Abstract
The Hepatitis C virus (HCV), a member of the family Flaviviridae, is a major cause of chronic liver disease. Patients are currently treated with alpha interferon (IFN-α) that is given alone or in combination with ribavirin. Unfortunately, this treatment is ineffective in eliminating the virus in a large proportion of individuals. IFN-induced antiviral activities have been intensively studied in the HCV replicon system. It was found that both IFN-α and IFN-γ inhibit HCV replicons, but the underlying mechanisms have not yet been identified. Of note is that nearly all of these studies were performed with the human hepatoma cell line Huh-7. Here, we report that genotypes 1b and 2a replicons also replicate in the human hepatoblastoma cell line HuH6. Similar to what has been described for Huh-7 cells, we observed that efficient HCV replication in HuH6 cells depends on the presence of cell culture-adaptive mutations and the permissiveness of the host cell. However, three major differences exist: in HuH6 cells, viral replication is (i) independent from ongoing cell proliferation, (ii) less sensitive to certain antiviral compounds, and (iii) highly resistant to IFN-γ. The latter is not due to a general defect in IFN signaling, as IFN-γ induces the nuclear translocation of signal transducer and activator of transcription 1 (STAT1), the enhanced transcription of several IFN-regulated genes, and the inhibition of unrelated viruses such as influenza A virus and Semliki Forest virus. Taken together, the results establish HuH6 replicon cells as a valuable tool for IFN studies and for the evaluation of antiviral compounds.
The Hepatitis C virus (HCV) belongs to the genus Hepacivirus within the family Flaviviridae (52). At least six different HCV genotypes exist which show specific geographical distributions. For each genotype, a series of more closely related subtypes have been described that differ from one another by 20 to 25% in their nucleotide sequence. HCV has an ∼9.6-kb single-stranded RNA genome of positive polarity (for a review, see reference 4). The genome is flanked by highly structured nontranslated regions (NTRs) important for both RNA translation and replication. Within the 5′ NTR, an internal ribosome entry site (IRES) has been identified that permits expression of the viral proteins in the absence of a cap structure (44). The HCV genome encodes a large, single polyprotein of approximately 3,000 amino acids that is co- and posttranslationally cleaved by cellular and viral proteinases into 10 polypeptides (core, E1, E2, p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B). The production of an additional viral protein by ribosomal frameshift has also been reported (53, 57), but its function remains to be defined.
Worldwide, about 170 million individuals have been infected with HCV; in >80% of all cases, the virus establishes a persistent infection (56) which frequently leads to chronic liver disease with liver fibrosis, cirrhosis, and eventually hepatocellular carcinoma (24, 50). The most advanced therapy for hepatitis C patients currently consists of polyethylene glycol-conjugated alpha interferon (IFN-α) and ribavirin, but this treatment has its limitations (37). One problem is the relatively poor response of patients infected with certain HCV genotypes. For example, only 50% of patients infected with genotype 1 mount a sustained viral response, whereas 80 to 90% of those infected with genotype 2 and genotype 3 viruses can do so. The correlation between therapy success and the infecting genotype suggests the involvement of viral factors, but the underlying molecular mechanism(s) is not yet understood.
IFNs are a rather diverse class of cytokines with key functions in the innate immune response to viruses (18, 43, 47). Two types of IFNs can be distinguished that have partially overlapping biological properties. Type I IFNs are secreted by most virus-infected cells and by a highly specialized leukocyte population, termed natural IFN-producing cells (8). The human genome contains several IFN genes including 12 IFN-α subtypes and IFN-β. In contrast, the expression of the only type II IFN, IFN-γ, is restricted to immune cells, such as activated T lymphocytes and natural killer (NK) cells. IFNs bind to highly specific cell surface receptors, which trigger the phosphorylation and nuclear translocation of certain latent transcription factors, known as signal transducers and activators of transcription (STATs). Type I IFNs bind to the IFN-α receptor, which leads to the formation of IFN-stimulated gene factor 3 (ISGF-3), a heterotrimer consisting of STAT1, STAT2, and IFN response factor 9/p48 that activates gene transcription via binding to the IFN-stimulated response element. A similar signaling pathway has been described for IFN-γ. In this case, the gamma activation factor, a phosphorylated STAT1 homodimer, is translocated to the nucleus where it enhances gene expression by binding to the gamma activation site (GAS). Besides these well-established pathways, alternative pathways have been described, but their contribution to the antiviral activity of IFN remains to be further elucidated (43).
Type I IFNs execute their antiviral activities through the induction of proteins such as the MxA guanosine triphosphatase, the double-stranded RNA-activated protein kinase (PKR), or the 2′-5′ oligoadenylate synthetase (OAS). These effector proteins may interfere with distinct steps in viral replication or trigger the degradation of viral RNAs. By contrast, IFN-γ predominantly induces the expression of proteins with systemic functions such as those involved in antigen processing and presentation (e.g., major histocompatibility complex class II). In addition, IFN-γ induces the expression and release of chemokines that activate and orchestrate the adaptive immune response (e.g., inducible protein 10 [IP10]). However, IFN-γ may also contribute to the establishment of an antiviral state by the induction of proteins with direct antiviral activities (19).
By using Huh-7 cells that contain selectable, self-replicating HCV RNAs (replicons), it has been shown that recombinant IFN-γ inhibits HCV replication (9, 14). The idea that IFN-γ enforces the critical first line of defense in the HCV-infected liver was further elaborated by Li and coworkers, who demonstrated in a coculture experiment that NK cells blocked HCV replication in Huh-7 cells through the secretion of IFN-γ (31). Clinical data are limited, and whether or not hepatitis C patients benefit from IFN-γ administration is still controversial (17). Nevertheless, it is interesting to note that type I and type II IFNs inhibit HCV RNA replication in Huh-7 cells in a highly synergistic manner (30, 42). Given the power of combination therapies in the treatment of persistent virus infections, it might be rewarding to elucidate the mechanism(s) responsible for the observed synergistic antiviral effects of IFN-α/β and IFN-γ, e.g., whether IFN-α and IFN-γ induce the expression of type-specific effector proteins that may interfere with different steps of the HCV life cycle.
Here, we report on a new host cell line for HCV replication. We show that the human hepatoblastoma cell line HuH6 supports persistent replication of Con1 (genotype 1b) and JFH-1 (genotype 2a) replicons. In line with previous studies in which Huh-7 cells were used as hosts for HCV replicons (5, 32, 34), we show that HCV replication in HuH6 cells depends on the presence of cell culture-adaptive mutations and host cell permissiveness. Compared with Huh-7 cells, however, HCV replication in HuH6 cells is independent from ongoing cell proliferation, less sensitive to certain NS5B-specific antiviral compounds, and highly resistant to IFN-γ. These differences make HuH6 cells an interesting new tool for HCV research.
MATERIALS AND METHODS
Cells and viruses.
The human liver cell lines HuH6 and Huh-7 were originally described by Doi (12) and Nakabayashi et al. (41), respectively. Some background information on these cell lines including the human leukocyte antigen (HLA) type is given in Table 1. Naive HuH6 cells were kindly provided by Brent E. Korba (Division of Molecular Virology and Immunology, Georgetown University Medical Center, Rockville, MD). Naive Huh-7 cells and the Huh-7 cell clone 9-13 (containing the subgenomic HCV Con1 replicon I377/NS3-3′) have been described previously (33). Cells were grown in Dulbecco's modified minimal essential medium (Life Technologies, Karlsruhe, Germany) supplemented with 10% fetal calf serum, 2 mM l-glutamine, nonessential amino acids, 100 U of penicillin G/ml, and 100 μg of streptomycin/ml. For cells with HCV replicons, the culture medium was additionally supplemented with 500- to 1,000-μg/ml G418 (Life Technologies).
TABLE 1.
Background information on HuH6 and Huh-7 cells
The mammalian cell-adapted variant FPV-B of the influenza A virus (FLUAV) strain A/FPV/Dobson/34 (H7/N7) has been described previously (25). The prototype strain of Semliki Forest virus (SFV) was kindly provided by Georg Kochs (Department of Virology, University of Freiburg, Freiburg, Germany). FLUAV and SFV stocks were grown on Vero cells and contained 2.0 × 107 PFU/ml and 2.0 × 108 50% tissue culture infective doses/ml, respectively (virus concentrations were determined with Vero cells).
The HCV replicons I341PI-Luc/NS3-3′/Con1/ET and I341PI-Luc/NS3-3′/Con1/GND and the corresponding pFK plasmids were described previously (32). The nucleotide exchanges T3472C, A4531G, C5474G, and C7754T were introduced into pFK-I341PI-Luc/NS3-3′/Con1/ET and pFK-I389neo/NS3-3′/Con1/ET by site-directed mutagenesis. JFH-1 replicons with a genome organization identical to that of the Con1 replicons I389Neo/NS3-3′/wt and I389Neo/NS3-3′/ΔGDD were transcribed by using plasmids pSGR-JFH1 and pSGR-JFH1/ΔGDD, respectively (26). Both plasmids were kindly provided by Takaji Wakita (Department of Microbiology, Tokyo Metropolitan Institute for Neuroscience, Tokyo, Japan). The newly constructed plasmids pFK-I341PI-Luc/NS3-3′/JFH1 and pFK-I341 PI-Luc/NS3-3′/JFH1/ΔGDD contain the T7-promoter sequence fused to the 5′ NTR of the JFH-1 consensus sequence, followed by the poliovirus IRES (PI), the firefly luciferase gene, the encephalomyocarditis virus IRES, the NS3-to-5B coding sequence and the 3′ NTR of JFH-1, the hepatitis delta virus genomic ribozyme, and the T7 terminator sequence. Plasmid pFK-I341PI-Luc/NS3-3′/JFH1 was constructed by replacing the Con1 and EMCV sequences of pFK-PI-Luc/NS3-3′/ET (32) by corresponding sequences derived from pSGR-JFH1. Plasmid pFK-I341PI-Luc/NS3-3′/JFH1/ΔGDD has an in-frame deletion of 10 amino acids (MLVCGDDLVV) encompassing the GDD motif of NS5B (underlined) and was generated by replacing the sequence between the KpnI and AscI restriction sites in pFK-I341PI-Luc/NS3-3′/JFH1 by a corresponding fragment derived from pSGR-JFH1/ΔGDD.
Interferons and other chemicals.
Recombinant human IFN-α2 and IFN-γ were purchased from Roche Molecular Biochemicals (Basel, Switzerland). Roscovitine was obtained from Sigma-Aldrich (Deisenhofen, Germany). Nucleosidic and nonnucleosidic NS5B-specific inhibitors were kindly provided by Gerhard Pürstinger (Institute of Pharmacy, University of Innsbruck, Innsbruck, Austria) and Piet Herdewijn and Johan Neyts (both from the Laboratories of Medicinal Chemistry and Virology, Rega Institute for Medical Research, Leuven, Belgium).
Generation and transfection of HCV RNAs.
The generation of HCV RNAs by in vitro transcription and conditions of electroporation have been described previously (33).
Virus plaque assay.
Cell monolayers in six-well microplates were infected for 1 h at 37°C with serial 10-fold dilutions of FLUAV and SFV stocks in medium containing 2% fetal calf serum and 20 mM HEPES (pH 7.3). The virus inoculum was removed, and medium containing 2% fetal calf serum, 20 mM HEPES (pH 7.3), and 0.4% SeaPlaque GTG agarose (BioWhittaker Molecular Applications, Walkersville, ME) was added. Plates were further incubated for 3 days, agarose was removed, and cells were stained with a solution of 1% crystal violet, 3.6% formaldehyde, 1% methanol, and 20% ethanol.
Western blot analysis.
Total cell extracts were prepared, samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and proteins were transferred to microporous polyvinylidene difluoride membranes (Perkin Elmer, Boston, MA). HCV nonstructural proteins were specifically immunostained by the use of polyclonal rabbit antisera directed against NS3 (2), NS4B (35), NS5A (2), and NS5B (3).
Northern blot analysis.
Details of this method have been described previously (34). Replicon and β-actin RNAs were detected by using a 32P-labeled negative-sense riboprobe complementary to the neo gene and a riboprobe complementary to β-actin mRNAs, respectively. HCV- and β-actin-specific signals were quantified, and HCV signals were corrected for total RNA amounts loaded in each lane of the gel.
Immunofluorescence analysis.
Cells grown on glass coverslips were fixed with 3% paraformaldehyde and permeabilized with 0.5% Triton X-100. Immunostaining was performed according to standard protocols. NS4B was detected by using a specific rabbit antiserum (35), and STAT1 was immunostained with a mouse monoclonal antibody recognizing the C terminus of the protein (BD Biosciences, San Diego, CA). Bound antibodies were visualized by using goat antibodies conjugated to the cyanine dye Alexa 488 (Molecular Probes, Eugene, OR). Cellular DNA was stained with 4,6-diamidino-2-phenylindol dihydrochloride (DAPI; Molecular Probes).
Luciferase assays.
Transient HCV RNA replication was determined by quantification of luciferase reporter activities as described previously (29). Briefly, 4 × 106 cells were transfected with 5 μg in vitro-transcribed RNA and resuspended in 12 ml culture medium, and 2-ml aliquots were seeded per well of a six-well plate for the determination of 4- and 24-h luciferase values. In case of 48- and 72-h values, only 1-ml aliquots were seeded to prevent the cells from becoming confluent before the time of harvest (subsequently, luciferase values were multiplied by a factor of 2).
RNA quantification by reverse transcription-PCR (RT-PCR).
The HCV Con1 and JFH-1 RNA concentrations were determined by using the ABI PRISM 7000 Sequence Detector system (Applied Biosystems, Foster City, CA) essentially as described previously by Vrolijk et al. (54) and Wakita et al. (55), respectively.
IFN-induced changes in the concentration of 15 different mRNA populations were quantified with the Rotor-Gene 2000 real-time amplification system (Corbett Research, Mortlake, Australia) and the QuantiTect SYBR Green RT-PCR kit (QIAGEN, Hilden, Germany). Briefly, total RNA was isolated by using the reagent Trizol (Invitrogen, Karlsruhe, Germany), following the manufacturer's protocol. RNA was further purified with the RNeasy Mini Kit and the RNase-free DNase Set (both from QIAGEN). For each PCR, 2 μl of total RNA (20 to 200 ng) was added to a 23-μl RT-PCR mixture containing a final primer and Mg2+ concentrations of 0.5 and 2.5 μM, respectively. Reactions were performed under the following conditions: 30 min at 50°C for reverse transcription; 15 min at 95°C for RT denaturation and activation of the Taq polymerase; 35 to 40 cycles, each consisting of 20 s at 95°C, 20 s at 55°C, 40 s at 72°C, and 15 s at 78°C. For each effector mRNA population, copy numbers were normalized to the number of β-actin transcripts. Note that treatment with IFN-α and IFN-γ did not change the activity of the β-actin gene in human hepatoma cells (data not shown).
Database accession numbers.
Effector gene-specific PCR primers were selected with the following database accession numbers: AI337069 (cig5), NM_005101.1 (G1P2), NM_022873.1 (G1P3), NM_002053.1 (GBP1),NM_004120.2 (GBP2), NM_005532.1 (IFI27), NM_001549.1 (IFIT4), NM_012420.1 (IFIT5), NM_ 001565.1 (IP10), NM_002462.1 (MxA), NM_002534.1 (OAS1), NM_002535.1 (OAS2), NM_002759.1 (PKR), NM_007315.1 (STAT1), and BC002704.1 (simSTAT1).
RESULTS
HuH6 cells support persistent HCV RNA replication.
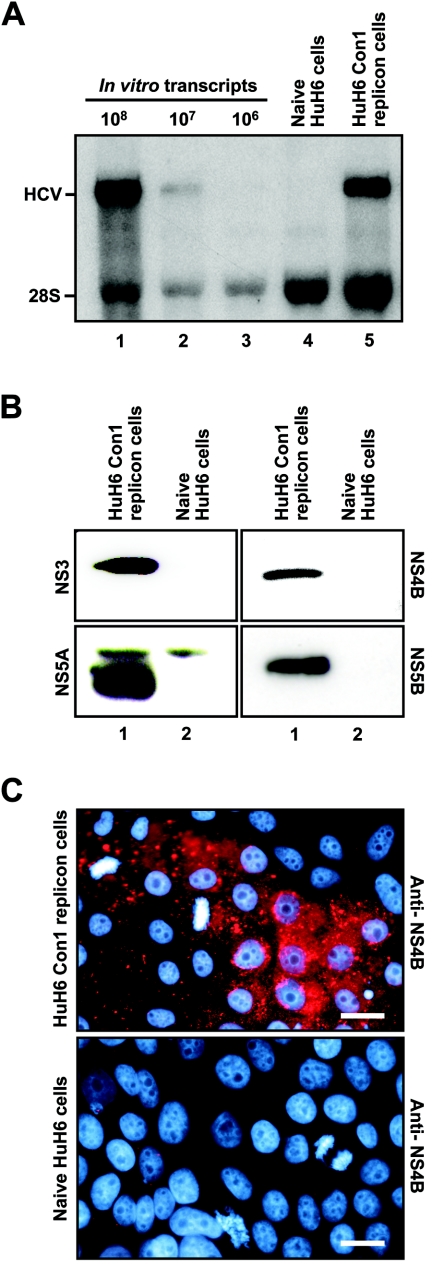
In an attempt to identify new host cells for HCV replication, we transfected Hep-3B, Hepa1-6, HepG2, HuH6, PLC/PRF/5, and HeLa cells with the following mixture of in vitro-transcribed Con1 replicons (cell culture-adaptive mutations are specified in parentheses): I389/NS3-3′/wt, I389/NS3-3′/ET (E1202G, T1280I, and K1846T), I389/NS3-3′/5.1 (E1202G, T1280I, and S2197P), and I389/NS3-3′/5B (R2884G). Although this approach was repeated several times, an increased number of G418-resistant colonies was only observed with HuH6 cells (for comparison, cells were also transfected with replication-defective HCV RNAs). Because we experienced difficulties in expanding G418-resistant HuH6 cell colonies, all cell clones (six clones) were pooled, further propagated, and subjected to Northern blot analysis. As shown in Fig. 1A, we detected positive-strand HCV RNAs of the expected size in HuH6 Con1 replicon cells. Subsequently, we used Western blotting to demonstrate the expression of NS3, NS4B, NS5A, and NS5B in these cells (Fig. 1B). Furthermore, the expression of NS3 and NS4B was confirmed by indirect immunofluorescence (data not shown and Fig. 1, respectively). Note that the punctate staining pattern of NS4B is very similar to what has previously been described as an indicator of the formation of the membranous web, which is the site of viral RNA replication (39). Taken together, these results demonstrate that HuH6 cells support persistent replication of HCV Con1 RNAs.
FIG.1.
HCV RNA and protein synthesis in HuH6 Con1 replicons cells. Naive HuH6 cells were transfected via electroporation with a mixture of subgenomic in vitro-transcribed Con1 replicons and subjected to G418 selection (for details, see the text). Six cell clones were pooled and further passaged. (A) HCV RNAs were detected by Northern hybridization to a neo gene-specific riboprobe (lane 5). Different amounts of in vitro transcripts were used as standards (lanes 1 to 3). RNA isolated from naive HuH6 cells served as a negative control (lane 4). The positions of replicon RNA and 28S rRNA are indicated. (B) Detection of HCV protein expression by Western blotting. Equal amounts of total cell lysates of HuH6 Con1 replicon cells (lane 1) and naive control cells (lane 2) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analyzed by using antibodies with given specificities. (C) Detection of NS4B protein expression by indirect immunofluorescence in HuH6 Con1 replicon cells (top) and naive control cells (bottom). Cells were seeded onto glass coverslips, cultured for 24 h, fixed, permeabilized, and stained for NS4B (red) and DNA (turquoise) by using a specific antibody and DAPI, respectively. Bars, 25 μm.
Cell culture-adaptive mutations enhance HCV Con1 replication in HuH6 cells.
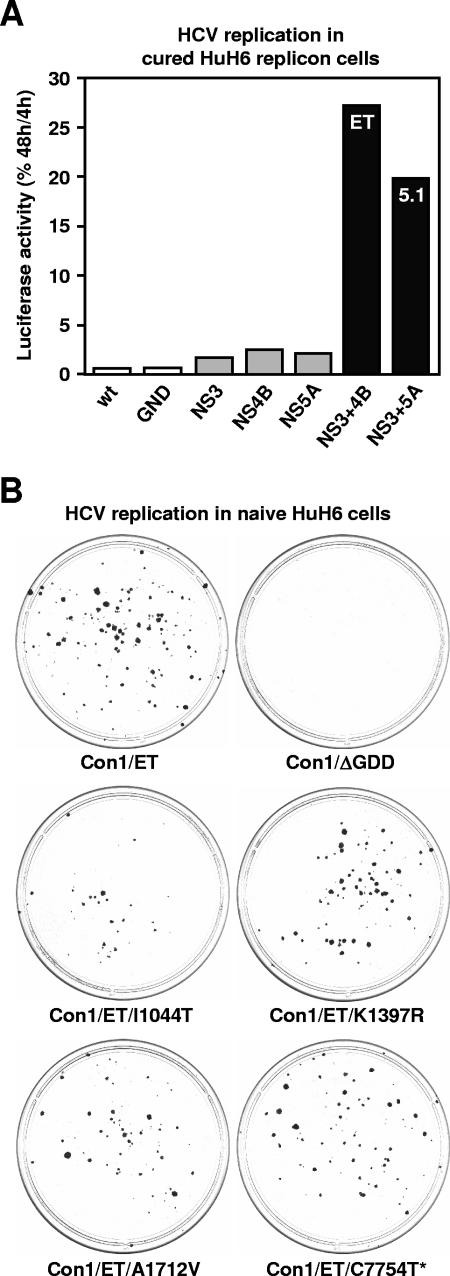
A mixture of four different Con1 replicons was used to establish the HuH6 Con1 replicon cell line. We reasoned that the replicon with the highest replication efficiency in HuH6 cells would become the dominant RNA species during colony formation and subsequent propagation of the cell pool. To address this hypothesis, total RNA was prepared from HuH6 Con1 replicon cells, and the HCV coding sequence was amplified by RT-PCR, cloned, and sequenced. All amplicons contained the so-called ET combination of cell culture-adaptive mutations (E1202G, T1280I, and K1846T) that has previously been identified to most efficiently enhance HCV Con1 replication in Huh-7 cells (32). This finding suggests that the ET mutations boost HCV RNA replication not only in Huh-7 but also in HuH6 cells. To verify this hypothesis, a transient replication assay was performed in which we analyzed a set of mutations known to enhance HCV replication in Huh-7 cells (32). To that end, we used HuH6 Con1 replicon cells that had been treated (cured) with IFN-α2 (for details, see below). As with Huh-7 cells, we found that individual cell culture-adaptive mutations and a combination of two mutations in the NS3 coding region only slightly enhanced HCV replication (Fig. 2A, gray columns), whereas certain combinatory substitutions in NS3 and NS4B (ET) or NS3 and NS5A (5.1) had a much stronger stimulatory effect (black columns). Interestingly, the order of efficiency by which these adaptive mutations enhance HCV replication in HuH6 cells exactly reflects the one previously observed with Huh-7 cells (32). To further corroborate our findings, we analyzed the efficiency by which ET and 5.1 mutations enhanced HuH6 cell colony formation under the selective pressure of G418. We calculated that the transfection of 5 μg of the Con1 replicon I389/NS3-3′/ET and I389/NS3-3′/5.1 resulted in the formation of approximately 75 to 150 and 50 to 100 cell colonies, respectively (data not shown). For comparison, we also transfected the cells with the JFH-1 replicon I389Neo/NS3-3′/wt, a genotype 2a replicon that has been shown to replicate in a variety of cell lines (see below). This time, the transfection of 1 μg of in vitro-transcribed replicon RNA yielded 600 to 700 colonies (data not shown), suggesting that as with Huh-7 cells, JFH-1 replicons replicate more efficiently than Con-1 replicons (26).
FIG. 2.
Effect of viral mutations on HCV RNA replication in HuH6 cells. (A) Effect of cell culture-adaptive mutations known to enhance Con1 replication in Huh-7 cells. Cured HuH6 replicon cells were transfected with in vitro-transcribed RNAs of I341PI-luc/NS3-3′ Con1 replicons and seeded into multiple cell culture dishes. After 4 and 48 h, cells were lysed and replication was quantified by measuring the luciferase activities. To correct for different transfection efficiencies, the 4-h values were set to 100%, and percentages of corresponding 48-h values were calculated. Column colors indicate the absence of cell culture-adaptive mutations (white), their presence in the coding sequence of a single (gray), or two different viral proteins (black). wt, wild-type Con1 sequence; GND, replicon with an inactivating mutation in the GDD motif of NS5B; NS3, replicon with E1202G and T1280I mutations in the protease domain of NS3; NS4B, replicon with K1846T mutation in the center of NS4B; NS5A, replicon with the S2204I mutation in NS5A; NS3+4B, replicon with the ET combination of mutations in NS3 and 4B (E1202G, T1280I, and K1846T); NS3+5A, replicon with the 5.1 combination of mutations in NS3 and 5A (E1202G, T1280I, and S2204I). The result of a single representative experiment is shown. (B) Colony formation assay to analyze the effect of conserved mutations that had been identified in HuH6 cells after the transfection of the Con1 replicon I389Neo/NS3-3′/ET. Naive HuH6 cells were electroporated with in vitro-transcribed replicons that contained either the ET combination of adaptive mutations, an inactivating mutation in the GDD motif of NS5B, or the ET combination of adaptive mutations plus one of the newly identified nucleotide substitutions: T3472C (I1044T in NS3), A4531G (K1397R in NS3), C5474G (A1712V in NS4A), or C7754U (silent mutation in NS5B). After the transfection, cells were seeded in 10-cm dishes, cultured for 4 weeks in the presence of 800-μg/ml G418, cultured for 2 additional weeks in the presence of 500-μg/ml G418, fixed, and stained with Coomassie blue. The positions of nucleotide/amino acid exchanges cited above refer to those in the Con1 consensus genome (EMBL database accession number AJ238799).
To analyze whether efficient HCV Con1 replication in HuH6 cells requires the accumulation of additional, cell-type-specific mutations, we cloned and partially sequenced replicons from HuH6 Con1 cells. However, owing to a very high degree of sequence diversity, no firm conclusions about adaptive mutations could be drawn (data not shown). Therefore, we cloned and sequenced replicons from a single HuH6 cell clone that had been selected after transfection of the Con1 replicon I389/NS3-3′/ET. This time, we encountered a much lower degree of sequence diversity, which enabled us to identify four conserved mutations within the coding sequence of NS3, NS4A, and NS5B. When individually tested by a colony formation assay (which is the most sensitive assay for evaluating the effect of viral mutations on HCV replication and the metabolism of the host cell), none of these mutations was found to significantly enhance HCV replication (Fig. 2B). On the contrary, the I1044T mutation slightly reduced HCV replication. Thus, these mutations seem not to represent true cell culture-adaptive mutations. We cannot, however, exclude cooperative effects because combinations of these mutations were not tested.
Increased permissiveness of cured HuH6 cells.
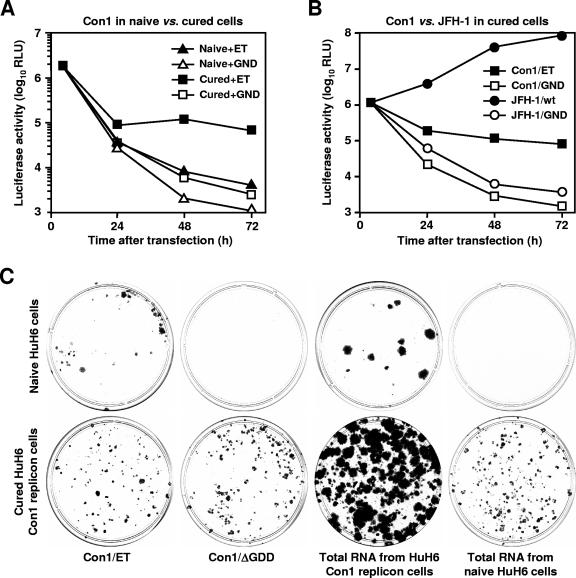
Huh-7 cells that once hosted HCV replicons but later lost them due to antiviral treatments may more readily support HCV replication than naive cells (6, 15, 40). This observation prompted us to propagate HuH6 Con1 replicon cells for a period of 4 weeks with 100-IU/ml IFN-α2, which was followed by an additional incubation period of 2 weeks without the cytokine (to enable the cells to restore normal gene expression). As a consequence, HCV RNA levels dropped below Northern hybridization detection limits (data not shown), but residual amounts of HCV RNAs could be detected by a more sensitive assay (see below). Nevertheless, we analyzed the permissiveness of cured HuH6 cells to newly transfected HCV RNAs in a series of experiments. We first performed transient replication assays. Cured HuH6 Con1 replicon cells and naive control cells were transfected with in vitro-transcribed Con1 replicons encoding a luciferase reporter, which allows a sensitive and precise quantification of HCV RNA replication (16). At several time points after transfection, cells were harvested and lysed, and luciferase assays were performed. Based on the data shown in Fig. 3A, we calculated the ratios between luciferase activities in cells with replication-competent and -incompetent replicons. Throughout the time course of the experiment, we found significantly higher values with cured HuH6 Con1 replicon cells than with naive control cells. Furthermore, significantly higher luciferase activities were measured with cured HuH6 cells after the transfection of replication-incompetent replicons than with naive control cells. Interestingly, this was also observed with Huh-7 cells that had been cured by IFN-α2 or an NS5B-specific antiviral compound (data not shown), suggesting that HCV RNAs are more readily translated and/or less rapidly degraded in cured cells.
FIG. 3.
Effect of host cell determinants on HCV RNA replication. (A) Transient Con1 replication in naive HuH6 cells and those cells that once contained Con1 replicons but later lost them due to a treatment with IFN-α2 (triangles and squares, respectively). Naive and cured cells were transfected with the replication-competent Con1 replicon I341PI-luc/NS3-3′/ET (closed symbols) or the inactive Con1 mutant I341PI-luc/NS3-3′/GND (open symbols). After 4, 24, 48, and 72 h, cells were lysed and luciferase activities were determined. Values were corrected for different transfection efficiencies by using the 4-h measurements. The result of a single representative experiment is shown. RLU, relative light units. (B) Transient replication of JFH-1 and Con1 replicons in cured HuH6 cells. Cells were transfected with the in vitro-transcribed JFH-1 replicons I389Neo/NS3-3′ and I389Neo/NS3-3′/ΔGDD (closed and open circles, respectively) or the Con1 replicons I341PI-luc/NS3-3′/ET and I341PI-luc/NS3-3′/GND (closed and open squares, respectively). The result of a single representative experiment is shown. RLU, relative light units. (C) Cell colony formation after transfection of Con1 replicons into naive and cured HuH6 cells (top and bottom, respectively). Cells were transfected with in vitro-transcribed RNAs of the Con1 replicons I389Neo/NS3-3′/ET and I389Neo/NS3-3′/ΔGDD or with total RNA of HuH6 Con1 cells containing the replicon I389Neo/NS3-3′/ET. In all cases, similar numbers of HCV RNAs were transfected (109 replicon molecules, as quantified by Northern hybridization). For comparison, cells were also transfected with total RNA from naive HuH6 cells. Transfected cells were seeded in 10-cm dishes, cultured for about 4 weeks in the presence of 500-μg/ml G418, fixed, and stained with Coomassie blue.
To corroborate our findings, we transfected cured HuH6 Con1 cells with a genotype 2a replicon that has been derived from the JFH-1 consensus sequence (28). Subgenomic JFH-1 replicons have previously been reported to replicate efficiently in a variety of cell lines including Huh-7, HepG2, IMY-N9, 293, and HeLa (10, 26, 27). To analyze the replication efficiency of JFH-1 replicons, we constructed a second generation of subgenomic JFH-1 replicons that contain the firefly luciferase gene under the control of the poliovirus IRES element (for details, see Materials and Methods) and transfected these replicons along with Con1 replicons of the same design into cured HuH6 Con1 cells. Based on the luciferase data shown in Fig. 3B, we calculated that JFH-1 replication in HuH6 cells is almost 3 orders of magnitude more efficient than that of the most adapted Con1 replicon, I341/PI-luc/NS3-3′/ET. For comparison, we also transfected naive HuH6 cells with the JFH-1 replicon and observed that this replicon replicated almost as efficiently in these cells as in cured HuH6 cells (data not shown). These results confirm previous findings, which indicate that the JFH-1 genome has an unusually high rate of replication. Furthermore, the results established cured HuH6 cells as a new host cell line for those HCV genomes that replicate less efficiently than the JFH-1 genome. It should, however, be noted that the increased susceptibility of cured cells to HCV replicons is a rather unstable phenotype which wears off with increasing passage numbers (data not shown).
Next, we performed a colony formation assay in which naive and cured HuH6 cells were transfected with in vitro-transcribed RNAs of the Con1 replicon I389Neo/NS3-3′/ET or a replication-incompetent mutant. Furthermore, cells were transfected with total RNA from HuH6 Con1 replicon cells (containing primarily I389Neo/NS3-3′/ET progenies). Note that in all cases, similar numbers of HCV RNA molecules were used. As a control, we also transfected equivalent amounts of RNA that were isolated from naive control cells. About 24 h after transfection, cells were subjected to G418 selection; 4 weeks later, colonies were fixed and stained (Fig. 3C). The unexpected finding that cured HuH6 replicon cells formed G418-resistent colonies after the transfection of replication-defective replicons and total RNA from naive HuH6 cells indicated the presence of HCV RNAs that had survived the previous treatment with IFN-α2. To exclude that the observed G418 resistance of cured HuH6 replicon cells was due to the integration of residual template DNA into the host cell genome, we performed a neo-specific PCR. In a pool of >200 G418-resistent HuH6 replicon cell colonies, we could not detect any neo DNA (data not shown), suggesting that G418 resistance in these cells was indeed conferred by surviving HCV replicons. This is remarkable because a similar IFN-α treatment was sufficient to almost completely cure Huh-7 cells from HCV RNAs (data not shown).
With naive HuH6 cells, we observed similar numbers of cell colonies after transfection of I389Neo/NS3-3′/ET replicons, irrespective of whether the replicon RNAs were synthesized in vitro or in replicon cells. The colony size, however, was significantly larger after the transfection of total RNA from HuH6 Con1 replicon cells. The same was observed with cured HuH6 Con1 replicon cells but not when HuH6 cells were transfected with total RNA from Huh-7 replicon cells (data not shown). The transfection of HepG2 cells with total RNA from HuH6 replicon cells also resulted in the formation of unusually large colonies (data not shown). It is therefore tempting to speculate that cotransfected host cell mRNAs present in HuH6 Con1 replicons cells but not (or to a much smaller number) in naive HuH6 cells may have caused this effect. Alternatively, cell-type-specific adaptive mutations may exist only in a small proportion of replicons that escaped our attempt to identify such mutations through the sequencing of cDNA clones (see above). Finally, we cannot exclude the possibility that the HCV replication machinery produced replicon RNAs which initiated viral replication more efficiently than in vitro-transcribed RNAs.
HCV RNA replication in HuH6 is not cell growth dependent.
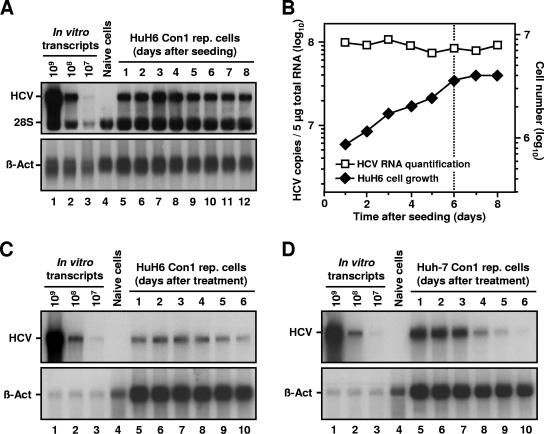
A peculiarity of Huh-7 replicon cells is that viral RNA concentrations drop dramatically when cells become confluent (20, 45), a phenomenon that has been ascribed to a cell cycle-dependent activation of the HCV IRES (22). To determine the efficiency of HCV replication in nondividing HuH6 cells, we performed a time course experiment in which we followed HCV RNA concentration in a growing cell culture over the point of confluence. The Northern blot analysis shown in Fig. 4A and B revealed that subconfluent and confluent HuH6 Con1 replicon cells contained similar amounts of HCV RNAs. In an additional experiment, we blocked cell growth by roscovitine, a potent and selective inhibitor of cdc2 and certain other cyclin-dependent kinases (11, 38). As expected, a treatment with 20 μM roscovitine largely blocked proliferation of both HuH6 and Huh-7 replicon cells (data not shown). Furthermore, we observed that HCV RNA levels in HuH6 cells did not change during the experiment, whereas roscovitine-treated Huh-7 cells quickly lost their replicons (Fig. 4C and D, respectively).
FIG. 4.
HCV RNA replication in nondividing HuH6 cells. (A) Detection of positive-stranded HCV RNAs in a growing culture of HuH6 Con1 replicon cells. Equal numbers of cells were seeded in multiple 10-cm cell culture dishes and harvested at given time points. Total RNA was prepared, and samples of 5 μg (each) were analyzed by Northern hybridization to a riboprobe complementary to the neo gene or to β-actin mRNAs (lanes 5 to 12). Different amounts of in vitro transcripts were used as standards (lanes 1 to 3). RNA isolated from naive HuH6 cells served as a negative control (lane 4). The positions of replicon RNA, 28S rRNA, and β-actin are indicated. (B) Comparison between the number of HCV RNAs (squares) and cell number at the time of harvest (diamonds). Prior to RNA preparation, HuH6 Con1 replicon cells were trypsinized and counted, and the number of cells per 10-cm cell culture dish was calculated. Hybridization signals of positive-stranded HCV RNAs and β-actin mRNAs shown in panel A were quantified and HCV RNA copy numbers were corrected for loading differences. Note that HCV copy and cell numbers are given in different log scales. The dotted line indicates the transition from the exponential to the stationary growth phase of the cell culture. (C and D) Detection of positive-stranded HCV RNAs in roscovitine-treated HuH6 Con1 cells and Huh-7 cells of clone 9-13, respectively. Equal numbers of cells were seeded in multiple 10-cm cell culture dishes, treated with 20 μM roscovitine, and harvested at given time points. Total RNA was prepared, and samples of 5 μg were analyzed by Northern hybridization as described above. The positions of replicon RNA and β-actin are indicated.
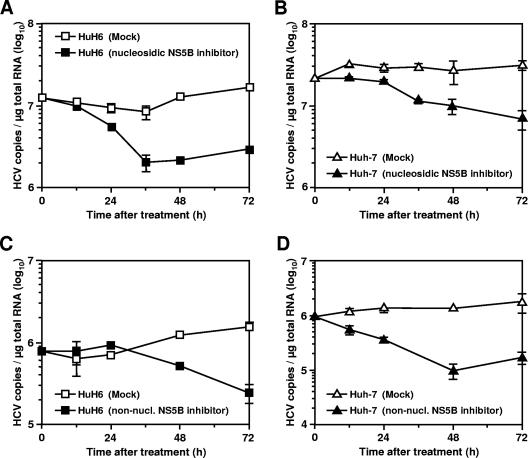
The findings suggest an unhindered HCV replication in nondividing HuH6 cells, an increased stability of HCV RNAs in HuH6 cells compared to Huh-7 cells, or a combination of both. To discriminate between these possibilities, we blocked HCV replication by using highly specific NS5B inhibitors and determined the effect of the antiviral treatment on RNA stability by quantitative, real-time RT-PCR. First, we incubated HuH6 and Huh-7 replicon cells with 2′-C-methyladenosine, a nucleoside analogue that functions as a chain terminator of newly synthesized HCV RNAs (7, 51). The results shown in Fig. 5A and B demonstrate that 1 μM of the drug inhibits HCV replication in both cell lines. As a consequence, we observed a decrease in the amount of replicon RNA in HuH6 and Huh-7 cells to approximately 18 and 22%, respectively (compared to the amount of viral RNA in mock-treated control cells). A similar inhibition profile was observed with higher drug concentrations (data not shown). The fact that this nucleosidic inhibitor was at least as potent in HuH6 cells as in Huh-7 cells indicates that HuH6 cells do not have an increased HCV RNA stability.
FIG. 5.
Inhibition of HCV RNA replication by antiviral compounds. (A and B) Effect of a nucleosidic, NS5B-specific inhibitor on the maintenance of HCV RNAs in HuH6 Con1 cells and Huh-7 cells of clone 9-13, respectively. Cells were seeded into multiple cell culture dishes; on the following day, the culture medium was supplemented with 1 μM 2′-C-methyladenosine. At given time points, cells were harvested, total RNA was prepared, and the number of HCV RNA copies was determined by quantitative, real-time RT-PCR. The result of a single representative experiment is shown. (C and D) Antiviral effect of a nonnucleosidic, NS5B-specific inhibitor of HCV replication. The experiment was performed and analyzed as described above.
To confirm that the half-life of HCV RNA is comparable in HuH6 and Huh-7 cells, we treated both replicon cell lines with a nonnucleosidic inhibitor of NS5B. Again, the antiviral treatment reduced the amount of replicon RNA in both cell lines, but this time, we observed a 24-h delay in HuH6 cells. Of note, a similar delay was found with another nonnucleosidic NS5B inhibitor (data not shown). We cannot exclude that the observed differences in the kinetic of virus inhibition are a consequence of differing drug uptake and/or metabolism, but it is also conceivable that these nonnucleosidic inhibitors interfere with the activity of newly formed replication complexes rather than affecting the activity of existing ones. This hypothesis is in line with recent findings of Ma et al., who reported that three different nonnucleosidic, NS5B-specific inhibitors failed to inhibit RNA synthesis by native replicase complexes isolated from replicon cells at concentrations 1,000-fold higher than the concentrations required for half-maximal inhibition of recombinant NS5B (36). Nevertheless, after a 24-h treatment with these nonnucleosidic inhibitors, the amount of replicon RNA decreased with a similar slope in both cell lines, which confirms our earlier finding that HCV RNAs have a comparable degree of stability in HuH6 and Huh-7. Taken together, these data support the idea of ongoing HCV RNA replication in nondividing HuH6 cells.
IFN-γ does not block HCV RNA replication in HuH6 cells.
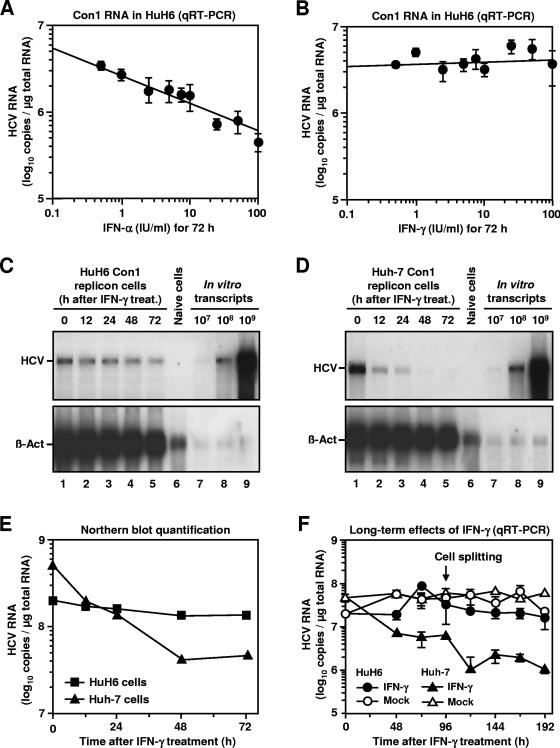
The fact that replicons could be removed from HuH6 replicon cells by IFN-α already implied that HCV replication in these cells is sensitive to type I IFNs. To further substantiate this finding, we determined the IFN-α sensitivity of HCV replication in HuH6 in comparison to Huh-7 cells. To that end, we incubated HuH6 Con1 replicon cells with various IFN-α2 concentrations ranging from 0.5 to 100 IU/ml. Three days later, total RNA was prepared and replicon RNA copy numbers were determined by quantitative, real-time RT-PCR. As expected, IFN-α2 reduced the amount of HCV RNAs in a dose-dependent manner with a 50% inhibitory concentration (IC50) of about 5 to 10 IU/ml (Fig. 6A), which is similar to what has previously been described for Huh-7 cells (5). Next, we treated HuH6 Con1 replicons cells with IFN-γ. Surprisingly, even doses of up to 100 U/ml did not affect the HCV RNA copy number (Fig. 6B). This observation is at variance to the strong inhibitory activity of IFN-γ in Huh-7 cells (9, 14). To determine whether higher doses of IFN-γ or longer incubation periods with the cytokine were required to block replication in HuH6 cells, we first performed a time course experiment. HuH6 Con1 replicon cells and Huh-7 cells of clone 9-13 (containing the Con1 replicon I377Neo/NS3-3′) were incubated with 1,000-IU/ml IFN-γ; at several time points after the start of treatment, the number of replicon RNAs was determined by Northern hybridization (Fig. 6C and D). A quantification of the hybridization signals revealed that the HCV RNA copy number in HuH6 cells hardly changed during the course of the experiment, whereas IFN-γ reduced viral RNA levels in Huh-7 cells by >90% (Fig. 6E). Analogous results were obtained when HCV RNA levels were quantified by real-time RT-PCR and cells were incubated for 96 h with IFN-γ (data not shown).
FIG. 6.
IFN-induced inhibition of HCV RNA replication in HuH6 cells. (A and B) Antiviral effect of different IFN-α and IFN-γ concentrations. HuH6 Con1 replicon cells were seeded into multiple cell culture dishes, cultured for 24 h, and incubated with different IFN concentrations (0, 0.5, 1, 2.5, 5, 7.5, 10, 25, 50, and 100 IU/ml). After 72 h, total RNA was prepared and the number of HCV RNA copies was determined by quantitative, real-time RT-PCR. Mean values and standard deviations of six different quantifications are given. (C and D) Northern blot analysis of HCV RNA levels in IFN-treated HuH6 Con1 replicon cells and Huh-7 cells of clone 9-13. Cells were seeded into multiple cell culture dishes, cultivated for 24 h in normal culture serum, and further cultivated in the presence of 1,000-IU/ml IFN-γ. Total RNA was prepared at given time points, and 5-μg samples (lanes 1 to 5) were analyzed by using a riboprobe complementary to the neo gene (top) or to β-actin mRNAs (bottom). Different amounts of in vitro transcripts were used as standards (lanes 7 to 9). RNA isolated from naive cells served as a negative control (lane 6). The positions of
Next, we addressed the question whether the observed IFN-γ resistance is a property of Con1 replicons only or holds true for replicons of different genotypes. A genotype 2a replicon cell pool was established after transfection of JFH-1 replicon I389Neo/NS3-3′/wt into naive HuH6 cells and the selection of ∼600 cell clones. As expected, an IFN-α treatment purged the majority of JFH-1 replicons from HuH6 cells (data not shown). However, when we incubated HuH6 JFH-1 replicon cells with 1,000-IU/ml IFN-γ, we found that this treatment did not result in significantly lower HCV RNA levels, even after a 192-h treatment (Fig. 6F). In contrast, an almost 2-log difference was observed between the HCV RNA copy numbers of Huh-7 replicon cells that had been treated with IFN-γ and those that had been mock treated. The results demonstrate that the resistance of HCV replicons in HuH6 to IFN-γ is not a short-term phenomenon and does not depend on the viral genotype.
Antiviral response to IFN-γ is not generally impaired in HuH6 cells.
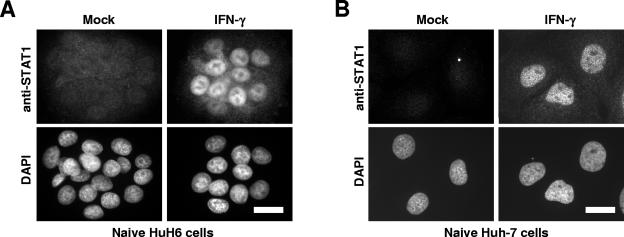
IFNs exert most of their biological functions through Jak/STAT signaling pathways. A key element in the pathway elicited by IFN-γ is the phosphorylation and nuclear translocation of STAT1. In line with this, we found that endoribonuclease-prepared small interfering RNA-mediated silencing of STAT1 expression restores HCV replication in Huh-7 cells treated with IFN-γ (U. Zeuge and M. Frese, unpublished data). Therefore, defective Jak/STAT signaling might well explain the incompetence of HuH6 cells in inhibiting HCV replication upon IFN-γ stimulation. To analyze the integrity of the Jak/STAT signaling pathway, naive HuH6 cells were treated for 24 h with 1,000-IU/ml IFN-γ, fixed, and analyzed by STAT1-specific immunofluorescence staining. Clearly, the IFN-γ treatment led to an enhanced expression of STAT1, accumulating almost exclusively in the nucleus (Fig. 7A), as was the case with Huh-7 cells (Fig. 7B). In a side-by-side experiment, we treated both replicon cell lines with 1,000-IU/ml IFN-α2, which in both cases also led to a nuclear accumulation of STAT1 (data not shown). These results indicate that both HuH6 and Huh-7 cells possess functional Jak/STAT signaling pathways.
FIG. 7.
Effect of IFN-γ on STAT1 expression and localization in HuH6 and Huh-7 cells. Naive HuH6 (A) and Huh-7 (B) cells were seeded onto glass coverslips, cultivated for 16 h in normal culture medium, and further incubated for 24 h in the absence of cytokines (left) or in the presence of 1,000-IU/ml IFN-γ (right). After fixation and permeabilization, cells were immunostained for STAT1. Bars, 25 μm.
A hallmark of IFNs is their ability to upregulate the expression of specific genes (18, 47). To check whether HuH6 cells respond to IFNs not only by the nuclear accumulation of STAT1 but also by a selective enhancement of gene expression, we stimulated the cells with 1,000-IU/ml of either IFN-α2 or IFN-γ for 24 h and measured the effect of this treatment on the transcript concentrations of 15 IFN-regulated genes by quantitative, real-time RT-PCR. Among these were genes encoding constituents of intracellular signaling pathways (e.g., STAT1), effector proteins with antiviral activities (e.g., MxA, OAS, and PKR), and a cytokine signaling at the interface between the innate and adaptive immune system (IP10). In agreement with the enhanced expression of STAT1 in IFN-treated HuH6 cells (Fig. 7), we found that both IFNs enhanced the synthesis of STAT1 mRNAs and those of several other IFN-regulated genes (Table 2). In addition, we analyzed Huh-7 cells for their cellular response to IFN-α and IFN-γ. A comparison of the IFN-induced changes in the gene expression of HuH6 and Huh-7 cells revealed that both cell lines showed a very similar response to IFNs, although some differences existed (Table 2). For example, the number of MxA and IP10 mRNAs was relatively low in HuH6 cells, whereas the same cells contained an increased number of IFI-27 transcripts. Given the different origin and history of both cell lines (Table 1), such differences are not unexpected and do not necessarily relate to the inability of HuH6 cells to block HCV replication in response to IFN-γ (see Discussion for further information).
TABLE 2.
Quantification of IFN-induced gene expression in naive HuH6 and Huh-7 cells
| Genea | Signalb,d
|
Fold changec,d
|
Signalb,e
|
Fold changec,e
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mock | IFN-α | IFN-γ | IFN-α | IFN-γ | Mock | IFN-α | IFN-γ | IFN-α | IFN-γ | |
| cig5 | 0.0 | 96 | 3.7 | 2,462 | 95 | 0.1 | 209 | 5.8 | 2,887 | 81 |
| G1P2 | 137 | 6,934 | 762 | 51 | 5.6 | 66 | 5021 | 914 | 77 | 14 |
| G1P3 | 2.9 | 657 | 41 | 224 | 14 | 0.9 | 945 | 75 | 1,043 | 84 |
| GBP1 | 0.8 | 5.2 | 1,152 | 6.2 | 1,362 | 9.3 | 184 | 4,046 | 20 | 435 |
| GBP2 | 12 | 13 | 1,082 | 1.1 | 89 | 390 | 665 | 2,672 | 1.7 | 6.9 |
| IFI-27 | 76 | 7,991 | 1,522 | 106 | 20 | 7.6 | 787 | 101 | 104 | 13 |
| IFI-T4 | 24 | 825 | 449 | 34 | 19 | 16 | 797 | 646 | 48 | 39 |
| IFI-T5 | 1.5 | 265 | 80 | 175 | 53 | 41 | 506 | 197 | 12 | 4.8 |
| IP10 | 0.3 | 3.6 | 256 | 11 | 755 | 67 | 1,089 | 8,702 | 16 | 131 |
| MxA | 4.2 | 340 | 20 | 80 | 4.6 | 46 | 24,896 | 2,763 | 540 | 60 |
| OAS1 | 0.76 | 983 | 21 | 1,288 | 27 | 7.0 | 1,391 | 102 | 198 | 15 |
| OAS2 | 2.4 | 2,384 | 180 | 987 | 75 | 1.9 | 1,154 | 97 | 599 | 50 |
| PKR | 3.3 | 9.6 | 4.6 | 2.9 | 1.4 | 5.1 | 12 | 10 | 2.4 | 2.0 |
| STAT1 | 176 | 1,667 | 2,749 | 10 | 16 | 180 | 2,151 | 1,695 | 12 | 9 |
| simSTAT1 | 16 | 209 | 293 | 13 | 18 | 19 | 253 | 242 | 14 | 13 |
Database accession numbers are given in the Materials and Methods.
mRNA copy number/100,000 β-actin mRNA copies 24 h after the IFN treatment.
Signal in IFN-treated cells per signal in mock-treated cells.
Values are for naive HuH6 cells.
Values are for naive Huh-7 cells.
The experiments described so far were performed only with naive cells. To exclude the possibility that HCV replicons compromise IFN signaling in HuH6 cells, we analyzed the IFN-induced gene expression in HuH6 Con1 and JFH-1 replicon cells. In this experiment, we followed the transcript levels of five genes that are preferentially induced either by IFN-α (MxA and OAS1), by IFN-γ (GBP1 and IP10), or by both IFN types (STAT1). Cells were treated with IFNs, and mRNA levels were quantified 24 h later as described in the previous paragraph. We observed that both replicon cell lines displayed an IFN response very similar to that of naive cells (Table 3). In HuH6 cells with HCV replicons, however, we measured a slightly increased baseline mRNA concentration for four out of five genes, indicating that the presence of virus RNA and/or proteins did not completely escape the attention of the cell's innate immune system. Since the concentrations of some IFN-induced mRNAs peaked earlier than 24 h after IFN stimulation, we performed an additional quantitative, real-time RT-PCR analysis in which we determined the concentration of GBP1, G1P2, IFI-T4, OAS1, and STAT1 mRNAs in HuH6 and Huh-7 cells with and without Con1 replicons. After 8 h of IFN treatment, we measured sometimes higher concentrations of IFN-induced mRNAs than we had detected after 24 h of treatment (for details, see the supplemental material). Nevertheless, the outcome of this experiment confirms that HuH6 cells possess intact IFN signaling pathways.
TABLE 3.
Quantification of IFN-induced gene expression in HuH6 cells with and without HCV replicons
| Genea and cell line | Signalb
|
Fold changec
|
|||
|---|---|---|---|---|---|
| Mock | IFN-α | IFN-γ | IFN-α | IFN-γ | |
| HuH6 Con1 replicon cells | |||||
| GBP1 | 2.4 | 18 | 933 | 7.6 | 395 |
| IP10 | 0.7 | 19 | 106 | 26 | 147 |
| MxA | 14 | 183 | 38 | 13 | 2.7 |
| OAS1 | 1.3 | 1,641 | 15 | 1,291 | 12 |
| STAT1 | 321 | 2,267 | 3,312 | 7.1 | 10 |
| HuH6 JFH-1 replicon cells | |||||
| GBP1 | 1.8 | 40 | 223 | 23 | 126 |
| IP10 | 0.5 | 330 | 39 | 648 | 77 |
| MxA | 18 | 3973 | 71 | 224 | 4.0 |
| OAS1 | 3.8 | 4553 | 37 | 1,211 | 9.7 |
| STAT1 | 222 | 4,278 | 1,366 | 19 | 6.2 |
| Naive HuH6 cells | |||||
| GBP1 | 0.8 | 3.7 | 586 | 4.9 | 768 |
| IP10 | 0.1 | 5.5 | 176 | 105 | 3,325 |
| MxA | 2.9 | 563 | 18 | 196 | 6.3 |
| OAS1 | 1.0 | 1,943 | 22 | 2,041 | 23 |
| STAT1 | 349 | 3,467 | 3,565 | 9.9 | 10 |
Database accession numbers are given in Materials and Methods.
mRNA copy number per 100,000 β-actin mRNA copies 24 h after IFN treatment.
Signal in IFN-treated cells per signal in mock-treated cells.
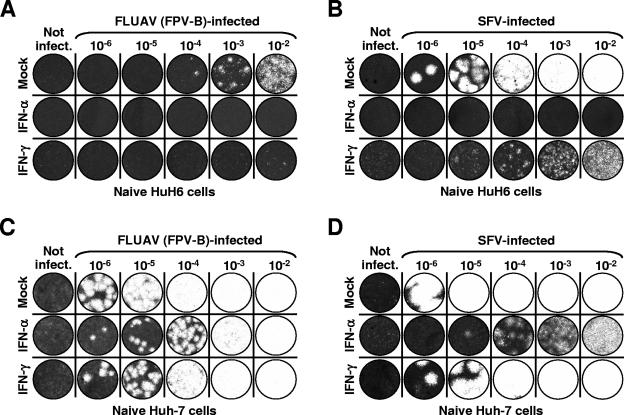
To test the capability of HuH6 cells of establishing a protective antiviral state in response to IFNs,we treated naive cells with either IFN-α2 or IFN-γ (each, 1,000 IU/ml) and challenged them with different concentrations of variant FPV-B of FLUAV (family Orthomyxoviridae) or SFV (family Togaviridae). Virus plaque inhibition assays revealed that both IFNs inhibited the multiplication of these viruses in HuH6 cells, with IFN-α2 being more potent than IFN-γ (Fig. 8A and B). Huh-7 cells that were also included in the experiment were found to be generally more susceptible to viral infections, and their IFN-induced antiviral response to viruses unrelated to HCV was less effective than that of HuH6 cells (Fig. 8C and D). To corroborate these findings, we analyzed the induction of an antiviral state by IFN-α2 and IFN-γ in HuH6 replicon cells. Similar to what has been described for naive cells, both IFNs largely inhibited the multiplication of SFV (data not shown). Taken together, the findings indicate a particular defect in the IFN-induced antiviral response of HuH6 cells, which allows an unhindered replication of HCV RNAs in the face of a seemingly otherwise-uncompromised antiviral state.
FIG. 8.
IFN-induced inhibition of FLUAV and SFV multiplication in HuH6 and Huh-7 cells. FLUAV (A) and SFV (B) plaque formation in HuH6 cells is shown. Semiconfluent monolayer cultures were incubated with 1,000-IU/ml IFN-α or IFN-γ or were left untreated. After 24 h, cells were infected with serial 10-fold dilutions of stock virus, plaques were allowed to develop for 72 h under soft agar, and remaining cells were fixed and stained with crystal violet. For FLUAV (C) and SFV (D) plaque formation in Huh-7 cells, the experiment was performed as described above.
DISCUSSION
Initially, only Huh-7 cells were found to support HCV RNA replication (5, 20, 33) but in subsequent work, other cell lines were identified that may also serve as suitable hosts for HCV RNAs. For example, Zhu et al. demonstrated persistent Con1 RNA replication in HeLa cells and in cells of the mouse hepatoma cell line Hepa1-6 (58). Furthermore, it has been shown that Con1 replicons replicate in 293 cells (1). By using a JFH-1 replicon that replicates efficiently in Huh-7 cells without the need for adaptive mutations, even more cell lines with persistent HCV replication could be established (10, 26, 27). In this study, we show that both Con1 and JFH-1 RNAs replicate in the human hepatoblastoma cell line HuH6 with an efficiency that is high enough to allow transient replication assays. This is a remarkable finding, because the detection of transient HCV replication in a cell line other than Huh-7 has not yet been reported.
In Huh-7 cells, adaptive mutations are an important prerequisite for efficient replication of Con1 RNAs (5, 34). Here, we show that the same is true for HuH6 cells. Wild-type Con1 replicons replicated poorly in HuH6 cells, but HCV replication could be enhanced by the introduction of mutations known to increase replication in Huh-7 cells. Even the order of efficiency by which these mutations enhanced HCV replication in HuH6 cells exactly reflects the one previously observed in Huh-7 cells (32). These findings suggest that HuH6 and Huh-7 cells provide a similar environment for HCV replication, a hypothesis that is in line with the fact that we failed to detect any cell-type-specific adaptive mutations in HuH6 Con1 cells.
On the other hand, remarkable differences exist in the ability of HuH6 and Huh-7 cells to support HCV replication in resting cells. It has previously been shown that HCV RNA levels rapidly decrease in confluent Huh-7 replicon cells (20, 45). In this paper, we report that subconfluent Huh-7 replicon cells which had been arrested in the G1 phase of the cell cycle also rapidly lost their HCV RNAs (Fig. 4C and D). These findings are consistent with the earlier observations of Honda and colleagues who reported that the HCV IRES activity in Huh-7 cells varied during the cell cycle (22). By using stably transfected cells that expressed a bicistronic reporter construct, the authors showed that HCV translation was greatest in the synthetic and mitotic phase of the cell cycle and lowest in the quiescent G0 phase of resting cells. These findings are in line with the more recent observation that an enhanced expression of the La protein or the polypyrimidine tract binding protein may not only correlate with regeneration of hepatocytes in chronic hepatitis C patients but may also stimulate HCV translation (23). Furthermore, it has been noted that confluent HeLa cells contain amounts of HCV RNAs similar to the amounts found in subconfluent ones (58), which has led to the hypothesis that HCV is able to replicate in resting cells. However, antiproliferating drugs have not been used to substantiate this speculation. Our finding that HCV RNA levels do not decrease in confluent HuH6 cells (which show a strong degree of contact inhibition) or those that have been arrested in G1 phase of the cell cycle strongly indicates that the persistence of HCV RNAs does not always require ongoing cell proliferation. It is generally assumed that hepatocytes are resting cells, raising the question of how HCV replication can be maintained if it is dependent on host cell growth. Since replication of HCV RNAs in HuH6 cells is cell growth independent, this cell line may reflect the in vivo situation more accurately than with Huh-7 cells. One might speculate that confluent HuH6 cells have larger nucleoside triphosphate pools than Huh-7 cells, which would imply that low nucleoside triphosphate concentrations restrict HCV RNA replication. It has indeed been demonstrated that high CTP and UTP levels are critical parameters for efficient HCV replication in Huh-7 cells (47a). Future experiments addressing the molecular basis for the observed difference between Huh-7 and HuH6 cells might reveal requirements for HCV RNA replication and maintenance that may provide us with new approaches for interviral intervention.
Another striking difference between HuH6 and Huh-7 cells relates to an apparent gap in the innate defense of HuH6 cells. It has previously been demonstrated that both IFN-α/β and IFN-γ efficiently terminate HCV replication in Huh-7 cells (5, 9, 13, 14, 42, 46). It is widely assumed that IFNs exert most, if not all, of their direct antiviral activities through the induction of effector proteins. Some of these are preferentially induced by IFN-α/β (e.g., Mx proteins), IFN-γ (e.g., GBP proteins), or by all IFNs (e.g., PKR). In several publications, it has been suggested that PKR plays a prominent role in IFN-induced inhibition of HCV replication, but these reports are controversial (4). Furthermore, it has been reported that HCV RNAs are sensitive to the OAS/RNase L degradation pathway (48) and to RNA editing performed by an adenosine deaminase acting on RNA (49), but details have not yet been elucidated.
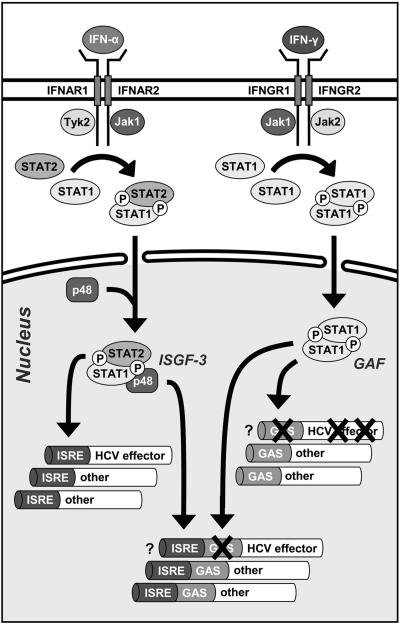
Based on the present finding that IFN-α but not IFN-γ inhibits HCV RNA replication in HuH6 cells, one might speculate that the innate immune response against HCV relies on more than one effector protein. According to the model depicted in Fig. 9, IFN-α/β and IFN-γ would induce the expression of different effector genes. While Huh-7 cells would possess a full set of functional effector genes, mutations in the genome of HuH6 cells may account for the inability of these cells to reduce the number of HCV RNAs in response to IFN-γ. For instance, mutations within a critical effector gene may have destroyed its promoter sequence, changed its coding sequence, or caused mRNA instability. In this context, it is interesting to note that IFN-γ induces >100 fold the number of MxA mRNAs in Huh-7 than in HuH6 cells (Table 3). MxA is a cytoplasmic GTPase with strong antiviral activities against a variety of positive- and negative-stranded RNA viruses (21). However, it has been shown that constitutive expression of MxA does not inhibit subgenomic HCV replicons and that the expression of a dominant-negative MxA mutant does not restore HCV replication during IFN-α treatment (13). These earlier observations are in line with the present finding that IFN-α inhibits HCV replication in Huh-7 and HuH6 cells with a similar IC50 (3 to 5 and 5 to 10 IU/ml, respectively), although the former synthesizes nearly 75-fold-more MxA mRNAs in response to IFN-α than the latter (Table 3).
FIG. 9.
Model for the IFN-induced effector gene expression of HuH6 cells. The binding of IFN-α and IFN-γ to their corresponding cell surface receptors (IFN-α receptor [IFNGR] and IFN-γ receptor [IFNAR], respectively) results in the phosphorylation and nuclear translocation of latent transcription factor complexes (IFN-stimulated gene factor 3 [ISGF-3] and gamma activation factor [GAF], respectively). In the case of IFN-α, a heterotrimer consisting of STAT1, STAT2, and p48 activates gene promoters containing one or more IFN-stimulated response elements ISREs. In the case of IFN-γ, a STAT1 homodimer activates gene transcription by binding to the gamma activation site (GAS). Crosses indicate potential mutations in the genome of HuH6 cells (see the text for further details).
Although we favor the idea that IFN-α and IFN-γ induce the expression of different effector proteins, it is also possible that the innate immune response in humans relies on a single HCV-specific effector that is expressed in response to both types of IFNs. In this case, however, it must be postulated that the genome of HuH6 cells carries a mutation in the promoter region of that effector gene which would specifically affect its responsiveness to IFN-γ. According to another model, HuH6 and Huh-7 cells may express a similar or even identical set of IFN-induced effector proteins, targeting at least two different steps of the viral life cycle. Such a scenario would imply that IFN-γ targets a viral replication step that somehow is less accessible to effector proteins. Interestingly, HuH6 replicon cells showed a delayed response to two different nonnucleosidic, NS5B-specific antiviral compounds, which might be a consequence of highly compact or otherwise shielded viral replication complexes. Further studies are required to determine why HuH6 cells cannot purge themselves from HCV RNAs in response to IFN-γ. To determine whether the cells are impaired in the expression of a pivotal IFN-induced effector gene, we performed a detailed transcriptome analysis by using cDNA microarrays. The data are currently being screened for effector transcripts that are differentially expressed in HuH6 and Huh-7 cells.
Taken together, HuH6 cells represent a valuable new host cell line for HCV replicons. Compared to Huh-7 cells, HCV RNA replication in HuH6 cells follows some unexpected rules. First, viral RNA levels do not decrease in confluent cells or in those that had been treated with a cell cycle inhibitor. Second, HCV replication is less sensitive to certain antiviral compounds, which may enable critical drug reevaluations. Third, IFN-γ does not inhibit HCV replication. Thus, HuH6 cells provide an environment in which HCV RNAs can persist despite drastic changes in host cell metabolism. In this context, it is interesting to mention that HuH6 cells are not the only ones unable to inhibit HCV replication in response to IFN-γ. We recently established persistent JFH-1 replication in HeLa, HepG2, and WRL-68 cells (WRL-68 cells are a human embryonic hepatocarcinoma cell line). In a first series of experiments, we found that HeLa and HepG2 cells were unable to inhibit HCV replication after IFN-γ stimulation, whereas WRL-68 cells behaved like Huh-7 cells and efficiently blocked HCV replication in a dose-dependent manner with an IC50 of approximately 1 IU/ml (unpublished data). Experiments to determine the molecular basis for the observed differences in the IFN-γ sensitivity of HCV replication in the different cell lines are under way.
Supplementary Material
Acknowledgments
We are grateful to Piet Herdewijn, Georg Kochs, Diana Koletzki, Brent E. Korba, Gerhard Pürstinger, Johan Neyts, and Takaji Wakita for reagents and advice; Christian Kleist for HLA genotyping; Bart Janssen for microsatellite analysis; Ulrike Herian, Nina Kezmic, Alexander Schuster, and Dirk Theile for technical assistance; and Kerry Mills for critical reading of the manuscript.
This work was supported by grants from the Bundesministerium für Bildung und Forschung (Kompetenznetz Hepatitis, Teilprojekt 13.2, 13.4, and 15.4) and the VIRGIL European Network of Excellence on Antiviral Drug Resistance (LSHM-CT-2004-503359).
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Ali, S., C. Pellerin, D. Lamarre, and G. Kukolj. 2004. Hepatitis C virus subgenomic replicons in the human embryonic kidney 293 cell line. J. Virol. 78:491-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartenschlager, R., L. Ahlborn-Laake, J. Mous, and H. Jacobsen. 1993. Nonstructural protein 3 of the hepatitis C virus encodes a serine-type proteinase required for cleavage at the NS3/4 and NS4/5 junctions. J. Virol. 67:3835-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartenschlager, R., L. Ahlborn-Laake, J. Mous, and H. Jacobsen. 1994. Kinetic and structural analyses of hepatitis C virus polyprotein processing. J. Virol. 68:5045-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartenschlager, R., M. Frese, and T. Pietschmann. 2004. Novel insights into hepatitis C virus replication and persistence. Adv. Virus Res. 63:71-180. [DOI] [PubMed] [Google Scholar]
- 5.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 6.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carroll, S. S., J. E. Tomassini, M. Bosserman, K. Getty, M. W. Stahlhut, A. B. Eldrup, B. Bhat, D. Hall, A. L. Simcoe, R. LaFemina, C. A. Rutkowski, B. Wolanski, Z. Yang, G. Migliaccio, R. De Francesco, L. C. Kuo, M. MacCoss, and D. B. Olsen. 2003. Inhibition of hepatitis C virus RNA replication by 2′-modified nucleoside analogs. J. Biol. Chem. 278:11979-11984. [DOI] [PubMed] [Google Scholar]
- 8.Cella, M., D. Jarrossay, F. Facchetti, O. Alebardi, H. Nakajima, A. Lanzavecchia, and M. Colonna. 1999. Plasmacytoid monocytes migrate to inflamed lymph nodes and produce large amounts of type I interferon. Nat. Med. 5:919-923. [DOI] [PubMed] [Google Scholar]
- 9.Cheney, I. W., V. C. Lai, W. Zhong, T. Brodhag, S. Dempsey, C. Lim, Z. Hong, J. Y. Lau, and R. C. Tam. 2002. Comparative analysis of anti-hepatitis C virus activity and gene expression mediated by alpha, beta, and gamma interferons. J. Virol. 76:11148-11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Date, T., T. Kato, M. Miyamoto, Z. Zhao, K. Yasui, M. Mizokami, and T. Wakita. 2004. Genotype 2a hepatitis C virus subgenomic replicon can replicate in HepG2 and IMY-N9 cells. J. Biol. Chem. 279:22371-22376. [DOI] [PubMed] [Google Scholar]
- 11.De Azevedo, W. F., S. Leclerc, L. Meijer, L. Havlicek, M. Strnad, and S. H. Kim. 1997. Inhibition of cyclin-dependent kinases by purine analogues: crystal structure of human cdk2 complexed with roscovitine. Eur. J. Biochem. 243:518-526. [DOI] [PubMed] [Google Scholar]
- 12.Doi, I. 1976. Establishment of a cell line and its clonal sublines from a patient with hepatoblastoma. Gann 67:1-10. [PubMed] [Google Scholar]
- 13.Frese, M., T. Pietschmann, D. Moradpour, O. Haller, and R. Bartenschlager. 2001. Interferon-α inhibits hepatitis C virus subgenomic RNA replication by an MxA-independent pathway. J. Gen. Virol. 82:723-733. [DOI] [PubMed] [Google Scholar]
- 14.Frese, M., V. Schwärzle, K. Barth, N. Krieger, V. Lohmann, S. Mihm, O. Haller, and R. Bartenschlager. 2002. Interferon-γ inhibits replication of subgenomic and genomic hepatitis C virus RNAs. Hepatology 35:694-703. [DOI] [PubMed] [Google Scholar]
- 15.Friebe, P., J. Boudet, J. P. Simorre, and R. Bartenschlager. 2005. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J. Virol. 79:380-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friebe, P., V. Lohmann, N. Krieger, and R. Bartenschlager. 2001. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 75:12047-12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao, B., F. Hong, and S. Radaeva. 2004. Host factors and failure of interferon-alpha treatment in hepatitis C virus. Hepatology 39:880-890. [DOI] [PubMed] [Google Scholar]
- 18.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 19.Guidotti, L. G., and F. V. Chisari. 2001. Noncytolytic control of viral infections by the innate and adaptive immune response. Annu. Rev. Immunol. 19:65-91. [DOI] [PubMed] [Google Scholar]
- 20.Guo, J.-T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haller, O., M. Frese, and G. Kochs. 1998. Mx proteins: mediators of innate resistance to RNA viruses. Rev. Sci. Tech. 17:220-230. [DOI] [PubMed] [Google Scholar]
- 22.Honda, M., S. Kaneko, T. Shimazaki, E. Matsushita, K. Kobayashi, L. H. Ping, H. C. Zhang, and S. M. Lemon. 2000. Hepatitis C virus core protein induces apoptosis and impairs cell-cycle regulation in stably transformed Chinese hamster ovary cells. Hepatology 31:1351-1359. [DOI] [PubMed] [Google Scholar]
- 23.Honda, M., T. Shimazaki, and S. Kaneko. 2005. La protein is a potent regulator of replication of hepatitis C virus in patients with chronic hepatitis C through internal ribosomal entry site-directed translation. Gastroenterology 128:449-462. [DOI] [PubMed] [Google Scholar]
- 24.Hoofnagle, J. H. 1997. Hepatitis C: the clinical spectrum of disease. Hepatology 26:15S-20S. [DOI] [PubMed] [Google Scholar]
- 25.Israel, A. 1979. Preliminary characterization of the particles from productive and abortive infections of L cells by fowl plague virus. Ann. Microbiol. (Paris) 130B:85-100. [PubMed] [Google Scholar]
- 26.Kato, T., T. Date, M. Miyamoto, A. Furusaka, K. Tokushige, M. Mizokami, and T. Wakita. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808-1817. [DOI] [PubMed] [Google Scholar]
- 27.Kato, T., T. Date, M. Miyamoto, Z. Zhao, M. Mizokami, and T. Wakita. 2005. Nonhepatic cell lines HeLa and 293 support efficient replication of the hepatitis C virus genotype 2a subgenomic replicon. J. Virol. 79:592-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kato, T., A. Furusaka, M. Miyamoto, T. Date, K. Yasui, J. Hiramoto, K. Nagayama, T. Tanaka, and T. Wakita. 2001. Sequence analysis of hepatitis C virus isolated from a fulminant hepatitis patient. J. Med. Virol. 64: 334-339. [DOI] [PubMed] [Google Scholar]
- 29.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larkin, J., L. Jin, M. Farmen, D. Venable, Y. Huang, S. L. Tan, and J. I. Glass. 2003. Synergistic antiviral activity of human interferon combinations in the hepatitis C virus replicon system. J. Interferon Cytokine Res. 23:247-257. [DOI] [PubMed] [Google Scholar]
- 31.Li, Y., T. Zhang, C. Ho, J. S. Orange, S. D. Douglas, and W. Z. Ho. 2004. Natural killercells inhibit hepatitis C virus expression. J. Leukoc. Biol. 76:1171-1179. [DOI] [PubMed] [Google Scholar]
- 32.Lohmann, V., S. Hoffmann, U. Herian, F. Penin, and R. Bartenschlager. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lohmann, V., J. O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 34.Lohmann, V., F. Körner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lohmann, V., F. Körner, U. Herian, and R. Bartenschlager. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma, H., V. Leveque, A. De Witte, W. Li, T. Hendricks, S. M. Clausen, N. Cammack, and K. Klumpp. 2005. Inhibition of native hepatitis C virus replicase by nucleotide and non-nucleoside inhibitors. Virology 332:8-15. [DOI] [PubMed] [Google Scholar]
- 37.McHutchison, J. G., and M. W. Fried. 2003. Current therapy for hepatitis C: pegylated interferon and ribavirin. Clin. Liver Dis. 7:149-161. [DOI] [PubMed] [Google Scholar]
- 38.Meijer, L., A. Borgne, O. Mulner, J. P. Chong, J. J. Blow, N. Inagaki, M. Inagaki, J. G. Delcros, and J. P. Moulinoux. 1997. Biochemical and cellular effects of roscovitine, a potent and selective inhibitor of the cyclin-dependent kinases cdc2, cdk2 and cdk5. Eur. J. Biochem. 243:527-536. [DOI] [PubMed] [Google Scholar]
- 39.Moradpour, D., R. Gosert, D. Egger, F. Penin, H. E. Blum, and K. Bienz. 2003. Membrane association of hepatitis C virus nonstructural proteins and identification of the membrane alteration that harbors the viral replication complex. Antiviral Res. 60:103-109. [DOI] [PubMed] [Google Scholar]
- 40.Murray, E. M., J. A. Grobler, E. J. Markel, M. F. Pagnoni, G. Paonessa, A. J. Simon, and O. A. Flores. 2003. Persistent replication of hepatitis C virus replicons expressing the beta-lactamase reporter in subpopulations of highly permissive Huh7 cells. J. Virol. 77:2928-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakabayashi, H., K. Taketa, K. Miyano, T. Yamane, and J. Sato. 1982. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 42:3858-3863. [PubMed] [Google Scholar]
- 42.Okuse, C., J. A. Rinaudo, K. Farrar, F. Wells, and B. E. Korba. 2005. Enhancement of antiviral activity against hepatitis C virus in vitro by interferon combination therapy. Antiviral Res. 65:23-34. [DOI] [PubMed] [Google Scholar]
- 43.Pestka, S., C. D. Krause, and M. R. Walter. 2004. Interferons, interferon-like cytokines, and their receptors. Immunol. Rev. 202:8-32. [DOI] [PubMed] [Google Scholar]
- 44.Pestova, T. V., I. N. Shatsky, S. P. Fletcher, R. J. Jackson, and C. U. Hellen. 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 12:67-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pietschmann, T., V. Lohmann, G. Rutter, K. Kurpanek, and R. Bartenschlager. 2001. Characterization of cell lines carrying self-replicating hepatitis C virus RNAs. J. Virol. 75:1252-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robek, M. D., B. S. Boyd, and F. V. Chisari. 2005. Lambda interferon inhibits hepatitis B and C virus replication. J. Virol. 79:3851-3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47a.Stuyver, L. J., T. R. McBrayer, P. M. Tharnish, A. E. A. Hassan, C. K. Chu, K. W. Pankiewicz, K. A. Watanabe, R. F. Schinazi, and M. J. Otto. 2003. Dynamics of subgenomic hepatitis C virus replicon RNA levels in Huh-7 cells after exposure to nucleoside antimetabolites. J Virol. 77:10689-10694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taguchi, T., M. Nagano-Fujii, M. Akutsu, H. Kadoya, S. Ohgimoto, S. Ishido, and H. Hotta. 2004. Hepatitis C virus NS5A protein interacts with 2′, 5′-oligoadenylate synthetase and inhibits antiviral activity of IFN in an IFN sensitivity-determining region-independent manner. J. Gen. Virol. 85:959-969. [DOI] [PubMed] [Google Scholar]
- 49.Taylor, D. R., M. Puig, M. E. Darnell, K. Mihalik, and S. M. Feinstone. 2005. New antiviral pathway that mediates hepatitis C virus replicon interferon sensitivity through ADAR1. J. Virol. 79:6291-6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Theodore, D., and M. W. Fried. 2000. Natural history and disease manifestations of hepatitis C infection. Curr. Top. Microbiol. 242:44-54. [DOI] [PubMed] [Google Scholar]
- 51.Tomassini, J. E., K. Getty, M. W. Stahlhut, S. Shim, B. Bhat, A. B. Eldrup, T. P. Prakash, S. S. Carroll, O. Flores, M. MacCoss, D. R. McMasters, G. Migliaccio, and D. B. Olsen. 2005. Inhibitory effect of 2′-substituted nucleosides on hepatitis C virus replication correlates with metabolic properties in replicon cells. Antimicrob. Agents Chemother. 49:2050-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Regenmortel, M. H. V., C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner. 2000. Virus taxonomy: the VIIth report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 53.Varaklioti, A., N. Vassilaki, U. Georgopoulou, and P. Mavromara. 2002. Alternate translation occurs within the core coding region of the hepatitis C viral genome. J. Biol. Chem. 277:17713-17721. [DOI] [PubMed] [Google Scholar]
- 54.Vrolijk, J. M., A. Kaul, B. E. Hansen, V. Lohmann, B. L. Haagmans, S. W. Schalm, and R. Bartenschlager. 2003. A replicon-based bioassay for the measurement of interferons in patients with chronic hepatitis C. J. Virol. Methods 110:201-209. [DOI] [PubMed] [Google Scholar]
- 55.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. K. Murthy, A. Habermann, H. G. Kräusslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.World Health Organization. 2000. Hepatitis C: global prevalence (update). Wkly. Epidemiol. Rec. 75:18-19. [PubMed] [Google Scholar]
- 57.Xu, Z., J. Choi, T. S. Yen, W. Lu, A. Strohecker, S. Govindarajan, D. Chien, M. J. Selby, and J. Ou. 2001. Synthesis of a novel hepatitis C virus protein by ribosomal frameshift. EMBO J. 20:3840-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu, Q., J. T. Guo, and C. Seeger. 2003. Replication of hepatitis C virus subgenomes in nonhepatic epithelial and mouse hepatoma cells. J. Virol. 77:9204-9210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.