNA viruses is associated with intracellular membranes (reviewed in references 17 and 59). Biochemical analyses of crude replicase complexes (CRCs) prepared from lysates of replicon cells provided deeper insights into the organization and structure of the viral replication complex (2, 5, 19, 36, 45, 57, 70); however, a detailed stoichiometric analysis of the HCV replication complex has not yet been carried out. The aim of our study was to determine the ratio of viral positive- and negative-strand RNA to proteins in replicon cells and CRCs and to analyze which portions are actively involved in viral RNA synthesis. We found that the nonstructural proteins are produced in large excess over viral RNA in replicon cells and that viral replication complexes contain a large number of nonstructural protein copies. These results suggest that the majority of HCV nonstructural proteins may serve some other function in the replication process apart from RNA synthesis such as formation or scaffolding of the viral replication complex.
MATERIALS AND METHODS
Cell cultures.
Cell monolayers of the human hepatoma cell line Huh-7 (60) were grown in Dulbecco modified Eagle medium (DMEM; Invitrogen, Karlsruhe, Germany) supplemented with 2 mM l-glutamine, nonessential amino acids, 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 10% fetal calf serum. G418 (Geneticin; Invitrogen) was added at a final concentration of 1 mg/ml in case of replicon cell clone 9-13 carrying a HCV replicon expressing neomycin phosphotransferase (53). In the case of cell clone 11-1 harboring a monocistronic replicon (24), the medium was supplemented with 25 μg of hygromycin (Invitrogen)/ml. Huh7-Lunet cells refer to a Huh-7 cell clone that was generated with a selectable replicon and cured from HCV by treatment with a specific inhibitor. Huh7-Lunet cells are more permissive for HCV replication than naive Huh-7 cells (26, 50).
In vitro transcription. In vitro transcripts of HCV positive and negative-strands were generated by using the protocol described recently (50). For transcription of positive-strand HCV RNAs, plasmid DNA was restricted with AseI and ScaI (New England Biolabs, Frankfurt/Main, Germany), in the case of negative-strand transcripts, DNA was restricted with PmeI (New England Biolabs). After restriction, DNA was extracted with phenol and chloroform, precipitated with ethanol, and dissolved in RNase-free water. In vitro transcription reactions contained 80 mM HEPES (pH 7.5), 12 mM MgCl2, 2 mM spermidine, 40 mM dithiothreitol, a 3.125 mM concentration of each nucleoside triphosphate, 1 U of RNasin (Promega, Mannheim, Germany)/μl, 0.05 μg of restricted plasmid DNA/μl, and 0.6 U of T7 RNA polymerase (Promega)/μl for positive-strand synthesis or T3 RNA polymerase for transcription of negative strands, respectively. After 2 h at 37°C, an additional 0.3 U of T7 or T3 RNA polymerase/μl was added, and the reaction was incubated for another 2 h. Transcription was terminated by the addition of 1 U of RNase-free DNase (Promega) per μg of plasmid DNA, followed by incubation for 30 min at 37°C. After extraction with acidic phenol and chloroform, RNA was precipitated with isopropanol and dissolved in RNase-free water. The concentration was determined by measurement of the optical density at 260 nm (OD260), and RNA integrity was checked by denaturing agarose gel electrophoresis.
Electroporation of HCV full-length genomes.
For electroporation, single-cell suspensions of Huh7-Lunet cells were prepared by trypsinization of monolayers, detaching the cells from the culture dish by a rinse with complete DMEM, followed by one wash with phosphate-buffered saline (PBS), counting, and resuspension at 107 cells per ml in Cytomix (76) containing 2 mM ATP and 5 mM glutathione. Then, 10 μg of in vitro-transcribed RNA was mixed with 400 μl of the cell suspension by pipetting, electroporated, and immediately transferred to 8 ml of complete DMEM. Electroporation conditions were 975 μF and 270 V using a Gene Pulser system (Bio-Rad, Munich, Germany) and a cuvette with a gap width of 0.4 cm (Bio-Rad). Cells of several electroporations were combined and seeded in aliquots. At 24, 48, 72, and 96 h after seeding, cells were treated with trypsin and counted. Aliquots of the cells were either lysed in protein sample buffer and subjected to immunoblot analysis or used for preparation of total RNA and Northern hybridization.
Preparation of total RNA and quantification of HCV RNA by Northern hybridization.
These methods have been described recently (50). In brief, total RNA from cells or CRCs was prepared by a single-step isolation method (15), denatured by treatment with 5.9% glyoxal in 50% dimethyl sulfoxide and 10 mM sodium phosphate buffer (pH 7.0), and analyzed after denaturing agarose gel electrophoresis by Northern hybridization. Prior to hybridization the membrane was stained with methylene blue and cut ∼1 cm below the 28S rRNA band. The upper strip containing the HCV replicon RNA was hybridized with a 32P-labeled negative-sense riboprobe complementary to NS4B-NS5A region (nt 5979 to 6699) to detect viral positive-strand RNA or a positive-sense riboprobe encompassing the same region for detection of HCV negative-strand RNA. The lower strip that was hybridized with a β-actin-specific antisense riboprobe was used to correct for total RNA amounts loaded in each lane of the gel. Specific bands were quantified by phosphorimaging with a Molecular Imager FX scanner (Bio-Rad), and the number of HCV molecules was determined by comparison with a serial dilution of in vitro transcripts corresponding to a known number of positive or negative strands of subgenomic replicons mixed with 2 μg of total RNA from naive Huh-7 cells loaded in parallel onto the gel. In vitro transcripts were checked by denaturing agarose gel electrophoresis to ensure that almost all RNAs are full length and quantified by measurement of the OD260.
Preparation of CRCs and in vitro replicase assay.
The protocol is adapted from a previously published procedure (73). In a standard CRC-preparation, 2 × 108 Huh-7 cells persistently harboring subgenomic replicons were washed with phosphate-buffered saline and then scraped in phosphate-buffered saline and pelleted by centrifugation at 800 × g for 10 min at 4°C. Cells were suspended to a density of 2.5 × 107 cells/ml in hypotonic buffer (10 mM Tris-HCl [pH 7.5], 10 mM KCl, 1.5 mM MgCl2, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 2 μg of aprotinin/ml) and lysed by 75 strokes with a Dounce homogenizer. Nuclei and unbroken cells were removed by centrifugation at 1,000 × g for 10 min at 4°C. The intracellular membranes in the resulting supernatant (S1) were then sedimented on 300 μl of 60% (wt/wt) sucrose in 10 mM Tris-HCl (pH 7.5)-10 mM KCl-1.5 mM MgCl2 in an ultracentrifuge at 68,500 × g for 1 h at 4°C. The resulting supernatant (S2) was carefully removed, and the membrane fraction containing the CRCs was resuspended in the sucrose cushion to obtain an ∼500-μl CRC fraction from 2 × 108 cells and directly subjected to proteinase K, S7 nuclease, and/or Triton X-100 treatment. The total protein concentrations of standard CRC preparations were in the range of 5 mg/ml. Alternatively, S1 obtained from 2 × 108 cells was directly pelleted for 1 h at 68,500 × g, resuspended in 200 μl of 10 mM Tris-HCl (pH 8.0)-10 mM NaCl-15% glycerol to obtain the CRC fraction, and stored in aliquots at −70°C.
HCV in vitro replicase activity was determined in a reaction mixture containing 20 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 5 mM dithiothreitol, 5 mM KCl, 40 μg of actinomycin D/ml, 20 μCi of [α-32P]CTP, 10 μM CTP, 1 mM concentrations each of ATP and UTP, 5 mM GTP, 2.5 mM phosphoenolpyruvate, 1 U of pyruvate kinase (Sigma, Taufkirchen, Germany), 1 U of RNasin, and a 4-μl sample fraction in a total volume of 10 μl at 35°C for 60 min. Reaction products were purified by phenol-chloroform extraction and isopropanol precipitation and then analyzed by denaturing glyoxal agarose gel electrophoresis, followed by autoradiography. A radioactively labeled in vitro transcript corresponding to the length of a replicon RNA was used to determine the size of the reaction products.
Proteinase K, S7 nuclease, and Triton X-100 treatment of CRCs.
To test the protease and nuclease resistance of the CRCs, different amounts of proteinase K (0.8- or 8-mg/ml final concentrations), nuclease (0.2- or 2-U/μl final concentrations), and/or 1% (vol/vol) Triton X-100 were directly added to 50 μl of freshly prepared CRCs, followed by incubation for 60 min at 25°C. After the incubation, proteinase K and S7 nuclease were inactivated by the addition of 1 μl of 100 mM PMSF and 1 μl of 200 mM EGTA, respectively. Then, 4 μl of CRCs was directly analyzed by in vitro replicase assays, and 2 μl was mixed with Laemmli sample buffer and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting. A 10-μl portion of CRCs was used for total RNA preparation. Total RNA was dissolved in 10 μl of water, and half of it was subjected to Northern hybridization analysis.
Fivefold-concentrated fractions of proteinase K-treated CRCs were generated by trichloroacetic acid precipitation.
Immunoblot analysis and silver staining of proteins.
For quantitative immunoblot analysis, cells or CRCs were directly lysed in Laemmli-sample buffer and proteins were separated by SDS-10% PAGE using standard techniques (68). After SDS-PAGE proteins were either stained with silver or transferred to a polyvinylidene difluoride membrane and detected with various primary antibodies and secondary antibodies conjugated with horseradish peroxidase (Sigma) and developed with an ECL plus Western blotting detection system according to the instructions of the manufacturer (Amersham Biosciences, Freiburg, Germany). Core protein was quantified by using a goat anti-rabbit secondary antibody conjugated with alkaline phosphatase (Dianova, Hamburg, Germany), an ECF substrate (Amersham Biosciences) and a FLA-3000 fluorescence scanner with a 570-nm filter (Fuji). For detection of Calnexin a rabbit polyclonal antibody (Stressgen) was used specific for the amino-terminal part, which is resident in the lumen of the endoplasmic reticulum (ER). Core-, NS3-, and NS4B-specific rabbit polyclonal antisera were raised against purified portions corresponding to a part of the core protein (amino acids [aa] 1 to 167 of the polyprotein), the helicase domain of NS3 (aa 1230 to 1526 of the polyprotein), or full-length NS4B of the HCV NC1 isolate (EMBL nucleotide sequence database accession no. AJ238800), respectively, differing only in a few residues to HCV Con1 (EMBL nucleotide sequence database accession no. AJ238799). Mouse monoclonal antibody 5B-3B1, specifically detecting a linear epitope encompassing aa 2791 to 2801 (58) was used to quantify NS5B.
For quantification of HCV proteins, bacterially expressed core protein and baculovirus-expressed full-length NS4B and NS5B proteins with C-terminal His6 tags were purified by Ni-nitrilotriacetic acid affinity chromatography according to the instructions of the manufacturer (QIAGEN, Hilden, Germany) or as described recently (52). NS4B-C-His and core-C-His was further purified by preparative SDS-PAGE. The purity of both proteins was >90%, and the protein concentration was determined by Bradford assay and analytical SDS-PAGE using a serial dilution of bovine serum albumin as a reference. Core, NS4B, and NS5B sequences correspond to the NC1 isolate (see above and reference 41). Samples were quantified by comparison of band intensities to signals obtained from the serially diluted purified proteins.
RESULTS
Quantification of the HCV RNA to protein ratio in Huh-7 cells.
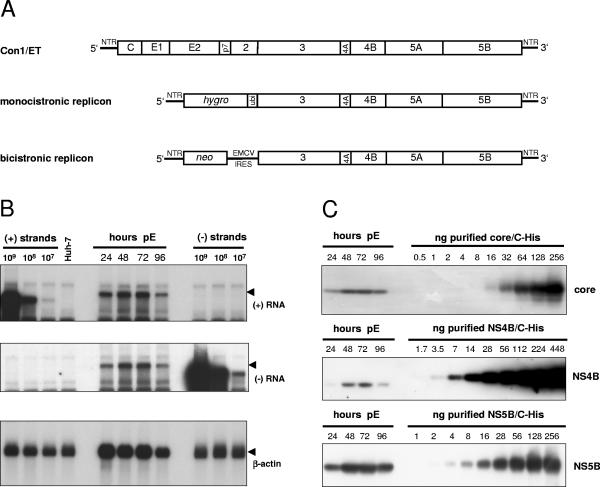
The first question addressed in our study was how the number of HCV positive- and negative-strand RNA molecules correlates with the amount of different HCV proteins in cells with productive HCV RNA replication. Therefore, we chose a full-length HCV genome with cell culture-adaptive mutations (Con1/ET, Fig. 1A) (64), which was transfected into Huh7-Lunet cells, a cured replicon cell clone exhibiting high permissiveness for HCV replication. Cells were seeded in aliquots after electroporation, harvested at various time points, counted, and analyzed for the amount of HCV RNA and proteins. Figure 1B shows a typical Northern blot analysis of such a transient replication assay by using known numbers of in vitro transcripts to determine the quantity of positive and negative-strand RNA in cells transfected with Con1/ET. RNA replication had already started 24 h after transfection (as judged by the clearly detectable negative-strand RNA signal), reached its maximum at 48 h and 72 h after transfection, and slightly decreased at 96 h, after the cells had reached confluence (Fig. 1B, top and middle panel). HCV core and nonstructural proteins 4B and 5B were quantified by Western blot analysis of cell lysates in comparison with defined amounts of purified proteins from the same HCV isolate (41, 52, 54) and by using antibodies raised against these particular antigens (Fig. 1C). The number of core, NS4B, and NS5B molecules in the transfected cells followed the same changes over time as the RNA, and the results obtained for the quantitative evaluation are summarized in Table 1. Previous analyses have shown that transfected RNA of replication-deficient genomes is degraded to trace amounts at 24 h after transfection and completely absent after 48 h (13, 64). To avoid any impact of transfected input RNA, the quantitative analysis was limited to the data obtained at 48 to 96 h. We found on average 40 copies of negative-strand RNA, a fivefold excess of positive-strand RNA and approximately one million copies of core, NS4B and NS5B per cell, indicating a vast excess of viral proteins to RNA molecules. Within the expected range of accuracy the ratio of the nonstructural proteins NS4B and NS5B was very similar. The three- to sixfold higher relative levels of core might be due to premature termination of translation leading to an overrepresentation of the amino-terminal portions of the HCV polyprotein. Nevertheless, the data were consistent in showing a tremendous excess of HCV proteins compared to RNA molecules.
FIG. 1.
Quantification of HCV RNA and nonstructural proteins in Huh-7 cells transfected with a full-length genome. (A) Structure of the HCV RNAs used in the present study. Con1/ET represents a full-length HCV genome harboring cell culture-adaptive mutations in NS3 and NS4B (64). The monocistronic replicon contains only a single ORF consisting of nt 342 to 389 of the core-coding region, the hygro gene (encoding the hygromycin phosphotransferase), the ubiquitin-encoding sequence (Ubi), and the HCV NS proteins NS3 to NS5B (24) and was used to select Huh-7 cell clone 11-1, which was analyzed in the present study (Table 1). The bicistronic replicon is composed of the first 377 nt of the HCV genome fused to the neo gene (encoding the neomycin phosphotransferase). Translation of the HCV NS proteins NS3-5B is initiated by the encephalomyocarditis virus-IRES. The bicistronic replicon was used to generate cell-clone 9-13 (53). (B) Time course of HCV positive- and negative-strand synthesis after transfection of Con1/ET RNA into Huh7-Lunet cells. Cells were harvested and counted at the time points indicated above the figure (pE, postelectroporation), and total RNA was prepared. A total of 5 μg of total RNA corresponding to 2.5 × 105, 2.1 × 105, 2.1 × 105, and 2.3 × 105 cells at 24, 48, 72, and 96 h, respectively, was subjected to Northern hybridization with radiolabeled riboprobes specific for the detection of HCV positive-strand (top panel), negative-strand (middle panel) or β-actin (lowest panel). Specific signals are indicated by arrowheads. Signals were quantified by phosphorimaging with known amounts of in vitro transcripts of positive or negative polarity corresponding to a subgenomic replicon and normalized for different loadings by the β-actin signal. Total RNA from naive Huh-7 cells was used as negative control (Huh-7). (C) Quantification of HCV core (upper panel), NS4B (middle panel), and NS5B (lowest panel) expression after transfection of Con1/ET RNA into Huh7-Lunet cells. An aliquot of the cells harvested for total RNA preparation at the time points given above each panel was lysed in Laemmli sample buffer and subjected to immunoblot analysis with monoclonal antibodies (NS5B) or polyclonal antisera (core, NS4B) with the specificities given on the right. Samples were quantified by comparison of signal intensities derived from known amounts of the respective antigens as indicated above each panel. The amount of loaded proteins correspond to 1.2 × 105, 1.8 × 105, 1.6 × 105, and 2.0 × 105 cells at 24, 48, 72, and 96 h, respectively.
TABLE 1.
Number of positive-strand RNA, negative-strand RNA, core, NS4B, and NS5B molecules per Huh-7 cell
| HCV sequence | Mean no. of molecules/cell ± SDa
|
||||
|---|---|---|---|---|---|
| Negative-strand | Positive-strand | Core | NS4B | NS5B | |
| Con1/ET (transient transfection)b | 40 ± 4 | (2.0 ± 0.3) × 102 | (4.9 ± 1.4) × 106 | (7.7 ± 1.8) × 105 | (1.5 ± 0.4) × 106 |
| Monocistronic replicon (cell clone 11-1) | 81 ± 11 | (4.4 ± 1.5) × 102 | NA | (8.9 ± 3.2) × 105 | (2.2 ± 0.8) × 106 |
| Bicistronic replicon (cell clone 9-13) | 94 ± 31 | (7.6 ± 2.9) × 102 | NA | (8.0 ± 0.4) × 105 | (1.2 ± 0.5) × 106 |
Data represent mean values and standard deviations of samples harvested at 48, 72, and 96 h after seeding. NA, not applicable.
Per-cell data was not normalized for transfection efficiency, which was routinely 50 to 80%.
Since we sought to determine the stoichiometry of RNA to protein in the HCV replication complex, we searched for the most appropriate biochemical equivalent. Every active replication complex must contain at least one negative-strand RNA molecule; therefore, the amount of negative-strand RNA gives the closest estimate of the maximal number of HCV replication complexes per cell. Based on this assumption, we found on average fewer than forty active replication complexes per cell, but each was accompanied by 20,000 to 40,000 copies of NS proteins.
Although the analysis of a HCV full-length genome should reflect the properties of viral translation and replication most closely, we wanted to confirm the data in a steady-state situation, which resembles a persistent infection. The most efficient and convenient systems to study persistent HCV RNA replication are Huh-7 cell clones with subgenomic replicons, keeping constant HCV RNA and protein levels over years, even in the absence of selective pressure (65). Therefore, we analyzed two different types of replicon cells to evaluate the data obtained with the full-length genome (Fig. 1A): (i) a Huh-7 cell clone designated 11-1, harboring a monocistronic replicon resembling closely the translational properties of a full-length genome (24) and (ii) a Huh-7 cell clone designated 9-13 (51, 53) with a bicistronic replicon representing the most efficient and most often used system to study persistent HCV replication. Cells harboring the respective replicons were seeded in aliquots and analyzed in the same way as the full-length genome at different time points after seeding. Despite the different architecture of the replicons and the assay format (transient replication versus stable replicon cell clones), we obtained very similar results (Table 1), with more than 10,000 NS4B or NS5B copies per negative-strand RNA molecule. This result indicated that the synthesis of a massive excess of NS-proteins over RNA seems to be an intrinsic property of HCV translation and replication in Huh-7 cells. Since the half-lives of the NS proteins (11 to 16 h) (65) were shown to be comparable to the half-life of RNA (about 11 h) (62) in replicon cells and HCV RNA and proteins are kept on similar steady-state levels over years of continuous passaging, HCV RNA and protein synthesis should also be quite constant. Therefore, based on the data shown in Table 1, each positive-strand RNA template is translated numerous times giving rise to 1,000 to 10,000 polyprotein copies. Another important conclusion was that the stoichiometry of HCV protein to RNA in replicon cell clones closely resembled the one found with full-length genomes. Therefore, replicon cell clones were an appropriate tool for further analyses.
Isolation of active replicase complexes from Huh-7 cell harboring subgenomic replicons.
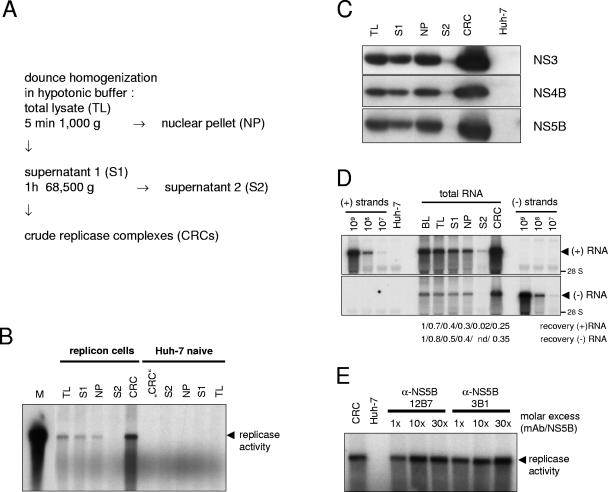
We wondered whether the massive surplus of nonstructural proteins really is directly involved in RNA synthesis or may serve some other function. To differentiate between these two possibilities, we isolated CRCs from replicon cells that could be further analyzed in vitro for their protein and RNA content. The method for the preparation of CRCs by pelleting heavy-membrane fractions from hypotonic cell lysates has been described for HCV and related viruses (5, 19, 36, 45, 73) and is shown schematically in Fig. 2A. To assay for in vitro replicase activity, cell lysates were incubated with radiolabeled nucleotides in the presence of actinomycin D and in the absence of exogenous template RNA. Reaction products were further analyzed by denaturing agarose gel electrophoresis (Fig. 2B). The dominant product of in vitro replication was a single band that corresponded in size to the full-length replicon RNA (arrowhead). HCV replicase activity was already detectable in the total hypotonic lysate of replicon cells but was enriched in CRCs that were obtained by pelleting the membranous material contained in supernatant 1. The resulting supernatant 2 did not contain detectable replicase activity. The distribution of the nonstructural proteins in different fractions of the CRC preparation is shown in Fig. 2C. Similar portions of NS3, NS4B, and NS5B were recovered in S1 and concentrated in parallel to the replicase activity in the CRC fraction, leaving only trace amounts in S2.
FIG. 2.
Preparation and characterization of CRCs from HCV replicon cells. (A) Schematic diagram of the CRC preparation protocol. (B) Analysis of in vitro replicase activity in total lysates (TL) and different subcellular fractions of replicon cells (left half) and naive Huh-7 cells (right half). In vitro replicase activity was determined in 4 μl of each fraction, and reaction products were analyzed by denaturing glyoxal-gel electrophoresis, followed by autoradiography of the dried gel. A radioactively labeled in vitro transcript identical in size to the replicon was loaded as a marker (M). The major reaction product of the in vitro replicase assay is indicated by an arrowhead. (C) Detection of NS3, NS4B, and NS5B in different fractions of the CRC preparation. The volume of the NP fraction was adjusted to the volume of S1 and 10 μl of each fraction were analyzed by immunoblot with a polyclonal antiserum raised against HCV NS3 (upper panel) or NS4B (middle panel) or monoclonal antibodies specific for NS5B (lower panel) and compared to 10 μl of “CRC” fraction from naive Huh-7 cells (Huh-7). (D) Fate of viral positive and negative strands during hypotonic lysis and CRC preparation. Total RNA was prepared from 50 μl of a replicon cell suspension before lysis (BL) and from the same volumes of TL, S1, NP (adjusted to the volume of S1), S2, and CRC and subjected to Northern hybridization analysis with the same controls and probes to detect HCV positive- (upper panel) and negative-strand RNA (lower panel) as in Fig. 1B. To calculate the recovery rate samples were analyzed by phosphorimaging and correlated with the value obtained before cell lysis (BL). The data of the CRC fraction were corrected for the difference in total volume. (E) Effect of NS5B-specific monoclonal antibodies on in vitro replicase activity. Two μl of a standard CRC preparation containing 40 ng of NS5B were preincubated 5 min on ice with 0.1, 1, or 3 μg of purified monoclonal antibodies as indicated in the top, resulting in a 1×, 10×, or 30× molar excess or incubated in the absence of antibodies (CRC) and analyzed for in vitro replicase activity. The same amount of “CRC” fraction from naive Huh-7 cells was used as a negative control (Huh-7). For further details refer to the text.
To exclude that the CRC fraction contains only a minor subpopulation of HCV replication complexes that might not be representative, we followed the fate of viral positive- and negative-strand RNA during CRC preparation (Fig. 2D). We found that 35% of the negative- and 25% of the positive-strand RNA present in the replicon cells before lysis were recovered in the CRC fraction. The remainder was either associated with the nuclear pellet (40% of negative-strand, 30% of positive-strand RNA) or was destroyed during cell lysis (20% of negative-strand, 30% of positive-strand RNA) or during centrifugation of CRCs (15% of negative-strand and positive-strand RNA, respectively); only traces of RNA were retained in S2. In consequence, about half of the positive-strand RNA and 25% of the negative-strand RNA were degraded, most likely by the action of cellular nucleases that were liberated during cell lysis. This fraction might include damaged replicase complexes and positive-strand HCV RNAs that were not integrated into the replicase complex but engaged in some other processes such as RNA translation. A varying amount of NS proteins, HCV RNA, and replicase activity always stayed associated with the nuclear pellet and could not be recovered even by vigorous dounce homogenization, most likely due to the accumulation of replication complexes in the perinuclear region (30) and due to the mild extraction conditions omitting any detergents.
HCV replicase complexes act selectively on endogenous templates in vitro and are blocked by an inhibitor of NS3 helicase (36), indicating that replicase activity is distinct from RNA synthesis exerted by isolated NS5B polymerase. In addition, RNA synthesis activity of the native replicase complex was inhibited by 3′-deoxy-CTP, a chain-terminating nucleotide analog, but not inhibited by non-nucleoside NS5B polymerase inhibitors interfering with the activity of purified NS5B (56; data not shown). To gain further insights into the properties of the HCV replication complex, we analyzed the impact of monoclonal antibody NS5B 12B7 on replicase activity in CRCs. It has previously been shown that this antibody inhibits HCV polymerase activity by binding to a conformational epitope in NS5B (58), in contrast to antibody NS5B 3B1, that did not interfere with polymerase activity. We found that none of the antibodies affected replicase activity in CRCs in an up to 30-fold molar excess (Fig. 2E), further substantiating that in vitro replicase activity exhibits features distinct from isolated HCV polymerase.
Taken together, CRCs most likely contained a representative fraction of HCV replication complexes in replicon cell clones and were therefore appropriate to study the stoichiometry of RNA to NS proteins required for RNA replication. Since clone 9-13, which harbors a bicistronic replicon, was the most efficient source for the preparation of active replicase complexes, we chose this cell clone for further experiments.
In vitro replicase activity and viral RNA are fully resistant to nuclease and protease treatment.
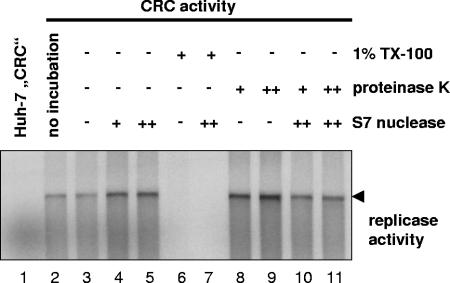
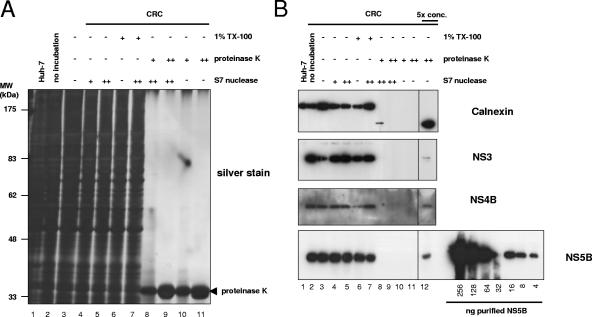
It has been shown previously that in vitro replicase activity is resistant to nuclease and protease treatment (2, 5, 19, 57). We exploited these results to evaluate which fraction of viral RNA and proteins is resistant to nuclease and protease, respectively, in order to determine the stoichiometry of the viral components of the HCV replication complex. Therefore, we treated CRCs with high concentrations of proteinase K (0.8 or 8 mg/ml) and/or S7 nuclease (200 or 2,000 U/ml), stopped the reaction by adding PMSF or EGTA, respectively, and analyzed an aliquot of the pretreated CRCs for in vitro replicase activity. As shown in Fig. 3, in vitro replicase activity was not affected by pretreatment with either S7 nuclease (lanes 4 and 5) or proteinase K (lanes 8 and 9) alone or in combination (lanes 10 and 11). Protease and nuclease resistance was not restricted to replicase activity contained in the CRC fraction but was also found for replicase activity in total cell lysate, the nuclear pellet, and supernatant 1 (data not shown), indicating that the replication complexes in the CRC fraction were not a selected subpopulation with distinct features. HCV replicase activity was only abolished by addition of 1% Triton X-100, in the absence (lane 6) or presence (lane 7) of additional nuclease, indicating that the resistance of HCV replicase to proteases and nucleases is mediated by detergent-sensitive structures. The full protease and nuclease resistance of in vitro replicase activity enabled us to analyze which portion of viral RNA and proteins were not affected by protease and nuclease and therefore were necessary and sufficient for RNA synthesis. To address this question, aliquots of the protease- and/or nuclease-treated CRCs were analyzed by Northern hybridization and immunoblotting to determine the ratio of HCV RNA and NS proteins involved in replication.
FIG. 3.
HCV replicase activity is completely resistant to protease and nuclease treatment. A total of 50 μl of CRCs prepared from replicon cell clone 9-13 was incubated for 60 min at 25°C in the presence of 1% Triton X-100 and/or 0.8 (+) or 8 (++) mg of proteinase K/ml and/or 0.2 (+) or 2 (++) U of S7 nuclease/μl, respectively, as indicated above each lane. After termination of the proteinase K and S7 nuclease digest by the addition of 1.4 mM PMSF and/or 2.75 mM EGTA, respectively, equal amounts of each sample were analyzed for in vitro replicase activity. Reaction products were separated by denaturing glyoxal agarose gel electrophoresis and autoradiography. Lane 1 and 2 represent control reactions with CRCs from naive Huh-7 or replicon cells in the absence of any preincubation. CRCs in lane 3 were mock incubated for 60 min at 25°C prior to the in vitro replicase assay.
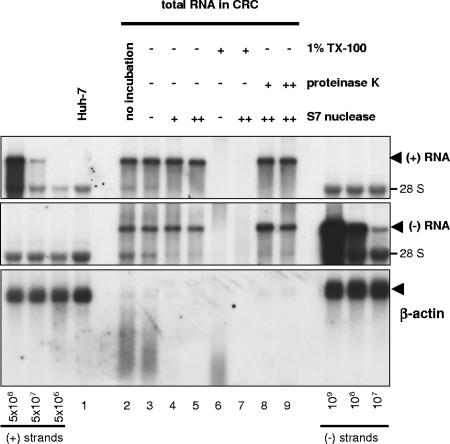
We first focused on the effect of S7 nuclease treatment on the fate of viral and cellular RNA (Fig. 4). Viral positive- and negative-strand RNAs were fully resistant to nuclease in CRCs, as shown by Northern blot analysis (Fig. 4, top and middle panels, compare lane 3 with lanes 4 and 5). The efficiency of the digest was indicated by the marked reduction of 28S rRNA in the S7 nuclease-treated samples (Fig. 4, top and middle panel, lanes 4, 5, 8, and 9). Protection from nucleases seemed not to be mediated by proteins, since further addition of proteinase K had no effect on RNA stability (lanes 8 and 9). In contrast, the addition of Triton X-100 resulted in complete degradation of both viral RNA species, even in the absence of exogenous nuclease (lanes 6 and 7), indicating that the RNA was protected by membranes rather than proteins and that the loss of replicase activity upon detergent treatment (Fig. 3) was primarily due to the destruction of template RNA by endogenous nucleases and S7 nuclease. In contrast to the viral RNAs, the cellular mRNA, exemplified by β-actin, was absent in CRCs because it was not attached to membranes or almost completely destroyed by the action of endogenous nucleases that were liberated during CRC-preparation (Fig. 4, lowest panel, lanes 2 and 3) and the remaining traces were fully accessible to S7 nuclease (lanes 4 and 5).
FIG. 4.
Quantification of nuclease-resistant HCV positive- and negative-strand RNA and β-actin mRNA in CRCs. Total RNA equivalent to 5 μl of CRCs treated with 1% Triton X-100, 0.8 (+) or 8 (++) mg of proteinase K/ml and/or 0.2 (+) or 2 (++) U of S7 nuclease/μl for 60 min at 25°C or from mock-treated CRCs, as indicated on top, was subjected to Northern hybridization analysis with a negative-strand riboprobe to detect viral positive-strand RNA (top panel), a positive-strand riboprobe for HCV negative-strand detection (middle panel), and a riboprobe specific for the detection of cellular β-actin mRNA. The positions of viral positive- and negative-strand RNA and β-actin are indicated by arrowheads at the right. For quantification, in vitro transcribed replicons corresponding to known amounts of viral positive- and negative-strand RNA were mixed with 2 μg of total cellular RNA from naive Huh-7 cells and loaded as indicated at the bottom of the figure. We used 4 μg of total RNA from naive Huh-7 cells as a negative control (Huh-7, lane 1). We quantified the viral RNAs by phosphorimaging and found ca. 3 × 106 negative-strand and 3 × 107 positive-strand RNAs per μl of CRCs in this particular experiment.
Taken together, our data indicate that the viral positive- and negative-strand RNA in the replication complex is completely protected from nucleases and that this protection is mediated by detergent sensitive membrane structures rather than by proteins.
Only a minor portion of the HCV NS proteins is resistant to protease treatment.
Since in vitro replicase activity was completely resistant to protease and nuclease treatment, the ratio of HCV NS proteins to RNA actively involved in RNA synthesis was determined by the number of nuclease resistant RNA molecules compared to protease-resistant protein molecules. To measure the fraction of viral NS proteins resistant to protease, we analyzed aliquots of the differently treated CRC samples for the impact of proteinase K incubation by Western blotting (Fig. 5B). Even with the lower protease concentrations, we found a massive degradation of almost all cellular proteins to nearly undetectable amounts (Fig. 5A, lanes 8 to 11), whereas detergent and nuclease treatment had, as expected, no significant effect (lanes 4 to 7). As an example for a cellular protein we chose calnexin for a closer analysis by Western blotting with an antiserum directed against its N-terminal part, which is located in the lumen of the endoplasmic reticulum and therefore should be protected from proteases in an intact ER structure, whereas the carboxy terminus was expected to be protease sensitive in a cell lysate (57). As shown in Fig. 5B (top panel), no full-length calnexin was detectable after protease treatment, and only traces of a C-terminally truncated fragment were visible with the lower amount of protease (lanes 8 to 11). Only after fivefold concentration of the sample were we able to obtain a clear signal (lane 12). The absence of full-length calnexin even in the concentrated samples indicated that the amount of proteinase K we used was sufficient for a complete digest. Since the majority of the ER-luminal N-terminal calnexin fragments were also accessible to protease (Fig. 5B, top panel, signal strength in lanes 1 to 7 compared to lanes 8 to 11), it seemed that most of the regular ER structures were not intact after CRC preparation and protease digest. The HCV replication complex therefore appears to represent a more rigid structure due to the complete protease and nuclease resistance of in vitro replicase activity. However, when we analyzed the viral nonstructural proteins 3, 4B, and 5B after protease digest (Fig. 5B, second, third, and lowest panels, respectively, lanes 8 to 12), we found detectable amounts again only after fivefold concentration of the samples (lane 12). The absence of lower-molecular-weight products indicated the completeness of digestion. Based on densitometric analysis and including the dilution factor, we calculated that roughly 2.5% of NS5B was resistant to proteinase K digest but accounted for the full replicase activity (Fig. 3), suggesting that the majority of viral nonstructural proteins was not directly involved in RNA synthesis at a given time point. The portions of protease-resistant NS3 and NS4B were similar, indicating a 1:1 stoichiometry of the NS proteins in the replication complex. A serial dilution of purified NS5B applied on the same Western blot allowed us to quantify the number of NS5B molecules resistant to protease digest (Fig. 5B, lowest panel), and we detected ca. 5 × 109 molecules NS5B per μl CRCs in this experiment. Compared to 3 × 106 negative strands and 3 × 107 positive strands (Fig. 4), the surprising result was that, although only 2.5% of the NS5B molecules were engaged in replicase activity at a given time, we still found a 1,600-fold excess of NS5B compared to negative-strand RNA and a >100-fold excess compared to positive-strand RNA in CRCs.
FIG. 5.
Effect of proteinase K digest on cellular proteins and quantification of protease-resistant HCV nonstructural proteins in CRCs. Equal amounts of CRCs prepared from replicon cell clone 9-13 were incubated for 60 min at 25°C in the presence or absence of 1% Triton X-100 and/or 0.8 (+) or 8 (++) mg of proteinase K/ml and/or 0.2 (+) or 2 (++) U of S7 nuclease/μl, respectively, as indicated above each lane. The reaction was stopped by the addition of 1.4 mM PMSF and 2.75 mM EGTA and boiled in sample buffer, and total protein equivalent to 2 μl of the CRCs was subjected to SDS-10% PAGE. In the fivefold-concentrated samples, the proteins were trichloroacetic acid precipitated, and the equivalent of 10 μl of CRCs was loaded. Proteins were either visualized by silver staining (A) or subjected to immunoblot analysis (B) with monoclonal antibodies specific for the ER luminal part of calnexin (upper panel), a polyclonal antiserum raised against HCV NS3 or NS4B (middle two panels), and monoclonal antibodies specific for HCV NS5B (bottom panel), as indicated at the right. The concentrated fractions (lane 12) are shown from identical expositions of the same blot. A serial dilution of purified NS5B was loaded in parallel for quantification as depicted at the bottom of the figure.
The HCV replication complex contains multiple copies of the NS proteins.
The data obtained from the experiment shown in Fig. 3 to 5 (experiment 1) and from an additional, independent experiment are summarized in Table 2. Both sets of data revealed very similar results. Viral positive- and negative-strand RNA was entirely resistant to nucleases in preparations of replication complexes compared to <3% of NS5B being protease resistant. The ratio of positive to negative-strand RNA varied and might be dependent on the physiological state of the cells at the time of harvest. Negative-strand RNA is the most limited component in HCV RNA replication and therefore the best indicator for the total number of active replication complexes. Given the data presented in Table 1 and assuming that each active replicase complex contains per definition at least one copy of negative-strand RNA, there were on average fewer than a hundred active replication complexes per replicon cell but more than a million polymerase molecules. After biochemical preparation of replicase complexes and excessive protease digest we still found more than 1,000 NS5B molecules per negative-strand RNA (Table 2) and similar amounts of NS4B and NS3, indicating that a huge excess of NS proteins is required to build up the viral replication complex.
TABLE 2.
Portions and ratios of nuclease-resistant positive-strand, negative-strand, and protease-resistant NS5B in CRCs
| Parametera | Expt 1 | Expt 2 |
|---|---|---|
| % Protease-resistant NS5B | 2.5 | 2.2 |
| % Nuclease-resistant positive-strand RNA | ∼100 | ∼100 |
| % Nuclease-resistant negative-strand RNA | ∼100 | ∼100 |
| Positive-strand/negative-strand RNA ratio | 12 ± 3 | 6 ± 2 |
| NS5B/negative-strand RNA ratio | 1,200 ± 400 | 700 ± 400 |
| NS5B/positive-strand RNA ratio | 100 ± 20 | 110 ± 50 |
Ratios are given as the mean and standard deviation of four nuclease-treated samples.
DISCUSSION
In the present study we analyzed the stoichiometry of HCV RNA and proteins in cells with ongoing HCV replication and found a massive excess of structural and nonstructural protein molecules over RNA, indicating that each positive-strand RNA molecule is excessively translated before a replication complex is formed and RNA synthesis is initiated. In agreement with the polyprotein nature of the major HCV ORF, we found all analyzed cleavage products in similar amounts. A ca. 1,000- to 10,000-fold excess of viral proteins to positive-strand RNA as observed in our study might fit well to the requirements for particle formation, since virions of other enveloped positive-strand RNA viruses such as alphaviruses, tick-borne encephalitis virus, or Dengue virus usually contain several hundred copies of structural proteins per particle (22, 28, 44, 61).
We wondered whether a similar excess of nonstructural proteins was required for RNA replication and addressed this question by studying crude replicase complexes isolated from cells harboring persistent HCV replicons. It has been shown previously that in vitro replicase activity is resistant to protease and nuclease treatment in CRCs (2, 5, 19) and that in digitonin-solubilized replicon cells only a minor portion of the HCV NS proteins is protease resistant and therefore seems to be involved in replication (57). Our data here are in good agreement with these earlier results; however, although we find as well that <5% of the NS proteins in CRCs are protease resistant and account for full replication activity in vitro, this still results in a ca. 1,000:1 stoichiometry of NS proteins to active replicase complexes, as estimated by the number of negative-strand RNA molecules.
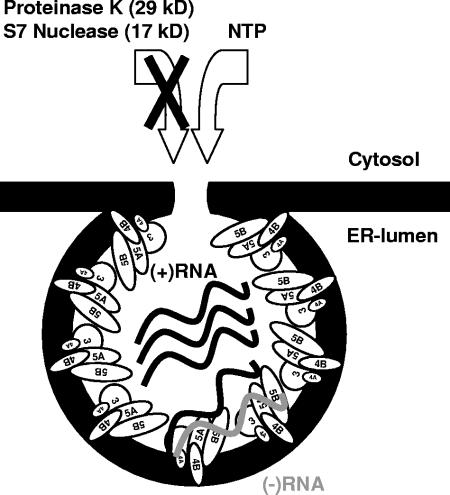
Based on our data and on previously published electron microscopic studies (18, 30), we propose a tentative model of the HCV replication complex (Fig. 6). This model is based on the well-supported assumption that polymerase activity as measured by CRC assays is a component of the replication complex and not of isolated enzyme. Multiple copies of HCV NS protein complexes encompassing NS3 to NS5B build up a vesicular membrane structure, which mediates the protection against nucleases and proteases and may hide double-strand RNA from detection by the host cells innate immune system. The existence of HCV NS protein complexes rather than individual proteins is indicated by similar fractions of NS3, NS4B, and NS5B being resistant to protease and seems plausible due to the numerous interactions within the NS proteins that have thus far been described (7, 16). Each vesicle should have a connection to the cytoplasm, allowing the constant supply with nucleotides for RNA synthesis, but preventing the access of molecules larger than 16 kDa, e.g., S7 nuclease and proteinase K. A number of these vesicles accumulate at distinct sites in the cytoplasm and form the membranous web, which was shown to be the site of RNA replication (30). Within every vesicle that contains an active replicase complex we find at least one negative-strand RNA, several positive-strand RNAs and up to 1,000 copies of each of the NS proteins. The precise stoichiometry of RNA to protein in the replicase complex is hard to tell, since a number of replication complexes might await initiation of negative-strand synthesis at a given time and therefore contain only NS proteins and positive-strand RNA. If this variant is frequent, ca. 100 to 200 copies of the nonstructural proteins per replicase complex would be a more realistic estimate. On the other hand, several replicase complexes might include more than one molecule of negative-strand RNA, and therefore the number of NS protein molecules per active replicase complex could even be higher. Finally, we cannot exclude the existence of vesicles without viral RNA, which still might render the included NS proteins protease resistant, because the induction of membrane alterations is an intrinsic property of NS4B (18), and we do not know whether the presence of RNA is necessary to induce protease resistance of the NS proteins.
FIG. 6.
Schematic model of the HCV replication complex. HCV NS proteins are indicated by ellipses; black and gray wavy lines represent viral positive- and negative-strand RNAs, respectively. Individual NS proteins and RNA are not drawn to scale. For closer explanations refer to the text.
Our picture of the HCV replication complex is very similar to a model suggested by Miyanari et al. (57); however, the data presented here provide the first experimental evidence that each HCV replication complex is composed of multiple copies of the viral nonstructural proteins. This resembles closely results obtained for brome mosaic virus (69). It has been shown that 400 copies of protein 1a form a spherular structure connected to the cytoplasm and containing viral RNA, together with a few copies of the viral polymerase 2a. In the case of HCV the majority of protease-resistant NS protein complexes might as well be required to build up the vesicular structure, whereas only a few complexes are required for polymerase activity. This calculation is based on the kinetics of RNA and protein synthesis in replicon cells: the intracellular levels of viral RNA and proteins remain relatively constant over years of continuous passaging (62), representing a steady-state situation at any given time point. Since the half-lives of viral NS proteins and viral positive-strand RNA in replicon cells have been shown to be 11 to 16 h (62, 65), the rate of newly synthesized RNA and protein per day should roughly be in the range we find in the steady-state situation (Table 1). Based on this knowledge and on our data, we estimate that only about 1,000 positive-strand RNA molecules are synthesized per day per cell by ca. 100 replicase complexes but more than 1,000,000 copies of NS proteins. In consequence, each newly synthesized positive strand has to be excessively translated to yield the ascertained surplus of proteins, whereas RNA synthesis is a rather rare event, which most likely is achieved only by a few NS protein complexes. Given our calculation, <0.1% of all NS5B molecules are required to be enzymatically active.
Many positive-strand RNA viruses have evolved strategies to regulate the amounts of active polymerase, either by expression from an independent cistron, like BMV (69); by the expression of a polyprotein containing rarely suppressed stop codons, like alphaviruses (47); or by producing stable precursor intermediates lacking polymerase activity, like poliovirus 3CD (10). In the case of HCV, polymerase activity could be regulated by different conformations of the NS protein complex, depending on its function. It has been shown that a carboxy-terminal region of NS5B, encompassing aa 545 to 562 inhibits polymerase activity (1) and that only a small fraction of purified NS5B containing this region is enzymatically active (14). Crystal structure analysis revealed that this carboxy-terminal domain protrudes into the RNA-binding cavity of NS5B and interferes with template binding (46). Deletion of this region increases polymerase activity, as well as RNA binding of NS5B (46); therefore, it is tempting to speculate that the majority of NS5B is inactive after translation by refolding of the carboxy terminus into the RNA binding cleft, whereas only those few molecules that stay bound to the viral RNA keep the template binding site in an open conformation and retain enzymatic activity. The ability to self-inactivate viral polymerase not required for RNA synthesis might represent an alternate strategy to deal with the stoichiometric constraints of a polyprotein.
In contrast, the majority of NS protein complexes not directly involved in RNA synthesis may be required, e.g., as a structural component of the membrane vesicle. The viral NS protein primarily involved in this process is NS4B, since it is able to induce vesicular structures even in the absence of other HCV proteins (18). The different roles of nonstructural protein complexes might be regulated by the association with individual host cell factors, such as hVAP-A, a cellular protein that has been shown to interact with NS5A and NS5B (20, 75). Since this protein is involved in intracellular vesicle trafficking, it seems to be a good candidate for a cofactor of membrane structure rearrangements and has already been suggested to be involved in HCV replication complex formation (2, 29). In addition, other cellular factors might be required for RNA synthesis. A detailed biochemical analysis of the protease resistant NS protein fraction might reveal some of the cellular proteins that are directly involved in the formation of the replication complex and in RNA synthesis.
The functions of the protease sensitive portion of the NS proteins, encompassing >95%, remain obscure. Since our analysis presents a snapshot at a given time point and we have no precise data on the dynamics of replication complex formation and degeneration, parts of these proteins might be in the process of vesicle generation and disintegration. The remainder could simply be a by-product of structural protein synthesis, which might be required in excess amounts for virus production. Miyanari et al. (57) suggested potential roles of the NS proteins in virion formation or indirect modulation of viral replication by interaction with host cell proteins. Along the same line, the NS3/4A moiety might be required to block the IRF-3 induced pathway of the host cells' innate immune system, which has recently been shown to play a critical role in HCV replication (23).
In the present study we have shown that during the replication cycle of HCV a massive excess of nonstructural proteins is produced due to extensive translation. An interesting but yet unanswered question is how the transition of translation to replication is regulated. A number of proteins have been shown to bind to the HCV 5′ NTR, which might be involved in this process (3, 4, 35, 40, 55); alternatively, the switch could rely on concentration-dependent inhibition of translation by an HCV protein. Interestingly, we found no significant differences in the ratio of HCV RNA and protein between an authentic HCV genome, a monocistronic replicon, and a bicistronic replicon (Table 1), indicating that the nature of the IRES-element directing translation of the NS proteins is not important for this stoichiometry. After translation, replication complex formation and RNA synthesis, the progeny RNA has to get back into the cytoplasm to enter a new round of translation/replication or packaging into particles. We currently do not know whether this process includes an active transport, as in the replication cycle of double-stranded RNA reoviruses (67), or if the progeny RNA accumulates in the replication complex and is released by disintegration of the vesicle.
An obvious limitation of our analysis is the fact that no viral particles are produced by the chosen replication systems (64). Since we do not know how many positive-strand RNA molecules would end up in virions and whether viral genomes undergo translation prior to packaging, it is very hard to predict the consequences of simultaneous RNA replication and particle morphogenesis on the ratio of RNA to proteins in infected cells. While the manuscript was in preparation, three independent studies demonstrated the production of infectious HCV particles in cell culture with the NS proteins of the JFH-1 HCV isolate (49, 77, 81). It will be interesting to evaluate our findings in this system covering the whole life cycle of HCV.
Acknowledgments
We thank Darius Moradpour for the monoclonal NS5B antibodies, Joanne Tomassini for initial help with the establishment of CRC preparation and in vitro replicase assays, and Thomas Pietschmann for critically reading the manuscript.
This study was funded in part by the Deutsche Forschungsgemeinschaft (SFB 638 Teilprojekt A5) and within the National Genome Research Network by the German Ministry for Research and Education.
REFERENCES
- 1.Adachi, T., H. Ago, N. Habuka, K. Okuda, M. Komatsu, S. Ikeda, and K. Yatsunami. 2002. The essential role of C-terminal residues in regulating the activity of hepatitis C virus RNA-dependent RNA polymerase. Biochim. Biophys. Acta 1601:38-48. [DOI] [PubMed] [Google Scholar]
- 2.Aizaki, H., K. J. Lee, V. M. Sung, H. Ishiko, and M. M. Lai. 2004. Characterization of the hepatitis C virus RNA replication complex associated with lipid rafts. Virology 324:450-461. [DOI] [PubMed] [Google Scholar]
- 3.Ali, N., and A. Siddiqui. 1995. Interaction of polypyrimidine tract-binding protein with the 5′ noncoding region of the hepatitis C virus RNA genome and its functional requirement in internal initiation of translation. J. Virol. 69:6367-6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ali, N., and A. Siddiqui. 1997. The La antigen binds 5′ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc. Natl. Acad. Sci. USA 94:2249-2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ali, N., K. D. Tardif, and A. Siddiqui. 2002. Cell-free replication of the hepatitis C virus subgenomic replicon. J. Virol. 76:12001-12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartenschlager, R., L. Ahlborn-Laake, J. Mous, and H. Jacobsen. 1993. Nonstructural protein 3 of the hepatitis C virus encodes a serine-type proteinase required for cleavage at the NS3/4 and NS4/5 junctions. J. Virol. 67: 3835-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartenschlager, R., M. Frese, and T. Pietschmann. 2004. Novel insights into hepatitis C virus replication and persistence. Adv. Virus Res. 63:71-180. [DOI] [PubMed] [Google Scholar]
- 8.Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. [DOI] [PubMed] [Google Scholar]
- 9.Bartenschlager, R., V. Lohmann, T. Wilkinson, and J. O. Koch. 1995. Complex formation between the NS3 serine-type proteinase of the hepatitis C virus and NS4A and its importance for polyprotein maturation. J. Virol. 69:7519-7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bedard, K. M., and B. L. Semler. 2004. Regulation of picornavirus gene expression. Microbes. Infect. 6:702-713. [DOI] [PubMed] [Google Scholar]
- 11.Behrens, S. E., L. Tomei, and R. De Francesco. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15:12-22. [PMC free article] [PubMed] [Google Scholar]
- 12.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 13.Bukh, J., T. Pietschmann, V. Lohmann, N. Krieger, K. Faulk, R. E. Engle, S. Govindarajan, M. Shapiro, M. St Claire, and R. Bartenschlager. 2002. Mutations that permit efficient replication of hepatitis C virus RNA in Huh-7 cells prevent productive replication in chimpanzees. Proc. Natl. Acad. Sci. USA 99:14416-14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carroll, S. S., V. Sardana, Z. Yang, A. R. Jacobs, C. Mizenko, D. Hall, L. Hill, J. Zugay-Murphy, and L. C. Kuo. 2000. Only a small fraction of purified hepatitis C RNA-dependent RNA polymerase is catalytically competent: implications for viral replication and in vitro assays. Biochemistry 39: 8243-8249. [DOI] [PubMed] [Google Scholar]
- 15.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 16.Dimitrova, M., I. Imbert, M. P. Kieny, and C. Schuster. 2003. Protein-protein interactions between hepatitis C virus nonstructural proteins. J. Virol. 77:5401-5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dubuisson, J., F. Penin, and D. Moradpour. 2002. Interaction of hepatitis C virus proteins with host cell membranes and lipids. Trends Cell Biol. 12: 517-523. [DOI] [PubMed] [Google Scholar]
- 18.Egger, D., B. Wolk, R. Gosert, L. Bianchi, H. E. Blum, D. Moradpour, and K. Bienz. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El Hage, N., and G. Luo. 2003. Replication of hepatitis C virus RNA occurs in a membrane-bound replication complex containing nonstructural viral proteins and RNA. J. Gen. Virol. 84:2761-2769. [DOI] [PubMed] [Google Scholar]
- 20.Evans, M. J., C. M. Rice, and S. P. Goff. 2004. Phosphorylation of hepatitis C virus nonstructural protein 5A modulates its protein interactions and viral RNA replication. Proc. Natl. Acad. Sci. USA 101:13038-13043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Failla, C., L. Tomei, and R. De Francesco. 1995. An amino-terminal domain of the hepatitis C virus NS3 protease is essential for interaction with NS4A. J. Virol. 69:1769-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferlenghi, I., M. Clarke, T. Ruttan, S. L. Allison, J. Schalich, F. X. Heinz, S. C. Harrison, F. A. Rey, and S. D. Fuller. 2001. Molecular organization of a recombinant subviral particle from tick-borne encephalitis virus. Mol. Cell 7:593-602. [DOI] [PubMed] [Google Scholar]
- 23.Foy, E., K. Li, C. Wang, R. Sumpter, Jr., M. Ikeda, S. M. Lemon, and M. Gale, Jr. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 300:1145-1148. [DOI] [PubMed] [Google Scholar]
- 24.Frese, M., V. Schwarzle, K. Barth, N. Krieger, V. Lohmann, S. Mihm, O. Haller, and R. Bartenschlager. 2002. Interferon-gamma inhibits replication of subgenomic and genomic hepatitis C virus RNAs. Hepatology 35:694-703. [DOI] [PubMed] [Google Scholar]
- 25.Friebe, P., and R. Bartenschlager. 2002. Genetic analysis of sequences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J. Virol. 76:5326-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friebe, P., J. Boudet, J. P. Simorre, and R. Bartenschlager. 2005. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J. Virol. 79:380-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friebe, P., V. Lohmann, N. Krieger, and R. Bartenschlager. 2001. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 75:12047-12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fuller, S. D. 1987. The T=4 envelope of Sindbis virus is organized by interactions with a complementary T=3 capsid. Cell 48:923-934. [DOI] [PubMed] [Google Scholar]
- 29.Gao, L., H. Aizaki, J. W. He, and M. M. Lai. 2004. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J. Virol. 78: 3480-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gosert, R., D. Egger, V. Lohmann, R. Bartenschlager, H. E. Blum, K. Bienz, and D. Moradpour. 2003. Identification of the hepatitis C virus RNA replication complex in huh-7 cells harboring subgenomic replicons. J. Virol. 77:5487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grakoui, A., D. W. McCourt, C. Wychowski, S. M. Feinstone, and C. M. Rice. 1993. A second hepatitis C virus-encoded proteinase. Proc. Natl. Acad. Sci. USA 90:10583-10587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grakoui, A., D. W. McCourt, C. Wychowski, S. M. Feinstone, and C. M. Rice. 1993. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J. Virol. 67:2832-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Griffin, S. D., L. P. Beales, D. S. Clarke, O. Worsfold, S. D. Evans, J. Jaeger, M. P. Harris, and D. J. Rowlands. 2003. The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, amantadine. FEBS Lett. 535:34-38. [DOI] [PubMed] [Google Scholar]
- 34.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hahm, B., Y. K. Kim, J. H. Kim, T. Y. Kim, and S. K. Jang. 1998. Heterogeneous nuclear ribonucleoprotein L interacts with the 3′ border of the internal ribosomal entry site of hepatitis C virus. J. Virol. 72:8782-8788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hardy, R. W., J. Marcotrigiano, K. J. Blight, J. E. Majors, and C. M. Rice. 2003. Hepatitis C virus RNA synthesis in a cell-free system isolated from replicon-containing hepatoma cells. J. Virol. 77:2029-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hijikata, M., N. Kato, Y. Ootsuyama, M. Nakagawa, and K. Shimotohno. 1991. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc. Natl. Acad. Sci. USA 88: 5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hijikata, M., H. Mizushima, T. Akagi, S. Mori, N. Kakiuchi, N. Kato, T. Tanaka, K. Kimura, and K. Shimotohno. 1993. Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J. Virol. 67:4665-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim, D. W., Y. Gwack, J. H. Han, and J. Choe. 1995. C-terminal domain of the hepatitis C virus NS3 protein contains an RNA helicase activity. Biochem. Biophys. Res. Commun. 215:160-166. [DOI] [PubMed] [Google Scholar]
- 40.Kim, J. H., K. Y. Paek, S. H. Ha, S. Cho, K. Choi, C. S. Kim, S. H. Ryu, and S. K. Jang. 2004. A cellular RNA-binding protein enhances internal ribosomal entry site-dependent translation through an interaction downstream of the hepatitis C virus polyprotein initiation codon. Mol. Cell. Biol. 24:7878-7890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koch, J. O., and R. Bartenschlager. 1999. Modulation of hepatitis C virus NS5A hyperphosphorylation by nonstructural proteins NS3, NS4A, and NS4B. J. Virol. 73:7138-7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuhn, R. J., W. Zhang, M. G. Rossmann, S. V. Pletnev, J. Corver, E. Lenches, C. T. Jones, S. Mukhopadhyay, P. R. Chipman, E. G. Strauss, T. S. Baker, and J. H. Strauss. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lai, V. C., S. Dempsey, J. Y. Lau, Z. Hong, and W. Zhong. 2003. In vitro RNA replication directed by replicase complexes isolated from the subgenomic replicon cells of hepatitis C virus. J. Virol. 77:2295-2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leveque, V. J., R. B. Johnson, S. Parsons, J. Ren, C. Xie, F. Zhang, and Q. M. Wang. 2003. Identification of a C-terminal regulatory motif in hepatitis C virus RNA-dependent RNA polymerase: structural and biochemical analysis. J. Virol. 77:9020-9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li, G. P., and C. M. Rice. 1989. Mutagenesis of the in-frame opal termination codon preceding nsP4 of Sindbis virus: studies of translational readthrough and its effect on virus replication. J. Virol. 63:1326-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin, C., J. A. Thomson, and C. M. Rice. 1995. A central region in the hepatitis C virus NS4A protein allows formation of an active NS3-NS4A serine proteinase complex in vivo and in vitro. J. Virol. 69:4373-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 50.Lohmann, V., S. Hoffmann, U. Herian, F. Penin, and R. Bartenschlager. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lohmann, V., F. Körner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lohmann, V., F. Korner, U. Herian, and R. Bartenschlager. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lohmann, V., F. Körner, J. O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 54.Lohmann, V., A. Roos, F. Korner, J. O. Koch, and R. Bartenschlager. 1998. Biochemical and kinetic analyses of NS5B RNA-dependent RNA polymerase of the hepatitis C virus. Virology 249:108-118. [DOI] [PubMed] [Google Scholar]
- 55.Lu, H., W. Li, W. S. Noble, D. Payan, and D. C. Anderson. 2004. Riboproteomics of the hepatitis C virus internal ribosomal entry site. J. Proteome Res. 3:949-957. [DOI] [PubMed] [Google Scholar]
- 56.Ma, H., V. Leveque, A. De Witte, W. Li, T. Hendricks, S. M. Clausen, N. Cammack, and K. Klumpp. 2005. Inhibition of native hepatitis C virus replicase by nucleotide and non-nucleoside inhibitors. Virology 332:8-15. [DOI] [PubMed] [Google Scholar]
- 57.Miyanari, Y., M. Hijikata, M. Yamaji, M. Hosaka, H. Takahashi, and K. Shimotohno. 2003. Hepatitis C virus nonstructural proteins in the probable membranous compartment function in viral genome replication. J. Biol. Chem. 278:50301-50308. [DOI] [PubMed] [Google Scholar]
- 58.Moradpour, D., E. Bieck, T. Hugle, W. Wels, J. Z. Wu, Z. Hong, H. E. Blum, and R. Bartenschlager. 2002. Functional properties of a monoclonal antibody inhibiting the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 277:593-601. [DOI] [PubMed] [Google Scholar]
- 59.Moradpour, D., R. Gosert, D. Egger, F. Penin, H. E. Blum, and K. Bienz. 2003. Membrane association of hepatitis C virus nonstructural proteins and identification of the membrane alteration that harbors the viral replication complex. Antivir. Res. 60:103-109. [DOI] [PubMed] [Google Scholar]
- 60.Nakabayashi, H., K. Taketa, K. Miyano, T. Yamane, and J. Sato. 1982. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 42:3858-3863. [PubMed] [Google Scholar]
- 61.Paredes, A. M., D. T. Brown, R. Rothnagel, W. Chiu, R. J. Schoepp, R. E. Johnston, and B. V. Prasad. 1993. Three-dimensional structure of a membrane-containing virus. Proc. Natl. Acad. Sci. USA 90:9095-9099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pause, A., G. Kukolj, M. Bailey, M. Brault, F. Do, T. Halmos, L. Lagace, R. Maurice, M. Marquis, G. McKercher, C. Pellerin, L. Pilote, D. Thibeault, and D. Lamarre. 2003. An NS3 serine protease inhibitor abrogates replication of subgenomic hepatitis C virus RNA. J. Biol. Chem. [DOI] [PubMed]
- 63.Pavlovic, D., D. C. Neville, O. Argaud, B. Blumberg, R. A. Dwek, W. B. Fischer, and N. Zitzmann. 2003. The hepatitis C virus p7 protein forms an ion channel that is inhibited by long-alkyl-chain iminosugar derivatives. Proc. Natl. Acad. Sci. USA 100:6104-6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pietschmann, T., V. Lohmann, A. Kaul, N. Krieger, G. Rinck, G. Rutter, D. Strand, and R. Bartenschlager. 2002. Persistent and transient replication of full-length hepatitis C virus genomes in cell culture. J. Virol. 76: 4008-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pietschmann, T., V. Lohmann, G. Rutter, K. Kurpanek, and R. Bartenschlager. 2001. Characterization of cell lines carrying self-replicating hepatitis C virus RNAs. J. Virol. 75:1252-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reed, K. E., and C. M. Rice. 2000. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr. Top. Microbiol. Immunol. 242:55-84. [DOI] [PubMed] [Google Scholar]
- 67.Reinisch, K. M., M. L. Nibert, and S. C. Harrison. 2000. Structure of the reovirus core at 3.6 Å resolution. Nature 404:960-967. [DOI] [PubMed] [Google Scholar]
- 68.Sambrook, J., Fritsch, E. F., and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 69.Schwartz, M., J. Chen, M. Janda, M. Sullivan, J. den Boon, and P. Ahlquist. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9:505-514. [DOI] [PubMed] [Google Scholar]
- 70.Shi, S. T., K. J. Lee, H. Aizaki, S. B. Hwang, and M. M. Lai. 2003. Hepatitis C virus RNA replication occurs on a detergent-resistant membrane that cofractionates with caveolin-2. J. Virol. 77:4160-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Suzich, J. A., J. K. Tamura, H. F. Palmer, P. Warrener, A. Grakoui, C. M. Rice, S. M. Feinstone, and M. S. Collett. 1993. Hepatitis C virus NS3 protein polynucleotide-stimulated nucleoside triphosphatase and comparison with the related pestivirus and flavivirus enzymes. J. Virol. 67:6152-6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tanji, Y., M. Hijikata, S. Satoh, T. Kaneko, and K. Shimotohno. 1995. Hepatitis C virus-encoded nonstructural protein NS4A has versatile functions in viral protein processing. J. Virol. 69:1575-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tomassini, J. E., E. Boots, L. Gan, P. Graham, V. Munshi, B. Wolanski, J. F. Fay, K. Getty, and R. LaFemina. 2003. An in vitro Flaviviridae replicase system capable of authentic RNA replication. Virology 313:274-285. [DOI] [PubMed] [Google Scholar]
- 74.Tsukiyama, K. K., N. Iizuka, M. Kohara, and A. Nomoto. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 66:1476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tu, H., L. Gao, S. T. Shi, D. R. Taylor, T. Yang, A. K. Mircheff, Y. M. Wen, A. E. Gorbalenya, S. B. Hwang, and M. C. Lai. 1999. Hepatitis C virus RNA polymerase and NS5A complex with a SNARE-like protein. Virology 263:30-41. [DOI] [PubMed] [Google Scholar]
- 76.van den Hoff, M. J., A. F. Moorman, and W. H. Lamers. 1992. Electroporation in “intracellular” buffer increases cell survival. Nucleic Acids Res. 20:2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Walewski, J. L., T. R. Keller, D. D. Stump, and A. D. Branch. 2001. Evidence for a new hepatitis C virus antigen encoded in an overlapping reading frame. RNA. 7:710-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xu, Z., J. Choi, T. S. Yen, W. Lu, A. Strohecker, S. Govindarajan, D. Chien, M. J. Selby, and J. Ou. 2001. Synthesis of a novel hepatitis C virus protein by ribosomal frameshift. EMBO J. 20:3840-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yanagi, M., C. M. St, S. U. Emerson, R. H. Purcell, and J. Bukh. 1999. In vivo analysis of the 3′ untranslated region of the hepatitis C virus after in vitro mutagenesis of an infectious cDNA clone. Proc. Natl. Acad. Sci. USA 96:2291-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102: 9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]