Abstract
Nonself recognition during vegetative growth in filamentous fungi is mediated by heterokaryon incompatibility (het) loci. In Neurospora crassa, het-c is one of 11 het loci. Three allelic specificity groups, termed het-cOR, het-cPA and het-cGR, exist in natural populations. Heterokaryons or partial diploids that contain het-c alleles of alternative specificity show severe growth inhibition, repression of conidiation and hyphal compartmentation and death (HCD). Using epitope-tagged HET-C, we show that nonself recognition is mediated by the presence of a heterocomplex composed of polypeptides encoded by het-c alleles of alternative specificity. The HET-C heterocomplex localized to the plasma membrane (PM); PM-bound HET-C heterocomplexes occurred in all three het-c incompatible allelic interactions. Strains containing het-c constructs deleted for a predicted signal peptide sequence formed HET-C heterocomplexes in the cytoplasm and showed a growth arrest phenotype. Our finding is a step towards understanding nonself recognition mechanisms that operate during vegetative growth in filamentous fungi, and provides a model for investigating relationships between recognition mechanisms and cell death.
Keywords: Neurospora crassa/nonself recognition/protein targeting/vegetative incompatibility
Introduction
Protein–protein interactions that mediate recognition of self and nonself have a fundamental role in many biological processes. Two distinct molecular mechanisms operate in self/nonself recognition. One mechanism operates via the recruitment of receptor ligands to accomplish allele-specific recognition. In flowering plants, gametophytic self-incompatibility is mediated by a receptor kinase encoded by the S locus (Nasrallah, 2000). At the mammalian major histocompatibility complex (MHC) loci, self and nonself MHC class I molecules are discriminated via MHC-antigen ligand and T-cell surface receptor (Jones et al., 1998). A second method for self/nonself discrimination involves combinatorial interactions of proteins to generate heterocomplexes with new regulatory functions. In fungi, combinatorial interactions between homeodomain proteins that function as transcriptional modulators have been described in several mating-type systems, such as the Mat a1p and Mat α2p in Saccharomyces cerevisiae (Ho et al., 1994), b locus gene-pair products in Ustilago maydis (Kämper et al., 1995) and the HD1 and HD2 polypeptides in Coprinus cinereus (Asante-Owusu et al., 1996).
In ascomycete fungi, nonself recognition during vegetative growth is mediated by vegetative (or heterokaryon) incompatibility. Different isolates are capable of undergoing hyphal fusion to form a heterokaryon, in which genetically different nuclei co-exist in a common cytoplasm. There are postulated benefits to heterokaryon formation, such as functional diploidy and mitotic recombination (Pontecorvo, 1956). The ability to form stable heterokaryons is regulated by heterokaryon (het) incompatibility loci (Glass et al., 2000; Saupe, 2000). Hyphal fusion between strains that differ in allelic specificity at a het locus generally results in rapid septal plugging and death of the fusion cell, a process referred to as hyphal compartmentation and death (HCD) (Garnjobst and Wilson, 1956; Jacobson et al., 1998; Wu and Glass, 2001). Nonself recognition mediated by het loci is thought to provide a protective mechanism to prevent transmission of infectious cytoplasmic elements, such as mycoviruses and senescence plasmids, and from exploitation by ‘aggressive’ genotypes (Debets and Griffiths, 1998; Cortesi et al., 2001).
In Neurospora crassa, 11 het loci have been genetically identified that regulate nonself recognition during vegetative growth (Perkins et al., 2000). At the het-c locus, isolates from natural populations fall into one of three het-c allelic specificity groups (Howlett et al., 1993; Saupe and Glass, 1997). These three allelic specificities are referred to as het-cOR, het-cPA and het-cGR (Saupe et al., 1996; Saupe and Glass, 1997). Transformants, heterokaryons or partial diploids containing het-c alleles of alternative specificity show severe growth inhibition, are aconidial and show HCD (Perkins, 1975; Howlett et al., 1993; Saupe and Glass, 1997; Jacobson et al., 1998; Wu and Glass, 2001). Phylogenetic analysis of het-c among different species and genera of filamentous fungi related to N.crassa shows that polymorphisms associated with het-c allelic specificity are subject to balancing selection (Wu et al., 1998). Thus, het-c displays an evolutionary pattern similar to other self/nonself recognition loci, such as the MHC loci and the S locus in plants (Klein et al., 1998), and the B mating-type locus of C.cinereus (May et al., 1999).
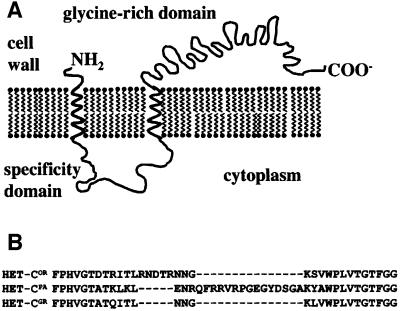
The het-c locus encodes an ∼960 amino acid protein (depending on the allele) with an N-terminal signal peptide and a C-terminal glycine-rich domain, and is predicted to reside in the plasma membrane (Saupe et al., 1996) (Figure 1A). The structure and size of het-c polypeptides that confer alternative het-c specificity (HET-COR, HET-CPA and HET-CGR) are very similar, with the exception of a short region (30–48 amino acids) that differs in insertion/deletion (indel) pattern (Figure 1B). Allelic specificity at het-c is dependent upon this indel: swapping of this region between alleles switches het-c allelic specificity (Saupe and Glass, 1997; Wu and Glass, 2001).
Fig. 1. Schematic diagram of HET-C. (A) HET-C is predicted to reside in the plasma membrane (http://psort.nibb.ac.jp/). (B) Amino acid sequence of the specificity domain of HET-C (Saupe and Glass, 1997). Allelic specificity (het-cOR, het-cPA, het-cGR) is based on characterization of laboratory strains (Howlett et al., 1993; Saupe and Glass, 1997).
In this study, we investigated the molecular mechanism of nonself recognition mediated by allelic differences at het-c. The simplest model describing het-c allelic recognition predicts either that heterocomplex formation between alternative het-c polypeptides is toxic to the cell or that its presence activates a pathway resulting in vegetative incompatibility. We show that nonself recognition is mediated by the physical interaction of HET-C polypeptides encoded by het-c alleles of alternative specificity. The HET-C heterocomplex specifically localizes to the plasma membrane of dead hyphal compartments. Deletion of a predicted signal peptide sequence in alternative het-c alleles led to cytoplasmic HET-C heterocomplex formation and a novel growth arrest phenotype. In all cases, co-expression of alternative het-c alleles resulted in stable HET-C heterocomplex formation.
Results
GFP- and HA-tagged versions of HET-C confer vegetative incompatibility
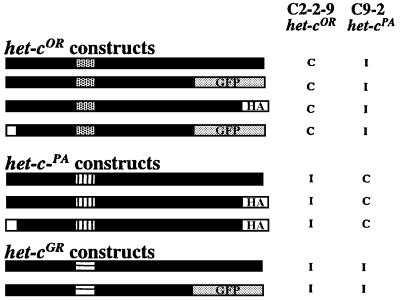
We used epitope-tagged het-c constructs in this study to assess HET-C localization and heterocomplex formation; alternative HET-C polypeptides (HET-COR, HET-CPA and HET-CGR) are 85–90% identical (Saupe and Glass, 1997). We used green fluorescent protein (GFP) (Cormack, 1998) and hemagglutinin (HA) (Forsburg and Sherman, 1997) as epitope tags (Figure 2). We confirmed the functionality of the epitope-tagged het-c constructs by introducing them into strains with different het-c allelic specificities (C9-2 het-cPA and C2-2-9 het-cOR) and via co-transformation into a het-c deletion strain, CJ44 (Wu and Glass, 2001). The phenotype displayed by transformants containing epitope-tagged het-c alleles (growth inhibition, repression of conidiation and ∼20% HCD) was indistinguishable from a typical het-c incompatibility phenotype mediated by untagged het-c alleles (Figure 2).
Fig. 2. Schematic diagram of epitope-tagged het-c constructs. N-terminal white boxes signify signal peptide deletion constructs. Patterned boxes indicate the het-c allelic specificity domain. GFP-tagged versions of het-cOR and het-cGR included the first 730 amino acids of HET-COR and 727 amino acids of HET-CGR, which are fully functional forms (Saupe et al., 1996). The het-cPA and het-cOR HA-tagged constructs included DNA sequences for the nine amino acid HA-epitope tag fused to the 3′ end of the het-c open reading frame (ORF). Constructs were introduced into standard het-c tester strains C2-2-9 (het-cOR) and C9-2 (het-cPA) (Saupe and Glass, 1997) to test for function. C, compatible transformants; I, incompatible transformants. Differences in phenotype or het-c allelic specificity were not observed when tagged, untagged or het-c signal peptide deletion constructs were introduced into C2-2-9 or C9-2, when using either the gpd promoter or het-c promoter (see Materials and methods).
HET-C localizes to the plasma membrane
HET-C is predicted to contain a signal peptide with a cleavage site at amino acids 29 and two transmembrane domains from amino acids 94–110 and 461–477. Thus, a mature, ∼100 kDa HET-C protein is predicted to reside in the plasma membrane with the glycine-rich domain extending to the outside of the cell (Figure 1A).
Initially, we tried to localize HET-C::GFP in living hyphae of CJ44 (het-cOR::GFP) compatible transformants by fluorescence microscopy. However, only high background fluorescence was detected (successful use of GFP for localization of fusion proteins in Neurospora has not been reported). We therefore chose the more sensitive analytical method of cell fractionation and western blot analysis to localize HET-C. The efficiency of our cell fractionation procedure was monitored using antibodies to the N.crassa plasma membrane H+-ATPase (Figure 3C) (Bowman et al., 1981), and anti-β-tubulin antibodies (Figure 3D), a cytoplasmic fraction marker. The cell wall fraction is from the processed pellet following homogenization of mycelia and low speed centrifugation (1000 g). We introduced het-cOR::GFP or het-cPA::HA into CJ44, isolated hygromycin-resistant transformants and subjected them to cell fractionation, immunoprecipitation with anti-GFP or anti-HA antibodies, followed by western blot analysis. An expected HET-COR::GFP or HET-CPA::HA polypeptide of ∼100 kDa was not detected in any of the cell fractions.
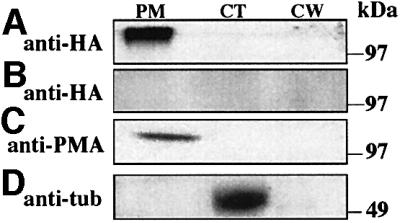
Fig. 3. HET-C is in the plasma membrane fraction. (A) CJ44 (het-cPA::HA) compatible transformants were subjected to cell fractionation and anti-HA-aminolinked column chromatography. Eluates from cellular fractions were probed by western analysis with a monoclonal anti-HA antibody. (B) CJ44 (het-cPA) compatible transformants treated identically to (A) show lack of cross-reacting material. (C) Western analysis using antibodies to the PM H+-ATPase (Bowman et al., 1981) show the predicted ∼100 kDa PM H+-ATPase band. (D) Western analysis using antibodies to β-tubulin, a cytoplasmic marker. MW markers are indicated. PM, plasma membrane fraction; CT, cytoplasmic fraction; CW, cell wall fraction.
Because of our inability to detect HET-C by cell fractionation and immunoprecipitation, we resorted to the still more definitive approach of aminolink chromatography to enrich for HET-C in cellular fractions. We introduced het-cPA::HA into CJ44, isolated transformants and subjected them to cell fractionation, as above. Each cellular fraction was passed through the anti-HA aminolinked column (see Materials and methods); bound proteins were eluted and subjected to western blot analysis using anti-HA antibodies. A protein with a molecular mass of ∼100 kDa was detected from the plasma membrane fraction of CJ44 (het-cPA::HA) transformants (Figure 3A). Cross-reacting proteins from untagged CJ44 (het-cPA) transformants were not observed (Figure 3B). Thus, HET-C localized to the plasma membrane fraction, consistent with computational analyses.
Alternative HET-C polypeptides form a heterocomplex
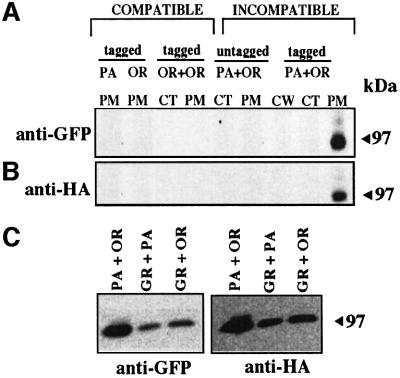
The simplest model for het-c-mediated nonself recognition is that a HET-C heterocomplex is perceived by the hyphal compartment, which subsequently triggers vegetative incompatibility. We therefore assessed HET-C heterocomplex formation by cell fractionation and co-immunoprecipitation assays. We co-transformed tagged (het-cOR:: GFP + het-cPA::HA) and untagged (het-cOR + het-cPA) alleles into CJ44, isolated incompatible transformants and subjected them to cell fractionation, immunoprecipitation using anti-HA antibodies, followed by western analysis using anti-GFP antibodies. In contrast to CJ44 (het-cOR:: GFP) or (het-cPA::HA) compatible transformants, a protein of estimated molecular mass of ∼100 kDa was detected in the plasma membrane fraction from CJ44 (het-cOR::GFP + het-cPA::HA) incompatible transformants by western analysis using anti-GFP antibodies (Figure 4A). The presence of HET-CPA::HA in the fraction was confirmed by stripping the immunoblot and re-probing with anti-HA antibody (Figure 4B). Reciprocal co-immunoprecipitation assays gave an identical result (see Supplementary data available at The EMBO Journal Online); a HET-C heterocomplex was identified specifically in the plasma membrane fraction from epitope-tagged incompatible transformants.
Fig. 4. HET-CPA::HA and HET-COR::GFP form a heterocomplex in incompatible transformants. (A) Anti-HA antibody immunoprecipitated cellular fractions from CJ44 (het-cOR::GFP + het-cOR::HA) compatible and (het-cOR::GFP + het-cPA::HA) incompatible transformants and their untagged counterparts were subjected to western analysis using anti-GFP antibodies. (B) The blot in (A) was stripped in stripping buffer (62.5 mM Tris–HCl pH 7.6, 100 mM 2-mercaptoethanol, 5% SDS) at 55°C for 30 min, rinsed in PBS, blocked and re-probed with anti-HA antibody. HET-C homocomplexes were undetectable in CJ44 (het-cOR::GFP + het-cOR::HA) transformants (OR+OR tagged). PM, plasma membrane fraction; CT, cytoplasmic fraction; CW, cell wall fraction. (C) HET-C heterocomplex formation was detected in three het-c allelic combinations by co-immunoprecipitation experiments. Plasma membrane fractions were isolated from CJ44 (het-cPA::HA + het-cOR::GFP), (het-cOR::HA + het-cOR::GFP) and (het-cOR::HA + het-cGR::GFP) incompatible transformants and subjected to co-immunoprecipitation, as above. MW markers are indicated.
We determined the ability of HET-COR to form a homocomplex by assessing cell fractions from CJ44 (het-cOR::GFP + het-cOR::HA) compatible transformants by co-immunoprecipitation assays. Unlike the HET-COR:: GFP + HET-CPA::HA heterocomplex, we were unable to detect HET-COR::GFP + HET-COR::HA homocomplex in any cellular fraction (Figure 4).
HET-C heterocomplex formation is independent of het-c allele type
Strains of one het-c specificity (het-cOR, het-cPA or het-cGR) are incompatible with strains carrying either of the other two het-c specificities (Howlett et al., 1993; Saupe and Glass, 1997; Wu and Glass, 2001). To address the question whether HET-C heterocomplex formation also occurs in other het-c allelic combinations [(het-cGR + het-cOR) and (het-cGR + het-cPA)], we constructed a tagged version of het-cGR [het-cGR::GFP; the predicted molecular weight (MW) of PM localized polypeptide is 99 kDa] (Figure 2). The het-cGR::GFP allele was co-transformed into CJ44 with het-cPA::HA or het-cOR::HA alleles and the resulting incompatible transformants were assessed for HET-C heterocomplex formation by cell fractionation and co-immunoprecipitation assays. A HET-C heterocomplex was detected in the plasma membrane fraction of CJ44 (het-cPA::HA + het-cGR::GFP) incompatible transformants (Figure 4C). Similarly, a HET-C heterocomplex was identified in the plasma membrane fraction of CJ44 (het-cOR::HA + het-cGR::GFP) incompatible transformants (Figure 4C). Reciprocal co-immunoprecipitations gave an identical result (our unpublished data). These data showed that all allelic combinations of het-c that cause vegetative incompatibility are capable of forming a plasma membrane-associated HET-C heterocomplex.
The HET-C heterocomplex is associated with the plasma membrane in hyphae
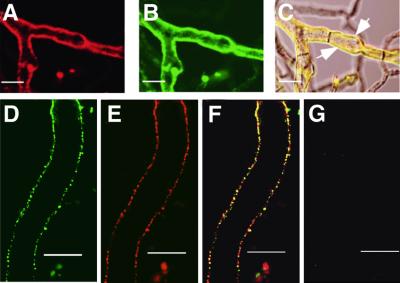
To assess the organization of the HET-C heterocomplex in hyphae, we performed immunolocalization of epitope-tagged HET-C using anti-GFP and/or anti-HA antibodies and laser scanning confocal microscopy (LSCM). Using anti-GFP antibodies and Cy5-conjugated streptavidin (Cy5), HET-COR::GFP localized to the plasma membrane region in CJ44 (het-cOR::GFP + het-cPA::HA) incompatible transformants (Figure 5A and B). Similarly, using anti-HA antibodies and Alexa Fluor 488 donkey anti-rabbit IgG (Alexa 488), HET-CPA::HA localized to the plasma membrane region in these same transformants (Figure 5C and D). HET-C showed an uneven distribution in hyphae of CJ44 (het-cOR::GFP + het-cPA::HA) incompatible transformants; only 20% of hyphal compartments have HET-C associated with their plasma membrane region. CJ44 transformants containing either het-cOR:: GFP and treated with anti-GFP (Cy5) (Figure 5G and H) or het-cPA::HA and treated with anti-HA antibodies (Alexa 488; data not shown) showed only background fluorescence, consistent with the difficulty in detecting HET-C in compatible transformants by immunoprecipitation and western blot analysis (see above).
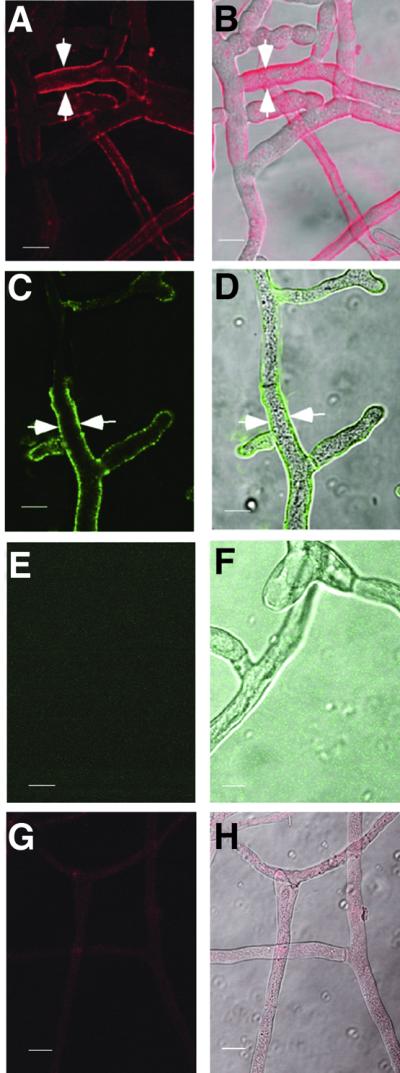
Fig. 5. HET-C heterocomplexes localize to the plasma membrane region by immunolocalization and LSCM. (A) A CJ44 (het-cOR:: GFP + het-cPA::HA) incompatible transformant labeled with anti-GFP antibodies (Cy5). (B) Image in (A) superimposed onto DIC. White arrows show plasma membrane labeling. (C) The same transformant labeled with anti-HA antibodies (Alexa 488). (D) Image in (C) superimposed onto DIC. White arrows show plasma membrane labeling. (E) A CJ44 (het-cOR + het-cPA) untagged incompatible transformant treated with anti-HA antibodies (Alexa 488). (F) Image in (E) superimposed onto DIC. (G) A CJ44 (het-cOR::GFP) compatible transformant treated with anti-GFP antibodies (Cy5). (H) The image shown in (G) superimposed onto DIC. (E–H) show a representative field; plasma membrane labeling was not observed across the entire colony. Scale bars = 10 µm.
Figure 6C shows the co-localization (in yellow) of HET-COR::GFP (Figure 6A) and HET-CPA::HA (Figure 6B) in a CJ44 (het-cOR::GFP + het-cPA::HA) incompatible transformant using anti-GFP (Alexa 633) and anti-HA (Alexa 488) antibodies; co-localization of HET-COR::GFP and HET-CPA::HA is apparent. For better resolution, we employed deconvolution microscopy at a higher magnification. Figure 6D and E shows individual green (Alexa 488) and red (Alexa 633) signals, indicative of HET-CPA::HA and HET-COR::GFP, respectively, in the plasma membrane region. Figure 6F shows a superimposed image of (D) and (E); co-localization in the plasma membrane region is indicated in yellow. Although the majority of HET-COR::GFP and HET-CPA::HA co-localized, separate green (Alexa 488) and red (Alexa 633) signals were observed, suggesting that the stoichiometry between HET-CPA::HA and HET-COR::GFP in these complexes may not be 1:1.
Fig. 6. HET-COR::GFP and HET-CPA::HA co-localize to the plasma membrane region. (A) Hyphae from a CJ44 (het-cOR::GFP + het-cPA::HA) incompatible transformant treated with anti-GFP (Alexa 633) and (B) anti-HA (Alexa 488) antibodies. (C) An image superimposed onto DIC showing co-localization of HET-COR::GFP and HET-CPA::HA in yellow (white arrows). (D) A deconvoluted image of a hypha from a CJ44 (het-cOR::GFP + het-cPA::HA) incompatible transformant in the green channel (Alexa 488; HET-CPA::HA). (E) The same hypha in the red channel (Cy5; HET-COR::GFP). (F) Co-localization of anti-HA (Alexa 488; HET-CPA::HA) and anti-GFP (Alexa 633; HET-COR::GFP) antibody labeling (yellow). (G) An untagged CJ44 (het-cOR + het-cPA) incompatible transformant treated with anti-GFP (Alexa 633) and anti-HA (Alexa 488) antibodies and viewed in green and red channels. Scale bars = 10 µm.
The distribution of HET-C heterocomplexes in the plasma membrane region by immunolocalization and LSCM is similar to that observed when vital dyes, such as Evans Blue, are used to stain incompatible hyphae (Jacobson et al., 1998; Wu and Glass, 2001): ∼20% of the hyphal compartments are dead. To determine whether HCD was correlated with HET-C heterocomplex formation, we took advantage of the fact that Evans Blue emits light at 611 nm and can be visualized using a rhodamine filter set. We stained hyphae of CJ44 (het-cOR::GFP + het-cPA::HA) incompatible transformants with Evans Blue prior to the process of fixation and then treated the samples with anti-HA antibodies (Alexa 488) to detect HET- CPA::HA. Figure 7A shows an image of Evans Blue fluorescence (red), while Figure 7B show plasma membrane labeling when the same sample was treated with anti-HA antibodies (Alexa 488; HET-CPA::HA). (HET-COR::GFP labeling could not be assessed because of overlap in fluorescence between Evans Blue and Cy5 or Alexa 633.) Figure 7C shows a merged image of Evans Blue fluorescence and anti-HA antibody (Alexa 488) labeling (see Supplementary data). The dead hyphal compartment (marked by a white arrow) was surrounded by plasma membrane labeling associated with HET-CPA::HA (Alexa 488; green).
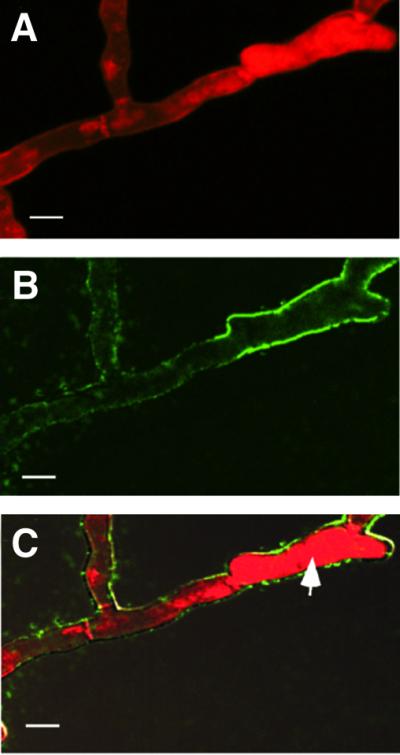
Fig. 7. HET-C heterocomplexes co-localize with dead hyphal compartments. (A) A CJ44 (het-cOR::GFP + het-cPA::HA) incompatible transformant was stained with Evans Blue and viewed with a rhodamine filter set (dead hyphal compartments show dark red). (B) An image of the same hyphae subsequently treated with anti-HA (Alexa 488) antibodies. (C) A merged image from (A) and (B). The white arrow indicates a dead cell. Scale bars = 10 µm.
Proper targeting of only one HET-C polypeptide is sufficient for vegetative incompatibility
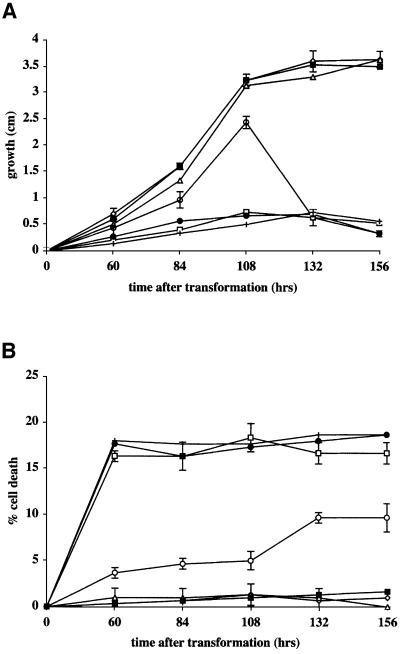
Targeting to the secretory system and the plasma membrane is accomplished by a signal peptide that promotes endoplasmic reticulum (ER)-associated translation. We therefore engineered het-cOR::GFP and het-cPA::HA constructs lacking signal peptides to determine whether localization to the plasma membrane was required for HET-C heterocomplex formation and vegetative incompatibility. The signal peptide deletion HET-C polypeptides are of nearly identical MW to HET-COR::GFP and HET-CPA::HA; the signal peptide is removed during ER-mediated translation. Surprisingly, the Δsp het-cOR::GFP and Δsp het-cPA::HA constructs triggered vegetative incompatibility in a manner that was indistinguishable from het-cOR::GFP and het-cPA::HA constructs when introduced into het-c tester strains C2-2-1 (het-cOR) and C9-2 (het-cPA) (Figure 2). Figure 8A and B shows the growth rate and percentage of HCD in CJ44 (Δsp het-cPA::HA + het-cOR::GFP) transformants; the morphological phenotype, growth inhibition and HCD rates (17–19%) were indistinguishable from CJ44 (het-cPA:: HA + het-cOR::GFP) incompatible transformants.
Fig. 8. Transformants containing alternative het-c alleles deleted for their signal peptide sequence show a novel phenotype. (A) Growth (in cm) was measured at 24 h intervals following transformation of CJ44 with different het-c constructs. Compatible transformants are CJ44 (het-cOR) (triangles), CJ44 (het-cPA::HA) (diamonds) and CJ44 (Δsp het-cPA::HA) (filled squares). Incompatible transformants are CJ44 (het-cPA + het-cOR) (open squares), CJ44 (het-cOR::GFP + het-cPA::HA) (crosses) and CJ44 (het-cOR::GFP + Δsp het-cPA::HA) (filled circles). The CJ44 (Δsp het-cOR::GFP + Δsp het-cPA::HA) (open circles) transformants showed a novel phenotype. Standard deviations are shown for transformants containing compatible, incompatible and double signal peptide het-c deleted constructs from three separate transformants. (B) Percentage of HCD in transformants shown in (A), which was calculated by counting an average of 100 hyphal compartments from three independent transformants after staining with Evans Blue (Gaff and Okong’O-Ogola, 1971).
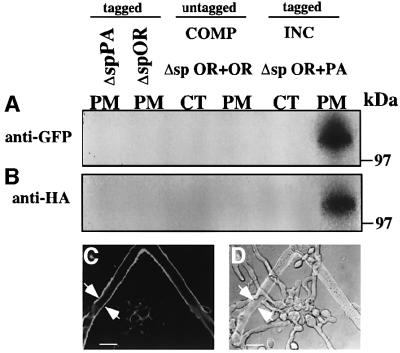
We localized HET-CPA::HA and HET-COR::GFP in CJ44 (spΔ het-cOR::GFP + het-cPA::HA) transformants by co-immunoprecipitation experiments. Immunoprecipit ation with anti-HA antibodies and subsequent immunoblot analyses with anti-GFP antibodies revealed ∼100 kDa HET-COR::GFP in the plasma membrane fraction (Figure 9A). Upon re-probing with anti-HA antibodies, an ∼100 kDa HET-CPA::HA protein was also detected in the plasma membrane fraction (Figure 9B). As with wild-type het-c constructs, HET-CPA::HA or HET-COR::GFP was not detected in cellular fractions from CJ44 (Δsp het-cPA::HA) or CJ44 (Δsp het-cOR::GFP) compatible transformants, respectively (Figure 9A and B).
Fig. 9. Transformants containing one Δsignal peptide het-c construct and a wild-type alternative het-c allele form a HET-C heterocomplex in the plasma membrane. (A) Cell-fractionated samples from CJ44 (Δsp het-cOR::GFP + het-cPA::HA) incompatible transformants, compatible transformants, CJ44 (Δsp het-cOR::GFP + het-cOR::HA), CJ44 (Δsp het-cOR::GFP) and CJ44 (Δsp het-cPA::HA) that were subjected to immunoprecipitation by anti-HA antibodies and western analysis using anti-GFP antibodies. (B) The same blot stripped and re-probed with anti-HA antibodies. CT, cytoplasmic fraction; PM, plasma membrane fraction. MW markers are indicated. (C) A confocal image of a CJ44 (Δsp het-cPA::HA + het-cOR::GFP) incompatible transformant treated with anti-HA antibodies (Alexa 488). (D) Image of (C) superimposed onto DIC. White arrows show plasma membrane labeling. Scale bars = 10 µm.
We performed LSCM on CJ44 (Δsp het-cPA::HA + het-cOR::GFP) incompatible transformants to assess HET-C heterocomplex localization within hyphae using anti-HA antibodies. HET-CPA::HA labeling (Alexa 488; green) was exclusively to the plasma membrane (Figure 9C and D; white arrows). In contrast, HET-CPA::HA was not detectable in CJ44 Δsp het-cPA::HA compatible transformants (data not shown). These data indicate that removal of the signal peptide from one of the alternative het-c alleles did not affect its localization to the plasma membrane, nor its ability to trigger vegetative incompatibility.
Mis-localization of alternative HET-C polypeptides results in cytoplasmic heterocomplexes
To determine whether mis-localization of HET-C occurs when both het-cPA and het-cOR are missing their signal peptides, we co-transformed Δsp het-cOR::GFP + Δsp het-cPA::HA into CJ44 and characterized the resulting transformants. The CJ44 (Δsp het-cOR::GFP + Δsp het-cPA::HA) transformants did not show typical het-c-mediated vegetative incompatibility. As shown in Figure 8A, the CJ44 (Δsp het-cOR::GFP + Δsp het-cPA::HA) transformants initially displayed an intermediate growth rate as compared with compatible and incompatible transformants, had growth characteristics that were more similar to wild type, and formed aerial hyphae and conidia. However, cessation of growth occurred between 108 and 132 h (4.5–5.5 days) in these transformants, with growth rates falling to <0.5 cm over a 24 h period. Figure 8B shows the relationship between HCD and growth rate cessation. Between 60 and 108 h post-transformation, the percentage of HCD was 5% in the CJ44 (Δsp het-cOR::GFP + Δsp het-cPA::HA) transformants, which increased to 10% at day 5. Phenotypic differences between CJ44 transformants carrying Δsp het-cOR::GFP + Δsp het-cPA::HA constructs driven either by the gpd promoter or the het-c promoter were not observed, consistent with results obtained with het-c constructs (Wu and Glass, 2001).
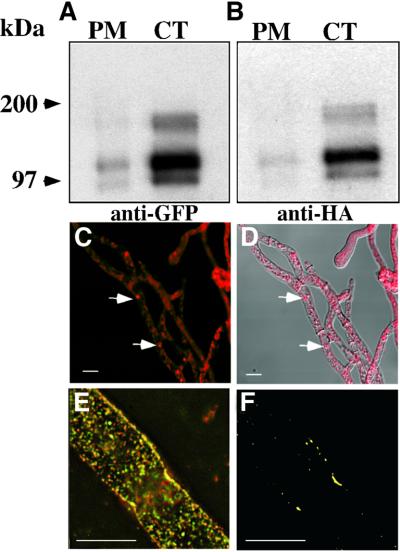
To determine whether HET-C heterocomplexes formed in CJ44 (Δsp het-cOR::GFP + Δsp het-cPA::HA) transformants, we performed cell fractionation and co-immunoprecipitation experiments. HET-CPA::HA + HET-COR:: GFP heterocomplexes were observed primarily in the cytoplasmic fraction (Figure 10A and B; CT), with a small fraction observed in the plasma membrane (Figure 10A and B; PM; Supplementary data). Both higher MW (∼190 kDa) and lower MW (∼90 kDa) bands were observed in both the cytoplasmic and plasma membrane fractions.
Fig. 10. HET-C heterocomplexes localize to the cytoplasm in transformants containing dual Δsignal peptide het-c constructs. (A) Cell fractions from a CJ44 (Δsp het-cPA::HA + Δsp het-cOR::GFP) transformant subjected to immunoprecipitation with anti-HA antibodies and western analysis using anti-GFP antibodies. (B) The same immunoblot after stripping and re-probing with anti-HA antibodies. PM, plasma membrane fraction; CT, cytoplasmic fraction. MW markers are indicated. (C) A confocal image of hyphae from a CJ44 (Δsp het-cPA::HA + Δsp het-cOR::GFP) transformant treated with anti-GFP (Alexa 633) antibodies. White arrows show cytoplasmic aggregates. (D) Image in (C) merged with DIC. (E) A deconvoluted image of a single hypha from a CJ44 (Δsp het-cPA::HA + Δsp het-cOR::GFP) transformant treated with both anti-HA (Alexa 488; green) and anti-GFP (Alexa 633; red) antibodies viewed with red and green channels. (F) Co-localization (yellow) of only HET-COR::GFP and HET-CPA::HA using the Bitplane co-localization program on the image in (E). Scale bars = 10 µm.
Confocal images of CJ44 (Δsp het-cOR::GFP + Δsp het-cPA::HA) transformants using anti-GFP antibodies (Alexa 633) showed cytoplasmic HET-C heterocomplexes (Figure 10C and D) as well as discrete cytoplasmic aggregates (white arrows). A similar pattern of labeling was observed when hyphae were treated with anti-HA antibodies (Alexa 488) (see Supplementary data). At a higher magnification, a deconvoluted image of a single hypha showed cytoplasmic aggregates (Figure 10E). Co-localization of anti-HA (Alexa 488) and anti-GFP (Alexa 633) antibody labeling is denoted in yellow (Figure 10F). Many aggregates do not show co-localization, suggesting that the stoichiometry between HET-CPA::HA and HET-COR::GFP in these complexes may not always be 1:1.
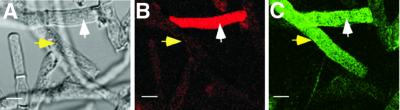
Are cytoplasmic HET-C heterocomplexes associated with HCD in the CJ44 (Δsp het-cOR::GFP + Δsp het-cPA:: HA) transformants? Figure 11 shows fluorescent images of hyphae from a CJ44 (Δsp het-cPA::HA + Δsp het-cOR:: GFP) transformant stained with Evans Blue prior to fixation and immunolocalization with anti-HA antibodies (Alexa 488). Co-localization of Evans Blue fluorescence (dead hyphal compartments are red; Figure 11B) and cytoplasmic HET-C heterocomplexes (green; Figure 11C) was observed (as shown by white arrows). However, HET-C heterocomplexes were common in the cytoplasm of hyphae not stained by Evans Blue, indicating that the formation of the cytoplasmic HET-C heterocomplexes may not always be associated with HCD (Figure 11A–C; yellow arrows).
Fig. 11. Cytoplasmic HET-C heterocomplexes do not always co- localize with dead hyphal compartments. (A) A DIC image of hyphae from a CJ44 (Δsp het-cPA::HA + Δsp het-cOR::GFP) incompatible transformant. (B) Transformant in (A) after staining with Evans Blue (rhodamine filter set). (C) The same field of hyphae showing labeling by anti-HA antibodies (HET-CPA::HA; Alexa 488). The white arrow in (A), (B) and (C) shows a dead hyphal compartment. The yellow arrow in (A), (B) and (C) shows a live hyphal compartment. Scale bars = 10 µm.
CJ44 transformants containing signal peptide deletion constructs of identical het-c specificity (Δsp het-cOR:: GFP + Δsp het-cOR::HA) were phenotypically similar to wild-type compatible transformants. We were unable to detect a ∼100 kDa HET-COR::HA or HET-COR::GFP polypeptide in any cellular fraction of CJ44 (Δsp het-cOR::GFP + Δsp het-cOR::HA) compatible transformants by co-immunoprecipitation, immunoprecipitation and western blot analyses (data not shown).
Discussion
The results of this study demonstrate that physical interaction of HET-C polypeptides encoded by het-c alleles of alternative specificity mediates nonself recognition during vegetative incompatibility in N.crassa and is a significant step in our understanding of how vegetative incompatibility is triggered. The presence of HET-C heterocomplexes in the plasma membrane is associated with the phenotypic aspects of vegetative incompatibility, namely, growth inhibition, repression of conidiation and HCD (Garnjobst and Wilson, 1956; Perkins, 1975; Jacobson et al., 1998). Nonself recognition mediated by HET-C heterocomplex formation was not restricted to one het-c allelic pair, but occurred in all combinations of het-c alleles that mediate vegetative incompatibility. In Podospora anserina, it has been shown using yeast two-hybrid experiments that both homo- and heterocomplexes are formed by proteins encoded by alternative specificities at the vegetative incompatibility locus, het-s (Coustou et al., 1997). Presumably, nonself recognition leading to vegetative incompatibility is also mediated by HET-S– HET-s heterocomplex formation. These observations suggest that heterocomplex formation between alternative HET proteins may be a common theme in nonself recognition during vegetative incompatibility.
HET-C heterocomplexes were abundant and readily detectable in the plasma membrane of incompatible transformants both by co-immunoprecipitation assays and by immunofluorescence using LSCM. However, under identical conditions, HET-C by itself or as a HET-C homocomplex was undetectable. We enriched for HET-C by using aminolink chromatography and cellular fractionation; HET-C was identified in the plasma membrane fraction. Similar differences in affinity between homo- and heterocomplexes have been described for Fos–Jun protein interactions in mammalian systems. Heterodimer formation is largely a thermodynamic consequence of Fos homodimer instability: Fos and Jun preferentially form a heterodimer (O’Shea et al., 1992). In U.maydis, heterodimer formation between homeodomain proteins encoded by different b alleles mediates nonself recognition (Kämper et al., 1995). Protein complexes between homeodomain proteins from the same b allele were not detectable. It is possible that the formation of a HET-C heterocomplex induces a conformational change in HET-C or causes the recruitment of other proteins to the HET-C heterocomplex that ‘mask’ protease degradation sites, thus rendering HET-C heterocomplexes more stable. Alternatively, regulation of HET-C protein stability may be determined by the activity of another protein, such as has been observed between the p53 tumor-suppressor protein and Mdm2: Mdm2 promotes the rapid degradation of p53 (Haupt et al., 1997).
According to topology predictions, the het-c specificity domain resides in the cytoplasm. The lack of the signal peptide in one of the HET-C polypeptides results in the formation of a HET-C heterocomplex that still localizes to the plasma membrane and is sufficient to cause a classical het-c vegetative incompatible phenotype. However, the signal peptide of het-c is essential for proper targeting because transformants containing alternative het-c alleles that are both deleted for their signal peptide resulted in primarily cytoplasmic localization of HET-C heterocomplexes.
Is the localization of HET-C heterocomplexes to the plasma membrane a prerequisite for vegetative incompatibility? Several lines of evidence suggest that it is. HET-C heterocomplexes are associated with the plasma membrane region of dead hyphal compartments. The mis-localization of alternative HET-C polypeptides resulted in the formation of HET-C heterocomplexes in the cytoplasm of both dead and live hyphal compartments. Transform ants containing cytoplasmic HET-C heterocomplexes initially grew at a rate near that of compatible transformants and formed aerial hyphae and conidia, but underwent growth arrest at ∼5 days of growth. Thus, localization of HET-C to the plasma membrane is required for typical phenotypic aspects of vegetative incompatibility, including repression of conidiation, HCD and growth inhibition. The formation of HET-C heterocomplexes, as well as their cellular localization is, we believe, one of the key factors in triggering vegetative incompatibility.
What is the nature of the HET-C heterocomplex? Deconvolution data of HET-C heterocomplexes in the plasma membrane and cytoplasm showed co-localization of HET-CPA::HA and HET-COR::GFP, but also complexes that appeared to be primarily HET-CPA::HA or HET- COR::GFP. Large aggregates were observed in the cytoplasm of transformants bearing het-c constructs deleted for their signal peptide sequence, suggesting that the HET-C heterocomplex can be of complex structure. The stoichiometric ratio of individual HET-C molecules in the heterocomplex is unclear. It has been shown that a het protein in a filamentous fungus, P.anserina, HET-s, forms protease-resistant aggregates (Coustou et al., 1997, 1999) and forms multimers even under denaturing conditions. HET-s has been show to function as a prion analog (Coustou et al., 1997; Wickner et al., 1999). In S.cerevisiae, oligopeptide repeats GGYGGT and SQPSYG in the N-terminal domain of Sup35p are involved in spontaneous propagation of novel [PSI+] prion elements (Nakayashiki et al., 2001). HET-C has eight repeats (partial or perfect) of the SQPSYG motif and six repeats of the GGYGGY motif from amino acids 708 to 951. However, deletion analysis of het-c showed that only part of the glycine-rich domain (from amino acids 610 to 730) is essential for vegetative incompatibility (Saupe et al., 1996; G.Iyer, unpublished results).
The indel motif in the het-c specificity domain (Figure 1) is sufficient to confer het-c allelic specificity (Saupe and Glass, 1997; Wu and Glass, 2001). Structural aspects of the het-c specificity domain must also regulate the capacity for alternative HET-C polypeptides to form a heterocomplex. It is unclear whether physical interactions between alternative HET-C polypeptides is mediated by the het-c specificity domain, or whether variations in this region affect the conformation of another region (such as the glycine-rich domain) that may be involved in physical interaction of alternative HET-C polypeptides. We are currently conducting experiments to determine the topology of HET-C in the plasma membrane, the region of physical interaction between alternative HET-C polypeptides and specific amino acid requirements of both the glycine-rich repeats and the specificity domain for HET-C heterocomplex formation.
How do plasma membrane-bound HET-C heterocomplexes mediate the pleiotropic phenotype associated with vegetative incompatibility? Two hypotheses are plausible. First, it is possible that HET-C heterocomplex formation acts as a signal to activate pathways associated with conidiation repression, growth inhibition and HCD. Activation of signal transduction pathways has been associated with programmed cell death in eukaryotes (Wilson, 1998). Alternatively, it is possible that the formation of HET-C heterocomplexes in the PM compromises plasma membrane function, thus destabilizing the hyphal compartment and causing death, perhaps by induction of a stress response pathway. It has been proposed that het genes encode products that form ‘poison’ heterocomplexes that lead to a lethal disorder (Bégueret et al., 1994). Septal plugging, a hallmark of the hyphal compartmentation during vegetative incompatibility, can also be induced by injury (Jedd and Chua, 2000). We have begun a genetic dissection of het-c-mediated vegetative incompatibility to differentiate these two hypotheses and to understand how the formation of HET-C heterocomplexes in the PM leads to the dramatic consequences of vegetative incompatibility.
Materials and methods
Strains and culture conditions
Escherichia coli DH5α (Bethesda Research Laboratories) was used for bacterial transformation (Sambrook et al., 1989) and plasmid maintenance. N.crassa strains used were C9-2 (thr-2 het-cPA a), C2-2-9 (thr-2 het-cOR A) (Saupe et al., 1996) and CJ44 (pan-2 arg-5 Δhet-c A) (Wu and Glass, 2001). Strains were grown on Vogel’s medium (VM) (Vogel, 1964) with required supplements.
het-c constructs
The het-cOR::GFP vector was constructed as described in Wu (2000). Constructs were cloned into either pOKE103 (gift from R.Metzenberg), which confers pantothenate prototrophy, or pCB1004 (Carroll et al., 1994), which confers hygromycin resistance. The het-cGR::GFP vector was constructed by ‘swapping’ a 588 bp EcoRV–StuI fragment containing the specificity domain of het-cGR into the het-cOR::GFP vector. The het-cPA::HA construct was made by amplifying a 3.2 kb het-cPA product using primers from ScaI 5′ SS.1 (AGTACTGCGCTACTAGCTACTAGA) and a 3′ primer SS.2 (GAGCTCGAACATCGGAGGTATTACCCTTATGATGTGCCAGATTATGCCTGA) with a SacI site, the het-cPA sequence from amino acids 970–974, plus 27 nucleotides of HA sequence followed by a stop codon. The HA sequence is in bold; restriction sites are underlined.
The 650 bp gpd promoter fragment (Punt et al., 1988) was amplified using primers SS.3 5′-GCATGCCATTAACCTAGGTA-3′ and SS.4 5′-AGATCTGTAACACTACAGACGATG-3′ as a SphI–XbaI fragment. The GFP ORF was ligated as a SacI–EcoR I fragment using primers SS.5 (GAGCTCATGGAGCAAGGGCGAGGAACT) and SS.6 (GAATTCACGAGCTGTACAAGTTGA). Primers containing sequences eliminating the signal peptide (amino acids 1–30) for het-cOR were used to amplify a 2.4 kb XbaI–SacI product, which was inserted into the gpd-GFP plasmid. Primers SS.7 (TCTAGAGCTGGCAACATTCCCTCCATC) and SS.8 (GAGCTCATCCTTCGCAGCCGTCATAT) contain XbaI and SacI sites, respectively. For the Δsp het-cPA::HA construct, a primer containing a XbaI site, SS.9 (TCTAGAGCTGCAGCCTTCGGTGCTGGC) was used with primer SS.2 to amplify a 3.1 kb het-cPA product. The het-c constructs were made by subcloning XbaI–SacI fragments from amino acid residues 2–730 and 2–974 of het-cOR::GFP and het-cPA::HA. Differences in the percentage of HCD, growth inhibition rate or other phenotypic characteristics associated with het-c incompatibility were not observed when the het-c promoter was replaced with the gpd promoter (this study and S.Sarkar, unpublished results). All constructs were verified by DNA sequencing and their functionality verified in vivo.
Neurospora transformation
Neurospora strains were transformed as described previously (Wu and Glass, 2001). For co-transformation experiments using pCB1004 and pOKE constructs, transformants were selected for both hygromycin resistance (250 µg/ml) and restoration of pantothenate prototrophy.
Microscopic analysis and cell death/growth inhibition assays
Growth of strains was measured at 24 h intervals using race tubes containing supplemented VM (Davis and De Serres, 1970). Evans Blue staining was carried out according to Jacobson et al. (1998). Evans Blue enters permeabilized cells and binds irreversibly to intracellular proteins (Gaff and Okong’O-Ogola, 1971). Samples were examined under bright field illumination on a Zeiss Axioskop or Zeiss LS 750 microscope for fluorescence with a rhodamine filter set. The percentage of dead hyphal compartments was determined by examining three independent compatible or incompatible transformants and calculating the number of dead cells per 100 hyphal compartments counted in a field.
Isolation and extraction of organelle fractions
Cell fractionation procedures using Neurospora were as described previously (Bowman and Bowman, 1988), in which separation of cellular fractions is performed by a series of centrifugation steps. To assess cell fractionation procedures, we used polyclonal antibodies to the N.crassa PM H+-ATPase (dilution 1:5000 as recommended; gift from the C.Slayman laboratory) (Bowman et al., 1981) and monoclonal antibody (clone TU27) against β-tubulin (dilution 1:2000; BAbCO), a cytoplasmic marker. The cell wall fraction is from a pellet following disruption of mycelia and centrifugation at 1000 g, which was subsequently resuspended in 50 mM HEPES pH 7.5, 10 mM NaCl, 0.1% NP-40, 0.05% sodium deoxycholate and homogenized. The resulting supernatant was concentrated prior to use, similar to other cellular fractions.
Protein extraction and affinity purification
An anti-HA aminolink column was prepared according to the manufacturer’s specifications (Pierce). Proteins from cytoplasmic (2.4 mg/ml), PM (0.5 mg/ml) and cell wall (1.1 mg/ml) fractions were loaded onto the anti-HA aminolink column, incubated and washed with seven columns of binding buffer. Eluates were subjected to immunoblotting with anti-HA antibodies. Immunodetection was carried out according to the manufacturer’s specifications (ECL kit; Amersham).
Co-immunoprecipitation experiments
Approximately 0.5 mg of protein extract from various cellular fractions were incubated at 4°C for 6 h with either anti-GFP (1:1500 dilution; Roche Biochemicals) or monoclonal anti-HA antibodies (clone 12CA5; 1:1000 dilution; Roche). Protein G–agarose beads (50 µl; Roche) were added; immunoprecipitation was according to the manufacturer’s specifications. Washing was repeated twice with a modified lysis buffer (50 mM HEPES pH 7.5, 750 mM NaCl, 0.1% NP-40, 0.05% sodium deoxycholate) with a final wash in the above buffer, but omitting NaCl. The resulting pellet was subjected to electrophoresis on an 8% SDS–PAGE gel and analyzed by immunoblotting, as above.
LSCM and immunofluorescence
Antibodies for LSCM were mouse anti-GFP mAb (Roche), biotinylated goat anti-mouse IgG (H+L) (Roche), Cy5-conjugated streptavidin (Jackson ImmunoResearch), goat anti-HA rabbit polyclonal antibody (Santa Cruz Biotechnologies), Alexa Fluor 488-conjugated donkey anti-rabbit IgG and Alexa Fluor 633-conjugated goat anti-mouse IgG (Molecular Probes).
Neurospora crassa hyphae were prepared for confocal microscopy as described in Tinsley et al. (1998), which involved cryofixation of hyphal tissue by plunging into liquid propane (purchased commercially). Incubation was for 4 h for the primary antibody (200 µg/ml) and for 2 h for the secondary antibody (5 µg/ml). During single-labeling experiments, the extent of red/green ‘bleed through’ was checked in the appropriate channels. Staining for co-localization of HET-COR::GFP and HET-CPA::HA was performed as follows: hyphae were incubated with both anti-GFP and anti-HA primary antibodies, followed by washing and incubation with the appropriate secondary antibodies, washed and mounted. Microscopes used were Zeiss LSM 510 for confocal with DIC optics (Plan-Apo 63×/1.25 N.A. objective) and API wide-field deconvolution microscope (Plan-Neo 100×/1.3 N.A. objective). Data were analyzed using Deltavision software (version 2.5) and the Imaris Bitplane co-localization (version 3.0) program. Alexa Fluor 488 was detected by excitation at 488 nm, emission at 505/550 nm. Cy5 and Alexa Fluor 633 were detected by excitation at 633 nm, emission long pass 650 nm.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We wish to thank Drs Steve Ruzin, Denise Schichnes (CNR Imaging Facility) and Kent Macdonald (Electron Microscopy Unit) for valuable assistance and technical support for microscopy. We acknowledge the help of Drs Sven Saupe and Richard Todd in making some of the het-c constructs. We are grateful to the C.Slayman laboratory for the gift of H+-ATPase antibodies. We thank Drs Sven Saupe and Bob Metzenberg for their critical reading of the manuscript. The work reported in this paper was supported by a grant from the National Institutes of Health (GM60468-01) to N.L.G.
References
- Asante-Owusu R.N., Banham,A.H., Böhnert,H.U., Mellor,E.J. and Casselton,L.A. (1996) Heterodimerization between two classes of homeodomain proteins in the mushroom Coprinus cinereus brings together potential DNA-binding and activation domains. Gene, 172, 25–31. [DOI] [PubMed] [Google Scholar]
- Bégueret J., Turcq,B. and Clave,C. (1994) Vegetative incompatibility in filamentous fungi—het genes begin to talk. Trends Genet., 10, 441–446. [DOI] [PubMed] [Google Scholar]
- Bowman B.J., Blasco,F. and Slayman,C.W. (1981) Purification and characterization of the plasma membrane ATPase of Neurospora crassa. J. Biol. Chem., 256, 12343–12349. [PubMed] [Google Scholar]
- Bowman E.J. and Bowman,B.J. (1988) Purification of vacuolar membranes, mitochondria and plasma membranes from Neurospora crassa and modes of discriminating among the different H+-ATPases. Methods Enzymol., 157, 562–573. [DOI] [PubMed] [Google Scholar]
- Carroll A.M., Sweigard,J.A. and Valent,B. (1994) Improved vectors for selecting resistance to hygromycin. Fungal Genet. Newsl., 41, 22. [Google Scholar]
- Cormack B. (1998) Green fluorescent protein as a reporter of transcription and protein localization in fungi. Curr. Opin. Microbiol., 1, 406–410. [DOI] [PubMed] [Google Scholar]
- Cortesi P., McCulloch,C.E., Song,H., Lin,H. and Milgroom,M.G. (2001) Genetic control of horizontal virus transmission in the chestnut blight fungus, Cryphonectria parasitica.Genetics, 159, 107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coustou V., Deleu,C., Saupe,S. and Bégueret,J. (1997) The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc. Natl Acad. Sci. USA, 94, 9773–9778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coustou V., Deleu,C., Saupe,S.J. and Bégueret,J. (1999) Mutational analysis of the [Het-s] prion analog of Podospora anserina. A short N-terminal peptide allows prion propagation. Genetics, 153, 1629–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R.H. and De Serres,F.J. (1970) Genetic and microbial research techniques for Neurospora crassa. Methods Enzymol., 17A, 79–143. [Google Scholar]
- Debets A.J.M. and Griffiths,A.J.F. (1998) Polymorphism of het-genes prevents resource plundering in Neurospora crassa.Mycol. Res., 102, 1343–1349. [Google Scholar]
- Forsburg S.L. and Sherman,D.A. (1997) General purpose tagging vectors for fission yeast. Gene, 191, 191–195. [DOI] [PubMed] [Google Scholar]
- Gaff D.F. and Okong’O-Ogola,O. (1971) The use of non-permeating pigments for testing the survival of cells. J. Exp. Bot., 22, 756–758. [Google Scholar]
- Garnjobst L. and Wilson,J.F. (1956) Heterocaryosis and protoplasmic incompatibility in Neurospora crassa. Proc. Natl Acad. Sci. USA, 42, 613–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass N.L., Jacobson,D.J. and Shiu,K.T. (2000) The genetics of hyphal fusion and vegetative incompatibility in filamentous ascomycetes. Annu. Rev. Genet., 34, 165–186. [DOI] [PubMed] [Google Scholar]
- Haupt Y., Maya,R., Kazaz,A. and Oren,M. (1997) Mdm2 promotes the rapid degradation of p53. Nature, 387, 296–299. [DOI] [PubMed] [Google Scholar]
- Ho C., Adamson,J.G., Hodges,R.S. and Smith,M. (1994) Hetero dimerization of the yeast MATa1 and Mata2 proteins is mediated by two leucine zipper-like coiled-coil motifs. EMBO J., 13, 1403–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett B., Leslie,J.F. and Perkins,D.D. (1993) Putative multiple alleles a the vegetative (heterokaryon) incompatibility loci het-c and het-8 in Neurospora crassa.Fungal Genet. Newsl., 40, 40–42. [Google Scholar]
- Jacobson D.J., Beurkens,K. and Klomparens,K.L. (1998) Microscopic and ultrastructural examination of vegetative incompatibility in partial diploids heterozygous at het loci in Neurospora crassa. Fungal Genet. Biol., 23, 45–56. [DOI] [PubMed] [Google Scholar]
- Jedd G. and Chua,N.H. (2000) A new self-assembled peroxisomal vesicle required for efficient resealing of the plasma membrane. Nat. Cell Biol., 2, 226–231. [DOI] [PubMed] [Google Scholar]
- Jones E.Y., Tormo,J., Reid,S.W. and Stuart,D.I. (1998) Recognition surfaces of MHC class I. Immunol. Rev., 163, 121–128. [DOI] [PubMed] [Google Scholar]
- Kämper J., Reichmann,M., Romeis,T., Bölker,M. and Kahmann,R. (1995) Multiallelic recognition: nonself-dependent dimerization of the bE and bW homeodomain proteins in Ustilago maydis. Cell, 81, 73–83. [DOI] [PubMed] [Google Scholar]
- Klein J., Sato,A., Nagl,S. and O’hUigin,C. (1998) Molecular trans-species polymorphism. Annu. Rev. Ecol. Syst., 29, 1–21. [Google Scholar]
- May G., Shaw,F., Badrane,H. and Vekemans,X. (1999) The signature of balancing selection: fungal mating compatibility gene evolution. Proc. Natl Acad. Sci. USA, 96, 9172–9177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayashiki T., Ebihara,K., Bannai,H. and Nakamura,Y. (2001) Yeast [PSI+] ‘prions’ that are crosstransmissible and susceptible beyond a species barrier through a quasi-prion state. Mol. Cell, 7, 1121–1130. [DOI] [PubMed] [Google Scholar]
- Nasrallah J.B. (2000) Cell–cell signaling in the self-incompatibility response. Curr. Opin. Plant Biol., 3, 368–373. [DOI] [PubMed] [Google Scholar]
- O’Shea E.K., Rutkowski,R. and Kim,P.S. (1992) Mechanism of specificity in the Fos–Jun oncoprotein heterodimer. Cell, 68, 699–708. [DOI] [PubMed] [Google Scholar]
- Perkins D.D. (1975) The use of duplication-generating rearrangements for studying heterokaryon incompatibility genes in Neurospora. Genetics, 80, 87–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins D.D., Radford,A. and Sachs,M.S. (2000) The Neurospora Compendium: Chromosomal Loci. Academic Press, San Diego, CA.
- Pontecorvo G. (1956) The parasexual cycle in fungi. Annu. Rev. Microbiol., 10, 393–400. [DOI] [PubMed] [Google Scholar]
- Punt P.J., Dingemanse,M.A., Jacobs-Meijsing,B.J., Pouwels,P.H. and van den Hondel,C.A. (1988) Isolation and characterization of the glyceraldehyde-3-phosphate dehydrogenase gene of Aspergillus nidulans. Gene, 69, 49–57. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Saupe S.J. (2000) Molecular genetics of heterokaryon incompatibility in filamentous ascomycetes. Microbiol. Mol. Biol. Rev., 64, 489–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saupe S.J. and Glass,N.L. (1997) Allelic specificity at the het-c heterokaryon incompatibility locus of Neurospora crassa is determined by a highly variable domain. Genetics, 146, 1299–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saupe S.J., Kuldau,G.A., Smith,M.L. and Glass,N.L. (1996) The product of the het-c heterokaryon incompatibility gene of Neurospora crassa has characteristics of a glycine-rich cell wall protein. Genetics, 143, 1589–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinsley J.H., Lee,I.H., Minke,P.F. and Plamann,M. (1998) Analysis of actin and actin-related protein 3 (ARP3) gene expression following induction of hyphal tip formation and apolar growth in Neurospora. Mol. Gen. Genet., 259, 601–609. [DOI] [PubMed] [Google Scholar]
- Vogel H.J. (1964) Distribution of lysine pathways among fungi: evolutionary implications. Am. Nat., 98, 435–446. [Google Scholar]
- Wickner R.B. Taylor,K.L., Edskes,H.K., Maddelein,M.L., Moriyama,H. and Roberts,B.T. (1999) Prions in Saccharomyces and Podospora spp.: protein-based inheritance. Microbiol. Mol. Biol. Rev., 63, 844–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M.R. (1998) Apoptotic signal transduction: emerging pathways. Biochem. Cell Biol., 76, 573–582. [DOI] [PubMed] [Google Scholar]
- Wu J. (2000) Non-self recognition in filamentous fungi—the het-c mediated vegetative incompatibility in Neurospora crassa. Ph.D. thesis, Botany Department and The Biotechnology Laboratory, University of British Columbia, Vancouver, Canada.
- Wu J. and Glass,N.L. (2001) Identification of specificity determinants and generation of alleles with novel specificity at the het-c heterokaryon incompatibility locus of Neurospora crassa. Mol. Cell. Biol., 21, 1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Saupe,S.J. and Glass,N.L. (1998) Evidence for balancing selection operating at the het-c heterokaryon incompatibility locus in a group of filamentous fungi. Proc. Natl Acad. Sci. USA, 95, 12398–12403. [DOI] [PMC free article] [PubMed] [Google Scholar]