Abstract
Actinobacillus actinomycetemcomitans causes periodontitis, a costly chronic infection that affects a large number of patients. The pathogenesis of this dental infection is a multifactorial process that results in a serious degenerative disease of the periodontium. Although significant progress has been achieved after the identification of this gram-negative bacterium as the etiological agent of this infection, much remains to be done to understand in detail the bacterial factors and host-pathogen interactions involved in the pathogenesis of this disease. Classical research approaches have resulted in the identification of important virulence factors and cellular processes, although they have provided a rather narrow picture of some of steps of this complex process. In contrast, a much wider picture could be obtained with the application of tools such as bioinformatics and genomics. These tools will provide global information regarding the differential expression of genes encoding factors and processes that lead to the pathogenesis of this disease. Furthermore, comparative genomics has the potential of helping to understand the emergence and evolution of this human pathogen. This genome-wide approach should provide a more complete picture of the pathogenesis process of this disease, and will facilitate the development of efficient diagnostic, preventive, and therapeutic measures for this disease.
Keywords: A. actinomycetemcomitans, dental pathogen, genomics, bioinformatics, iron metabolism
THE HUMAN ORAL CAVITY AS A MICROBIAL MODEL
The human oral cavity is a complex biological system in which members of a diverse microbial community interact with themselves as well as different host structures and components. It has been reported that this cavity is colonized by approximately 500 bacterial species (Kroes et al., 1999; Paster et al., 2001), most of which are still poorly characterized. The bacterial flora of this human cavity includes a wide range of bacterial cell types as well as important fungi such as Candida albicans, most of which are considered to be commensal while some of them are classified as opportunistic pathogens capable of causing severe infections in compromised patients. Although complex, the interactions among the members of the oral flora develop according to reproducible temporal and spatial patterns, a process that results in coaggregation-mediated interactions and formation of multi-species microbial biofilms known as dental plaques (Rickard et al., 2003). These cell arrangements allow microbial cells to prosper in a hostile environment, which is the case for significant clinical bacterial pathogens that prosper in the human host (Donlan and Costerton, 2002). These cell structures are the result of specific microbial-microbial and microbial-host interactions, which play a role in the ability of microbial cells to evade the host response and resist antimicrobial agents (Stewart and Costerton, 2001). Furthermore, there is evidence that biofilms play a significant role in the horizontal transfer of genetic material among members of these cell aggregates (Ghigo, 2001), a process that facilitates their adaptation to different ecological niches.
Most of the knowledge available to date with regard to the microbial and host factors and processes involved in these interactions has been obtained with classical approaches based on the utilization of molecular genetics and molecular biology. Although these approaches have produced important and novel information, they have provided a rather narrow picture of these complex interactions. In contrast, a holistic view of the genetic blueprint of a bacterial cell can be readily obtained once its entire genome is sequenced with automated methods and annotated using computer analysis of nucleotide sequences. This genome-wide approach, which uses tools such as genomics and bioinformatics, is based mainly in the comparative analysis of gene content and structure among different bacterial cells and gene expression under different physiological conditions using DNA arrays. Whole-genomic comparisons and transcriptome analysis have the benefit of providing wider and more comprehensive information regarding the genome structures and gene expression patterns that will facilitate the understanding of the evolution and adaptation of bacterial pathogens, and the complex mechanisms by which they interact with their hosts. For example, the application of DNA microarrays and global transcriptional analysis has facilitated the study of the genetic composition and the analysis of gene expression under different environmental conditions, respectively, of important bacterial pathogens such as Escherichia coli (Perna et al., 2001) and Pseudomonas aeruginosa (Ochsner et al., 2002; Schuster et al., 2003; Wagner et al., 2003). These studies have provided novel and important information with regard to lateral gene transfer and the emergence of pathogens with new or different virulence attributes, the adaptation of bacteria to particular hosts and environmental niches through genome rearrangements, the evolution of pathogens that share the same host niche, and potential bacterial targets for the development of new therapeutic agents and the design of vaccines to prevent microbial diseases. Similar approaches could be applied to dental pathogens once their genomes are completely sequenced and annotated, a process that is near completion for several of them. However, there are investigators who have not waited until the genomes of oral bacteria were completely annotated and published, and have already started these types of analyses with some oral pathogens (Duncan, 2003).
GENOME SEQUENCING OF DENTAL MICROBES
As mentioned above, genome-wide analysis could be applied more efficiently to dental commensals and pathogens once their genomes are completely sequenced and annotated. Most of these efforts are supported by funding from federal agencies, mainly the National Institute of Dental and Craniofacial Research (NIDCR). Currently, this NIH institute supports the genome sequencing of Actinobacillus actinomycetemcomitans, Actinomyces naeslundii, Bacteroides forsythus, Candida albicans, Fusobacterium nucleatum, Porphyromonas gingivalis, Prevotella intermedia, Streptococcus gordonii, Streptococcus mitis, Streptococcus mutans, Streptococcus sanguis, Streptococcus sobrinus, and Treponema denticola (http://www.nidcr.nih.gov/research/research_resources.asp). NIDCR has also established the specialized Oral Pathogen Sequence Databases in collaboration with Los Alamos National Laboratory with the purpose of providing information pertaining to the genomes of oral pathogens to the scientific community (http://www.oralgen.lanl.gov/). The Genomes OnLine Database (GOLD, http://igweb.integratedgenomics.com/GOLD/) list 16 genome sequencing projects related to oral bacteria, with those of A . actinomycetemcomitans HK1651, P. gingivalis W83, F. nucleatum 25586, and S. mutants UA159 shown as completed projects. The genomic data of the two latter strains were published last year (Ajdic et al., 2002; Kapatral et al., 2002). It is interesting to note that Dr. M. Duncan from the Forsyth Institute (http://www.forsyth.org/re/re_i_duncan.htm) was using the genomic data of P. gingivalis W83, even before this genome project was finished and published, to analyze the gene composition of this oral bacterium and compare it with those of other bacterial genomes (Duncan, 2003). Now, her study can be extended even further since the complete genome sequence and analysis of this oral pathogenic bacterium was published very recently (Nelson et al, 2003). Previous information such as the phylogeny and taxonomy of P. gingivalis W83 was confirmed by comparative genomics between this oral bacterium and that of other completely sequenced bacterial genomes. On the other hand, the computer analysis of the genome of this strain revealed the presence of pathways and virulence determinants, some of which are novel, that can explain the particular biology and virulence properties of this bacterium, and the mechanisms involved in the development of periodontal diseases caused by this oral pathogen. It is clear from the data reported in this publication that this revolutionary approach is already having a profound impact on basic and applied research, and will advance the understanding of basic biological process that are central to microbial physiology, genetics, and virulence.
It is evident that it would be more tedious and difficult to obtain this type of global information in such a short time using classical molecular genetics and molecular biology methods. However, these tools, which include gene cloning, the generation of isogenic mutants by random and site-directed mutagenesis, and genetic complementation tests and biological assays, should be used together with genomics and bioinformatics to confirm and assess some of the hypotheses and predictions made with global methods.
A. actinomycetemcomitans AS A MODEL FOR A GENOME-WIDE APPROACH TO STUDY A BACTERIAL DENTAL PATHOGEN
Actinobacillus actinomycetemcomitans is the major causative agent of Localized Juvenile Periodontitis (LJP), which has been named Localized Aggressive Periodontitis (LAP), as well as one of the microorganisms responsible for adult periodontitis. Periodontitis is the most prevalent chronic inflammatory diseases in humans, and it is a major cause of tooth loss (Slots and Genco, 1984; Zambon, 1985; Meyer and Fives-Taylor, 1997). The genome of the clinical isolate HK1651 has been completely sequenced and is in the final stages of the annotation process (http://www.genome.ou.edu/act.html). According to the information provided by Dr. Dyer, the genome size of this strain is 2,105,503 bp and contains 2,345 predicted open reading frames. Table 1 shows some information that was obtained after the initial automated annotation of this bacterial genome was completed. It is interesting to note that about half of the genes of this dental pathogen encode proteins that were classified either as hypothetical, unclassified, or of unknown function. This information demonstrates that much remains to be done to understand the basic biology of this pathogen and the functions it expresses during the interaction with the host’s cells and systems.
Table 1.
Classification of A. actinomycetemcomitans HK1651 predicted genes according to their cellular role.
| Cellular main role | No. of predicted genes |
|---|---|
| Amino acid | 65 |
| Biosynthesis of cofactors, prosthetic groups, and carriers | 74 |
| Cell envelope | 97 |
| Cellular processes | 64 |
| Central intermediary metabolism | 28 |
| DNA metabolism | 79 |
| Energy metabolism | 184 |
| Fatty acid and phospholipid metabolism | 40 |
| Hypothetical proteins | 671 |
| Other categories | 16 |
| Protein fate | 77 |
| Protein synthesis | 124 |
| Purines, pyrimidines, nucleosides, and nucleotides | 42 |
| Regulatory functions | 54 |
| Signal transduction | 7 |
| Transcription | 34 |
| Transport and binding proteins | 181 |
| Unclassified | 460 |
| Unknown function | 67 |
We and other investigators have been unable to use genomics and bioinformatics to study A. actinomycetemcomitans because, although closed (http://www.genome.ou.edu/act.html), the genome of this bacterium has not been completely annotated and the results made public by Dr. Dyer’s research group. However, the information already available at their web site has been instrumental to design classical molecular genetic and molecular biology experiments to test the presence and expression of genes potentially involved in iron acquisition. Analysis of the HK1651 genome with BLAST (Altschul et al., 1997) revealed that the genome of this strain contains genes encoding functions involved in iron acquisition and iron regulation of gene expression. PCR amplification of genomic DNA isolated from other A . actinomycetemcomitans strains, that have been used by other investigators to study different biological aspects of this oral pathogen, proved that the afe and afu genes, which code for periplasmic-binding protein-dependent transport (PBT) systems, are present in all strains tested (Table 2). The presence of TonB, which provides the energy required for the transport process, and HgpA, which could be involved in the acquisition of iron from human hemoglobin, were also detected in these strains. This analysis also showed that the HK1651 strain lacks genes required for the expression of siderophore-mediated iron uptake systems, an observation that agrees with the data we have obtained experimentally (unpublished observations). However, it seems that this oral pathogen has the potential to express transport systems that could use siderophore compounds produced by other bacterial species. This possibility could be confirmed once the annotation of the genome is completed and a more complete picture can be drawn with regard to its physiological and virulence properties. Another important observation we have made related to the expression of a transferrin-binding system is that the HK1651 genome contains sequences encoding some components of this system that plays a role in other pathogens such as Haemophilus influenzae and Neisseria that obtain iron via siderophore-independent iron acquisition systems (Chen et al., 1993; Sanders et al., 1994). However, the predicted open reading frame for the transferrin-binding protein A present in the HK1651 genome contains a point mutation that impairs the expression of this activity. This finding was confirmed experimentally in our laboratory and is in agreement with the data reported in a recent publication (Hayashida et al., 2002).
Table 2.
Detection of genes related to iron acquisition and gene regulation in different strains of A. actinomycetemcomitans.
| Genes | HK1651 | Strains Y4 | SUNY465 | CU1000 |
|---|---|---|---|---|
| afeA | +a | + | + | + |
| afeB | + | + | + | + |
| afeC | + | + | + | + |
| afeD | + | + | + | + |
| afuA | + | + | + | + |
| afuB | + | + | + | + |
| afuC | + | + | + | + |
| tonB | + | + | + | + |
| hgpA | + | NDb | ND | + |
| fur | + | + | + | + |
The amplicons obtained with each template were validated by automated DNA sequencing
Not determined
We have also used the genomic information already available to confirm the transcriptional expression of the afe and afu genes, which have been reported to be associated with siderophore-independent iron acquisition in bacteria, in the strains HK1651, SUNY465, and CU1000. In addition, we proved that in the case of HK1651, these loci are polycistronic as predicted by the computer analysis of their nucleotide sequence. RT-PCR experiments also showed that all three strains express the tonB iron transport gene and the fur iron regulatory gene. The results obtained with the latter gene are in agreement with our previous report describing the presence and expression of fur and iron regulated proteins in A. actinomycetemcomitans Y4 (Graber et al., 1998) as well as in the SUNY465 strain (unpublished observation).
We have been using the presence and expression of fur and the genomic data available to initiate the identification and characterization of A. actinomycetemcomitans genes that belong to the Fur and iron regulons using a genetic approach that to some extent provides genome-wide information. For this purpose, we have used the Fur titration assay (FURTA) (Stojiljkovic et al., 1994) with a genomic library constructed by cloning ~1 kb Sau3AI-digested DNA fragments from HK1651 into the BamHI site of the multi-copy plasmid vector pUC18 (Fig. 1). This approach resulted in the isolation of several clones that produced red colonies on Fe-containing MacConkey agar plates because the presence of multiple copies of Fur boxes allows the expression of the reporter fusion fhu::lacZ under iron-rich conditions. Sequence analysis of several positive clones showed that some of them contain either promoter elements or intergenic regions that appear to have sequences that resemble the Fur consensus binding site found in the promoter regions of bacterial Fur-regulated genes (Calderwood and Mekalanos, 1988). Computer analysis of the nucleotide sequence of some clones showed that they contain sequences related to genes encoding transferrin and hemoglobin binding proteins, ferritin, formate dehydrogenase, a transmembrane protein, and some proteins with no significant matches in the GenBank database. We are currently confirming these results by subcloning the promoter regions of these genes to test their expression under different conditions, and their ability to bind the A. actinomycetemcomitans HK1651 Fur repressor protein, which was overexpressed and purified by affinity chromatography. We are also in the process of testing the expression of these putative Fur-regulated genes by using real-time RT-PCR with RNA samples isolated from HK1651 cells cultured in the presence of either heme or inorganic iron. We are also using the available genomic information to generate isogenic derivatives to test the role of some of the loci and genes that are predicted to be involved in iron acquisition by computer analysis of their nucleotide sequences and test their differential expression under different experimental conditions.
Figure 1.
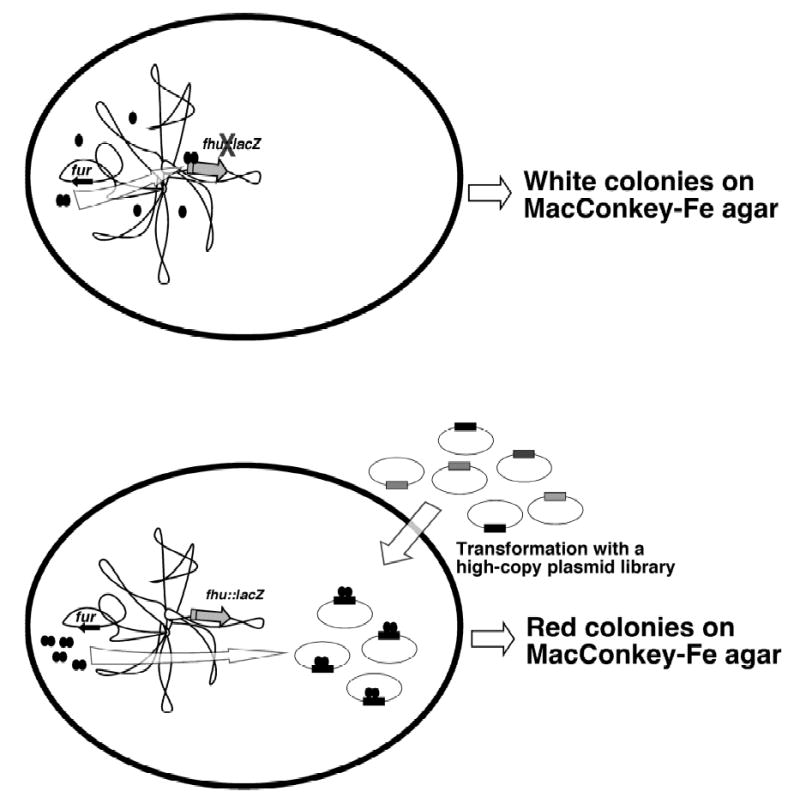
Diagram depicting the basis of FURTA used to clone Fur-regulated genes from the A. actinomycetemcomitans HK1651 strain. The grey and black straight arrows represent the fhu-lacZ genetic fusion and the fur gene, respectively, located in the chromosome of the E. coli indicator strain H1717. The black ovals represent the Fur repressor protein, and the black boxes in the plasmids indicate the presence of Furbinding sequences cloned from the genomic DNA of HK1651. Titration of the Fur repressor protein by the cloned Fur boxes derepresses the expression of the fhu-lacZ genetic fusion, which results in the formation of red colonies on MacConkey agar supplemented with inorganic iron.
WHAT IS IN THE FUTURE?
From the standpoint of genomics and bioinformatics, a bright and promising future is ahead for dental microbes since the genomes of several of them were completed or are near completion. The scientific community is already benefiting from bacterial genomes that have been completed and published, like that of S. mutans UA159 which is providing basic global information such as that related to gene composition, the presence of genetic elements potentially involved in genome evolution, and the mechanisms and elements involved in horizontal gene transfer (Ajdic et al., 2002). The genomic information has also allowed the in silico construction of the metabolic pathways of this bacterium (Ajdic et al., 2002), a practical and convenient result that will aid in the understanding of basic biological aspects of this member of the oral flora. For instance, the analysis derived from the nucleotide sequence of this cariogenic pathogen demonstrates that it contains a large number of membrane transport systems, some of which could be involved in iron acquisition. This analysis also showed that this bacterium lacks some important genes required for the expression of siderophore-mediated iron acquisition systems in other bacteria, an observation that is in agreement with the idea that the main mechanism by which this bacterium acquires iron is either via the direct interaction of host iron-binding proteins, such as lactoferrin or transferrin, or the expression of iron reductase activity that allows the transport of ferrous iron across the bacterial cell wall. Certainly, the S. mutans UA159 genomic information will be used to construct DNA microarrays (DNA chips) containing probes for each open reading frame found in the genome of this bacterium. Hybridization of these chips with labeled total DNA obtained from other pathogens or clinical isolates of the same pathogen should provide data regarding the genetic components that are common and unique among them, information that should give insights into the genetic variations among bacteria without the burden of sequencing the entire genome of a large number of clinical isolates. On the other hand, hybridization of DNA chips with labeled cDNA probes generated from RNA samples obtained from cells grown under different conditions could be used to study differential gene expression and understand the host-pathogen interactions at the molecular level. This type of approach has been used recently to study the effects of iron limitation (Ochsner et al., 2002), quorum-sensing and the extracellular environment (Schuster et al., 2003; Wagner et al., 2003) in P. aeruginosa, conditions and systems that are known to play a role in the pathogenesis of significant human infectious diseases.
The sequenced genomes could be used to search for the presence and expression of classical as well as novel global regulators that could play significant roles in differential gene expression, and the consequent adaptation of bacteria to different environments. Small RNAs is one of the latter elements that is being investigated intensively in different bacterial systems after the initial observation that it plays a regulatory role in gene expression in E. coli (Gottesman, 2002), particularly in the expression of genes that are involved in iron metabolism (Masse and Gottesman, 2002). A similar regulatory mechanism has been found recently in P. aeruginosa (Wilderman and Vasil, 103rd General Meeting of the American Society for Microbiology, abstract B404) which was detected after the genome of this bacterium was searched with algorithms designed to find intergenic chromosomal regions that can produce transcripts with structures similar to that described for the E. coli small RNA RyhB (Masse and Gottesman, 2002). This, in conjunction with the appropriate bioinformatic tools, will most likely produce novel and important information that should facilitate the analysis and understanding of central cellular process in bacteria.
The examples described above demonstrate how the experimental approaches to study the molecular biology, genetics, physiology, and genome evolution of microbial cells has been revolutionized with the development and application of bioinformatics and genomics. It is evident that the same approach will be applied to study different aspects of the microbial community found in the human oral cavity. With regard to the particular case of A. actinomycetemcomitans, it is possible to predict that significant advances in basic and clinical research will be attained once the complete genome of this oral pathogen is published. Furthermore, the application of genomics and bioinformatics together with the utilization of the animal model recently described (Schreiner et al., 2003), in which lesions similar to those found in human patients were observed after inoculating Sprague-Dawley rats with food containing this oral pathogen, should provide a more comprehensive picture of the pathogenesis of the periodontal diseases. For instance, it will be possible to identify and characterize genes that are differentially expressed in the host when compared to laboratory conditions used to maintain and propagate A. actinomycetemcomitans cells, an approach that will provide information on the factors involved in the host-pathogen interactions that are central to microbial pathogenicity and infectious diseases.
In our particular case, the availability of the complete genome and DNA microarrays will facilitate our work in the identification of genes that belong to the iron and Fur regulon, which are differentially expressed under iron-limiting and iron-rich conditions. Another benefit would be the utilization of comparative genomics, an approach that will facilitate the comparison of the complete genomes of different dental pathogens. The information obtained by these means will allow investigators to understand different processes, such as bacterial evolution and adaptation, species relationships, and gene transfer by different means among members of a complex microbial community at the molecular and genetic levels. These achievements will bring Oral Microbiology to levels of sophistication and advancement comparable to those already achieved with other bacterial pathogens that have different targets in the human host.
Acknowledgments
Miami University research funds and Public Health Grants AI44776-01A1 and DE13657-02 supported part of the research work presented in this manuscript. The development of this manuscript was supported in part by award 1R13DE014611-01 from the National Institute of Dental and Craniofacial Research/National Library of Medicine. We are grateful to Dr. D. Dyer, University of Oklahoma health Science Center, for making available to us unpublished data related to the genome of A. actinomycetemcomitans HK1651. We also thank Dr. D. Figurski, College of Physicians & Surgeons of Columbia University, for giving us the CU1000 strain of A . actinomycetemcomitans and some isogenic derivatives that have been used in our work.
References
- Ajdic D, McShan WM, McLaughlin RE, Savic G, Chang J, Carson MB, et al. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci USA. 2002;99:14434–14439. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood SB, Mekalanos J. Confirmation of the fur operator site by insertion of a synthetic oligonucleotide into an operon fusion plasmid. J Bacteriol. 1988;170:1015–1017. doi: 10.1128/jb.170.2.1015-1017.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-Y, Berish SA, Morse SA, Mietzner TA. The ferric-iron binding protein of pathogenic Neisseria spp. functions as a periplasmic transport protein in iron acquisition from human transferrin. Mol Microbiol. 1993;10:311–318. doi: 10.1111/j.1365-2958.1993.tb01957.x. [DOI] [PubMed] [Google Scholar]
- Donlan RM, Costerton JW. Biofilms: Survival mechanisms of clinically relevant microorganisms. J Clin Microbiol Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan MJ. Genomics of oral bacteria. Crit Rev Oral Biol Med. 2003;14:175–187. doi: 10.1177/154411130301400303. [DOI] [PubMed] [Google Scholar]
- Ghigo JM. Natural conjugative plasmids induce bacterial biofilm development. Nature. 2001;412:442–445. doi: 10.1038/35086581. [DOI] [PubMed] [Google Scholar]
- Gottesman S. Stealth regulation: biological circuits with small RNA switches. Genes Dev. 2002;16:2829–2842. doi: 10.1101/gad.1030302. [DOI] [PubMed] [Google Scholar]
- Graber K, Smoot LM, Actis LA. Expression of iron binding proteins and hemin binding activity in the dental pathogen Actinobacillus actinomycetemcomitans. FEMS Microbiol Lett. 1998;163:135–142. doi: 10.1111/j.1574-6968.1998.tb13037.x. [DOI] [PubMed] [Google Scholar]
- Hayashida H, Poulsen K, Kilian M. Differences in iron acquisition from human haemoglobin among strains of Actinobacillus actinomycetemcomitans. Microbiology. 2002;148:3993–4001. doi: 10.1099/00221287-148-12-3993. [DOI] [PubMed] [Google Scholar]
- Kapatral V, Anderson I, Ivanova N, Reznik G, Los T, Lykidis A, et al. Genome sequence and analysis of the oral bacterium Fusobacterium nucleatum strain ATCC 25586. J Bacteriol. 2002;184:2005–2018. doi: 10.1128/JB.184.7.2005-2018.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroes I, Lepp PW, Relman DA. Bacterial diversity within the human subgingival crevice. Proc Natl Acad Sci USA. 1999;96:14547–14552. doi: 10.1073/pnas.96.25.14547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci USA. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer DD, Fives-Taylor PM. The role of Actinobacillus actinomycetemcomitans in the pathogenesis of periodontal disease. Trends Microbiol. 1997;6:224–228. doi: 10.1016/S0966-842X(97)01055-X. [DOI] [PubMed] [Google Scholar]
- Nelson KE, Fleischmann RD, DeBoy RT, Paulsen IT, Fouts DE, Eisen JA, et al. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis Strain W83. J Bacteriol. 2003;185:5591–601. doi: 10.1128/JB.185.18.5591-5601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsner UA, Wilderman PJ, Vasil AI, Vasil ML. GeneChip expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol Microbiol. 2002;45:1277–1287. doi: 10.1046/j.1365-2958.2002.03084.x. [DOI] [PubMed] [Google Scholar]
- Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, et al. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perna NT, Plunkett GI, Burland V, Mau B, Glasner JD, Rose DJ, et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- Rickard AH, Gilbert P, High NJ, Kolenbrander PE, Handley PS. Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol. 2003;11:94–100. doi: 10.1016/s0966-842x(02)00034-3. [DOI] [PubMed] [Google Scholar]
- Sanders JD, Cole LD, Hansen EJ. Identification of a locus involved in the utilization of iron by Haemophilus influenzae. Infect Immun. 1994;62:4515–4525. doi: 10.1128/iai.62.10.4515-4525.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner HC, Sinatra K, Kaplan JB, Furgang D, Kachlany SC, Planet PJ, et al. Tight-adherence genes of Actinobacillus actinomycetemcomitans are required for virulence in a rat model. Proc Natl Acad Sci USA. 2003;100:7295–7300. doi: 10.1073/pnas.1237223100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M, Lostroh CP, Ogi T, Greenberg EP. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J Bacteriol. 2003;185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slots J, Genco RJ. Black-pigmented Bacteroides species, Capnocytophaga species, and Actinobacillus actinomycetemcomitans in human periodontal disease: virulence factors in colonization, survival, and tissue destruction. J Dent Res. 1984;63:412–421. doi: 10.1177/00220345840630031101. [DOI] [PubMed] [Google Scholar]
- Stewart PS, Costerton JW. Antibiotic resistance of bacteria in biofilms. Lancet. 2001;358:135–138. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- Stojiljkovic I, Baumler AJ, Hantke K. Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a Fur titration assay. J Mol Biol. 1994;236:531–545. doi: 10.1006/jmbi.1994.1163. [DOI] [PubMed] [Google Scholar]
- Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J Bacteriol. 2003;185:2080–2095. doi: 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambon JJ. Actinobacillus actinomycetemcomitans in human periodontal disease. J Clin Periodontol. 1985;12:1–20. doi: 10.1111/j.1600-051x.1985.tb01348.x. [DOI] [PubMed] [Google Scholar]


