Abstract
The two major constituents of coronavirus virions are the membrane (M) and nucleocapsid (N) proteins. The M protein is anchored in the viral envelope by three transmembrane segments flanked by a short amino-terminal ectodomain and a large carboxy-terminal endodomain. The M endodomain interacts with the viral nucleocapsid, which consists of the positive-strand RNA genome helically encapsidated by N protein monomers. In previous work with the coronavirus mouse hepatitis virus (MHV), a highly defective M protein mutant, MΔ2, was constructed. This mutant contained a 2-amino-acid carboxy-terminal truncation of the M protein. Analysis of second-site revertants of MΔ2 revealed mutations in the carboxy-terminal region of the N protein that compensated for the defect in the M protein. To seek further genetic evidence corroborating this interaction, we generated a comprehensive set of clustered charged-to-alanine mutants in the carboxy-terminal domain 3 of N protein. One of these mutants, CCA4, had a highly defective phenotype similar to that of MΔ2. Transfer of the CCA4 mutation into a partially diploid MHV genome showed that CCA4 was a loss-of-function mutation rather than a dominant-negative mutation. Analysis of multiple second-site revertants of CCA4 revealed mutations in both the M protein and the N protein that could compensate for the original lesion in N. These data more precisely define the region of the N protein that interacts with the M protein. Further, we found that fusion of domain 3 of the N protein to the carboxy terminus of a heterologous protein caused it to be incorporated into MHV virions.
Coronavirus assembly results from an accumulation of interactions among four structural proteins, the positive-sense RNA genome, and a host membrane envelope obtained from the site of budding, which is the endoplasmic reticulum-Golgi intermediate compartment. Three of the four structural proteins are embedded in the virion envelope: the spike protein (S), the membrane protein (M), and the small envelope protein (E). The fourth, the nucleocapsid protein (N), resides in the virion interior, wrapping the positive-strand RNA genome into a helical nucleocapsid.
The most abundant viral constituent is M, a 25-kDa protein containing three transmembrane segments flanked by a short amino-terminal ectodomain and a large carboxy-terminal endodomain. The M protein is the central organizer of assembly, in that it self-associates (8, 10, 21), captures S for virion incorporation (36, 37), and selectively packages the fraction of N protein that is bound to genomic RNA (32-34). The S protein is responsible for viral attachment to host cell receptors and for the membrane fusion event that initiates infection. This type I membrane protein, shortly after its synthesis, folding, and oligomerization, forms complexes with M protein in the endoplasmic reticulum (36, 37). The M-interacting domain of the S protein was localized to the transmembrane-endodomain region of the molecule, based on the assembly of chimeric S proteins into virus-like particles (VLPs) (16) or virions (18, 23). More recently, this determinant was further localized to the endodomain of S, which could confer virion assembly competence if transferred to a heterologous transmembrane protein (48).
The general (1, 3, 4, 31, 45), but not universal (20), consensus from studies of VLPs is that formation of coronaviruses is mediated by just the M and the E proteins, and that neither S protein nor the nucleocapsid plays an obligate role in virion morphogenesis. The precise role of E protein in this process is enigmatic, with some evidence pointing to a direct interaction between E and M (1, 5) and other observations suggesting that E acts independently of M in the budding compartment (4, 26, 41). A very recent finding that may shed light on the workings of the E protein is the demonstration that the E protein of severe acute respiratory syndrome coronavirus has the properties of a cation-selective ion channel (47). The construction of E protein mutants of mouse hepatitis virus (MHV) has confirmed the critical role of E in viral assembly (14). Surprisingly, however, MHV remains viable, although severely impaired, following deletion of the E gene (25), whereas disruption of the E gene of porcine transmissible gastroenteritis virus (TGEV) is lethal (6, 38).
The interaction between M protein and N protein has been previously explored by biochemical and molecular biological methods for both MHV (33, 34, 44) and TGEV (12). In a genetic approach, we constructed and analyzed a highly defective MHV mutant with a carboxy-terminal truncated M protein, MΔ2, and we identified suppressors of this mutation that mapped to the carboxy terminus of the N protein (24). This led us to more closely examine which amino acids in N protein determine its recognition by M protein. Based on interstrain sequence comparisons, the MHV N protein has been proposed to comprise three conserved domains, separated by two highly divergent spacer regions (A and B) (39). Domains 1 and 2, which make up most of the molecule, are very basic, with the RNA-binding property of N mapping to domain 2 (27, 35). In contrast, domain 3, the carboxy-terminal 45 amino acids of N, has an excess of acidic residues. In the present study, we carried out a systematic mutagenesis of domain 3, and we identified an adjacent pair of functionally essential aspartates, analysis of which revealed further genetic cross talk between the carboxy termini of the N and M proteins. In addition, we found that fusion of N protein domain 3 to the carboxy terminus of a heterologous protein caused it to be selectively incorporated into MHV virions.
MATERIALS AND METHODS
Cells and viruses.
Wild-type MHV strain A59 and mutants were propagated in mouse 17 clone 1 (17Cl1) or L2 cells; plaque assays and plaque purifications of mutant recombinants and revertants were carried out in mouse L2 cells. The interspecies chimeric virus fMHV.v2 (17) was grown in feline FCWF cells or AK-D fetal lung cells.
Plasmid constructs.
Clustered charged-to-alanine (CCA) mutants 1 through 6 were originally constructed in pB36 (29), which is a donor RNA transcription vector that encodes an RNA comprising the 5′ end of the MHV genome (467 nucleotides [nt]) fused (via a 49-nt linker) to the 3′ end of the genome, beginning at the start codon of the N gene. Mutations were generated by splicing overlap extension-PCR (19) and were inserted into pB36 by exchange of the segment between the AccI site in spacer B and the BstEII site in the 3′ untranslated region (3′ UTR) (29), producing vectors pBL56 through pBL61, respectively. Of note for subsequently obtained results, the CCA4 mutations in pBL59 changed N gene codons 440 and 441 from GAU (D) to GCU (A). In CCA1 plasmid pBL56, an additional silent mutation was inadvertently generated, changing N gene codon 447 from GGG to GGA.
All other mutants were constructed in pMH54 (23) or pSG6 (17), which are donor RNA transcription vectors that encode RNAs comprising the 5′ end of the MHV genome (467 nt) fused (via a 73-nt linker) to the 3′ end of the genome, beginning at codon 28 of the HE pseudogene. Plasmid pSG6 is identical to pMH54, except for a coding-silent BspEI site that spans codons 444 to 446 of the N gene (17). The CCA4 mutations of pBL59 were transferred to pMH54, by exchange of the segment between the NheI site in domain 2 of the N gene and the BclI site in the 3′ UTR, to produce vector pLK94. For the purpose of reconstructing intragenic CCA4 revertants, a reverse transcription-PCR (RT-PCR) product, generated from revertant 2 and containing the I438S, D440A, and D441A mutations, was cloned between the NheI and BclI sites of pSG6 to produce vector pKRH5. Similarly, an RT-PCR product generated from revertant 17 and containing the Q437L, I438S, D440A, and D441A mutations was cloned between the NheI and BclI sites of pSG6. For the purpose of reconstructing intergenic CCA4 revertants, individual RT-PCR products generated from chosen revertants were incorporated into pKRH5 via unique restriction sites. For the M gene mutations I128T (revertant 1), Y143H (revertant 9), and Y156H (revertant 5), an RT-PCR product was cloned between the EagI and BssHII sites occurring in the M gene ectodomain and endodomain, respectively. For the M gene mutation V202I (revertant 7), an RT-PCR product was cloned between the M gene BssHII and N gene NheI sites.
Transcription vector pA112, containing a duplication of the N gene, was constructed as a precursor to the previously described pA122 (11), which contains a single copy of the N gene transposed to the position occupied by gene 4 in the wild-type MHV genome. The sequences of the upstream and downstream boundaries of the transposed N gene are given as junctions 5 and 6 in Fig. 3 of reference 11. In the upstream copy of the N gene in pA112, the hemagglutinin (HA) epitope tag, YPYDVPDYA, replaces amino acids 386 to 394 of spacer B, and a His6 tag is appended to the carboxy terminus. Details of the construction of pA112 and deletion derivatives will be reported elsewhere (C. A. Koetzner and P. S. Masters, unpublished results). CCA4 mutations in the duplicated copy of the N gene were created in a subclone by PCR mutagenesis, with simultaneous elimination of the His6 tag, and these were shuttled into pA112 via unique Sse8387I and XbaI sites flanking the upstream copy of the N gene.
FIG. 3.
Reconstruction of viruses containing the N gene adaptive mutation I438S and M gene reverting mutations of the CCA4 lesion. Derivative plasmids were constructed from the wild-type plasmid pSG6 (17), or from pSG6 containing the N gene CCA4 mutations (D440A and D441A) and the adaptive mutation I438S, in the presence or absence of individual candidate M gene reverting mutations. Donor RNAs transcribed from these vectors were used in targeted recombination experiments with fMHV.v2, as shown in the schematic in Fig. 1B, and progeny viruses were titrated directly on mouse L2 cells at 37°C. Monolayers were stained with neutral red at 72 h postinfection and were photographed 18 h later. In each case, plaques from two independent infection-transfections were subsequently purified by two rounds of plaque titration, and the presence of the expected mutant or wild-type sequences in the M and N genes was confirmed by direct sequencing of RT-PCR products prepared from RNA purified from virus-infected cells.
Donor RNA transcription vectors encoding derivatives of the gene for green fluorescent protein (GFP) replacing most of gene 4 of MHV were constructed from pMH54GFP (7) (generously provided by Jayasri Das Sarma and Susan Weiss, University of Pennsylvania School of Medicine). N protein domain 3 (in construct pMH54GFP-d3) or N protein spacer B plus domain 3 (in construct pMH54GFP-Bd3) was fused to the GFP open reading frame (ORF) by three-way ligations making use of the unique SalI and NotI sites bounding the GFP gene in pMH54GFP (7) as well as a BsrGI site near the carboxy terminus of the GFP ORF. In each case, the BsrGI-NotI fragment incorporating the N gene segment was generated by PCR, and a two-amino-acid spacer (SG), creating a unique BspEI site, was added between the GFP ORF and the start of the N gene segment. Derivatives of pMH54GFP-Bd3 containing the CCA4 mutations were then made by PCR mutagenesis, replacing the BspEI-NotI fragment encompassing the N gene segment.
To bacterially produce maltose binding protein fused to the carboxy terminus of the N protein, the region of the N gene encoding spacer B plus domain 3 was cloned between the BamHI and HindIII sites of vector pMAL-p2 (New England Biolabs). The fusion protein (designated MBP-Bd3) and control maltose binding protein were inducibly expressed in Rosetta(DE3)pLysS cells (Novagen).
All manipulations of DNA were carried out by standard methods (42). The compositions of all plasmid constructs were initially verified by restriction analysis. Then all cloned cDNA precursors, PCR-generated segments, and newly reconstructed or created junctions of each plasmid were confirmed by automated DNA sequencing.
Mutant construction by targeted RNA recombination.
Mutants CCA1, CCA2, CCA3, CCA5, and CCA6 were created by targeted recombination between donor RNAs from pB36-derived transcription vectors and the recipient virus Alb4 (22), as described previously (29). All other viral mutants were obtained from targeted recombination using the interspecies chimera fMHV.v2 as the recipient virus, as described in detail previously (17, 23-25). In brief, monolayers of feline AK-D or FCWF cells were infected with fMHV.v2 and were subsequently transfected with capped synthetic donor RNA by electroporation (Gene PulserII; Bio-Rad). Donor RNAs were generated by transcription with T7 RNA polymerase (mMessage mMachine; Ambion) using PacI-truncated plasmid templates. The infected and transfected feline cells either were overlaid onto murine cell monolayers or else were directly plated into 10-cm2 wells. Released progeny virus was harvested at 24 to 48 h postinfection at 37°C. Recombinant candidates were selected and purified by two rounds of plaque titration on murine L2 cell monolayers at 37°C.
For analysis of each recombinant candidate, total RNA was extracted from infected 17Cl1 cell monolayers (Ultraspec reagent; Biotecx), and reverse transcription of RNA was carried out with a random hexanucleotide primer and avian myeloblastosis virus reverse transcriptase (Life Sciences). To ascertain the presence of incorporated mutations or genes, PCR amplifications of cDNAs were performed with AmpliTaq polymerase (Roche) using primer pairs flanking the relevant regions of the genome. RT-PCR products were analyzed directly by agarose gel electrophoresis and were purified with Quantum-prep columns (Bio-Rad) prior to automated sequencing.
Western blotting.
For preparation of lysates, confluent monolayers (10 to 25 cm2) of 17Cl1 cells either were mock infected or were infected with wild-type MHV or constructed mutants; cells were then incubated at 33°C or 37°C. At 6 to 18 h postinfection, monolayers were washed twice with phosphate-buffered saline and then lysed by addition of 600 μl of 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1.0% Nonidet P-40, 0.7 μg/ml pepstatin, 1.0 μg/ml leupeptin, 1.0 μg/ml aprotinin, and 0.5 mg/ml Pefabloc SC (Roche). Lysates were held for 5 to 15 min on ice and were then clarified by centrifugation. For analysis of proteins assembled into MHV virions, viruses were purified by polyethylene glycol precipitation followed by two cycles of equilibrium centrifugation on potassium tartrate-glycerol gradients, as described in detail previously (48). Samples of either infected-cell lysates or purified virions were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis through 10% or 12% polyacrylamide gels and then transferred to a polyvinylidene difluoride membrane. Blots were probed with one of the following antibodies: anti-N monoclonal antibody (MAb) J.3.3, anti-M MAb J.1.3 (MAbs J.3.3 and J.1.3 were generously provided by John Fleming, University of Wisconsin, Madison), an anti-HA epitope tag MAb (MAb 12CA5; Roche), a polyclonal rabbit antiserum raised against a bacterially expressed maltose binding protein-MHV N protein fusion, or an anti-GFP MAb (BD Biosciences). Bound primary antibodies were visualized using a chemiluminescent detection system (ECL; Amersham).
RESULTS
Isolation of mutants in domain 3 of the N protein.
In previous work our laboratory constructed a highly defective M protein mutant, MΔ2, in which the carboxy-terminal two amino acids of M protein were truncated (24). Analysis of second-site revertants of MΔ2 demonstrated that certain mutations in the carboxy-terminal region of the N protein could compensate for the M protein truncation, suggesting a direct interaction between these two protein domains. To further examine the function of domain 3 of the MHV N protein, we generated a comprehensive set of six CCA mutants spanning domain 3, generally following the algorithm of Wertman and coworkers (46). In each case where two or more charged amino acids occurred within a sliding window of five or six residues, a mutant was designed that replaced all charged residues with alanines (Fig. 1A). This mutagenesis strategy tests the assumption that linear clusters of charged amino acids are found mainly on the surfaces of proteins, where they can potentially make strong contributions to protein-protein interactions.
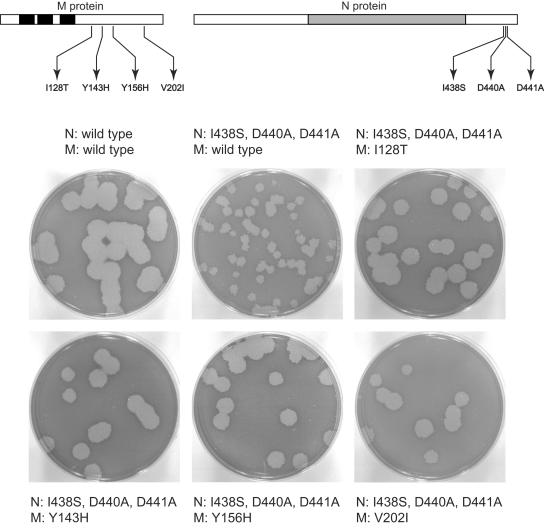
FIG. 1.
Construction of a comprehensive set of CCA mutants in domain 3 of the N protein of MHV. (A) Schematic of the proposed structure of MHV N protein, with three domains separated by two short spacers (designated A and B) (22, 29, 39, 40). The expanded region of amino acid sequence shows the carboxy terminus of N protein, including part of spacer B and all of domain 3. Residue numbers and charged residues are indicated above the wild-type sequence; for CCA mutants 1 through 6, only those residues that differ from the wild type are shown. (B) Strategy for selection of mutant CCA4 by targeted RNA recombination between the interspecies chimera fMHV.v2 (17) and donor RNA transcribed from a derivative of plasmid pMH54 (23). fMHV.v2 contains the ectodomain-encoding region of the FIPV S gene (shaded rectangle) and grows in feline cells but not in murine cells. A single crossover within the HE gene should generate a recombinant that has simultaneously reacquired the MHV S ectodomain and the ability to grow in murine cells and has also incorporated the mutations in the N gene (star). The rearranged order of genes following the S gene in fMHV.v2 precludes the occurrence of a secondary crossover event downstream of the S gene (17). (C) Plaques of purified mutant CCA4 (Alb334) compared to the reconstructed wild type (Alb240) (25). Plaque titrations were carried out on mouse L2 cells at 37°C. Monolayers were stained with neutral red at 72 h postinfection and were photographed 18 h later.
The six CCA mutants were isolated by targeted RNA recombination, a method that is used for the site-specific introduction of mutations into coronaviruses via recombination between a synthetic donor RNA and a recipient virus that can be counterselected (28, 30). CCA mutants 1, 2, 3, 5, and 6 were obtained by an earlier version of targeted recombination that made use of selection against a thermolabile recipient virus, Alb4 (22, 29). These mutants were phenotypically indistinguishable from the wild type, with the exception of mutant CCA3, which formed somewhat smaller plaques than the wild type at all temperatures tested (data not shown). Despite repeated attempts, however, we could not obtain mutant CCA4 from Alb4. Additional attempts were made to obtain CCA4 following the development of a host-range-based selective system for targeted RNA recombination. This system makes use of an interspecies chimera, named fMHV, which has the spike ectodomain of feline infectious peritonitis virus (FIPV) in place of that of MHV. As a result, fMHV grows in feline cells but not in mouse cells. This tropism is reversed by targeted recombination with a donor RNA that restores the MHV S ectodomain and simultaneously transduces additional constructed mutations into the MHV genome. Resulting mutants are selected on the basis of the restoration of the ability of the virus to grow in mouse cells (23, 24). In a recent version of this system, the genes downstream of S have been rearranged in the recipient virus fMHV.v2 (Fig. 1B), thereby effectively eliminating the possibility of downstream crossover events that could exclude the mutation(s) of interest (17).
A total of 12 independent targeted recombination experiments with donor RNA containing the CCA4 mutations were conducted in three separate sets, in which wild-type donor RNA yielded robust numbers of control recombinants. In only one of these trials were we able to recover three tiny-plaque siblings, designated Alb334, Alb335, and Alb340. All three of these mutant candidates were confirmed to have the two point mutations creating the residue changes D440A and D441A expected for CCA4. However, all three also contained a nearby mutation, I438S, resulting from an additional single-nucleotide change. This low frequency of success, as well as our failure to obtain any recombinant solely harboring the CCA4 mutations, suggested that the additional mutation is required for the viability of the CCA4 mutant.
Isolation and mapping of revertants of CCA4.
The CCA4 mutant exhibited an extremely defective phenotype, reminiscent of that of the previously studied MΔ2 mutant (24). It produced much smaller plaques than the wild type (Fig. 1C), and it grew to maximal titers that were orders of magnitude below those of the wild type. Even more severely than for the MΔ2 mutant, stocks of the CCA4 mutant could accumulate revertants within a single passage, because of the strong selective pressure imposed by the growth defect of the mutant. To examine the molecular basis for reversion, we isolated and characterized a total of 28 revertants (Fig. 2). Revertants 3, 15, 26, and 28 were identified as large-plaque isolates from the original passage-3 stocks of Alb334 and Alb335 and are independent, since they differ from one another. Each of the other revertants was individually obtained as a large plaque isolated from a passage-3 stock begun from an independent tiny plaque of either Alb334, Alb335, or Alb340. Most revertants had plaques that were wild-type sized, although some had sizes intermediate between those of the mutant and the wild type; all revertants isolated were purified through two rounds of plaque titration.
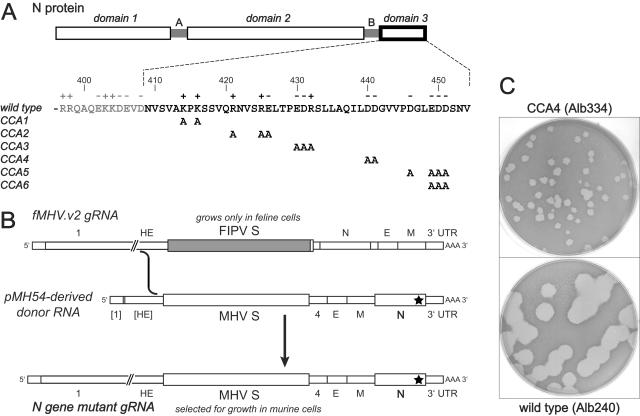
FIG. 2.
Summary of the sequence analysis of the E, M, and N genes of revertants of CCA4 sibling isolates Alb334, Alb335, and Alb340. Revertants 1, 4 to 6, 10, 12, 13, 15, 25, and 26 were obtained from Alb334; revertants 2, 3, 7 to 9, 11, 14, 16, 27, and 28 were from Alb335; revertants 17 to 24 were from Alb340. The E, M, and N proteins are represented linearly at the top. Solid rectangles indicate the membrane-bound domains of the E and M proteins; the gray rectangle indicates the RNA-binding domain of the N protein (27, 35). Positions within the M and N proteins at which potential reverting mutations were found are connected by arrows to the wild-type residue, under which the changes found in each revertant are given. All coding changes are shown; the only other changes were silent mutations at codon 8 of the M protein and at codon 212 of the N protein of revertants 11 and 9, respectively. No changes were found in the E gene of any of the revertants. Boxed residues are those that were chosen for further analysis. At the bottom are shown the carboxy-terminal sequences of the N proteins of revertants 25 to 28 in comparison to those of the wild type and the CCA4 mutant. Amino acid residues that differ from those of the wild type, including insertions, are underlined; arrowheads indicate the positions of deletions.
To map possible genetic changes that could counteract the CCA4 lesion, we sequenced RT-PCR products from each revertant spanning the entirety of the E, M, and N genes. The results of this analysis, summarized in Fig. 2, allow two general inferences. First, with one potential exception, all reverting mutations were second-site. This possibly means that a single change of either A440 or A441 back to aspartate is insufficient to restore wild-type function to the N protein. Second, most reverting mutations were found more than once, implying that the revertant search was saturating. In light of this, it is noteworthy that, as with revertants of the MΔ2 mutant (24), no change was found in the E gene of any of the 28 independent revertants of CCA4.
Significantly, nine revertants were mapped to the M gene, identifying four residue changes, I128T, Y143H, Y156H, and V202I, that potentially could individually suppress the CCA4 mutation in the N gene (Fig. 2). This intergenic compensation, although spread over a wider region than that of the intragenic revertants of the MΔ2 mutant (24), provided further evidence for an interaction between the M protein endodomain and domain 3 of the N protein. To establish whether the mutations found in M were indeed responsible for reversion of the N CCA4 mutant, we independently reconstructed each of these revertants. Initially, however, it was necessary to clarify the role of the I438S mutation in N protein. To accomplish this, we directly examined the progeny of targeted recombinations resulting from transfection of fMHV.v2-infected cells with donor RNA containing the mutations of interest. This direct approach was taken to avoid selection of additional reverting mutations that might accumulate upon purification and passage of reconstructed candidate revertants. When targeted recombination was carried out with donor RNA containing the I438S mutation in addition to the original CCA4 mutations (D440A and D441A), tiny-plaque recombinants were obtained at a high frequency (Fig. 3). This showed both that I438S is an adaptive mutation that is required for recovery of the CCA4 mutant and also that the original D440A and D441A mutations alone are apparently lethal. When targeted recombination was carried out with donor RNA containing each one of the candidate M protein reverting mutations combined with all three CCA4 mutations, only large-plaque recombinants were obtained, and these were the same size as reconstructed wild-type recombinants (Fig. 3). This demonstrated that all four of the mutations identified in the M protein were individually able to compensate for the defect in the CCA4 mutant. It incidentally also showed that the N protein mutation P199S in revertant 6 is extraneous (Fig. 2), since the M protein Y156H mutation is sufficient for reversion.
The other 19 revertants mapped to the N gene (Fig. 2). Thirteen of these contained the same single-residue change, Q437L. Remarkably, this identical mutation had previously been shown to be an intergenic suppressor of the MΔ2 mutation (24). Two other point mutations, D386G and S438C (the latter of which changes the adaptive mutation I438S), also appeared to account for reversion in revertants 24 and 10, respectively. The remaining four intragenic revertants had more extensive changes brought about by insertions or deletions. In revertant 25, a 15-nt insertion resulted in the replacement of V407 by ELKYYL at the boundary of spacer B and domain 3 (Fig. 2). The 15 inserted nucleotides (5′-AACUGAAAUACUACC-3′) exactly match a tract of negative-strand RNA in the S1 region of the S gene. In revertant 26, a 40-nt insertion resulted in the replacement of the carboxy-terminal four amino acids of the N molecule by 13 heterologous residues. Curiously, this insertion (5′-AAUGCUGCAACACUUUCAUUUAUUGGAUGGUCAUCAUCCU-3′) may also have arisen by nonhomologous recombination with the S gene, since 31 of its bases form a gapped match to the positive strand of a different part of the S1 region. Conversely, the other revertants contained in-frame deletions: of a single amino acid, Q437 (revertant 28), and of 17 amino acids at the boundary of spacer B and domain 3 (revertant 27). Additionally, revertant 27 had a point mutation, A440V (changing the original D440A mutation). For the most frequent intragenic reverting mutation, Q437L, we demonstrated by reconstruction that this single change was in fact responsible for the suppression of the CCA4 phenotype (data not shown). At this time, however, the other N protein revertants have not been reconstructed. Although it remains formally possible that undiscovered mutations elsewhere in the genome actually caused the observed reversion, we think this is unlikely, because the putative reverting mutations were the only changes found in the entire E, M, and N genes, and they were located very close to the primary mutations.
Rescue of the CCA4 mutant by wild-type N protein.
To examine whether N protein harboring the CCA4 mutations could interfere with the function of wild-type N protein, we examined the phenotype of a partially diploid MHV containing both a wild-type and a mutant copy of the N gene. It has been shown previously that the N gene, with its intact transcription-regulating sequence, can be transposed to an upstream position, replacing the nonessential gene 4 (11). This genomic rearrangement has little effect on the level of N protein expression and no detectable effect on the viral phenotype in tissue culture. Moreover, the placement of an HA epitope tag within spacer B plus a His6 tag at the carboxy terminus has no effect on the function of the transposed N gene (Koetzner and Masters, unpublished). We have also constructed mutants in which the transposed copy of the N gene (designated N[2]) is present in addition to the N gene in its normal genomic position (designated N[1]) (Fig. 4A). This duplication is stable because, although recombination can occur between the two copies of the N gene, such recombinants do not propagate due to deletion of the intervening M and E genes (Koetzner and Masters, unpublished). To test the effect of the CCA4 mutations in such a context, we constructed N duplication mutants in which the N[2] gene contained the D440A and D441A mutations in either the presence (Alb429) or the absence (Alb430) of the adaptive mutation I438S (Fig. 4A). Both Alb429 and Alb430 formed plaques that were indistinguishable from those of Alb284, which has a wild-type N[2] gene (Fig. 4B). In turn, plaques of all three of these duplication mutants were the same size as those of the wild-type virus, Alb240, and all mutants and the wild type grew to comparable titers. This result shows that the CCA4 lesion is not strongly dominant-negative, which is consistent with the notion that the CCA4 mutations act by attenuating or abolishing an essential interaction of N protein rather than by creating a novel, deleterious interaction.
FIG. 4.
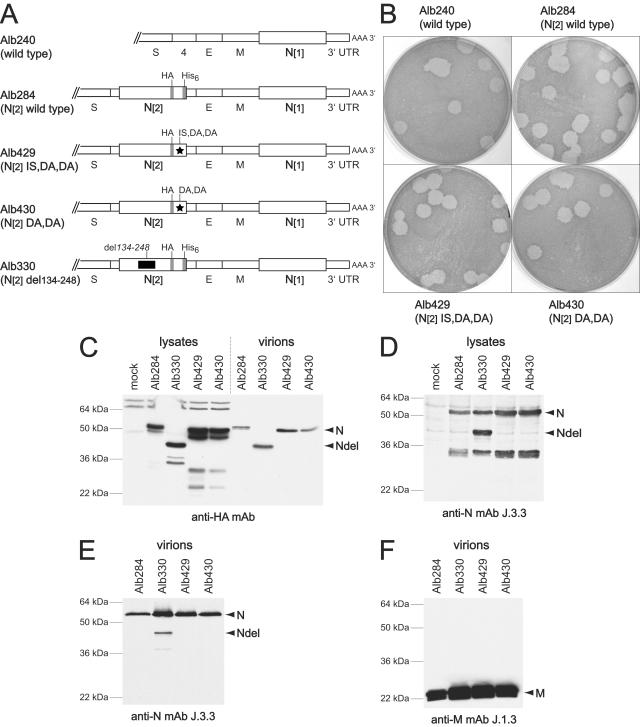
Rescue of the CCA4 mutant by wild-type N protein. (A) Schematic of the downstream end of the genome of the wild type (Alb240) and four recombinants in which the N gene is duplicated. In each, the wild-type copy of the N gene, in its normal genomic locus, is designated N[1]. In Alb284, the second copy of the N gene, designated N[2], is wild type. In Alb429 and Alb430, N[2] contains the CCA4 mutations D440A and D441A, and N[2] of Alb429 additionally contains the adaptive mutation I438S. In Alb330, N[2] contains a deletion of amino acids 134 to 248 within domain 2 of the molecule. The HA and His6 epitope tags are indicated by gray rectangles. (B) Plaques of the wild type (Alb240) and the N duplication mutants Alb284, Alb429, and Alb430. Plaque titrations were carried out on mouse L2 cells at 37°C. Monolayers were stained with neutral red at 72 h postinfection and were photographed 18 h later. (C to F) Western blots of lysates from cells infected with N duplication mutant Alb284, Alb330, Alb429, or Alb430 (C and D) or of purified virions (C, E, and F) probed with an antibody for the HA epitope tag (C), N protein (D and E), or M protein (F).
To ascertain whether the N[2] protein was expressed in the duplication mutants, Western blot analysis was performed with infected-cell lysates and with purified virions. As shown in Fig. 4C, an anti-HA antibody detected expression of similar amounts of N[2] protein in cells infected with Alb284, Alb429, and Alb430. To demonstrate the specificity of the anti-HA tag, we included, as an additional control, a duplication mutant, Alb330, in which N[2] contains an extensive deletion in domain 2. For Alb330-infected cells, a band of the expected molecular mass for N[2], 39 kDa, was detected by the anti-HA antibody (Fig. 4C), whereas anti-N MAb J.3.3 detected both the 39-kDa N[2] and the 50-kDa N[1] proteins (Fig. 4D). Figure 4D also shows that, at least for Alb330, the N[2] and N[1] proteins were expressed in comparable amounts. We also found that N[2] protein from each of the CCA4 duplication mutants Alb429 and Alb430 was incorporated into purified virions in amounts similar to that of the wild-type N[2] from Alb284 (Fig. 4C). Control blots probed with anti-N and anti-M MAbs showed that equivalent amounts of virions were analyzed (Fig. 4E and F). Taken together, these results suggest that the CCA4 lesion is a specific defect rather than one creating gross misfolding or degradation, since sufficient N-N or N-RNA interactions were retained to allow incorporation of the mutant N protein into virions in the presence of wild-type N protein. Somewhat surprisingly, a fraction of the N[2] protein of Alb330 was also incorporated into virions. Owing to the large deletion in domain 2, we would expect this mutant N protein to have lost RNA-binding ability, but it apparently still retained either N-M or N-N interactions, or both.
Overlap of the CCA4 mutations and the epitope for MAb J.3.3.
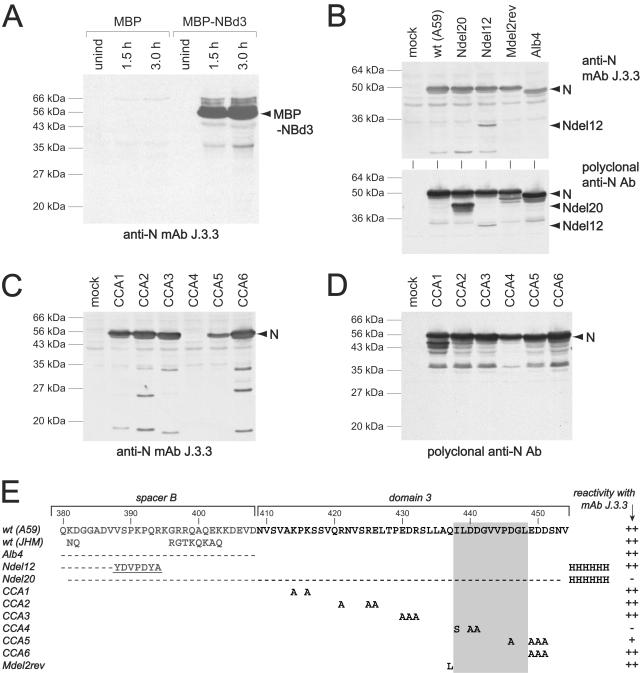
The anti-N MAb J.3.3 is a widely used reagent in studies of MHV. This antibody was raised against MHV strain JHM, but it is broadly cross-reactive with the N proteins of a number of MHV strains, including A59 (15). During the course of experiments related to the present work, we found that MAb J.3.3 recognized an expressed (MHV-A59) N protein fragment, comprising just spacer B and domain 3 (amino acids 380 through 454), fused to the maltose binding protein (Fig. 5A). This observation was consistent with a previous report that MAb J.3.3 failed to react with expressed MHV-JHM N protein that had been truncated from amino acid 374 through 455 (43). These results prompted us to attempt to further localize the epitope for MAb J.3.3 with various N protein mutants. For this purpose, we used two N duplication mutants with large deletions in N[2], similar to Alb330 (Fig. 4C): N[2] of Ndel12 has a deletion spanning parts of domain 2 and spacer B, and N[2] of Ndel20 has a deletion spanning all of spacer B and domain 3. We also examined the classical N protein deletion mutant, Alb4, which lacks spacer B (22), and an intergenic revertant of the MΔ2 mutant that has a Q437L mutation in N protein (and no other change in spacer B or domain 3) (24). In Western blots of lysates from cells infected with each of these viruses, both MAb J.3.3 and polyclonal anti-N antisera detected all expected N proteins for wild-type MHV-A59 (50 kDa), the MΔ2 revertant (50 kDa), Alb4 (47 kDa), and Ndel12 (50 kDa for N[1] and 29 kDa for N[2] (Fig. 5B). For Ndel20, however, only N[1] was recognized by MAb J.3.3, while a polyclonal anti-N antibody detected both the 50-kDa N[1] and the 43-kDa N[2]. These observations, together with the known N protein sequence variation between the A59 and JHM strains (39), both confirmed the mapping of the MAb J.3.3 epitope to the carboxy terminus of the molecule and ruled out spacer B (Fig. 5E). We next examined lysates from cells infected with each of the six CCA mutants. In this case, MAb J.3.3 was found to be fully reactive with N proteins from mutants CCA1, CCA2, CCA3, and CCA6, but it failed to recognize the CCA4 N protein at all and only weakly detected the CCA5 N protein (Fig. 5C and D). Based on these results, we infer that the epitope for MAb J.3.3 is contained within amino acids 438 through 448 of the N protein (Fig. 5E). It must be noted, however, that since we have not shown that this 11-amino-acid segment can be recognized in the context of an entirely heterologous protein, we cannot yet rule out the possibility that other residues in domain 3 contribute to the ability to react with MAb J.3.3. Interestingly, the locus of residues 438 through 448 precisely overlaps the major determinant of the N-M interaction as defined by the CCA4 mutant, suggesting that this region of the N molecule is displayed on a surface easily accessible for protein-protein contacts, either with antibody or with M protein.
FIG. 5.
Localization of the epitope for anti-N MAb J.3.3 (15). (A) Western blots of lysates from bacterial cells that inducibly expressed either maltose binding protein (MBP) or maltose binding protein with a carboxy-terminal fusion of spacer B and domain 3 of the N protein (MBP-NBd3). Bacterial cultures were harvested prior to induction (unind) or after 1.5 or 3.0 h of induction. (B) Western blots of lysates from cells that were either mock infected or else infected with wild-type MHV-A59, N duplication mutant Ndel20 (Alb354) or Ndel12 (Alb381), or Mdel2rev (Alb302). The genomic arrangement of the N duplication mutants is similar to that of Alb330 (see Fig. 4). In Ndel20, the N[2] gene contains a deletion of amino acids 381 to 453 in spacer B and domain 3; in Ndel12, the N[2] gene contains a deletion of amino acids 188 to 387 in domain 2 and spacer B. Both Ndel20 and Ndel12 contain the His6 epitope tag; Ndel12 also retains a fragment of the HA epitope tag, while Ndel20 lacks it entirely. Mdel2rev is an intergenic revertant of the MΔ2 mutant that has the suppressor mutation Q437L in the N gene. (C and D) Western blots of lysates from cells that were mock infected or infected with CCA mutants 1 to 6. Blots were probed with MAb J.3.3 (A, top panel of B, and C) or polyclonal anti-N antibodies (bottom panel of B and D). (E) Summary of the abilities of various strains and mutants to cross-react with MAb J.3.3. The sequence of the carboxy terminus of the wild-type MHV-A59 N protein is shown at the top. For all other viruses, only residues that differ from the wild type (A59) are shown; dashes indicate deleted residues. In Alb4 and Ndel20, the entire deletion is shown. In Ndel12 only the downstream end of the deletion is represented, and the remnant of the HA tag is underlined. The gray rectangle indicates the region to which the MAb J.3.3 epitope maps.
N protein domain 3 is sufficient for incorporation of a foreign protein into MHV virions.
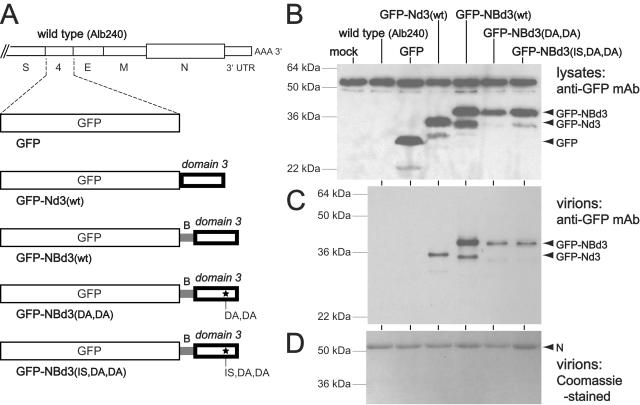
To determine whether the putative M-interacting region of N can be transferred to another protein, we created a series of recombinants expressing variants of GFP. For this purpose, we used the same donor RNA vector, pMHV54GFP, that had previously been reported by Das Sarma and coworkers to optimally express the gene for enhanced GFP in place of the nonessential gene 4 (7). In addition to a recombinant expressing native (enhanced) GFP, we constructed viruses expressing GFP with a carboxy-terminal extension of N protein domain 3, GFP-Nd3(wt), or expressing GFP with both spacer B and domain 3, GFP-NBd3(wt) (Fig. 6A). Two further recombinants were also tested: GFP-NBd3(DA,DA) and GFP-NBd3(IS,DA,DA), in which the GFP-NBd3 fusion protein contained the CCA4 mutations D440A and D441A, without or with the adaptive mutation I438S. Western blot analysis of lysates from cells infected with these and wild-type control viruses revealed abundant expression of the expected set of proteins, as identified by the anti-GFP antibody (Fig. 6B). As noted previously (7, 13), GFP and derivative proteins had slower mobilities by sodium dodecyl sulfate-polyacrylamide gel electrophoresis than would be expected for proteins of their molecular masses (27 kDa for GFP, 32 kDa for GFP-Nd3, and 35 kDa for GFP-NBd3). We also detected a cross-reacting protein of roughly 55 kDa in all lysates, including mock-infected cells and cells infected with wild-type MHV; however, this nonspecific band was easily differentiated from those of the lower-molecular-weight GFP constructs.
FIG. 6.
Transfer of the M-interacting region of N protein to a heterologous protein. (A) Schematic of the downstream end of the genome of the wild type (Alb240) and five recombinants in which the GFP gene replaces the nonessential gene 4. Mutant GFP is identical to the MHV-A59 EGFP recombinant described previously (7). Mutant GFP-Nd3(wt) contains the wild-type domain 3 of the N protein linked to the carboxy terminus of GFP. Mutant GFP-NBd3(wt) contains the wild-type spacer B and domain 3 of the N protein. Mutants GFP-NBd3(DA,DA) and GFP-NBd3(IS,DA,DA) both contain spacer B and domain 3 with the CCA4 mutations D440A and D441A; GFP-NBd3(IS,DA,DA) additionally contains the adaptive mutation I438S. (B and C) Western blots of lysates from cells infected with the wild type or GFP-expressing mutants (B), or of purified virions (C), probed with an antibody for GFP. The amounts of virions analyzed in each lane of panel C were normalized to contain equivalent amounts of N protein, as determined by staining with Coomassie blue (D).
When purified virions of the same set of recombinant viruses were analyzed in Western blots, only GFP constructs that contained fusions of N protein domain 3 were detected (Fig. 6C). The GFP-NBd3(wt) protein was incorporated into virions at a markedly higher level than the GFP-Nd3(wt) protein, possibly because spacer B provided the necessary distance and flexibility for the functioning of domain 3 outside of its native context. Notably, the CCA4 mutations in GFP-NBd3(DA,DA) and GFP-NBd3(IS,DA,DA) severely reduced, but did not abolish, the incorporation of GFP-NBd3 into virions. Although domain 3 clearly enabled the specific incorporation of GFP into MHV virions, the amount of GFP incorporation, on a molar basis, was well below that of the N protein. This was apparent from Coomassie blue staining of gels of electrophoresed purified virions, which detected N protein but not GFP derivatives (Fig. 6D). The lower molar incorporation of GFP-NBd3 or GFP-Nd3 into virions was likely due to the monovalent nature of the GFP fusion constructs, in contrast to the highly multivalent setting of N domain 3 in the nucleocapsid. We would expect the latter to have a far higher avidity for its interacting partner.
DISCUSSION
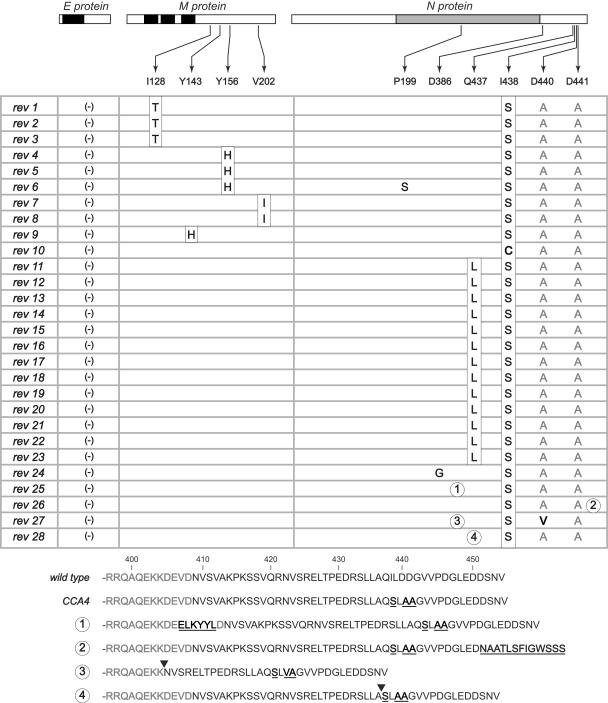
In previous work it was shown that truncation of just the two carboxy-terminal residues of the MHV M protein, R227 and T228, produced a severely impaired mutant, designated MΔ2 (24). The MΔ2 virus exhibited a tiny-plaque phenotype and a low growth yield, and it gave rise to spontaneous large-plaque revertants upon being passaged. These revertants were all found to be second-site, mapping either to nearby residues of the M protein or else to a locus near the carboxy terminus of the N protein that included residue Q437 (Fig. 7). In the present study, we constructed a comprehensive set of all six possible clustered charged-to-alanine mutants in the carboxy-terminal domain 3 of the N protein. Each of these mutants had a wild-type or nearly wild type phenotype, except for one, CCA4, which had severely defective characteristics very similar to those of MΔ2, including the formation of tiny plaques.
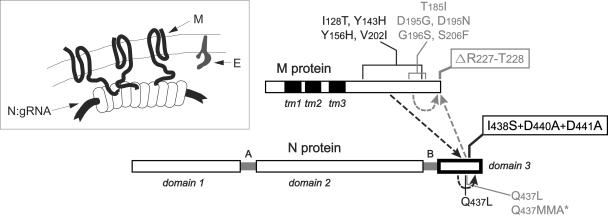
FIG. 7.
Genetic cross talk between the N and M proteins. The domain structure of N protein is shown linearly, as in Fig. 1. For the M protein, solid rectangles indicate the three transmembrane (tm) domains. Primary mutations that disrupt N-M interactions (the original lesions of the MΔ2 and CCA4 mutants) are boxed. The locations of second-site mutations, each of which individually suppresses one of the primary mutations, are indicated by lines or brackets; the asterisk represents a stop codon. The MΔ2 primary and reverting mutations (24) are in gray; the CCA4 primary and reverting mutations (this study) are in black. Arrows indicate compensating intramolecular and intermolecular interactions. (Inset) Schematic of the relative topologies and interfaces of the M, N, and E proteins at or within the virion envelope.
The originally engineered mutations of the CCA4 mutant, D440A and D441A, were most likely lethal, as evidenced by the fact that they could be recovered only in the company of an additional nearby mutation, I438S. We found that, in an MHV construct containing a duplication of the N gene, wild-type N protein was able to rescue N proteins containing either the two charged-to-alanine mutations or all three mutations. This showed that the CCA4 mutations are loss-of-function mutations rather than dominant-negative mutations. Moreover, in the context of the N duplication construct, both of the mutant N proteins were expressed at levels comparable to those of the wild-type copy of the N protein (Fig. 4). This suggested that the mutant N proteins folded normally and that the CCA4 phenotype was due to failure to form an essential interaction and did not result from extensive degradation of mutant N protein.
Analysis of multiple independent large-plaque-forming revertants of the CCA4 mutant has resulted in a very clear convergence with the MΔ2 mutant results (Fig. 7). Like the MΔ2 revertants (24), revertants of the CCA4 mutant retained the original lesion and exhibited second-site mutations that were either intragenic (in spacer B or domain 3 of the N protein) or intergenic (in the carboxy-terminal tail of the M protein) (Fig. 2). The presence of the latter class of revertants leads us to believe that we have now mapped here the reciprocal of the M protein-N protein interaction that was previously revealed by the MΔ2 mutant. One picture that emerges from our results is that a major component of the M protein-N protein interface may well be a salt bridge between R227 of M protein and D440 and/or D441 of N protein, as was formerly hypothesized (24). However, other observations argue that the M-N interaction is more complex. Arginine side chains not only form electrostatic interactions but also are capable of participation in hydrogen bond networks and in hydrophobic contacts (2). In addition, we were not able to simply exchange R227 of the M protein and D440 and D441 of the N protein (data not shown). Surprisingly, it was possible to construct a viable mutant in which R227 of M protein was replaced with a pair of aspartate residues. However, the replacement of both D440 and D441 of N protein with a single arginine was lethal, whether in the background of a wild-type M protein or in that of an R227DD mutant M protein.
Remarkably, 13 of 19 independent intragenic reversion events in the CCA4 mutant were caused by a single mutation, Q437L (Fig. 2), which had previously been identified as responsible for second-site reversion of the MΔ2 mutation (24). As with the MΔ2 revertants, this may be explained by the substitution of hydrophobic interactions between L437 of N protein and L225 and/or L226 of M protein, replacing lost electrostatic interactions. Other N protein intragenic reverting mutations are more difficult to account for at this time. Notably, four of these contain insertions or deletions in spacer B or domain 3 (Fig. 2, revertants 25 to 28). The net effect of these reverting events may be to change the phasing of the protein secondary structure, making alternative residues available for electrostatic or hydrophobic interactions with the M protein. At the RNA level, these revertants show that extreme selective pressure can reveal unusual nonhomologous rearrangements that are likely to be occurring constantly at a low frequency during MHV RNA synthesis. In particular, the insertions in revertants 25 and 26 appear to be scavenged fragments of ORFs from other parts of the genome.
Consideration of the combined results from the MΔ2 and CCA4 mutants suggests that the interacting regions of the N and M proteins can be sharply localized in the N protein primary sequence but not in the M protein primary sequence (Fig. 7). In the N protein, our CCA mutation set spanned all charged residues in domain 3. Among these mutants, only the CCA4 mutant had a noteworthy phenotype, and the CCA4 mutations fall precisely adjacent to the locus of the intergenic revertants of MΔ2 (24). By contrast, in the M protein, intergenic revertants of the CCA4 mutant cover a region of some 75 residues of the cytoplasmic tail, partially overlapping the sites of intragenic revertants of the MΔ2 mutant. If residue R227 is the major point of M protein interaction with the N protein, then many other residues, some quite distant in the primary sequence, presumably alter the way in which R227 is made accessible to N domain 3. This is likely a reflection of the complexity of the folding of the M cytoplasmic endodomain. Additionally, there must be limitations on allowable changes in the M sequence that could compensate for the CCA4 lesion while not adversely affecting any of the other functions of M, since mutations in many parts of this protein can alter critical interactions with other M molecules as well as with the S protein (9, 10).
The more defined localization of the interacting portion of the N protein allowed us to establish that attachment of domain 3 of N was sufficient for the selective incorporation of a heterologous molecule into MHV virions (Fig. 6). Incorporation of a GFP-N domain 3 chimera into virions was enhanced by the inclusion of spacer B of the N protein, an effect that is most likely attributable to the provision of sufficient separation of domain 3 from the GFP molecule. We do not think that spacer B plays a specific role in interacting with M, because the sequence of spacer B is highly divergent among different MHV strains (39), and it can also be engineered to contain various foreign motifs with no consequence to viral phenotype (40) (unpublished data). The monovalent GFP-NBd3 construct, however, was not able to mimic the cooperative M binding of the multivalent nucleocapsid. Consequently, GFP-NBd3 was not incorporated into virions at levels comparable to those of native N protein. The attachment of N domain 3 to a self-associating heterologous protein might produce a chimera capable of competing with the nucleocapsid.
It should be noted that we cannot yet conclusively rule out the possibility that other parts of the N molecule play a role in association with M protein or that more than one type of N-M interaction exists. In this regard, it remains to be shown how or whether our genetically defined interaction correlates with results from biochemical and molecular biological assays that have addressed the same question. For MHV, an early study using virion fractionation procedures uncovered a temperature-dependent interaction between nonionic detergent-solubilized M protein and the viral nucleocapsid (44). Some of the experiments in that work suggested that the M protein-nucleocapsid association was mediated by M binding to viral RNA rather than to N protein. More recently, it was found that N protein could be coimmunoprecipitated from MHV-infected cells by MAbs specific for M protein. Significantly, although N was shown to be associated with all viral RNAs, the M protein selected only those complexes of N molecules that were bound to genomic RNA (33). This selectivity was subsequently determined to depend on the presence of the genomic RNA packaging signal, which could be transferred to allow the incorporation of heterologous RNAs into virions (34). Further related work with coexpressed MHV proteins and RNAs attributed the selection of packaging signal RNA to the M protein (32). Thus, VLPs composed of M and E but not N protein were found to selectively incorporate RNA containing the MHV packaging signal. The interaction that we have mapped appears to be independent of RNA in that it can be transferred to a heterologous protein, GFP, which is not known to bind to RNA. In addition, domain 3 of the MHV N protein has been shown to be dispensable for RNA binding (27, 35). Nevertheless, it is possible that, within the native N protein, domain 3 becomes accessible for interaction with M only after binding to particular RNA substrates. Such an RNA-dependent binding of N to M has been proposed for the TGEV N protein (38). For this group 1 coronavirus, the M-N interaction was measured by binding of in vitro-translated M protein to immobilized nucleocapsid purified from virions. It was reported that purified N protein alone was not bound by the M protein, suggesting that the N protein that was complexed with RNA in the viral nucleocapsid had an altered conformation compared with free N protein. Analysis of a series of in vitro-translated deletion mutants of the TGEV M protein localized that protein's N-interacting region to amino acids 237 to 252 (38). This segment corresponds to amino acids 205 to 220 of the MHV M protein, which overlap with only one of the critical residues that we have identified in the MHV M protein by genetic means (Fig. 7). However, this region of the two M sequences is very poorly conserved, and the apparent discord between the MHV and TGEV results may stem from differences in the respective folds of the two M proteins, or differences in how individual residues influence those folds. These relationships should be clarified when crystal structures of either, or both, of the interacting domains of M and N become available.
Acknowledgments
We are grateful to John Fleming for providing MAbs J.3.3 and J.1.3 and to Jayasri Das Sarma and Susan Weiss for pMH54GFP. We thank the Molecular Genetics Core Facility of the Wadsworth Center for oligonucleotide synthesis and DNA sequencing.
This work was supported by Public Health Service grants AI 39544 and AI 64603 from the National Institutes of Health.
REFERENCES
- 1.Baudoux, P., C. Carrat, L. Besnardeau, B. Charley, and H. Laude. 1998. Coronavirus pseudoparticles formed with recombinant M and E proteins induce alpha interferon synthesis by leukocytes. J. Virol. 72:8636-8643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bogan, A. A., and K. S. Thorn. 1998. Anatomy of hot spots in protein interfaces. J. Mol. Biol. 280:1-9. [DOI] [PubMed] [Google Scholar]
- 3.Bos, E. C. W., W. Luytjes, H. van der Meulen, H. K. Koerten, and W. J. M. Spaan. 1996. The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology 218:52-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corse, E., and C. E. Machamer. 2000. Infectious bronchitis virus E protein is targeted to the Golgi complex and directs release of virus-like particles. J. Virol. 74:4319-4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corse, E., and C. E. Machamer. 2003. The cytoplasmic tails of infectious bronchitis virus E and M proteins mediate their interaction. Virology 312:25-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curtis, K. M., B. Yount, and R. S. Baric. 2002. Heterologous gene expression from transmissible gastroenteritis virus replicon particles. J. Virol. 76:1422-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das Sarma, J., E. Scheen, S. H. Seo, M. Koval, and S. R. Weiss. 2002. Enhanced green fluorescent protein expression may be used to monitor murine coronavirus spread in vitro and in the mouse central nervous system. J. Neurovirol. 8:381-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Haan, C. A. M., L. Kuo, P. S. Masters, H. Vennema, and P. J. M. Rottier. 1998. Coronavirus particle assembly: primary structure requirements of the membrane protein. J. Virol. 72:6838-6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Haan, C. A. M., M. Smeets, F. Vernooij, H. Vennema, and P. J. M. Rottier. 1999. Mapping of the coronavirus membrane protein domains involved in interaction with the spike protein. J. Virol. 73:7441-7452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Haan, C. A. M., H. Vennema, and P. J. M. Rottier. 2000. Assembly of the coronavirus envelope: homotypic interactions between the M proteins. J. Virol. 74:4967-4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Haan, C. A. M., H. Volders, C. A. Koetzner, P. S. Masters, and P. J. M. Rottier. 2002. Coronaviruses maintain viability despite dramatic rearrangements of the strictly conserved genome organization. J. Virol. 76:12491-12493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Escors, D., J. Ortego, H. Laude, and L. Enjuanes. 2001. The membrane M protein carboxy terminus binds to transmissible gastroenteritis coronavirus core and contributes to core stability. J. Virol. 75:1312-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer, F., C. F. Stegen, C. A. Koetzner, and P. S. Masters. 1997. Analysis of a recombinant mouse hepatitis virus expressing a foreign gene reveals a novel aspect of coronavirus transcription. J. Virol. 71:5148-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer, F., C. F. Stegen, P. S. Masters, and W. A. Samsonoff. 1998. Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J. Virol. 72:7885-7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fleming, J. O., S. A. Stohlman, R. C. Harmon, M. M. C. Lai, J. A. Frelinger, and L. P. Weiner. 1983. Antigenic relationships of murine coronaviruses: analysis using monoclonal antibodies to JHM (MHV-4) virus. Virology 131:296-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Godeke, G. J., C. A. de Haan, J. W. Rossen, H. Vennema, and P. J. M. Rottier. 2000. Assembly of spikes into coronavirus particles is mediated by the carboxy-terminal domain of the spike protein. J. Virol. 74:1566-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goebel, S. J., B. Hsue, T. F. Dombrowski, and P. S. Masters. 2004. Characterization of the RNA components of a putative molecular switch in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 78:669-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haijema, B. J., H. Volders, and P. J. M. Rottier. 2003. Switching species tropism: an effective way to manipulate the feline coronavirus genome. J. Virol. 77:4528-4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horton, R. M., and L. R. Pease. 1991. Recombination and mutagenesis of DNA sequences using PCR, p. 217-247. In M. J. McPherson (ed.), Directed mutagenesis, a practical approach. IRL Press, New York, N.Y.
- 20.Huang, Y., Z. Y. Yang, W. P. Kong, and G. J. Nabel. 2004. Generation of synthetic severe acute respiratory syndrome coronavirus pseudoparticles: implications for assembly and vaccine production. J. Virol. 78:12557-12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klumperman, J., J. Krijnse Locker, A. Meijer, M. C. Horzinek, H. J. Geuze, and P. J. M. Rottier. 1994. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J. Virol. 68:6523-6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koetzner, C. A., M. M. Parker, C. S. Ricard, L. S. Sturman, and P. S. Masters. 1992. Repair and mutagenesis of the genome of a deletion mutant of the coronavirus mouse hepatitis virus by targeted RNA recombination. J. Virol. 66:1841-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuo, L., G.-J. Godeke, M. J. B. Raamsman, P. S. Masters, and P. J. M. Rottier. 2000. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 74:1393-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo, L., and P. S. Masters. 2002. Genetic evidence for a structural interaction between the carboxy termini of the membrane and nucleocapsid proteins of mouse hepatitis virus. J. Virol. 76:4987-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuo, L., and P. S. Masters. 2003. The small envelope protein E is not essential for murine coronavirus replication. J. Virol. 77:4597-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maeda, J., A. Maeda, and S. Makino. 1999. Release of E protein in membrane vesicles from virus-infected cells and E protein-expressing cells. Virology 263:265-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masters, P. S. 1992. Localization of an RNA-binding domain in the nucleocapsid protein of the coronavirus mouse hepatitis virus. Arch. Virol. 125:141-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masters, P. S. 1999. Reverse genetics of the largest RNA viruses. Adv. Virus Res. 53:245-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masters, P. S., C. A. Koetzner, C. A. Kerr, and Y. Heo. 1994. Optimization of targeted RNA recombination and mapping of a novel nucleocapsid gene mutation in the coronavirus mouse hepatitis virus. J. Virol. 68:328-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masters, P. S., and P. J. M. Rottier. 2005. Coronavirus reverse genetics by targeted RNA recombination. Curr. Top. Microbiol. Immunol. 287:133-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mortola, E., and P. Roy. 2004. Efficient assembly and release of SARS coronavirus-like particles by a heterologous expression system. FEBS Lett. 576:174-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narayanan, K., C. J. Chen, J. Maeda, and S. Makino. 2003. Nucleocapsid-independent specific viral RNA packaging via viral envelope protein and viral RNA signal. J. Virol. 77:2922-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Narayanan, K., A. Maeda, J. Maeda, and S. Makino. 2000. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J. Virol. 74:8127-8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narayanan, K., and S. Makino. 2001. Cooperation of an RNA packaging signal and a viral envelope protein in coronavirus RNA packaging. J. Virol. 75:9059-9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nelson, G. W., and S. A. Stohlman. 1993. Localization of the RNA-binding domain of mouse hepatitis virus nucleocapsid protein. J. Gen. Virol. 74:1975-1979. [DOI] [PubMed] [Google Scholar]
- 36.Nguyen, V.-P., and B. Hogue. 1997. Protein interactions during coronavirus assembly. J. Virol. 71:9278-9284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Opstelten, D.-J. E., M. J. B. Raamsman, K. Wolfs, M. C. Horzinek, and P. J. M. Rottier. 1995. Envelope glycoprotein interactions in coronavirus assembly. J. Cell Biol. 131:339-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ortego, J., D. Escors, H. Laude, and L. Enjuanes. 2002. Generation of a replication-competent, propagation-deficient virus vector based on the transmissible gastroenteritis coronavirus genome. J. Virol. 76:11518-11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parker, M. M., and P. S. Masters. 1990. Sequence comparison of the N genes of five strains of the coronavirus mouse hepatitis virus suggests a three domain structure for the nucleocapsid protein. Virology 179:463-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng, D., C. A. Koetzner, T. McMahon, Y. Zhu, and P. S. Masters. 1995. Construction of murine coronavirus mutants containing interspecies chimeric nucleocapsid proteins. J. Virol. 69:5475-5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raamsman, M. J. B., J. Krijnse Locker, A. de Hooge, A. A. F. de Vries, G. Griffiths, H. Vennema, and P. J. M. Rottier. 2000. Characterization of the coronavirus mouse hepatitis virus strain A59 small membrane protein E. J. Virol. 74:2333-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Stohlman, S. A., C. Bergmann, D. Cua, H. Wege, and R. van der Veen. 1994. Location of antibody epitopes within the mouse hepatitis virus nucleocapsid protein. Virology 202:146-153. [DOI] [PubMed] [Google Scholar]
- 44.Sturman, L. S., K. V. Holmes, and J. Behnke. 1980. Isolation of coronavirus envelope glycoproteins and interaction with the viral nucleocapsid. J. Virol. 33:449-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vennema, H., G.-J. Godeke, J. W. A. Rossen, W. F. Voorhout, M. C. Horzinek, D.-J. E. Opstelten, and P. J. M. Rottier. 1996. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 15:2020-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wertman, K. F., D. G. Drubin, and D. Botstein. 1992. Systematic mutational analysis of the yeast ACT1 gene. Genetics 132:337-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilson, L., C. McKinlay, P. Gage, and G. Ewart. 2004. SARS coronavirus E protein forms cation-selective ion channels. Virology 330:322-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ye, R., C. Montalto-Morrison, and P. S. Masters. 2004. Genetic analysis of determinants for spike glycoprotein assembly into murine coronavirus virions: distinct roles for charge-rich and cysteine-rich regions of the endodomain. J. Virol. 78:9904-9917. [DOI] [PMC free article] [PubMed] [Google Scholar]