Abstract
Background
Human herpesvirus-8 (HHV-8) is linked to the pathogenesis of Kaposi's sarcoma (KS), and the HHV-8 DNA load in peripheral blood mononuclear cells (PBMC) is associated with the clinical stage of KS. To examine the expression of HHV-8 in PBMC, four HHV-8 mRNA specific NASBA assays were developed
Methods
We have developed four quantitative nucleic acid sequence-based amplification assays (NASBA-QT) specifically to detect mRNA coding for ORF 73 (latency-associated nuclear antigen, LANA), vGCR (a membrane receptor), vBcl-2 (a viral inhibitor of apoptosis) and vIL-6 (a viral growth factor). The NASBA technique amplifies nucleic acids without thermocycling and mRNA can be amplified in a dsDNA background. A molecular beacon is used during amplification to enable real-time detection of the product. The assays were tested on PBMC samples of two AIDS-KS patients from the Amsterdam Cohort.
Results
For all four assays, the limit of detection (LOD) of 50 molecules and the limit of quantification (LOQ) of 100 molecules were determined using in vitro transcribed RNA. The linear dynamic range was 50 to 107 molecules of HHV-8 mRNA. We found HHV-8 mRNA expression in 9 out of the 10 tested samples.
Conclusion
These real-time NASBA assays with beacon detection provide tools for further study of HHV-8 expression in patient material.
Background
Homosexual men infected with HIV-1 are at increased risk for developing AIDS-related Kaposi's sarcoma (AIDS-KS) due to co-infection with HHV-8, a gamma herpes virus. Also known as Kaposi's sarcoma associated herpes virus, HHV-8 was first discovered in 1994 in KS-affected tissue [1]. Since then it also has been associated with the development of multicentric Castleman's disease and primary effusion lymphoma (PEL) [2-5]. HHV-8 DNA has been detected in both tissues and bodily fluids [6,7]. Studies initially focused on HHV-8 and its association with KS, transmission of HHV-8, and the natural history of HHV-8 infection. As shown by Renwick et al. and Jacobsen et al. [8,9] seroconversion for HHV-8 during HIV infection increases the risk of developing KS, implying that HIV-1 has an impact on HHV-8. Laboratory assays developed for the detection of HHV-8 include serologic assays such as immunofluorescence antibody assays [10-13], immunoblot assays [10], and various enzyme immunoassays [14-16]. PCR-based assays have also been used to detect HHV-8 DNA [1]. With the serologic assays, sensitivity and specificity are always problematic. No single assay is completely sensitive and specific [17].
We would prefer a method using viral RNA or DNA as a marker for the presence of HHV-8. Though DNA is an obvious choice because HHV-8 is a DNA virus, we have chosen RNA, because it indicates activity of the virus and may thus be a better prognostic marker. Active infected cells produce more RNA than DNA. In situ hybridisation has shown that most HHV-8 infected cells in KS lesions are latently infected [18,19]. but lytic HHV-8 infection is clearly present in a small proportion of cells [20-22].
A method for the detection of RNA is RT-PCR. To increase sensitivity an additional amplification step can follow the RT-PCR. This method is time consuming and also increases the likelihood of false positive reactions due to cross-contamination. Nucleic acid sequence-based amplification (NASBA) is a single-step isothermal RNA-specific amplification process [23] that amplifies mRNA in a dsDNA background [24]. NASBA has proved successful in the detection of various mRNAs [24-26] and in the detection of both viral [23] and bacterial [27] RNA in clinical samples.
By combining the standard NASBA technology [28] with a molecular beacon that anneals during amplification to the target sequence, a real-time detection system is generated [29]. This paper describes our development of real-time NASBA assays to quantify four functionally different genes of HHV-8: ORF 73, a latent gene, vGCR, a constitutively active gene, and the lytic genes vBcl-2 and vIL-6. To normalise the input mRNA we developed a fifth NASBA for a constitutively expressed mRNA, U1A mRNA, and standardised the specific HHV8 mRNAs to the expression of U1A mRNA.
Methods
HHV-8 genes
Our first assay was developed for ORF 73 and is based on the amplification of a 192 bp fragment situated within the gene. ORF 73 encodes the major immunogenic latency-associated nuclear antigen (LANA) of HHV-8. LANA is essential for maintenance of this virus in latently infected cells [30,31]. It is expressed only in such cells and thus gives a good indication of latent HHV-8 infection. In addition, LANA interacts with the tumour suppressor protein p53 and represses its transcriptional activity [32].
Our second assay was developed for vGCR, G-protein coupled receptor, and amplifies a 168 bp fragment. A receptor that binds several CXC and CC chemokines, vGCR appears to be constitutively active [33], although some studies indicate that it is expressed only during lytic replication [34]. It may contribute to the pathogenesis of KS by increasing the release of cellular growth factors such as VEGF [33]. It transforms rodent fibroblasts and induces expression of angiogenic factors [35].
The final two assays were developed for the genes vBcl-2 and vIL-6. The assay for vBcl-2 amplifies a 216-bp fragment, and for vIL-6 the fragment is 186 bp. vBcl-2 and vIL-6 are both early lytic genes, as their expression increases within 10 hours after induction [9].
vBcl-2 is a member of the Bcl-2 family, and functional studies indicate that vBcl-2 prevents Bax-mediated apoptosis and thus is an anti-apoptotic protein [36,37]. It is primarily active during lytic replication and its expression pattern suggests that it may function to prolong the survival of cells in which lytic infection is present.
vIL-6 is a secreted cytokine that maintains proliferation of IL-6-dependent mouse and human mycelia cell lines [21,38,39] and prevents apoptosis [21,38,40].
NASBA
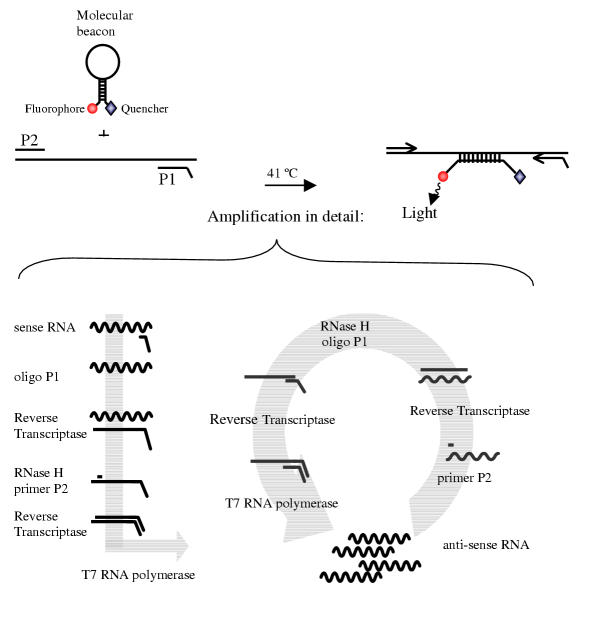
A NASBA reaction is based on the simultaneous activity of avian myeloblastosis virus (AMV) reverse transcriptase (RT), RNase H and T7 RNA polymerase with two oligonucleotide primers to produce amplification of the desired fragment more than 1012 fold in 90 to 120 minutes [23,41]. In a NASBA reaction, nucleic acids are a template for the amplification reaction only if they are single stranded and located in the primer-binding region. Because the NASBA reaction is isothermal (41°C), specific amplification of ssRNA is possible if denaturation of dsDNA is prevented in the sample preparation procedure. It is thus possible to pickup mRNA in a dsDNA background without getting false positive results caused by genomic dsDNA, in contrast with RT-PCR. In figure 1 a schematic presentation of the NASBA amplification is given. NASBA is achieved with the P1 (antisense) – P2 (sense) oligonucleotide set. The overhang on P1 encodes the promoter sequence for the T7 RNA polymerase. By using molecular beacons with the NASBA amplification a real-time detection system can be generated. Molecular beacons are stem-and-loop-structured oligonucleotides with a fluorescent label at the 5' end and a universal quencher at the 3' end. They are highly specific for their target and hybridise with their amplified target RNA when present in a NASBA amplification reaction to form a stable hybrid [42] at a relative low temperature (41°C).
Figure 1.
NASBA amplification reaction NASBA amplification reaction with the P1 (anti-sense) – P2 (sense) oligonucleotide primer set. The overhang on the P1 encodes the promoter sequence for the T7 RNA polymerase. A molecular beacon with a fluorophore and a quencer with the NASBA amplification reaction generates a real-time detection system.
Standard RNA
The NASBA quantification method is based on a standard curve with a known input of RNA. For this input we used in vitro RNA that was transcribed from four different plasmids. These plasmids were generated by cloning a specific PCR product for each of the four target genes, using primer sets located on the borders of the different genes. Primer sequences are shown in table 1. Input for the PCR was the nucleic acids of the BCP-1 cell line (a cell line containing HHV-8). The sizes of the four different PCR fragments were 595 bp, 926 bp, 473 bp, and 589 bp for ORF 73, vGCR, vBcl-2 and vIL-6, respectively (table 1). Each PCR fragment was cloned into the TOPO-TA plasmid (Invitrogen/ Lifetechnologies, Carlsbad, USA), and after all inserts were verified by sequencing, the fragments were cloned in a plasmide, pBluescript SK (Stratagene, La Jolla, USA). In vitro RNA was generated from these plasmids using T7 or T3 RNA polymerase, depending on the orientation of the fragment, and was treated with DNase to remove the plasmid. The Q-RNA was purified by RNeasy (Qiagen, Hilden, Germany), quantified spectrophotometrically, and checked by eye on agarose gel. The resulting in vitro RNA was used for the four different standard curves.
Table 1.
Primers for PCR fragment in plasmids
| 5' primer | 3' primer | PCR fragment size | |
| ORF 73 | agcccaccaggagataataca | tcatttcctgtggagagtccc | 595 bp |
| vGCR | gcggatatgactactctggaaact | gaggctttggaagagaccgt | 926 bp |
| vBcl-2 | atggacgaggacgttttgcct | cccaatagcgctgtcattct | 473 bp |
| vIL-6 | ggttcaagttgtggtctctctt | ggagtcacgtctgggatagagt | 589 bp |
Real-time amplification system
NASBA amplification requires a sense and an anti-sense primer, and for real-time detection a unique beacon is added to the reaction. We developed molecular beacons that could hybridise with the known sequences of the four HHV-8 genes, as shown in table 2. All the beacons have 6-fluorescine (6-FAM) as the fluorescent label at the 5' end and a (4-(dimethylamino) phenyl) azo) benzoic acid (Dabcyl) as universal quencher at the 3' end. A BLAST analysis revealed that in each case, neither the primers nor the beacon shared significant homology with any known nucleotide sequences other than the target gene. The quality of the different beacons was tested with a melting curve (from 80°C to 20°C) and acryl amide gel electrophoresis.
Table 2.
Sequences of the primers and beacons for each of the assays
| Primer P1 Anti-sense primer | Primer P2 Sense primer | Beacon Stem-loop in lowercase italics | |
| ORF73 | AG AGA CAA TAC ACA TAT ACA CAA TAA G | GAA AGG ATG GAA GAC GAG ATC CA | gca cgc AGG AGT AAA GGC AGG CCC CGT GTC gc g tgc |
| vGCR | AA CGA GGT TAC TGC CAG ACC CAC GT | CAG GCG GAA GGT AAG GGG GGT GA | gca cgc TGA TTG TTG CTG TGG TGC TGC T gcg tcg |
| vBcl-2 | AA GCG AAA CCA CTG GGG TCC GAT TG | GTG AGA TTT CAC AGC ACC ACC GGT A | gca cgc TGA CCT TTG GCA GTT TTG TGG CC gcg tgc |
| vIL-6 | AG AAC ATA AAA CGA AGC AAA GTG TCT CA | GGA AAA TCA GTG ATA AAC GTG GA | gca cgc GGA AGA GCT CAA TAA GCT GAC TA gcg tgc |
| U1A | AG AGG CCC GGC ATG TGG TGC ATA A | CAG TAT GCC AAG ACC GAC TCA GA | cgt acg AGA AGA GGA AGC CCA AGA GCC A cgt acg |
The 3' anti-sense primer is elongated with T7-promotor recognition sequence: AAT TCT AAT ACG ACT CAC TAT AGG G
Each reaction consisted of 5 μl of the sample RNA and 10 μl of NASBA reaction mix. This mix consisted of 80 mM Tris-HCl [pH 8.5], 24 mM MgCl2, 140 mM KCl, 1.0 mM DTT, 2.0 mM of each dNTP, 4.0 mM each of ATP, UTP and CTP, 3.0 mM GTP, and 1.0 mM ITP in 30% DMSO. This solution also contained the anti-sense and sense primers for amplification and the molecular beacons used for detection. The final concentration for the primes was 0.1 μM and for the beacons 40 nM.
The reaction mixtures were incubated at 65°C for 5 min, and after cooling to 41°C for 5 min to allow for primer annealing, 5 μl of enzyme mix was added by pipetting into the lids of the microtubes and subsequently spinning down. This mix contained, per reaction, 375 mM sorbitol, 2.1 μg BSA, 0.08 U RNase H, 32 U T7 RNA polymerase and 6.4 U AMV reverse transcriptase. Reactions were incubated at 41°C for 120 min in a fluorometer (Cytofluor 4000; Perkin-Elmer, Wellesley, Mass.). The RNA amplicons generated in the NASBA process are detected by molecular beacons [29]. These beacons generate a fluorescent signal during amplification when hybridising with their target. The fluorescence is measured continuously during amplification for real-time monitoring (i.e., as the reaction proceeds). A threshold is determined by setting the fluorescence emitted in the first 5 measure points as a background. The time-point at which a reaction rises above this threshold of detection and becomes positive is determined. This time-to-positivity (TTP) principle is similar to real-time PCR [43]. A calibration curve with 50, 102, 103, 104, 105, 106 and 107 molecules HHV-8 mRNA was included in each experiment. The number of mRNA copies per sample input can be extrapolated from the standard curve. De Baar et al showed that it is possible to use ttp values for quantification without the use of an internal calibrator [44].
RNA quantification
For standardisation of the amount of RNA input the U1A assay was used in each experiment. U1A mRNA encodes for one of the proteins of the U1 snRNP [45,46]. Because U1A is constitutively expressed, the amount of U1A mRNA measured in the samples is an indication of total RNA input. The mean of 6 assays (variation 0.6 log) was taken as the input RNA amount of the sample, and sample input RNA was normalised accordingly. The NASBA conditions for the U1A assay were the same as those for the HHV-8 assay, with the exception of the final concentration of the primer (0.2 μM) and the beacon (50 nM).
HHV-8 DNA assay
The DNA fractions of the PBMC samples were tested for the presence of HHV-8 DNA using a nested PCR for ORF 73 described by Goudsmit et al. [47]. As a quantification of the amount of DNA input, a limited dilution PCR for the cellular CCR5 gene was used. With limited dilution, an estimation of copies of HHV-8 DNA was made in relation to the amount of cells. For both the ORF 73 and the CCR5 nested PCR, the detection level was 5 molecules input per reaction.
Patient samples
To test our assays in vivo, we selected PBMC samples from two participants of the Amsterdam Cohort Studies. Described elsewhere in detail [48,49], they were both HIV-1-infected men with Kaposi's sarcoma but had very different disease development. Patient 1 was a 30-year-old man who was demonstrated to be HIV-1-seropositive in 1995. In July 1996 he was diagnosed with KS. He did not respond to anti-retroviral or chemotherapy and died in January of 1997 as a result of severe infiltration of KS in both lungs. Patient 2 is a 36-year-old man who visited our outpatient clinic in 1992 with an increasing number of KS-related skin lesions. After he started anti-retroviral therapy several lesions disappeared and complete remission was gradually reached over the course of two years. These two patients were chosen in large part for the diversity in their course of Kaposi's sarcoma. We tested three samples for patient 1 and seven samples for patient 2 collected over time, all taken after diagnosis of KS.
After the frozen PBMC samples were thawed the cells were collected and resuspended in Trizol™ buffer to isolate RNA and DNA simultaneously, according to the manufacturer's recommendations. Precipitated RNA was redissolved in 50 μl H2O. For each time-point, a known amount of cells (7.6–11.1 * 106 cells) were isolated. With high input of total RNA we found that background RNA can disturb the amplification if present in large amounts and it was necessary to dilute the samples. To have a broad range in the dilution for the RNA quantification in the NASBA experiments, the samples were diluted 10,100, and 1000 times. Of these diluted samples 5 μl was used per reaction, and 10 μl of reaction mix was added. U1A mRNA was used as an external standard to compare quantification signals among the different samples.
Results
Sensitivity and specificity
These newly developed assays can be used quantitatively by testing a standard curve of samples with a known amount of mRNA molecules within the same experiment as the unknown samples, then extrapolating the results to the standard curve. Typical amplification curves could be plotted in which an increase of fluorescence was observed until most of the molecular beacon had hybridised with the synthesised amplicons and the fluorescence reached a maximum level.
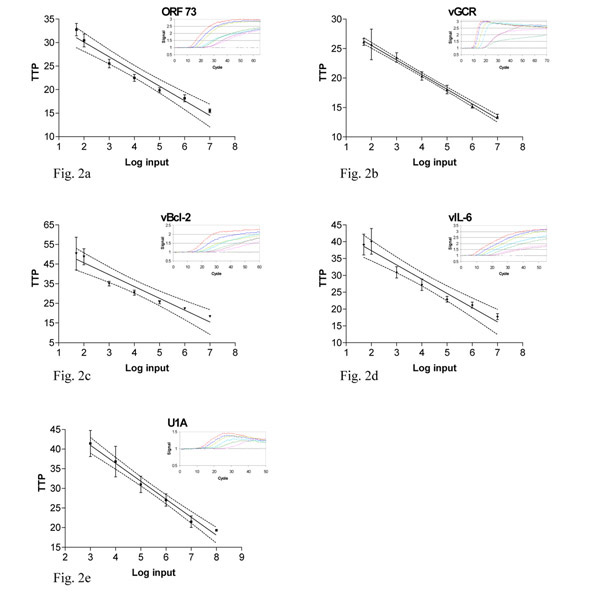
Such an amplification of in vitro synthesised ORF 73 RNA is shown in the insert in figure 2a. The time-point at which the fluorescence signal became detectable over the background i.e., time-to-positivity (ttp), was linear over a range of at least six orders of magnitude of input RNA molecules, as shown by the dilution series of in vitro synthesised RNA. In figure 2a the relationship between ttp values of the standard curve for ORF 73 and the log input of in vitro RNA is shown. This relationship extended from 107 to 50 molecules input.
Figure 2.
Standard curves in vitro RNA Figure 2 a-d. Relationship of time-to-positivity (TTP) to HHV-8 mRNA copy number. The number of mRNA molecules present in the reaction is indicated on the x axis and the ttp value in indicated on the y axis. For the data obtained with 50, 102 to 107 molecules, the values of TTP are the mean of five replicates of independent experiments. For the U1A the range of RNA input was 103 to 108. Error bars indicate the standard deviation for the values of ttp. The solid line was obtained by linear regression analysis of the data from 50 to 107 molecules, and the dotted lines indicate the 95% confidence intervals for the regression. Inserts a-d: Amplification plot of a 10-fold dilution serial dilution of in vitro RNA for ORF 73, vGCR, vBcl-2 and vIL-6. The amount of input RNA 1 * 107, 1 * 106, 1 * 105, 1 * 104, 1 * 103, 1 * 102, 50 and 0 molecules. Insert e: Amplification plot of a 10-fold dilution serial dilution of U1A in vitro RNA, the amount of input RNA 1 * 108, 1 * 107, 1 * 106, 1 * 105, 1 * 104, 1 * 103 and 0 molecules.
The dilution series of in vitro RNA showed a detection level of 50 molecules input. For accurate quantification the required level is 102 molecules input per assay. The line is calculated with linear regression analysis. Besides the limit of detection (LOD), the limit of quantification (LOQ) can be determined. For ORF 73, the LOD is 50 molecules input (84% pos.) while the LOQ is 102 molecules per reaction. The linear quantification of the ORF 73 assay was determined to be between 102 to 107 copies RNA per reaction (R 2 = 0.96, p < 0.001, fig. 2a). The sensitivity of all four assays was studied using five serial dilutions. The linear regression analyses for the other assays are shown in figure 2b,2c,2d,2e. The assay for vGCR has a very small standard deviation, and the amplification is linear from 107 to 50 molecules input per reaction (R 2 > 0.99, p < 0.001, fig. 2b). The LOD is 50 molecules (55% pos.) and the LOQ is 102 molecules per reaction. The assays for both vBcl-2 and vIL-6 are weaker but still within a workable range. As above, the linear quantification was between 102 to 107 copies RNA per reaction (R 2 = 0.94 and R 2 = 0.95 respectively, p < 0.001, fig. 2c,2d). For both assays, the LOD is 50 molecules (72% and 50% pos. respectively) and the LOQ is 102 molecules per reaction. U1A has a linear quantification range of 103 to 108 molecules (R2 = 0.99, p < 0.001, fig 2e). The assay for U1A had a sensitivity of only 103 copies input/reaction, which is feasible due to the high expression level of U1A.
The specificity of the five assays was tested by spiking each reaction with 106 molecules of in vitro RNA that the primers and beacon were not designed for. For example, ORF 73 in vitro RNA did not give a signal with the vGCR assay (data not shown).
Robustness
In replicate independent experiments, the time-to-positivity (TTP) value had a linear relationship with the logarithm of the amount of in vitro RNA added in the reaction. Subsequent experiments used serial 10-fold dilutions of in vitro RNA (from 107 to 102 and 50 molecules per reaction) as a standard curve to determine the amount of HHV-8 RNA in samples. The experiments were done on different days and by different people with no significant variation in results, which confirmed the robustness of the assays and also their ease of performance.
Patient samples
To address the functionality and quality of the HHV-8 assays in vivo, they were tested on PBMC samples collected after KS diagnosis from two participants in the Amsterdam Cohort Studies. In total, 10 frozen samples were analysed. As a positive control, all samples were tested for the presence of HHV-8 DNA with a nested PCR and were found to be positive (see Materials and Methods). The copies of HHV-8 DNA varied between 2 and 2 * 102 per 103 cells (table 3). A relation has been shown between the HHV-8 DNA levels in PBMC and the extent of Kaposi's sarcoma [50]. A possible correlation between DNA levels and mRNA expression should be investigated.
Table 3.
The ratio mRNA of (HHV-8 / U1A) × 107
| ORF 73 | vGCR | vBcl-2 | vIL-6 | HHV-8 DNA copies per 103 cells | |
| 1.1 | 6,3 × 105 | 3,1 | 3,1 | 62 | 20 |
| 1.2 | 92 | 71 | 71 | - | 20 |
| 1.3 | 2,2 × 102 | 9,2 | 3,3 × 103 | 1,8 × 102 | 200 |
| 2.1 | 2,9 × 102 | - | 3,9 × 104 | - | 20 |
| 2.2 | 2,7 × 102 | - | 2,7 × 102 | 2,2 × 102 | 2 |
| 2.3 | 5,0 × 102 | 2,6 × 105 | 3,2 × 103 | 25 | 2 |
| 2.4 | 3,0 × 102 | 8,5 × 103 | 5,7 × 105 | 3,0 × 103 | 2 |
| 2.5 | 2,6 ×102 | - | 4,0 × 102 | - | 2 |
| 2.6 | 41 | - | 4,1 × 103 | 2,6 × 104 | 2 |
| 2.7 | - | - | - | - | 2 |
Negative samples are shown as -. Samples from patient 1 are numbered 1.1–1.3; samples from patient 2: 2.1–2.7.
The results of the HHV-8 assays are shown in table 3. Of the 10 samples, 9 were positive for ORF 73, whereas only half were positive for vGCR. While vBcl-2 was measured in 9 out of 10 samples, vIL-6 was detected in approximately half of the samples. All the samples that were positive for ORF 73 were also positive for vBcl-2. Surprisingly the samples were positive for a latent and a lytic gene at the same time. The variation in expression levels is large both among the different assays and among the samples. No clear trend in the expression of HHV-8-specific mRNA can be distinguished, based on HHV-8 DNA levels, patient, or disease status.
Discussion
In general the NASBA assay has several advantages over the PCR technique [51]. The amplification procedure of the NASBA assay is entirely isothermal and is conducted at 41°C; it does not require thermal cycling instrumentation. The NASBA product is single-stranded and can therefore be readily detected by hybridisation analysis.
Our four real-time NASBA assays offer these benefits and also the five features that define validation of any assay: accuracy, sensitivity, precision, robustness, and specificity. With the use of standard curves in each experiment, accuracy is achieved. The sensitivity is determined with a linear standard curve. The limit of detection (LOD) and the limit of quantification (LOQ) are determined. For all the HHV-8 assays, the LOD is 50 molecules per reaction and LOQ was 100 molecules per reaction. To achieve linear amplification requires at least 5 concentrations on the linear portion of the curve. For the HHV-8 assays, the linear range spans 7 concentrations and for U1A it spans 6 concentrations. Quantification was achieved within this linear range of at least six orders of magnitude (102 to 107 molecules per reaction). Within this linear range quantification of at least six orders of magnitude (102 to 107 molecules per reaction) was achieved.
The precision of the assays was measured as the standard deviation and the confidence interval (fig. 2). We used 5 replicates of 7 different concentrations of the standard curve. Robustness was determined by having different people perform the experiments on different days. Specificity was shown with spiking experiments. The lower detection level of the assays was approximately 50 copies per reaction. The assays work equally well in vivo as in vitro e.g., for quantification of HHV-8 RNA in PBMCs.
The onset of KS is associated with a high HHV-8 load in PBMC [19] and this indicates that HHV-8 infected circulating cells play an important role in KS development [20,52,53]. HHV-8 can infect circulating B cells, monocytes/macrophages, T cells and KS-like spindle cell progenitors [54-56]. In KS lesions B cells are rare or absent, but monocytes macrophages are abundantly present [52], this suggest that circulating monocytes may recruit the virus into tissues or transmit the virus to other cells. Since lymphocytes and monocytes infiltrating KS lesions can be productively infected by HHV-8 [20,22], it is important to look at active infection in the PBMC. PBMCs of two AIDS-KS patients were selected and analysed for the presence of HHV-8 mRNA.
In 90% of our samples, ORF 73 mRNA and vBcl-2 mRNA were detected, whereas for vIL-6 mRNA and vGCR mRNA were present in 60% and 50%, respectively. The expression levels of all four genes varied widely. Comparing the different expression levels of the four HHV-8 genes within one sample or comparing expression levels of one gene in one patient did not show an apparent tendency. No clear trend can be distinguished based on the HHV-8 DNA levels, patient or disease status. Nevertheless the finding of expression of different HHV8 specific genes in PBMCs is notable in itself, as it has not been described before. HHV-8 RNA has been found in tissues, endothelial cells and spindle cells [18,57,58]. In 90% of fresh KS-affected tissues, HHV-8 RNA has been detected with RT-PCR [59]. In situ hybridisation and LANA immunohistochemistry have confirmed HHV-8 gene expression in KS spindle cells within KS lesions [18,58,60-62]. Expression of lytic HHV-8 genes has also been documented in monocytes and macrophages in KS lesions [20].
Conclusion
The assays that were developed work well for the detection and quantification of HHV-8 specific mRNA. Although no clear trend in the expression pattern could be distinguished the finding of expression of different HHV8 specific genes in PBMCs is important in itself. As this suggests that HHV-8 replication takes place in the PBMC. The PBMCs we used were obtained from two patients. To draw conclusions about the clinical and prognostic utility of these assays more patients and patient samples need to be analysed. However, as the assays are able to quantify HHV-8 mRNA in PBMC samples we feel that these real-time NASBA assays with beacon detection provide tools for further study of HHV-8 expression in patient material.
Competing interests
None declared.
Authors' contributions
AP carried out the assay development and validation, preformed the NASBA experiments and drafted the manuscript. JG participated in the study design. MC participated in the study design and carried out its supervision.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Acknowledgments
Acknowledgements
We thank Remco van den Burg, Margreet Bakker and Eveline Timmermans for their technical assistance, Bob van Gemen for his advice on assay design and Lucy Phillips for editorial review.
Contributor Information
Abeltje M Polstra, Email: a.m.polstra@amc.uva.nl.
J Goudsmit, Email: j.goudsmit@amc.uva.nl.
M Cornelissen, Email: m.i.cornelissen@amc.uva.nl.
References
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. New England Journal of Medicine. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- Cesarman E, Moore PS, Rao PH, Inghirami G, Knowles DM, Chang Y. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood. 1995;86:2708–2714. [PubMed] [Google Scholar]
- Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, D'Agay M-F, Clauvel J-P, Raphael M, Degos L, et al. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- Rettig MB, Ma HJ, Vescio RA, Pold M, Schiller G, Belson D, Savage A, Nishikubo C, Wu C, et al. Kaposi's sarcoma-associated herpesvirus infection of bone marrow dendritic cells from multiple myeloma patients. Science. 1997;276:1851–1854. doi: 10.1126/science.276.5320.1851. [DOI] [PubMed] [Google Scholar]
- LaDuca JR, Love JL, Abbott LZ, Dube S, Freidman-Kien AE, Poiesz BJ. Detection of human herpesvirus 8 DNA sequences in tissues and bodily fluids. Journal of Infectious Diseases. 1998;178:1610–1615. doi: 10.1086/314514. [DOI] [PubMed] [Google Scholar]
- Stamey FR, Patel MM, Holloway BP, Pellett PE. Quantitative, fluorogenic probe pcr assay for detection of human herpesvirus 8 dna in clinical specimens. J Clin Microbiol. 2001;39:3537–3540. doi: 10.1128/JCM.39.10.3537-3540.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renwick N, Halaby T, Weverling GJ, Dukers NHTM, Simpson GR, Coutinho RA, Lange LMA, Schulz TF, Goudsmit J. Seroconversion for human herpesvirus 8 during HIV infection is highly predictive of Kaposi's sarcoma. AIDS. 1998;12:2481–2488. doi: 10.1097/00002030-199818000-00018. [DOI] [PubMed] [Google Scholar]
- Jenner RG, Alba MM, Boshoff C, Kellam P. Kaposi's sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. Journal of Virology. 2001;75:891–902. doi: 10.1128/JVI.75.2.891-902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao SJ, Kingsley L, Li M, Zheng W, Parravicini C, Ziegler J, Newton R, Rinaldo CR, Saah A, Phair J, et al. KSHV antibodies among Americans, Italians and Ugandans with and without Kaposi's sarcoma. Nat Med. 1996;2:925–928. doi: 10.1038/nm0896-925. [DOI] [PubMed] [Google Scholar]
- Kedes DH, Operskalski E, Busch M, Kohn R, Flood J, Ganem D. The seroepidemiology of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus): distribution of infection in KS risk groups and evidence for sexual transmission. Nat Med. 1996;2:918–924. doi: 10.1038/nm0896-918. [DOI] [PubMed] [Google Scholar]
- Lennette ET, Blackbourn DJ, Levy JA. Antibodies to human herpesvirus type 8 in the general population and in Kaposi's sarcoma patients. Lancet. 1996;348:858–861. doi: 10.1016/S0140-6736(96)03240-0. [DOI] [PubMed] [Google Scholar]
- Koelle DM, Huang ML, Chandran B, Vieira J, Piepkorn M, Corey L. Frequent detection of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) DNA in saliva of human immunodeficiency virus-infected men: clinical and immunologic correlates. J Infect Dis. 1997;176:94–102. doi: 10.1086/514045. [DOI] [PubMed] [Google Scholar]
- Chatlynne LG, Lapps W, Handy M, Huang YQ, Masood R, Hamilton AS, Said JW, Koeffler HP, Kaplan MH, Friedman-Kien A, et al. Detection and titration of human herpesvirus-8-specific antibodies in sera from blood donors, acquired immunodeficiency syndrome patients, and Kaposi's sarcoma patients using a whole virus enzyme-linked immunosorbent assay. Blood. 1998;92:53–58. [PubMed] [Google Scholar]
- Davis DA, Humphrey RW, Newcomb FM, O'Brien TR, Goedert JJ, Straus SE, Yarchoan R. Detection of serum antibodies to a Kaposi's sarcoma-associated herpesvirus-specific peptide. J Infect Dis. 1997;175:1071–1079. doi: 10.1086/516444. [DOI] [PubMed] [Google Scholar]
- Smith MS, Bloomer C, Horvat R, Goldstein E, Casparian JM, Chandran B. Detection of human herpesvirus 8 DNA in Kaposi's sarcoma lesions and peripheral blood of human immunodeficiency virus-positive patients and correlation with serologic measurements. J Infect Dis. 1997;176:84–93. doi: 10.1086/514043. [DOI] [PubMed] [Google Scholar]
- Spira TJ, Lam L, Dollard SC, Meng YX, Pau CP, Black JB, Burns D, Cooper B, Hamid M, Huong J, et al. Comparison of serologic assays and PCR for diagnosis of human herpesvirus 8 infection. J Clin Microbiol. 2000;38:2174–2180. doi: 10.1128/jcm.38.6.2174-2180.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staskus KA, Zhong W, Gebhard K, Herndier B, Wang H, Renne R, Beneke J, Pudney J, Anderson DJ, Ganem D, et al. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. Journal of Virology. 1997;71:715–719. doi: 10.1128/jvi.71.1.715-719.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker LL, Shankar P, Khan G, Freeman RB, Dezube BJ, Lieberman J, Thorley DA-Lawson. The Kaposi sarcoma-associated herpesvirus (KSHV) is present as an intact latent genome in KS tissue but replicates in the peripheral blood mononuclear cells of KS patients. Journal of Experimental Medicine. 1996;184:283–288. doi: 10.1084/jem.184.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasig C, Zietz C, Haar B, Neipel F, Esser S, Brockmeyer NH, Tschachler E, Colombini S, Ensoli B, Sturzl M. Monocytes in Kaposi's sarcoma lesions are productively infected by human herpesvirus 8. Journal of Virology. 1997;71:7963–7968. doi: 10.1128/jvi.71.10.7963-7968.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PS, Boshoff C, Weiss RA, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- Orenstein JM, Alkan S, Blauvelt A, Jeang K-T, Weinstein MD, Ganem D, Herndier B. Visualization of human herpesvirus type 8 in Kaposi's sarcoma by light and transmission electron microscopy. AIDS. 1997;11:F35–F45. doi: 10.1097/00002030-199705000-00001. [DOI] [PubMed] [Google Scholar]
- Kievits T, Van Gemen B, Van Strijp D, Schukkink R, Dircks M, Adriaanse H, Malek L, Sooknanan R, Lens P. NASBA(TM) isothermal enzymatic in vitro nucleic acid amplification optimized for the diagnosis of HIV-1 infection. Journal of Virological Methods. 1991;35:273–286. doi: 10.1016/0166-0934(91)90069-C. [DOI] [PubMed] [Google Scholar]
- Heim A, Grumbach IM, Zeuke S, Top B. Highly sensitive detection of gene expression of an intronless gene: Amplification of mRNA, but not genomic DNA by nucleic acid sequence based amplification (NASBA). Nucleic Acids Research. 1998;26:2250–2251. doi: 10.1093/nar/26.9.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blok MJ, Goossens VJ, Vanherle SJV, Top B, Tacken N, Middeldorp JM, Christiaans MHL, Van Hooff JP, Bruggeman CA. Diagnostic value of monitoring human cytomegalovirus late pp67 mRNA expression in renal-allograft recipients by nucleic acid sequence-based amplification. Journal of Clinical Microbiology. 1998;36:1341–1346. doi: 10.1128/jcm.36.5.1341-1346.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darke BM, Jackson SK, Hanna SM, Fox JD. Detection of human TNF-alpha mRNA by NASBA(TM). Journal of Immunological Methods. 1998;212:19–28. doi: 10.1016/S0022-1759(97)00211-1. [DOI] [PubMed] [Google Scholar]
- Morre SA, Sillekens P, Jacobs MV, van Aarle P, de Blok S, Van Gemen B, Walboomers JM, Meijer CJ, van den Brule AJ. RNA amplification by nucleic acid sequence-based amplification with an internal standard enables reliable detection of Chlamydia trachomatis in cervical scrapings and urine samples. J Clin Microbiol. 1996;34:3108–3114. doi: 10.1128/jcm.34.12.3108-3114.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gemen B, Wiel VP, Van Beuningen R, Sillekens P, Jurriaans S, Dries C, Schoones R, Kievits T. The one-tube quantitative HIV-1 RNA NASBA: Precision, accuracy, and application. PCR Methods & Applications. 1995;4:S177–S184. doi: 10.1101/gr.4.4.s177. [DOI] [PubMed] [Google Scholar]
- Leone G, Van Schijndel H, Van Gemen B, Kramer FR, Schoen CD. Molecular beacon probes combined with amplification by NASBA enable homogeneous, real-time detection of RNA. Nucleic Acids Research. 1998;26:2150–2155. doi: 10.1093/nar/26.9.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballestas ME, Chatis PA, Kaye KM. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science. 1999;284:641–644. doi: 10.1126/science.284.5414.641. [DOI] [PubMed] [Google Scholar]
- Cotter MA, Robertson ES. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology. 1999;264:254–264. doi: 10.1006/viro.1999.9999. [DOI] [PubMed] [Google Scholar]
- Friborg J Jr, Kong W, Hottiger MO, Nabel GJ. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature. 1999;402:889–894. doi: 10.1038/47266. [DOI] [PubMed] [Google Scholar]
- Arvanitakis L, Geras-Raaka E, Varma A, Gershengorn MC, Cesarman E. Human herpesvirus KSHV encodes a constitutively active G-protein-coupled receptor linked to cell proliferation. Nature. 1997;385:347–349. doi: 10.1038/385347a0. [DOI] [PubMed] [Google Scholar]
- Kirshner JR, Staskus K, Haase A, Lagunoff M, Ganem D. Expression of the open reading frame 74 (G-protein-coupled receptor) gene of Kaposi's sarcoma (KS)-associated herpesvirus: Implications for KS pathogenesis. Journal of Virology. 1999;73:6006–6014. doi: 10.1128/jvi.73.7.6006-6014.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bais C, Santomasso B, Coso O, Arvanitakis L, Raaka EG, Gutkind JS, Asch AS, Cesarman E, Gerhengorn MC, Mesri EA. G-protein-coupled receptor of Kaposi's sarcoma-associated herpesvirus is a viral oncogene and angiogenesis activator. Nature. 1998;391:86–89. doi: 10.1038/34193. [DOI] [PubMed] [Google Scholar]
- Sarid R, Sato T, Bohenzky RA, Russo JJ, Chang Y. Kaposi's sarcoma-associated herpesvirus encodes a functional Bcl-2 homologue. Nature Medicine. 1997;3:293–298. doi: 10.1038/nm0397-293. [DOI] [PubMed] [Google Scholar]
- Cheng EHY, Nicholas J, Bellows DS, Hayward GS, Guo H-G, Reitz MS, Hardwick JM. A Bcl-2 homolog encoded by Kaposi sarcoma-associated virus, human herpesvirus 8, inhibits apoptosis but does not heterodimerize with Bax or Bak. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:690–694. doi: 10.1073/pnas.94.2.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas J, Ruvolo VR, Burns WH, Sandford G, Wan X, Ciufo D, Hendrickson SB, Guo H-G, Hayward GS, Reitz MS. Kaposi's sarcoma-associated human herpesvirus-8 encodes homologues of macrophage inflammatory protein-1 and interleukin-6. Nature Medicine. 1997;3:287–292. doi: 10.1038/nm0397-287. [DOI] [PubMed] [Google Scholar]
- Burger R, Neipel F, Fleckenstein B, Savino R, Ciliberto G, Kalden JR, Gramatzki M. Human herpesvirus type 8 interleukin-6 homologue is functionally active on human myeloma cells. Blood. 1998;91:1858–1863. [PubMed] [Google Scholar]
- Neipel F, Albrecht J-C, Ensser A, Huang Y-Q, Jian J, Li , Kien AE, Fleckenstein B. Human herpesvirus 8 encodes a homolog of interleukin-6. Journal of Virology. 1997;71:839–842. doi: 10.1128/jvi.71.1.839-842.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton J. Nucleic acid sequence-based amplification. Nature. 1991;350:91–92. doi: 10.1038/350091a0. [DOI] [PubMed] [Google Scholar]
- Tyagi S, Kramer FR. Molecular beacons: Probes that fluoresce upon hybridization. Nature Biotechnology. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- Higuchi R, Fockler C, Dollinger G, Watson R. Kinetic PCR analysis: Real-time monitoring of DNA amplification reactions. Bio-Technology. 1993;11:1026–1030. doi: 10.1038/nbt0993-1026. [DOI] [PubMed] [Google Scholar]
- De Baar MP, van Dooren MW, de Rooij E, Bakker M, Van Gemen B, Goudsmit J, De Ronde A. Single rapid real-time monitored isothermal RNA amplification assay for quantification of human immunodeficiency virus type 1 isolates from groups M, N, and O. J Clin Microbiol. 2001;39:1378–1384. doi: 10.1128/JCM.39.4.1378-1384.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Reed R. The role of small nuclear ribonucleoprotein particles in pre-mRNA splicing. Nature. 1987;325:673–678. doi: 10.1038/325673a0. [DOI] [PubMed] [Google Scholar]
- Maxwell ES, Fournier MJ. The small nucleolar RNAs. Annu Rev Biochem. 1995;64:897–934. doi: 10.1146/annurev.biochem.64.1.897. [DOI] [PubMed] [Google Scholar]
- Goudsmit J, Renwick N, Dukers NHTM, Coutinho RA, Heisterkamp S, Bakker M, Schulz TF, Cornelissen M, Weverling GJ. Human herpesvirus 8 infections in the Amsterdam Cohort Studies (1984–1997): Analysis seroconversions to ORF65 and ORF73. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:4838–4843. doi: 10.1073/pnas.97.9.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins JM, Sol CJA, Renwick N, Goudsmit J, Veenstra J, Reiss P. Favourable effect of chemotherapy on clinical symptoms and human herpesvirus-8 DNA load in a patient with Kaposi's sarcoma presenting with fever and anemia. European Journal of Clinical Microbiology & Infectious Diseases. 1999;18:499–502. doi: 10.1007/s100960050331. [DOI] [PubMed] [Google Scholar]
- Wit FWN, Sol CJA, Renwick N, Roos MTL, Pals Van ST, Leeuwen R, Goudsmit J, Reiss P. Regression of AIDS-related Kaposi's sarcoma associated with clearance of human herpesvirus-8 from peripheral blood mononuclear cells following initiation of antiretroviral therapy [2]. AIDS. 1998;12:218–219. [PubMed] [Google Scholar]
- Campbell TB, Borok M, Gwanzura L, MaWhinney S, White IE, Ndemera B, Gudza I, Fitzpatrick L, Schooley RT. Relationship of human herpesvirus 8 peripheral blood virus load and Kaposi's sarcoma clinical stage. AIDS. 2000;14:2109–2116. doi: 10.1097/00002030-200009290-00006. [DOI] [PubMed] [Google Scholar]
- Romano JW, Shurtliff RN, Dobratz E, Gibson A, Hickman K, Markham PD, Pal R. Quantitative evaluation of simian immunodeficiency virus infection using NASBA technology. J Virol Methods. 2000;86:61–70. doi: 10.1016/S0166-0934(99)00184-6. [DOI] [PubMed] [Google Scholar]
- Fiorelli V, Gendelman R, Sirianni MC, Chang HK, Colombini S, Markham PD, Monini P, Sonnabend J, Pintus A, Gallo RC, et al. gamma-Interferon produced by CD8+ T cells infiltrating Kaposi's sarcoma induces spindle cells with angiogenic phenotype and synergy with human immunodeficiency virus-1 Tat protein: an immune response to human herpesvirus-8 infection? Blood. 1998;91:956–967. [PubMed] [Google Scholar]
- Sirianni MC, Vincenzi L, Fiorelli V, Topino S, Scala E, Uccini S, Angeloni A, Faggioni A, Cerimele D, Cottoni F, et al. gamma-Interferon production in peripheral blood mononuclear cells and tumor infiltrating lymphocytes from Kaposi's sarcoma patients: correlation with the presence of human herpesvirus-8 in peripheral blood mononuclear cells and lesional macrophages. Blood. 1998;91:968–976. [PubMed] [Google Scholar]
- Mesri EA, Cesarman E, Arvanitakis L, Rafii S, Moore MA, Posnett DN, Knowles DM, Asch AS. Human herpesvirus-8/Kaposi's sarcoma-associated herpesvirus is a new transmissible virus that infects B cells. J Exp Med. 1996;183:2385–2390. doi: 10.1084/jem.183.5.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirianni MC, Vincenzi L, Topino S, Scala E, Angeloni A, Gonnella R, Uccini S, Faggioni A. Human herpesvirus 8 DNA sequences in CD8+ T cells. J Infect Dis. 1997;176:541. doi: 10.1086/514072. [DOI] [PubMed] [Google Scholar]
- Sirianni MC, Uccini S, Angeloni A, Faggioni A, Cottoni F, Ensoli B. Circulating spindle cells: correlation with human herpesvirus-8 (HHV-8) infection and Kaposi's sarcoma. Lancet. 1997;349:255. doi: 10.1016/s0140-6736(05)64866-0. [DOI] [PubMed] [Google Scholar]
- Ascherl G, Hohenadl C, Monini P, Zietz C, Browning PJ, Ensoli B, Sturzl M. Expression of human herpesvirus-8 (HHV-8) encoded pathogenic genes in Kaposi's sarcoma (KS) primary lesions. Adv Enzyme Regul. 1999;39:331–339. doi: 10.1016/S0065-2571(98)00019-3. [DOI] [PubMed] [Google Scholar]
- Sturzl M, Blasig C, Schreier A, Neipel F, Hohenadl C, Cornali E, Ascherl G, Esser S, Brockmeyer NH, Ekman M, et al. Expression of HHV-8 latency-associated T0.7 RNA in spindle cells and endothelial cells of AIDS-associated, classical and African Kaposi's sarcoma. Int J Cancer. 1997;72:68–71. doi: 10.1002/(SICI)1097-0215(19970703)72:1<68::AID-IJC10>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Huang YQ, Li JJ, Zhang WG, Feiner D, Friedman-Kien AE. Transcription of human herpesvirus-like agent (HHV-8) in Kaposi's sarcoma. J Clin Invest. 1996;97:2803–2806. doi: 10.1172/JCI118735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturzl M, Wunderlich A, Ascherl G, Hohenadl C, Monini P, Zietz C, Browning PJ, Neipel F, Biberfeld P, Ensoli B. Human herpesvirus-8 (HHV-8) gene expression in Kaposi's sarcoma (KS) primary lesions: an in situ hybridization study. Leukemia. 1999;13:S110–S112. doi: 10.1038/sj.leu.2401323. [DOI] [PubMed] [Google Scholar]
- Davis MA, Sturzl MA, Blasig C, Schreier A, Guo HG, Reitz M, Opalenik SR, Browning PJ. Expression of human herpesvirus 8-encoded cyclin D in Kaposi's sarcoma spindle cells. J Natl Cancer Inst. 1997;89:1868–1874. doi: 10.1093/jnci/89.24.1868. [DOI] [PubMed] [Google Scholar]
- Luppi M, Barozzi P, Maiorana A, Trovato R, Marasca R, Morselli M, Cagossi K, Torelli G. Expression of cell-homologous genes of human herpesvirus-8 in human immunodeficiency virus-negative lymphoproliferative diseases. Blood. 1999;94:2931–2933. [PubMed] [Google Scholar]