Abstract
Moloney murine leukemia virus (MLV) particles contain both viral genomic RNA and an assortment of host cell RNAs. Packaging of virus-encoded RNA is selective, with virions virtually devoid of spliced env mRNA and highly enriched for unspliced genome. Except for primer tRNA, it is unclear whether packaged host RNAs are randomly sampled from the cell or specifically encapsidated. To address possible biases in host RNA sampling, the relative abundances of several host RNAs in MLV particles and in producer cells were compared. Using 7SL RNA as a standard, some cellular RNAs, such as those of the Ro RNP, were found to be enriched in MLV particles in that their ratios relative to 7SL differed little, if at all, from their ratios in cells. Some RNAs were underrepresented, with ratios relative to 7SL several orders of magnitude lower in virions than in cells, while others displayed intermediate values. At least some enriched RNAs were encapsidated by genome-defective nucleocapsid mutants. Virion RNAs were not a random sample of the cytosol as a whole, since some cytoplasmic RNAs like tRNAMet were vastly underrepresented, while U6 spliceosomal RNA, which functions in the nucleus, was enriched. Real-time PCR demonstrated that env mRNA, although several orders of magnitude less abundant than unspliced viral RNA, was slightly enriched relative to actin mRNA in virions. These data demonstrate that certain host RNAs are nearly as enriched in virions as genomic RNA and suggest that Ψ− mRNAs and some other host RNAs may be specifically excluded from assembly sites.
Host cell RNA packaging has been observed in several viruses, including DNA viruses such as human cytomegalovirus (54) and herpes simplex virus type 1 (50), RNA viruses such as flock house virus (49), and several retroviruses (9, 11, 15, 16, 20). In the herpesviruses, cellular mRNAs are reportedly packaged in a random fashion, most likely representative of their intracellular abundance (50, 54). For retroviruses, the primer tRNA is specifically recruited, and mRNAs are thought to a represent a random sampling (37, 41), but it is unclear whether encapsidation is random among most remaining groups of cellular RNAs.
From the mixture of viral and cellular transcripts in the infected cell, two copies of genomic RNA are selected for incorporation into each retroviral particle (7, 36). Virus-encoded RNAs within a Moloney murine leukemia virus (MLV)-infected cell include the capped and polyadenylated unspliced primary transcript as well as spliced env mRNA. Unspliced MLV RNAs contain a ∼350-nucleotide (nt) region near their 5′ ends termed the packaging signal, or Ψ (38), which lies downstream of the splice donor site and upstream of the gag start codon. MLV RNAs that contain Ψ are packaged at least 100-fold more efficiently than RNAs that lack Ψ (38). Although other viral sequences and motifs have been implicated in facilitating the specific recruitment of RNAs for packaging (6, 7, 42, 57), the selective encapsidation of MLV and other retroviral genomes is largely attributed to the presence of Ψ (1, 3, 24, 38).
In addition to the unspliced viral genomic RNA, retroviruses package RNAs derived from the infected host cell (2, 3, 7), and estimates suggest that up to 30% of RNAs within a retrovirus originate from the host (36). Even in the absence of viral genomic RNA, retroviral particles contain RNA, and it has been shown both in vitro (13) and in cell culture (40) that RNA, although not necessarily of viral origin, is required for virion assembly. Previous studies of the RNA content of retroviral particles have demonstrated packaging of short RNA species, initially designated 4S to 7S RNAs based on their sedimentation properties, in virions of avian myeloblastosis virus (55), equine infectious anemia virus (15), feline leukemia virus (11), Visna virus (35), Rous sarcoma virus (RSV) (9), MLV (20, 33, 34, 43), and human immunodeficiency virus type 1 (27, 28, 31). Early work on the RNA content of RSV particles demonstrated that along with the dimer of viral RNA, 7S RNA (now known as 7SL or SRP RNA, the RNA component of the signal recognition particle [29]), 4S RNA, 5S rRNA, and trace amounts of 28S and 18S ribosomal RNAs were also present (9, 21, 36, 46). Furthermore, in RSV, the packaging of cellular mRNA has been suggested (2), and a recent report showed that cellular spliceosomal RNAs, especially U6 snRNA, are also packaged (22).
The best-understood host RNAs that are packaged into virions are the cellular tRNAs. Retroviruses are known to contain uncharged tRNAs (37) and are enriched for primer tRNAs required to initiate minus-strand DNA synthesis. Both simple and complex retroviruses selectively package their own particular primer tRNA (14), with estimates of up to 8 copies of primer tRNA associated with the retroviral genome within each particle (34). Primer tRNA packaging in retroviruses depends on the presence of reverse transcriptase (RT) (32, 34) and in at least some cases involves tRNA synthetases, but apparently not for MLV (14).
In this work, we examined host cell RNA packaging by MLV. We confirmed the packaging of some previously described host cell RNAs, including 7SL RNA, primer tRNA, and 5S rRNA, and examined the packaging of several additional host RNAs. Virion and cellular RNAs were directly compared by Northern hybridization, RNase protection assays, and quantitative RT-PCR. The results showed that host cell RNAs in MLV were not packaged in proportion to their intracellular abundance but rather revealed differential enrichment of some and exclusion of other RNAs.
MATERIALS AND METHODS
Cells and virus.
NIH 3T3 and derivative cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% bovine serum (Invitrogen). ET cells (a 293T derivative that constitutively expresses murine ecotropic envelope) (44) were maintained in DMEM plus 10% fetal bovine serum (Gemini). Wild-type MLV particles were collected from the medium of NIH 3T3 cells chronically infected with wild-type MLV (NIH 3T3-MLV). Genome-defective particles were obtained from PG13 cell medium. PG13 cells are an NIH 3T3-based packaging cell line that expresses MLV Gag and Pol and gibbon ape leukemia virus Env (39). All cell lines were grown at 37°C under 5% CO2. Virus-containing media were filtered using 0.2-μm filters and stored at −70°C prior to use.
Cells that expressed nucleocapsid (NC) mutant virions were generated as follows: the NC region of pMLV GPP (previously known as pGPP, which encodes MLV Gag and Pol, but instead of env, it has an simian virus 40-driven puromycin expression cassette) (44) was mutated to bear each of three previously described packaging-defective NC alleles, including single-amino-acid substitutions in the zinc finger (H34C or C39H; referred to in Fig. 3 as CCCC and CCHH, respectively), and a three-substitution change (R16L-R17S-R18S, also known as ELSS) in the basic region N terminal to the zinc finger (23, 25). ET cells were transfected with the resulting plasmids using Fugene-6 transfection reagent (Roche). Virions were harvested and used to infect NIH 3T3 cells in the presence of 0.8 μg/ml hexadimethrine bromide (Polybrene; Sigma). Infected cells were selected in 6 μg/ml puromycin (Sigma) for 14 days. Surviving colonies were pooled and expanded, and virus was harvested from the media for analysis.
FIG. 3.
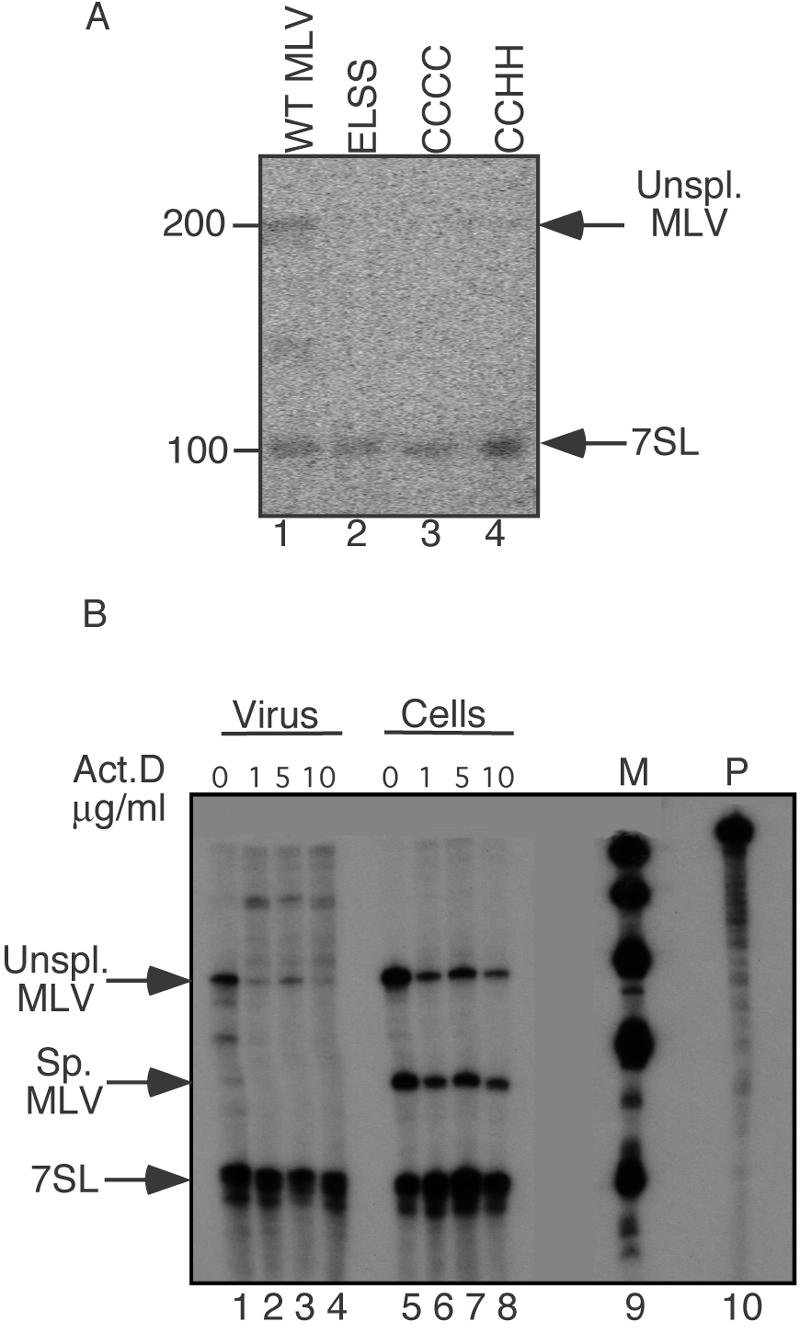
7SL packaging determinants (RNase protection assays). (A) Virion RNAs from wild-type (WT) or NC mutant MLV. Lanes: 1, wild type; 2, ELSS basic region NC mutant (R16L-R17S-R18S); 3, CCCC zinc finger NC mutant (H34C); 4, CCHH zinc finger NC mutant (C39H). 7SL RNA- and MLV genomic RNA-protected product bands are indicated by the arrows. (B) Virion and cellular RNAs from actinomycin D (Act.D)-treated MLV-producing cells. Lanes: 1 to 4, viral RNAs; 5 to 8, cellular RNAs; 9, marker (M); 10, probe (P). Lanes 1 and 5, untreated control; lanes 2 and 6, cells treated with 1 μg/ml actinomycin D; lanes 3 and 7, 5 μg/ml actinomycin D; lanes 4 and 8, 10 μg/ml actinomycin D. 7SL RNA and MLV genomic RNA bands are indicated by the arrows. Note that products migrating between genome and 7SL products in lane 1 are spurious products of RNase digestion.
For experiments that involved actinomycin-treated cells, NIH 3T3 cells chronically infected with wild-type MLV were treated with 1, 5, or 10 μg actinomycin D (Sigma)/ml in DMEM plus 10% fetal bovine serum for 24 h. Virion-containing media and cells were then collected, and RNAs were extracted and analyzed by RNase protection assay as described below.
Viral and cellular RNA extraction.
Tissue culture medium was harvested at 8- to 16-h intervals from chronically infected NIH 3T3-MLV cells, PG13 cells, and uninfected NIH 3T3 cells at comparable confluence. Virus-containing medium was centrifuged at 25,000 rpm for 90 min using the AH629 rotor in a Sorvall Discovery ultracentrifuge at 4°C. Viral pellets were resuspended in Trizol (Invitrogen), and RNA was extracted according to the manufacturer's instructions. Total cell RNA was extracted from confluent 6- or 10-cm dishes of cells using 1 or 3 ml of Trizol.
Cellular fractionation.
Wild-type MLV-infected cells, genome-defective virion-expressing (PG13) cells, and uninfected NIH 3T3 cells were harvested using phosphate-buffered saline (PBS)-5 mM EDTA and pelleted by spinning at 1,500 rpm for 5 min at 4°C. The cell pellet was suspended in 200 μl cytoplasmic lysis buffer (20% glycerol-0.01 M Tris [pH 8.4]-1.5 mM MgCl2-0.14 M NaCl-0.5% NP-40) and incubated for 3 min on ice, followed by a 3-min 6,000-rpm spin at 4°C in a bench-top microcentrifuge. The supernatant was designated the cytoplasmic fraction; the pellet was resuspended in 200 μl of the same lysis buffer and incubated for 3 additional minutes on ice and then spun as before. The supernatant from this second spin was discarded, and the pellet was designated the nuclear fraction. RNA was extracted from each fraction using Trizol per the manufacturer's instructions.
Gradient purification of Moloney MLV.
Pelleted virions were suspended in DMEM containing 10% calf serum. Gradient steps were prepared by diluting 60% (wt/vol) iodixanol (Optiprep; Nycomed Pharma) in PBS. One milliliter of each gradient step was layered at 1.2% intervals ranging from 18% iodixanol at the bottom of the tube to 6% at the top, for a total of 11 steps. The gradient was then topped off with 1 ml of concentrated virus, thereby forming the 12th step. Virus was spun through the gradient at 4°C using a Beckman SW41 rotor in the Sorvall Discovery ultracentrifuge at 27,500 rpm for 1 h. After centrifugation, 0.5 ml of each gradient step was collected, generating a total of 24 fractions. Each fraction was divided into 4 aliquots. Two aliquots were treated with 40 U micrococcal nuclease (Takara Biotech) at 37°C for 30 min. Of the micrococcal nuclease-treated samples, one set was digested in a detergent-free buffer (a 150-μl reaction mixture comprised of virions in iodixanol plus micrococcal nuclease mix) at a final concentration of 20 mM Tris-HCl (pH 8.0)-50 mM NaCl-2.5 mM CaCl2 in 0.5× PBS plus residual iodixanol. The other set of gradient aliquots were digested in the same buffer but in the presence of 0.1% Triton X-100. Reactions were stopped by the addition of 1 mM EDTA-500 μl Trizol, and the samples were stored at −70°C until further processing. The other 2 aliquots were left untreated and stored at −70°C. Viral RNA was extracted from thawed samples using Trizol according to the manufacturer's instructions.
RNase protection assay.
RNase protection assays used a chimeric riboprobe that hybridized to both the 5′ untranslated region (UTR) of MLV genomic RNA and part of 7SL RNA. The probe overlapped the splice donor site within the MLV 5′ UTR, making it possible to differentiate spliced and unspliced viral RNAs. To generate this riboprobe, the MLV 5′ UTR and murine 7SL sequences were amplified separately with primers containing linker sequences. The two primary PCR products were then used as templates in a secondary PCR to generate a single hybrid product which was subcloned into the EcoRV site of pBluescript SK(+) (Stratagene). The resulting plasmid was linearized with HindIII, and T3 RNA polymerase (Promega) was used to generate an antisense transcript of approximately 400 nt. The riboprobe was radiolabeled by including [α-32P]rCTP (Perkin-Elmer) in the in vitro transcription reaction mixture. The in vitro transcription reaction mixture was subjected to two phenol-chloroform extractions followed by one chloroform extraction, ethanol precipitated, and resuspended in 1× S1 hybridization buffer [80% formamide-40 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (pH 6.4)-400 mM NaCl-1 mM EDTA].
RNA samples were hybridized with riboprobe by first denaturing samples at 85°C for 5 min followed by a 16-h incubation at 60°C and then slow cooling to room temperature. Samples were digested by RNases A and T1 and separated on a 5% polyacrylamide-8 M urea gel in 1× Tris-borate-EDTA (TBE) buffer at 500 V for 1 h. Gels were dried and exposed to film or PhosphorImager screens (GE Biosciences). The riboprobe protected 200 nt of unspliced MLV viral RNA, 150 nt of spliced viral RNA, and 100 nt of 7SL RNA. Molar ratios of each band were determined by normalizing for the number of radiolabeled Cs in each protected fragment.
Northern blotting.
Viral and/or cellular RNAs were separated by 5% polyacrylamide-8 M urea gel electrophoresis in 1× TBE buffer at 500 V for 1 h and then transferred by electroblotting onto Zeta-probe GT nylon membranes (Bio-Rad) in 0.5× TBE buffer. After transfer, membranes were air dried and UV cross-linked (Stratalinker; Stratagene). Prehybridization was performed at 45°C for 2 h in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-5× Denhardt's solution-0.5% sodium dodecyl sulfate (SDS)-0.025 M sodium phosphate-625 μg/ml of denatured salmon sperm DNA. Oligonucleotide probes were denatured by boiling in a 100°C water bath for 10 min before being added to the membranes, and hybridization was performed at 45°C for 16 h. After hybridization, blots were washed in 2× SSC-0.1% SDS at 25°C for 15 min and then transferred to 0.5× SSC-0.1% SDS at 25°C for another 15 min. Damp blots were wrapped in plastic wrap and exposed to PhosphorImager screens and/or film. For reprobing, blots were stripped by at least two washes in 0.1% SDS at 80°C and then prehybridized and probed as described above.
Oligonucleotide probes.
All oligonucleotides were synthesized by Invitrogen. A total of 10 pmol of the oligonucleotides complementary to the RNAs of interest was 5′-end labeled using [γ-32P]ATP (Perkin-Elmer) and T4 polynucleotide kinase (NEB). Labeled oligonucleotides were separated from unincorporated nucleotides by purification on G-25 Sephadex columns (Roche) according to the manufacturer's instructions. The following oligonucleotides were used to probe Northern blots: 5′-TGCTCCGTTTCCGACCTGGGCCGGTTCACCCCTCCTT-3′ (7SL), 5′-CGAGATCGGGCGCGTTCAGGGTGGTATGGCCGTA-3′ (5S), 5′-GGGCCGCAAGTGCGTTCGAAGTGTCGATGATCAATGTG-3′ (5.8S), 5′-GAAAACATTCTTGGCAAATGCTTTCGCTC-3′ (18S), 5′-CACCCCAAGCGAGAATCATACCCCTAGACCAACG-3′ (tRNAPro), 5′-CGGGAAGGATTTAAACCAACGTTTTCGGGG-3′ (tRNAMet), 5′-CTGACTGTGAACAATCAATTGAGATAACTCACTAC-3′ (Y1), 5′-TAACTGGTTGTGATCAATTAGTTGTAAACACCACTACTC-3′ (Y3), 5′-CAGGCTGGCCTCGAACTCAGAAATCCGCCTGCCT-3′ (B1), 5′-CGTGTCATCCTTGCGCAGGGGCCATGCTAATCTTCTCTGT-3′ (U6), 5′-TCGTATACCCTTGACCGAAGACCGGTCCTC-3′ (7SK), 5′-GGATAAACCTCGCCCTGGGAAAACCACCTTCGTGATCATG-3′ (U1), and 5′-GCCGAAAGATTTATACGATCTGAAGAGAAACCAGAGT-3′ (U5).
Quantitative RT-PCR.
cDNA transcripts were generated from DNase-treated RNA samples derived from supernatants and whole-cell lysates of uninfected and Moloney MLV-infected cells. RNAs were DNased as follows. Samples were incubated in 100 μl of 25 mM Tris-Cl (pH 8)-50 mM MgCl2-5 mM dithiothreitol-100 U of RNase-free DNase (Roche)/ml-400 U of RNasin (Promega)/ml for 15 min at 37°C. DNase treatment was stopped by the addition of 25 μl of 50 mM EDTA-1.5 M sodium acetate (pH 5.2)-1% SDS. Samples were phenol-chloroform extracted, ethanol precipitated, air dried, and resuspended in diethylpyrocarbonate (Sigma)-treated water. A total of 150 ng random hexamers (Invitrogen) was annealed to RNA samples in the presence of a 1 mM mix of deoxynucleoside triphosphates (Invitrogen). Reverse transcriptase mix (1× first-strand buffer [20 mM Tris-HCl {pH 8.4}-50 mM KCl]-10 mM dithiothreitol- 5 mM MgCl2-40 U RNasin [Promega]) was added, and samples were incubated at 25°C for 2 min. MLV RNase H-RT (Superscript; Invitrogen) was then added, samples were incubated at 25°C for 10 min and transferred to 42°C, and after an additional 50 min, reactions were terminated by incubating at 70°C for 15 min. To degrade the RNA template, 20 U Escherichia coli RNase H was added to each sample and incubated at 37°C for 30 min. Samples were diluted in distilled water, with 50 μg/ml of herring DNA as a carrier, and stored at −20°C prior to use.
Quantitative RT-PCR was performed using a TaqMan probe-based assay system and the ABI 7700 sequence detection system (Perkin-Elmer). The mouse β-actin sequence was amplified and measured using the following primers and fluorescent-tagged probe: 5′-GGCCAACCGTGAAAA GATGA-3′, 5′-GACCAGAGGCATACAGGGACA-3′, and 5′-6-carboxyfluorescein (FAM)-TTCAACACCCCAGCCATGTACGTAGCC-6-carboxytetramethylrhodamine (TAMRA)-3′. To amplify and measure spliced MLV cDNAs, the following primers and probe were used: 5′-GGCTCGTCCGGGATCG-3′, 5′-TGGACTAAGTAGAGAGCCTGTAAGTGA-3′, and 5′-FAM- CTGCCCAGGGACCACCGACCCACCACCG-TAMRA-3′. To analyze unspliced MLV cDNAs, the following primers and probes were used: 5′-GATGGGAATCTCAGGACAATTGA, 5′-TCAAACAGGGTGGGACTGTTT-3, and 5′-FAM- CTGGACCAGACTCCCACAGGGTTTCA-TAMRA-3′. Standard curves were generated by plotting the threshold cycle when PCR product was detected versus the log of the input template copy number. This response was linear over a wide range of copy numbers, achieving correlation coefficients (r) of >0.93. Results within the linear range were used for all calculations. The ratio of the unspliced MLV transcript to β-actin transcript in cells was compared to the same ratio in virions, yielding the severalfold enrichment of the unspliced MLV RNA over β-actin mRNA in virions. Using the same approach, the severalfold enrichment of the unspliced MLV RNA over the spliced MLV RNA as well as the severalfold enrichment of the spliced MLV RNA over β-actin mRNA in virions were determined.
RESULTS
Confirmation that 7SL RNA and unspliced MLV RNA, but not spliced env mRNA, are enriched in virions.
The RNA component of the host signal recognition particle, 7SL RNA, is found among RNAs prepared from retroviral particles (16). Here, the presence of the approximately 300-nucleotide 7SL RNA within uninfected and chronically infected cells as well as its presence in RNA extracted from the medium of infected cells, but its absence from an equal volume of uninfected cell medium, were confirmed by Northern blot (Fig. 1A).
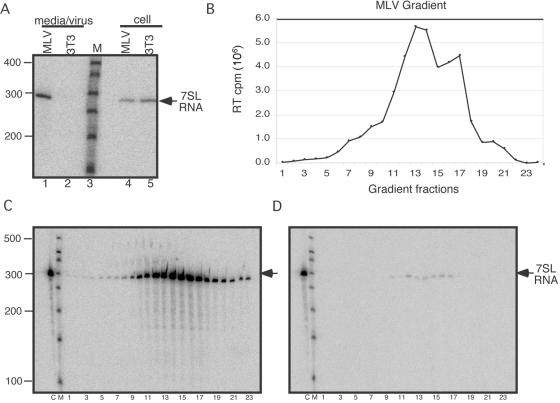
FIG. 1.
Analysis of 7SL RNA in MLV virions. (A) Northern blot to detect 7SL RNA packaging in MLV using RNAs derived from cells or medium/virus of MLV-infected or uninfected cells. Lanes: 1, RNA from supernatant of MLV-infected cells; 2, RNA from supernatant of uninfected cells; 3, size markers; 4, RNA from MLV-infected cells; 5, RNA from uninfected cells. (B) RT assay of gradient-purified MLV. (C and D) Northern blots of RNA from gradient-purified MLV probed for 7SL RNA. (C) Micrococcal nuclease-treated samples. (D) micrococcal nuclease plus Triton X-100. On both gradient blots, lanes are labeled as follows: C, cellular RNA; M, size markers; 1 to 23, gradient fractions. The position of 7SL RNA is indicated by the arrows.
To verify that 7SL RNA was associated with virions, virion preparations were purified on iodixanol gradients, and the sedimentation of MLV in the gradient was determined by assaying aliquots of each fraction for RT activity (Fig. 1B). Parallel aliquots of each fraction were digested by treatment with micrococcal nuclease, with (Fig. 1D) or without (Fig. 1C) Triton X-100, prior to RNA extraction, and the presence of 7SL RNA in each fraction was examined on Northern blots. For fractions digested in the absence of detergent, the nuclease-protected 7SL RNA peak corresponded to the virion peak determined by RT assay. Treatment of gradient fractions with detergent rendered 7SL RNA susceptible to nuclease digestion. The observations that 7SL RNA was within detergent-sensitive particles that cosedimented with MLV RT activity support the notion that 7SL RNA is encapsidated in virions. Although not all 7SL RNA was removed with micrococcal nuclease treatment, comparative PhosphorImager analysis of cell and viral samples (Fig. 1C and D, compare lanes 12 to 17) indicated that >90% of the RNA in each fraction was degraded in the presence of Triton X-100. These initial experiments confirmed previous observations and established the approaches used throughout the current study.
The molar ratio of 7SL RNA to unspliced viral genomic RNA in virions was compared to ratios within cells using RNase protection assays (Fig. 2). RNA samples were extracted from total cell lysates or pelleted virions and hybridized to a chimeric riboprobe comprised of a region that hybridized to MLV RNA linked to a 7SL-complementary region (Fig. 2A). By using a single riboprobe to detect both RNAs, the stoichiometry of probes to 7SL and to MLV RNAs remained equal. The MLV-specific portion of the probe overlapped the splice donor site within the MLV 5′ UTR so that a single probe could differentiate between spliced and unspliced viral RNA. As expected, spliced and unspliced MLV RNA as well as 7SL RNA products were detected in the cellular RNA (Fig. 2B, lane 5), while only unspliced viral RNA and 7SL RNA were observed in RNA samples from infected cell medium (lane 8). Neither product was detectable in an equal volume of medium from uninfected cells (Fig. 2B, lane 7). From the PG13 packaging cell line, a shorter unspliced MLV band (corresponding to a 5′ Ψ deletion in the viral protein expression construct) and 7SL RNA were detected in cells (Fig. 2B, lane 3), and only 7SL RNA was detected in the virions (lane 6).
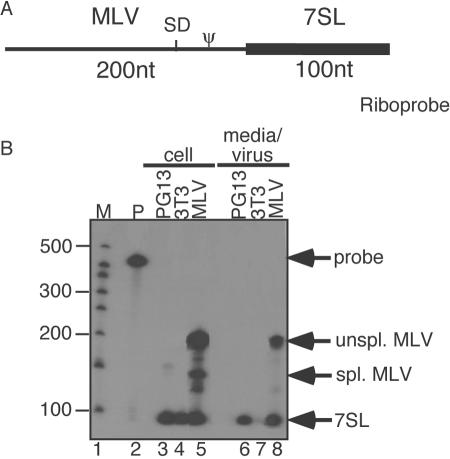
FIG. 2.
Packaging of 7SL and unspliced, but not spliced, MLV RNAs (RNase protection assay). (A) Riboprobe used. Ψ, packaging signal; SD, splice donor site; thin line, MLV-protected portion of riboprobe; boldface line, 7SL-protected region of riboprobe. (B) Viral and cellular RNAs from uninfected and MLV-infected cells using the 7SL-MLV riboprobe. The positions of the undigested probe, unspliced MLV RNA, spliced MLV RNA, and 7SL RNA bands are indicated on the right (probe, unspl. MLV, spl. MLV, and 7SL, respectively). Lanes: 1, size markers (M); 2, undigested probe; 3, RNA from packaging cells; 4, uninfected cells; 5, infected cells; 6, packaging cell supernatants; 7, uninfected cell supernatants; 8, infected cell supernatants. Bands were quantified by PhosphorImager analysis and normalized for the number of Cs in each protected probe fragment.
Quantification by PhosphorImager, after corrections for protected probe lengths, revealed that 7SL RNA was present in a three- to fourfold molar excess over unspliced MLV RNA in wild-type MLV virions. Thus, for every dimer of viral genomic RNA, there were 6 to 8 copies of 7SL RNA (Fig. 2B). This value is consistent with previous calculations (9).
7SL packaging determinants differ from those for the MLV genome.
The above-described results demonstrated that 7SL is packaged in particles produced by both Ψ+ and Ψ− MLV, which is consistent with findings from previous work (16). To address whether domains of viral NC protein that are necessary for genome packaging contribute to 7SL recruitment, the 7SL content of virions produced by NC mutants was examined (Fig. 3). The RNA content of three previously described assembly-competent MLV NC mutants that are severely impaired for genome packaging, including both basic flank and zinc finger mutants (23, 25), was examined by RNase protection assay (Fig. 3) using the same MLV-7SL chimeric riboprobe described in the legend of Fig. 2. As in Fig. 2, wild-type MLV virion RNA displayed both MLV genome and 7SL protection products (Fig. 3A, lane 1), whereas only 7SL RNA was detected among NC mutants' RNA (Fig. 3A, lanes 2 through 4). As an alternate way of addressing possible genome dependence of 7SL packaging, virus-producing cells were treated with actinomycin D under conditions known to deplete MLV particles of genomic RNA (19, 33), and 7SL and MLV genomic RNAs were examined by RNase protection assay (Fig. 3B). The results confirmed that this treatment vastly reduced MLV RNA in virions, but it did not affect 7SL RNA packaging (Fig. 3B). Virions generated by env-deficient proviruses also retained 7SL (data not shown). These latter conditions were examined because Env is the only MLV protein known to traffic through the secretory pathway and because Env has previously been suggested to affect MLV RNA trafficking (5). The presence of 7SL RNA in the genome-defective NC mutant particles indicated that the NC mechanisms involved in genome selection are not the determinants of 7SL RNA packaging, nor is 7SL encapsidation inhibited by some conditions that alter MLV genome trafficking.
MLV packages specific tRNA and rRNA subsets.
Northern blots were used to quantify the enrichment of additional small host RNAs in MLV particles by using 7SL RNA as a standard (Table 1). The question these experiments sought to address was whether or not the spectra and relative abundance of various host RNAs in virions were similar to those in cells, as would be predicted if virions contained a random sample of total host cell RNA. This experimental approach allowed us to calculate the relative abundance of each tested RNA (that is, whether the RNA was packaged in proportion to 7SL or if it was packaged more or less efficiently) but did not address these RNAs' molar quantities.
TABLE 1.
Relative abundance of host cell RNAs in MLV virions
| Host cell RNA species | Approx fold underrepresentation relative to 7SL RNA |
|---|---|
| 7SL | 1a |
| tRNAPro | 3 |
| tRNAMet | >180 |
| tRNAVal | >250 |
| 5S RNA | 3 |
| 5.8S RNA | 90 |
| 18S RNA | 140 |
| Y1 RNA | 1 |
| Y3 RNA | 1 |
| B1 RNA | 2 |
| U1 snRNA | 700 |
| U2 snRNA | 50 |
| U4 snRNA | 180 |
| U5 snRNA | 170 |
| U6 snRNA | 5 |
| 7SK RNA | 20 |
Values for 7SL RNA were set to 1. Values for the other RNAs were determined using Northern blots of cell and viral samples coprobed for each RNA plus 7SL, as described in the text.
To quantify each host RNA's relative abundance, filters with cellular and virion RNAs were probed with a mixture of two oligonucleotide probes: one specific to 7SL and the other specific to a second test RNA. Signals were quantified by PhosphorImager analysis, with test RNA signals normalized by the 7SL signal for each lane. A single Northern blot was repeatedly stripped and reprobed in these experiments, with the cellular RNA sample lane (for example, see Fig. 4, lane C) serving as a reference for the intracellular ratios of the compared RNAs. Note that the two probes used in each hybridization differed from one another in specific activity. For less-abundant test RNAs, the 7SL probe was deliberately radiolabeled to a low specific activity to facilitate simultaneous detection of both 7SL and the other host RNA. Thus, the intensities of bands on the Northern blots do not represent the coprobed RNAs' molar ratios.
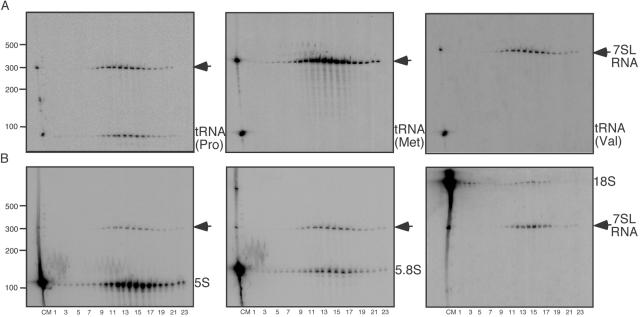
FIG. 4.
Northern blots to assess virion enrichment of ribosomal RNAs and tRNAs. (A) Northern blots using RNA from gradient-purified, micrococcal nuclease-treated MLV probed for 7SL RNA and tRNAs. Left, probed for tRNAPro; middle, probed for tRNAMet; right, probed for tRNAVal. On all blots, lanes loaded right to left are cellular RNA, size markers, and gradient fractions 1 to 23, as indicated below panel B. (B) Northern blots using RNA from gradient-purified, micrococcal nuclease-treated MLV probed for 7SL RNA and rRNAs. Left, probed for 5S rRNA; middle, probed for 5.8S rRNA; right, probed for 18S rRNA. On all blots, lanes are labeled as follows: C, cellular RNA; M, size markers; 1 to 23, gradient fractions. The position of 7SL RNA is indicated by the arrows.
It is known that retroviruses preferentially package the tRNA required for priming reverse transcription (37). To investigate the magnitude of discrimination in tRNA packaging in MLV, Northern blots of RNA from gradient-purified, micrococcal nuclease-treated virions were probed for the MLV primer tRNAPro and separately for two nonprimer tRNAs: tRNAMet and tRNAVal (Fig. 4A [note that the leftmost lanes of each blot contain a sample of total cell RNA]). The packaging of these three tRNAs differed strikingly: primer tRNAPro was present in virions, and its proportion in the cell and in virions relative to the 7SL standard differed by only threefold (Fig. 4A, left, compare peak gradient fractions, lanes 12 to 17, with cell RNA sample, lane C; see also Table 1). Although tRNAMet and tRNAVal were readily detectable in the cellular RNA lane, unlike the primer tRNAPro, they were not detectable in viral RNA preparations by the approaches used here. Both tRNAVal and tRNAMet were present below the threshold of detection in their respective assays and were at 200-fold less enriched than primer tRNAPro (Fig. 4A, middle and right panels).
The rRNA content of MLV virions was also examined (Fig. 4B), and the results confirmed the enrichment of a distinct subset of ribosomal RNAs. Again, using 7SL RNA as a standard, Northern blots of gradient-purified, micrococcal nuclease-treated virions were probed for 5S, 5.8S, and 18S rRNAs (Fig. 4B). Although all three RNAs were detected in the gradient-fractionated virion RNA and thus their encapsidation was not as biased as tRNA packaging, a comparison of the viral and cellular samples showed that 5S rRNA was at least 30-fold more enriched in MLV particles than were 5.8S or 18S rRNAs (Fig. 4B and Table 1).
MLV packaging of cytoplasmic and nuclear host RNAs.
At least for mRNAs, the prevailing model for host RNA encapsidation is random incorporation at assembly sites, and it has been suggested that the packaging of most classes of host RNAs occurs by accident (41). Because particle assembly is first detected at the plasma membrane, this suggests that virions should contain a random collection of cytoplasmic RNAs. To address the extent to which cytoplasmic RNAs were encapsidated by MLV, the packaging of abundant cytoplasmic RNAs such as Y1 and Y3 was examined (Fig. 5A, left and middle panels). Y1 and Y3 are small cytoplasmic RNAs of uncertain function that are components of the Ro RNP complex in association with Ro protein (18, 52). Here, it was observed that both Y1 and Y3 RNAs were abundant in virions and that their ratios relative to 7SL RNA were similar to the ratios in cells (Fig. 5A and Table 1).
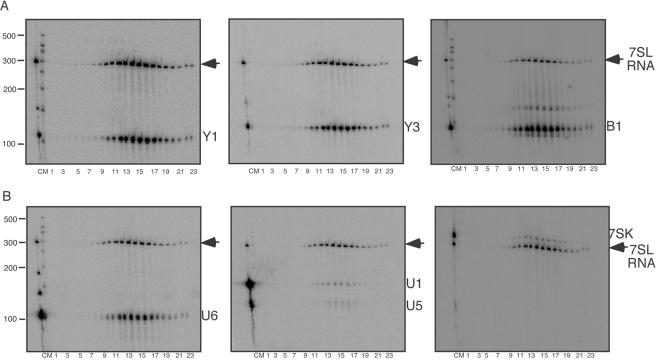
FIG. 5.
Northern blots to assess virion enrichment of some other small host RNAs. (A) Northern blots using RNA from gradient-purified, micrococcal nuclease-treated MLV probed for 7SL RNA and cytoplasmic RNAs. Left, probed for Y1 RNA; middle, probed for Y3 RNA; right, probed for B1 RNA. (B) Northern blots using RNA from gradient-purified, micrococcal nuclease-treated MLV probed for 7SL RNA and nuclear RNAs. Left, probed for U6 snRNA; middle, probed for U1 and U5 snRNAs; right, probed for 7SK RNA. On all blots, lanes are labeled as follows: C, cellular RNA; M, size markers; 1 to 23, gradient fractions. The position of 7SL RNA is indicated by the arrows.
B1 RNA is a murine SINE RNA abundantly expressed in the ∼110-nucleotide form in NIH 3T3 cells that is structurally related to a portion of 7SL RNA (48). The enrichment of B1 RNA was examined (Fig. 5A, right, and Table 1). Like Y1 and Y3, the virion enrichment of this cytoplasmic RNA was similar to that of 7SL.
Packaging of some nuclear small RNAs also was examined. A recent report demonstrated that Rous sarcoma virus particles contain cellular spliceosomal RNAs, with U6 snRNA being the most abundant (22). Here, MLV Northern blots were probed for U1, U2, U4, U5, and U6 spliceosomal RNAs and for 7SK small nuclear RNA, a 330-nt-long RNA that acts as a negative regulator of the transcription factor p-TEFb (17) (Fig. 5B and Table 1). The results demonstrated that U6 snRNA was encapsidated by MLV (Fig. 5B, left) and was only slightly less enriched in virions than 7SL RNA. Under conditions where U1, U2, U4, and U5 snRNAs were readily detectable in total cell RNA, their signals in the viral RNA samples were very low, indicating a significant underrepresentation of most spliceosomal components in virions (Table 1, Fig. 5B, middle, and data not shown). 7SK RNA, although more highly represented in virions than U1 and U5, was less enriched in wild-type MLV than U6 snRNA (Fig. 5B, right).
To confirm that biases in RNA packaging did not mirror, at least on a gross level, the intracellular partitioning of these RNAs into different cellular compartments, we isolated RNAs from cytoplasmic and nuclear fractions from three cell types: those chronically infected with MLV, the MLV-based packaging cell line PG13 (39), and uninfected NIH 3T3 cells, which are the parent line to both PG13 and the chronically infected cells used here. RNA content of whole cells and the crude nuclear and cytoplasmic fractions was compared to that of pelleted virions by Northern blotting (Fig. 6). Some nuclear leakage during cell fractionation was evident from the detection of U6 in cytoplasmic fractions (Fig. 6C, lanes 10 to 12). However, nearly all of the 7SL, which is cytoplasmic except for a minor nucleolar component (45), was detected in the cytoplasmic fraction, and the majority of U5, which is nuclear most of its lifetime, was detected in the nuclear fraction (Fig. 6A, lanes 7 to 12). Subsequent probing of this blot for 7SL and Y1 (Fig. 6B) revealed that although similar ratios of 7SL to Y1 were present in the cytoplasmic fractions of all three cell types, these RNAs exhibited differential packaging, with particles shed by PG13 cells showing diminished packaging of Y1 RNA, as determined by 7SL per Y1 ratios in the virion lanes (Fig. 6B).
FIG. 6.
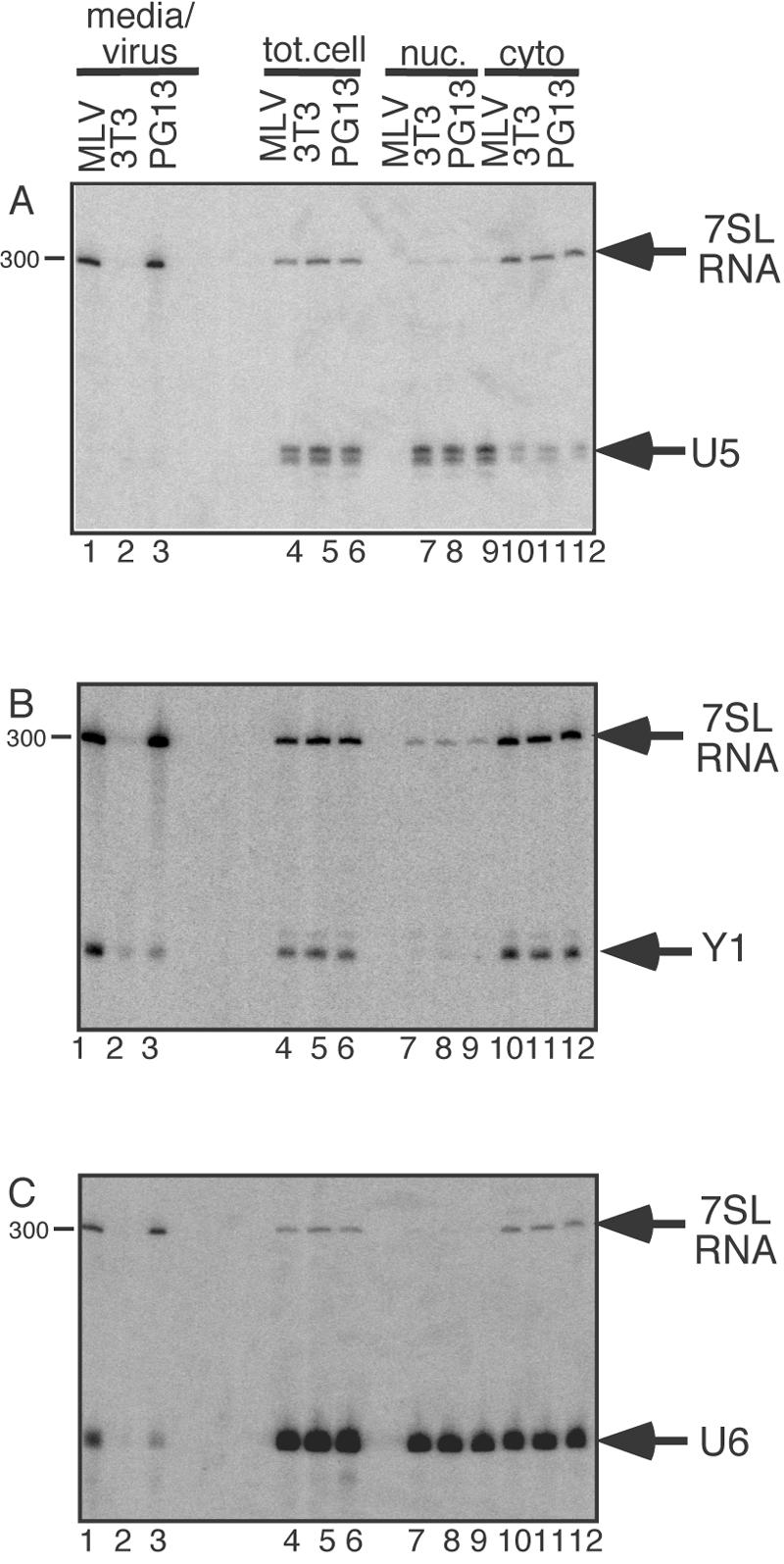
Northern blots of viral, total cell (tot.cell), nuclear (nuc.), and cytoplasmic (cyto) RNAs. Top, probed for 7SL RNA and U5 snRNA; middle, probed for 7SL RNA and Y1 RNA; bottom, probed for 7SL RNA and U6 snRNA. On all blots, lanes 1 to 3 are supernatant/viral RNA, lanes 4 to 6 are total cell RNA, lanes 7 to 9 are nuclear RNA, and lanes 10 to 12 are cytoplasmic RNA.
This same blot was reprobed for 7SL and U6 (Fig. 6C). The presence of U6 in cytoplasmic fractions was pronounced, presumably due to differential cytoplasmic leakage during cell fractionation. Examination of the virion RNAs revealed that, as was the case for Y1, U6 RNA was less enriched in particles produced by PG13 packaging cells than it was in wild- type MLV. The greater enrichment of Y1 and U6 RNAs in wild-type virions than in PG13-produced particles was observed consistently in repeated experiments with independently harvested and processed virion RNA preparations (data not shown).
mRNA and genome packaging.
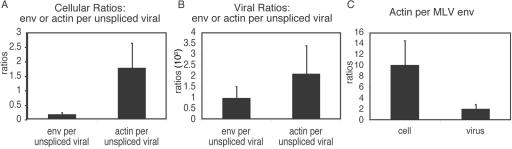
Small amounts of host mRNA are believed to become encapsidated at random, while the enrichment of viral genomic RNA in virions is well established (7). To examine both their molar amounts and their levels of enrichment in MLV particles, the encapsidation of env mRNA, unspliced viral RNA, and host actin mRNA were quantified by RT-PCR (Fig. 7). The data indicated that whereas there was about twice as much actin mRNA as unspliced viral RNA in cells, there was only about one actin mRNA per 500 viral genomic RNAs in virions, and thus, the viral genome monomer was more than 1,000-fold enriched over actin mRNA in virions. After accounting for the fact that cells contained more actin mRNA than env mRNA but that the amounts of env and actin mRNAs were more similar in virions (Fig. 7B and C), the data indicated that env mRNA was three- to fivefold more enriched in virions than was actin mRNA.
FIG. 7.
Histograms of quantitative RT-PCR data. (A) cellular ratios of unspliced MLV genome cDNA versus actin cDNA and spliced MLV env cDNA; (B) viral ratios of unspliced MLV genome cDNA versus actin cDNA and spliced MLV env cDNA; (C) actin cDNA versus MLV env cDNA. x axis, source of RNA for cDNA synthesis; y axis, ratio of actin to MLV env.
DISCUSSION
MLV selectively packages its own genome, but virions also contain host RNAs (7). Here, we identified and quantified several host RNAs in MLV, including the Y1 and Y3 RNA components of the Ro ribonucleoprotein complex, which had not been previously been reported in retroviral particles. In humans, autoantibodies to the Ro complex develop in association with systemic lupus erythematosus and Sjögren's syndrome (30), and accumulating evidence suggests that Ro may be involved in RNA quality control (52). The data here indicated that the abundance of 7SL and several other small host RNAs, relative to unspliced viral RNA, was similar in virions and in cells, arguing that if genomic RNA is considered to be enriched in virions, these cellular RNAs should also be considered enriched. In addition, previously unrecognized host RNAs with lengths similar to those of tRNAs, such as Y1 and Y3, may help explain the cluster of 4S RNAs that is consistently observed in virions and suggest that the MLV virion 4S RNA population may not be entirely, or even principally, composed of tRNAs.
Host RNAs readily detected in MLV included 7SL RNA, primer tRNAPro, U6 snRNA, 5S rRNA, B1 retroelement RNA, and Y1 and Y3 Ro RNP RNAs. Nuclear 7SK RNA and U2 snRNA showed intermediate levels of enrichment, while tRNAMet; tRNAVal; actin and env mRNAs; U1, U4, and U5 snRNAs; and 5.8S and 18S rRNAs were significantly less enriched. The exclusion of cytoplasmic RNAs such as tRNAMet and tRNAVal, the relative enrichment in virions of nuclear RNAs U6 and 7SK, and the differential packaging of Y1 and U6 in particles formed by Ψ+ and Ψ− proviruses all attest to the nonrandomness of host RNA encapsidation by MLV.
Other than the primer tRNA, the consequences of incorporating host RNAs into retroviral particles are largely unknown. In rare instances, host RNA sequences have been transduced by retroviruses, presumably as a consequence of encapsidation (53). Reverse transcription products of 7SL RNA and U6 snRNA have been detected among RSV endogenous reverse transcription products (16, 22), indicating that nonviral RNAs can function as templates for reverse transcription. Based on these observations and the high prevalence of U6 pseudogenes (12), it has been postulated that retroviral reverse transcription and integration of cellular RNAs may contribute to the pseudogenes observed in host genomes (22).
The specific determinants governing the recruitment and incorporation of host RNA packaging remain undefined, although previously published observations and those described here provide a foundation for speculation. Nonretroviral RNAs engineered to contain Ψ sequences can be packaged into virions and are encapsidated as dimers (24). Thus, it is possible that some enriched host RNAs may contain sequences that are functionally equivalent to Ψ. As a first attempt at identifying such motifs, the highly enriched host RNAs (7SL, U6, tRNAPro, Y1, and Y3 RNAs) were aligned using MegAlign from DNAStar. However, besides previously described patches of identity between 5S and 7SL RNA (58), there were no significant homologies (data not shown). Although this analysis did not address possible contributions of these RNAs' folded structures, it showed that these RNAs do not have any obvious sequences in common that may mediate their recruitment. Similar analysis demonstrated the absence of significant complementarity between the host RNAs and the MLV genome.
Some RNAs may be largely excluded from packaging due to molecular interactions that confine them. For example, tRNAs and their synthetases are normally compartmentalized in a way that relies on the integrity of the actin cytoskeleton, and tRNAs are never free of the translation machinery once they associate with it (26, 51). Some host RNAs may become passively incorporated into virions due to their presence at assembly or subassembly sites. If so, trafficking pathways employed by different host RNAs and/or RNPs and the subcellular locales where viral protein precursors are synthesized or recruited may contribute to differential RNA packaging.
Finding nuclear RNAs in MLV was surprising. We first detected U6 snRNA in MLV when a U6 probe was used as a control for cell fractionation. Although leakage of U6 into cytoplasmic fractions is a common technical problem (10), as was the case in the fractionation presented here, the entire U6 biosynthetic process takes place in the nucleus, and U6 is not known to have a cytoplasmic presence (10). While this work was in progress, U6 encapsidation by Rous sarcoma virus was reported by Giles et al. (22), who determined that RSV packages approximately 1 copy of U6 snRNA per virion. One copy per virion was also the value we determined for U6 packaging by infectious MLV (data not shown). Giles et al. (22) demonstrated an association between U6 snRNA and RSV genomic RNA, which may explain the reduced levels of U6 observed here for PG13 particles that lacked genomic RNA. How nuclear RNAs are recruited to virions that form at the plasma membrane is unclear. However, observations that avian sarcoma virus Gag enters the nucleus prior to assembly and that this replication step may be required for genome maturation challenge the classic view that late stages of retroviral replication occur solely in the cytoplasm and leave open the possibility that the nuclear RNAs in retroviral particles were recruited from the nucleus (47).
The current work confirmed widely reported differences in spliced and unspliced virus-encoded RNA packaging and determined that the extent to which spliced MLV RNA was excluded from packaging was not greater than the exclusion of actin mRNA. This may indicate a difference between MLV and certain avian retroviruses, for which env RNAs possess sequences that contribute to their exclusion from packaging (4), and a possible similarity to human immunodeficiency virus type 1, in which Ψ− viral RNAs are more enriched than cell mRNAs (8). The generally low level of mRNA virion enrichment, compared to the packaging of certain other host RNAs, may simply reflect sequestration of mRNAs at polysomes. Alternately, it might be that RNA splicing is incompatible with recruitment for packaging, as appears to be the case for avian leukosis virus spliced viral RNAs which contain Ψ but are not packaged (4).
Potential roles for most host RNAs within retroviruses are as yet unknown. Because RNA is involved in assembly but virions assemble efficiently in the absence of viral RNA, host RNAs probably contribute to the assembly process. However, whether any kind of host RNA can serve in assembly or if a specific species plays this role remains unknown. One highly speculative hypothesis is that some host RNAs may contribute to assembly by playing roles analogous to those they ordinarily play in the cell. For instance, 7SL RNA may act as scaffolding around which virus particles are formed. This hypothesis has no experimental support but is suggested by the fact that 7SL RNA fills a scaffolding role for the signal recognition particle (29). Consistent with this hypothesis, numerous attempts in this laboratory to identify conditions under which particles can bud in the absence of 7SL RNA have yet to bear results (our unpublished results).
It seems likely that the majority of the small RNAs observed in virions here are not required for replication. It has previously been reported that the encapsidated small RNAs vary by cell type without affecting infectivity (28). Although we calculated the relative and not the molar amounts of cellular RNAs in virions, the data here suggest that many of these RNAs are present at far less than 1 copy per virion. For example, the level of 7SK RNA enrichment, paired with estimates of 7SK molecules per cell (56), suggests that roughly 1 in 100 MLV particles contains a molecule of 7SK RNA. Thus, barring the possibility that their presence may influence the relatively low ratio of infectious units per particle in retroviruses, these low-abundance RNAs cannot be necessary for replication.
One interesting observation from the current study was an apparent bias toward the packaging of RNA polymerase III (Pol III) transcripts. All RNAs we found enriched in MLV particles (7SL RNA, primer tRNA, 5S rRNA, Ro RNP RNAs, and U6 snRNA) were RNA polymerase III transcripts, while small Pol II transcripts, such as the core spliceosomal RNAs other than U6, were largely excluded from packaging. This observation raises the possibility that some common feature or consequence of Pol III transcription might contribute to the packageability of the enriched RNAs. Ultimately, an understanding of the causes and effects of these RNAs' encapsidation may help unravel the RNA-trafficking pathways employed by MLV or other aspects of early assembly.
Acknowledgments
This work was supported by an American Heart Association predoctoral fellowship to A.O.-N. and by NIH grant number CA69300 to A.T.
We thank Sharon Marr, Mallika Krishnan, and Crystal Pacut for generating the NC mutants, Nicole Robson-Dixon for designing the RNase protection probe, Sandra Wolin and Joan Steitz for valuable advice, and Kathy Spindler and David Miller for critical readings of the manuscript.
REFERENCES
- 1.Adam, M. A., and A. D. Miller. 1988. Identification of a signal in a murine retrovirus that is sufficient for packaging of nonretroviral RNA into virions. J. Virol. 62:3802-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adkins, B., and T. Hunter. 1981. Identification of packaged cellular mRNA in virions of Rous sarcoma virus. J. Virol. 39:471-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aronoff, R., and M. Linial. 1991. Specificity of retroviral RNA packaging. J. Virol. 65:71-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banks, J. D., B. O. Kealoha, and M. L. Linial. 1999. An MΨ-containing heterologous RNA, but not env mRNA, is efficiently packaged into avian retroviral particles. J. Virol. 73:8926-8933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basyuk, E., T. Galli, M. Mougel, J.-M. Blanchard, M. Sitbon, and E. Bertrand. 2003. Retroviral genomic RNAs are transported to the plasma membrane by endosomal vesicles. Dev. Cell 5:161-174. [DOI] [PubMed] [Google Scholar]
- 6.Beasley, B. E., and W. S. Hu. 2002. cis-Acting elements important for retroviral RNA packaging specificity. J. Virol. 76:4950-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berkowitz, R., J. Fisher, and S. P. Goff. 1996. RNA packaging. Curr. Top. Microbiol. Immunol. 214:177-218. [DOI] [PubMed] [Google Scholar]
- 8.Berkowitz, R. D., M. L. Hammarskjold, C. Helga-Maria, D. Rekosh, and S. P. Goff. 1995. 5′ regions of HIV-1 RNAs are not sufficient for encapsidation: implications for the HIV-1 packaging signal. Virology 212:718-723. [DOI] [PubMed] [Google Scholar]
- 9.Bishop, J. M., W. E. Levinson, D. Sullivan, L. Fanshier, N. Quintrell, and J. Jackson. 1970. The low molecular weight RNAs of Rous sarcoma virus. II. The 7S RNA. Virology 42:927-937. [DOI] [PubMed] [Google Scholar]
- 10.Boelens, W. C., I. Palacios, and I. W. Mattaj. 1995. Nuclear retention of RNA as a mechanism for localization. RNA 1:273-283. [PMC free article] [PubMed] [Google Scholar]
- 11.Brian, D. A., A. R. Thomason, F. M. Rottman, and L. F. Velicer. 1975. Properties of feline leukemia virus. III. Analysis of the RNA. J. Virol. 16:535-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buzdin, A., S. Ustyugova, E. Gogvadze, T. Vinogradova, Y. Lebedev, and E. Sverdlov. 2002. A new family of chimeric retrotranscripts formed by a full copy of U6 small nuclear RNA fused to the 3′ terminus of L1. Genomics 80:402-406. [DOI] [PubMed] [Google Scholar]
- 13.Campbell, S., and V. M. Vogt. 1995. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 69:6487-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cen, S., H. Javanbakht, S. Kim, K. Shiba, R. Craven, A. Rein, K. Ewalt, P. Schimmel, K. Musier-Forsyth, and L. Kleiman. 2002. Retrovirus-specific packaging of aminoacyl-tRNA synthetases with cognate primer tRNAs. J. Virol. 76:13111-13115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheevers, W. P., B. G. Archer, and T. B. Crawford. 1977. Characterization of RNA from equine infectious anemia virus. J. Virol. 24:489-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, P. J., A. Cywinski, and J. M. Taylor. 1985. Reverse transcription of 7S L RNA by an avian retrovirus. J. Virol. 54:278-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen, R., Z. Yang, and Q. Zhou. 2004. Phosphorylated positive transcription elongation factor b (P-TEFb) is tagged for inhibition through association with 7SK snRNA. J. Biol. Chem. 279:4153-4160. [DOI] [PubMed] [Google Scholar]
- 18.Chen, X., and S. L. Wolin. 2004. The Ro 60 kDa autoantigen: insights into cellular function and role in autoimmunity. J. Mol. Med. 82:232-239. [DOI] [PubMed] [Google Scholar]
- 19.Dorman, N., and A. Lever. 2000. Comparison of viral genomic RNA sorting mechanisms in human immunodeficiency virus type 1 (HIV-1), HIV-2, and Moloney murine leukemia virus. J. Virol. 74:11413-11417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duesberg, P. H., and W. S. Robinson. 1966. Nucleic acid and proteins isolated from the Rauscher mouse leukemia virus (MLV). Proc. Natl. Acad. Sci. USA 55:219-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faras, A. J., A. C. Garapin, W. E. Levinson, J. M. Bishop, and H. M. Goodman. 1973. Characterization of the low-molecular-weight RNAs associated with the 70S RNA of Rous sarcoma virus. J. Virol. 12:334-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giles, K. E., M. Caputi, and K. L. Beemon. 2004. Packaging and reverse transcription of snRNAs by retroviruses may generate pseudogenes. RNA 10:299-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorelick, R. J., D. J. Chabot, D. E. Ott, T. D. Gagliardi, A. Rein, L. E. Henderson, and L. O. Arthur. 1996. Genetic analysis of the zinc finger in the Moloney murine leukemia virus nucleocapsid domain: replacement of zinc-coordinating residues with other zinc-coordinating residues yields noninfectious particles containing genomic RNA. J. Virol. 70:2593-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hibbert, C. S., J. Mirro, and A. Rein. 2004. mRNA molecules containing murine leukemia virus packaging signals are encapsidated as dimers. J. Virol. 78:10927-10938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Housset, V., H. De Rocquigny, B. P. Roques, and J. L. Darlix. 1993. Basic amino acids flanking the zinc finger of Moloney murine leukemia virus nucleocapsid protein NCp10 are critical for virus infectivity. J. Virol. 67:2537-2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hudder, A., L. Nathanson, and M. P. Deutscher. 2003. Organization of mammalian cytoplasm. Mol. Cell. Biol. 23:9318-9326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang, M., J. Mak, A. Ladha, E. Cohen, M. Klein, B. Rovinski, and L. Kleiman. 1993. Identification of tRNAs incorporated into wild-type and mutant human immunodeficiency virus type 1. J. Virol. 67:3246-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang, M., J. Mak, M. A. Wainberg, M. A. Parniak, E. Cohen, and L. Kleiman. 1992. Variable tRNA content in HIV-1IIIB. Biochem. Biophys. Res. Commun. 185:1005-1015. [DOI] [PubMed] [Google Scholar]
- 29.Keenan, R. J., D. M. Freymann, R. M. Stroud, and P. Walter. 2001. The signal recognition particle. Annu. Rev. Biochem. 70:755-775. [DOI] [PubMed] [Google Scholar]
- 30.Keene, J. D. 1996. RNA recognition by autoantigens and autoantibodies. Mol. Biol. Rep. 23:173-181. [DOI] [PubMed] [Google Scholar]
- 31.Kleiman, L. 2002. tRNA(Lys3): the primer tRNA for reverse transcription in HIV-1. IUBMB Life 53:107-114. [DOI] [PubMed] [Google Scholar]
- 32.Levin, J. G., and J. G. Seidman. 1981. Effect of polymerase mutations on packaging of primer tRNAPro during murine leukemia virus assembly. J. Virol. 38:403-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levin, J. G., P. M. Grimley, J. M. Ramseur, and I. K. Berezesky. 1974. Deficiency of 60 to 70S RNA in murine leukemia virus particles assembled in cells treated with actinomycin D. J. Virol. 14:152-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levin, J. G., and J. G. Seidman. 1979. Selective packaging of host tRNA's by murine leukemia virus particles does not require genomic RNA. J. Virol. 29:328-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin, F. H., and H. Thormar. 1971. Characterization of ribonucleic acid from visna virus. J. Virol. 7:582-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Linial, M. L., and A. D. Miller. 1990. Retroviral RNA packaging: sequence requirements and implications. Curr. Top. Microbiol. Immunol. 157:125-152. [DOI] [PubMed] [Google Scholar]
- 37.Mak, J., and L. Kleiman. 1997. Primer tRNAs for reverse transcription. J. Virol. 71:8087-8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mann, R., R. C. Mulligan, and D. Baltimore. 1983. Construction of a retrovirus packaging mutant and its use to produce helper-free defective retrovirus. Cell 33:153-159. [DOI] [PubMed] [Google Scholar]
- 39.Miller, A. D., J. V. Garcia, N. von Suhr, C. M. Lynch, C. Wilson, and M. V. Eiden. 1991. Construction and properties of retrovirus packaging cells based on gibbon ape leukemia virus. J. Virol. 65:2220-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muriaux, D., J. Mirro, D. Harvin, and A. Rein. 2001. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. USA 98:5246-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muriaux, D., J. Mirro, K. Nagashima, D. Harvin, and A. Rein. 2002. Murine leukemia virus nucleocapsid mutant particles lacking viral RNA encapsidate ribosomes. J. Virol. 76:11405-11413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pambalk, K., C. Hohenadl, B. Salmons, W. H. Gunzburg, and M. Renner. 2002. Specific packaging of spliced retroviral vector transcripts lacking the Psi-region. Biochem. Biophys. Res. Comm. 293:239-246. [DOI] [PubMed] [Google Scholar]
- 43.Peters, G., F. Harada, J. E. Dahlberg, A. Panet, W. A. Haseltine, and D. Baltimore. 1977. Low-molecular-weight RNAs of Moloney murine leukemia virus: identification of the primer for RNA-directed DNA synthesis. J. Virol. 21:1031-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pfeiffer, J. K., R. S. Topping, N. H. Shin, and A. Telesnitsky. 1999. Altering the intracellular environment increases the frequency of tandem repeat deletion during Moloney murine leukemia virus reverse transcription. J. Virol. 73:8441-8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Politz, J. C., S. Yarovoi, S. M. Kilroy, K. Gowda, C. Zwieb, and T. Pederson. 2000. Signal recognition particle components in the nucleolus. Proc. Natl. Acad. Sci. USA 97:55-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sawyer, R. C., and J. E. Dahlberg. 1973. Small RNAs of Rous sarcoma virus: characterization by two-dimensional polyacrylamide gel electrophoresis and fingerprint analysis. J. Virol. 12:1226-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scheifele, L. Z., R. A. Garbitt, J. D. Rhoads, and L. J. Parent. 2002. Nuclear entry and CRM1-dependent nuclear export of the Rous sarcoma virus Gag polyprotein. Proc. Natl. Acad. Sci. USA 99:3944-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmid, C. W. 1998. Does SINE evolution preclude Alu function? Nucleic Acids Res. 26:4541-4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schneemann, A., and D. Marshall. 1998. Specific encapsidation of nodavirus RNAs is mediated through the C terminus of capsid precursor protein alpha. J. Virol. 72:8738-8746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sciortino, M. T., M. Suzuki, B. Taddeo, and B. Roizman. 2001. RNAs extracted from herpes simplex virus 1 virions: apparent selectivity of viral but not cellular RNAs packaged in virions. J. Virol. 75:8105-8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stapulionis, R., and M. P. Deutscher. 1995. A channeled tRNA cycle during mammalian protein synthesis. Proc. Natl. Acad. Sci. USA 92:7158-7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stein, A. J., G. Fuchs, C. Fu, S. L. Wolin, and K. M. Reinisch. 2005. Structural insights into RNA quality control: the Ro autoantigen binds misfolded RNAs via its central cavity. Cell 121:529-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sugden, B. 1993. How some retroviruses got their oncogenes. Trends Biochem. Sci. 18:233-235. [DOI] [PubMed] [Google Scholar]
- 54.Terhune, S. S., J. Schroer, and T. Shenk. 2004. RNAs are packaged into human cytomegalovirus virions in proportion to their intracellular concentration. J. Virol. 78:10390-10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Waters, L. C., B. C. Mullin, E. G. Bailiff, and R. A. Popp. 1975. tRNA's associated with the 70S RNA of avian myeloblastosis virus. J. Virol. 16:1608-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu, Y.-T., E. Scharl, C. Smith, and J. A. Steitz. 1998. The growing world of small nuclear ribonucleoproteins, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 57.Zhang, Y., and E. Barklis. 1995. Nucleocapsid protein effects on the specificity of retrovirus RNA encapsidation. J. Virol. 69:5716-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zwieb, C. 1985. The secondary structure of the 7SL RNA in the signal recognition particle: functional implications. Nucleic Acids Res. 13:6105-6124. [DOI] [PMC free article] [PubMed] [Google Scholar]