Abstract
In ClpXP protease complexes, hexameric rings of the ATP-dependent ClpX chaperone stack on one or both faces of the double-heptameric rings of ClpP. We used electron microscopy to record the initial binding of protein substrates to ClpXP and their accumulation inside proteolytically inactive ClpP. Proteins with N- or C-terminal recognition motifs bound to complexes at the distal surface of ClpX and, upon addition of ATP, were translocated to ClpP. With a partially translocated substrate, the non-translocated portion remained on the surface of ClpX, aligned with the central axis of the complex, confirming that translocation proceeds through the axial channel of ClpXP. Starting with substrate bound on both ends, most complexes translocated substrate from only one end, and rarely (<5%) from both ends. We propose that translocation from one side is favored for two reasons: initiation of translocation is infrequent, making the probability of simultaneous initiation low; and, further, the presence of protein within the cis side translocation channel or within ClpP generates an inhibitory signal blocking translocation from the trans side.
Keywords: AAA-ATPase/ATP-dependent proteolysis/chaperone/ClpXP protease/electron microscopy
Introduction
Protein quality control in cells is mediated by molecular chaperones and proteases. Chaperones promote proper protein folding and prevent aggregation, while ATP-dependent proteases eliminate irremediably misfolded, damaged or foreign proteins (Gottesman and Maurizi, 1992; Wickner et al., 1999). The ATP-dependent proteases are large multi-subunit assemblies that share certain key features: they have proteolytic active sites sequestered in internal chambers, thereby protecting normal cellular proteins, and they possess ATP-dependent chaperone components, which recognize substrates and deliver them to the digestion chambers (Lupas et al., 1997; Zwickl et al., 2000).
In Escherichia coli, the Clp family of ATP-dependent proteases comprises ClpXP, ClpAP and ClpYQ (HslUV). ClpP, the peptidase component of both ClpXP and ClpAP, has two stacked heptameric rings of 21 kDa subunits enclosing a chamber with 14 proteolytic sites (Wang et al., 1997). The chaperones ClpX (subunit Mr 47 K) and ClpA (subunit Mr 84 K) form hexameric rings in the presence of ATP or a compatible analog and mount coaxially on one or both faces of ClpP, producing 1:1 or 2:1 complexes (Kessel et al., 1995; Grimaud et al., 1998). Because the only access to the digestion chamber of ClpP is via a narrow axial channel (Wang et al., 1997), the chaperone rings control access to the proteolytic sites. The molecular architecture of ClpYQ (Bochtler et al., 2000; Ishikawa et al., 2000; Sousa et al., 2000; Wang et al., 2001a) is generally similar, although the fold of the ClpQ subunit differs from ClpP (Bochtler et al., 1997), and both ClpQ and ClpY assemble into hexameric rings (Kessel et al., 1996; Rohrwild et al., 1997). A complex of hexameric ClpY bound on both sides of the double hexamer of ClpQ has been crystallized (Bochtler et al., 2000; Sousa et al., 2000; Wang et al., 2001a), demonstrating that symmetry-matched ATP-dependent proteolytic complexes are also stable and functional.
Interaction of ClpXP and ClpAP with protein substrates has been studied at both a biochemical (Weber-Ban et al., 1999; Kim et al., 2000; Singh et al., 2000, 2001; Burton et al., 2001) and a structural level (Ortega et al., 2000; Ishikawa et al., 2001). This process involves three steps: first, substrates initially bind to specific sites on the distal surface of the ATPase; secondly, they are then unfolded and translocated along a pathway that appears, for ClpAP, to be axial (Ishikawa et al., 2001); and finally, degradation of the protein and dispersal of the resulting oligopeptides take place. These steps can be decoupled and studied separately in vitro. The third step, degradation, can be blocked by inhibitors or mutations affecting the activity of ClpP, in which case, non-degraded substrate accumulates in the digestion chamber. ATPγS supports the assembly of complexes and the binding of substrate, allowing stable substrate complexes with ClpX and ClpA to be isolated. In the case of ClpXP with λO as a substrate, subsequent translocation and degradation steps proceed at a slow rate in the presence of ATPγS (J.Rozycki and M.R.Maurizi, manuscript in preparation).
A 2:1 ClpXP complex is formally bipolar, with the translocation pathways from both ends converging in the digestion chamber. Bipolarity is shared with other ATP-dependent proteases and with other molecular machines such as group I chaperones, e.g. GroEL (Grantcharova et al., 2001), and Group II chaperones, e.g. the thermosome (Klumpp and Baumeister, 1998). Like ClpP, GroEL is composed of two apposed heptameric rings. GroEL rings act in concert to fold a single substrate, with the cycle being completed by the binding of ATP to one ring of GroEL and release of the folded or folding-competent protein from the other end (Fenton and Horwich, 1997). Kinetic studies have revealed negative cooperativity between the rings in binding of substrates or GroES (Burston et al., 1995), as well as positive cooperativity in binding and hydrolysis of ATP (Bochkareva et al., 1992).
Here we used electron microscopy to address several aspects of substrate binding and translocation by ClpXP. To approach the question of whether substrates bearing different degradation tags follow the same pathway, we examined the processing of green fluorescent protein C-terminally tagged with the SsrA peptide (GFP–SsrA; Weber-Ban et al., 1999; Kim et al., 2000; Singh et al., 2000) and compared it with λO protein, whose recognition motif is near its N-terminus (Gonciarz-Swiatek et al., 1999), and with a doubly tagged chimera consisting of the N-terminal portion of λO fused to GFP–SsrA (λO162– GFP–SsrA). We then investigated the translocation of these substrates in the presence of ATPγS or ATP and addressed the functional polarity of ClpXP complexes, specifically asking whether substrate translocation proceeds simultaneously from both ends or by an alternating mechanism, first from one end, then from the other. We used conditions that gave mostly 2:1 ClpXP complexes with substrate bound at both ends and performed long incubations to allow time for multiple rounds of translocation to occur. By correlating the appearance of density within ClpP with the decrease in density at one or both ends of the complex, we were able to evaluate the relative frequencies with which substrates were translocated from either end.
Results
Binding of substrates with N- and C-terminal tags to ClpX and ClpXP
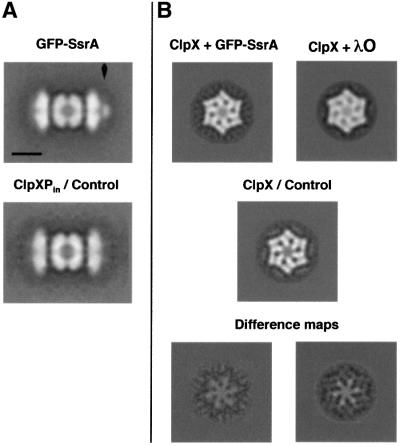
Previous studies with λO showed that this substrate binds to the distal surface of ClpX in ClpXP complexes (Ortega et al., 2000). To ascertain whether substrates with C-terminal recognition sites bind to the same surface, we formed complexes of ClpXP with a recombinant fusion protein, GFP–SsrA. SsrA is a C-terminal motif (AAN DENYALAA) added co-translationally to incomplete proteins to target them for degradation by E.coli ClpXP (Keiler et al., 1996; Gottesman et al., 1998). GFP–SsrA bound to ClpXP was directly visible by negative stain electron microscopy (EM; data not shown). The averaged image shows the increased density due to GFP–SsrA at the distal surface of ClpXP (Figure 1A, upper versus lower). At low ratios of substrate to ClpXP, only one substrate is bound and the difference between the ends with and without bound substrate is evident. When excess substrate was added, identical densities were visible at both ends of the complexes (Figure 2A, upper left). Compared with the images of λO bound to ClpXP (Figure 2A, middle left), the GFP–SsrA density is more condensed and more focused on the central axis of the complex.
Fig. 1. GFP–SsrA and λO protein binding to ClpXP and ClpX. (A) The ClpXP complexes were formed in the presence of ATPγS, substrate proteins were added in slight molar excess, and samples prepared by negative staining within 2 min. Averaged side-views of 2:1 ClpXP complexes with GFP–SsrA bound at one end (arrow, top panel) and without substrate (bottom). The complexes in individual images in the data set are viewed at various angular settings around their central axis, so that these images, obtained by averaging after translational alignment, represent cylindrically averaged structures. (B) ClpX hexamers were assembled in ATPγS, and negatively stained images were obtained with and without bound substrates. ClpX hexamers are viewed with the six-fold symmetry axis parallel to the grid. The upper row shows ClpX with GFP–SsrA (left) and λO bound (right). Substrate bound to the surface of ClpX hexamers is evident from the decreased staining of the center channel and the six petal-like projections on the surface of ClpX. The latter may represent surface depressions where substrates may bind and where stain can accumulate in the absence of substrate. Decrease in stain penetration upon binding is further illustrated in difference images (bottom row, central and right panel). Each average image combined ∼700 particles for resolutions between 25 (top views of ClpX) and 31 Å (side views of ClpXP). The scale bar represents 100 Å.
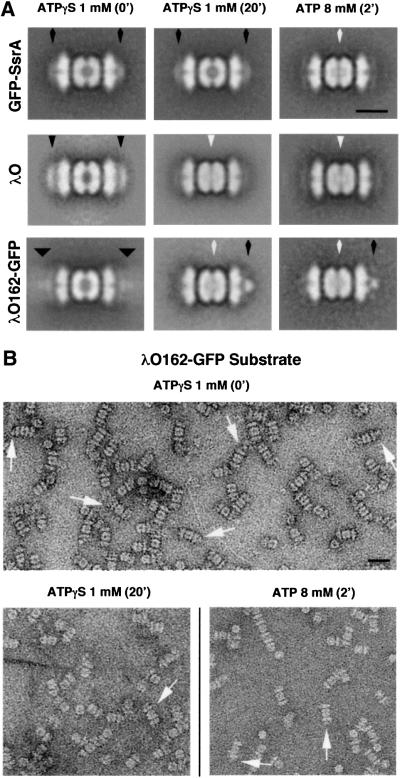
Fig. 2. Translocation of substrates into ClpP in the presence of ATPγS. (A) Complexes of GFP–SsrA protein bound to ClpXPin in the presence of 1 mM ATPγS were immediately applied to a grid and negatively stained (top, left). Grids were prepared after incubation of the same sample at room temperature for 20 min (top, middle). An aliquot of the sample assembled in ATPγS was treated with 8 mM ATP for 2 min at room temperature before applying to a grid and staining (top, right). Complexes of λO or λO162–GFP with ClpXPin were assembled and treated as above. Averages for complexes at zero time in ATPγS (middle and bottom left, respectively); complexes after 20 min with ATPγS (middle and bottom center, respectively) and complexes after 2 min with ATP (middle and bottom right, respectively) were obtained. The average images of ClpXPin complexes with λO162–GFP show that under both conditions of translocation a portion of the substrate has been translocated into ClpP (white arrow), which is no longer stain-penetrable, and the remaining non-translocated domain (GFP) has condensed nearer the opening to the axial channel of ClpX suggesting that the substrate remaining on the surface of ClpX (black arrow) is apparently compact and well folded. Each average image combined ∼300 particles for resolutions of 31 Å. (B) The upper panel shows negative stained electron micrographs of ClpXPin complexes with λO162–GFP (white arrows) in the presence of ATPγS. The bottom panels show ClpXPin complexes after either ATPγS (left, white arrows) or ATP (right, white arrows) translocation. Scale bars represent 100 Å in the averages, and 200 Å for negative-stained electron micrographs.
We also compared ‘top views’ of ClpX hexamers (Figure 1B, middle) with top views of ClpX with bound GFP–SsrA (Figure 1B, upper left) or λO (Figure 1B, upper right). The presence of bound protein resulted in decreased staining of the central channel of ClpX and of the surrounding six stain-accumulating indentations. Differ ence images allowed the distribution of the extra density to be more easily visualized (Figure 1B, lower panels). In both cases, one sees a 6-fold motif of positive density that extends ∼70% of the way to the outer edges of the hexamer. A ClpX hexamer can bind only one λO dimer or one GFP–SsrA monomer (J.Rozycki and M.R.Maurizi, unpublished data), but the signal-to-noise ratio of the original images precluded alignment of particles according to the site occupied by substrate. Averaging over single substrate molecules randomly distributed among six symmetry-related binding sites effects an apparent six-fold symmetry on the bound substrate, and renders the difference density fainter because of fractional occupancy (∼1/6). Substrate density overlaps only in the center of the difference images, possibly marking a subsite common to all six substrate-binding sites, or simply indicating a region of potential steric clash between molecules attempting to bind at two different symmetry-related sites on the same ClpX ring. By either mechanism, one bound molecule would impede binding of additional substrate molecules at the other symmetry-related sites. The six-fold density motif is similar between the difference maps for λO and GFP–SsrA (Figure 1B, lower center and right), suggesting that there is significant overlap in the sites occupied by these substrates.
Translocation of λO in the presence of ATPγS
Earlier, we showed that addition of ATP to pre-assembled ClpXPin–λO complexes resulted in the disappearance of protein bound to the distal surface of ClpX and the appearance of density in the center of ClpPin (Ortega et al., 2000). To follow the kinetics of translocation, we took advantage of the ability of ClpXP to degrade λO in the presence of ATPγS at ∼1% of the rate seen with ATP (J.Rozycki and M.R.Maurizi, unpublished results). Negatively stained grids were prepared immediately after adding λO to ClpXPin with 1 mM ATPγS, and again after incubation for 20 min. In complexes left with ATPγS for 20 min, the λO had migrated from its initial binding site at the ends of ClpXPin (Figure 2A, middle left) to the digestion chamber of ClpPin (Figure 2A, middle center). The final product had the same appearance as that formed when ATP was added (Figure 2A, middle right). The time course of ATPγS-mediated translocation is considered further below. GFP–SsrA was not translocated in the presence of ATPγS (Figure 2A, upper center), although ATP supported its complete translocation into ClpXPin (Figure 2A, upper right). This result is consistent with biochemical experiments showing that ATPγS does not efficiently promote unfolding or degradation of GFP–SsrA (Singh et al., 2000; data not shown).
Binding and translocation of λO162–GFP
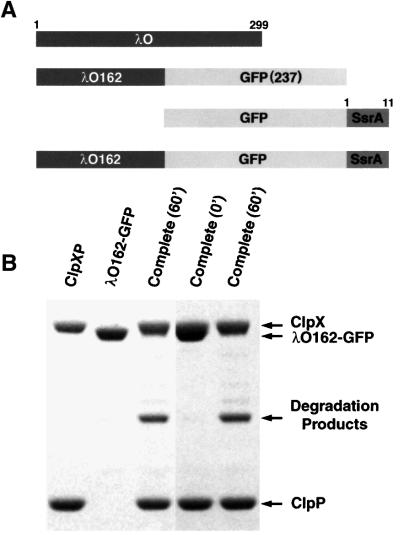
In order to visualize intermediate stages of translocation, we wanted a substrate that was translocated either very slowly or only partially. We found that a fusion of the N-terminal half of λO with GFP (λO162–GFP; Mr ∼ 45 K; Figure 3A) undergoes limited proteolysis in the presence of ATP or ATPγS. Most of the λO moiety was degraded, but the GFP plus a short N-terminal extension remained intact and retained its fluorescence (H.S.Lee and M.R.Maurizi, manuscript in preparation). Figure 3B shows a typical result of incubating λO162–GFP with ClpXP in the presence of ATP. After 60 min, the original fusion protein was gone, and a 30 kDa product accumulated. λO162–GFP was thus a good candidate for visualizing partially translocated substrate complexes with ClpXPin.
Fig. 3. Limited proteolysis of λO162–GFP by ClpXP. (A) Schematic diagram of the various substrates used in the experiments. (B) λO162–GFP was incubated with or without ClpXP in the presence of 8 mM ATP. Samples were run on an SDS gel and stained with Coomassie Blue. Lanes 1–3, samples incubated for 60 min. Lane 1, ClpXP alone; lane 2, λO162–GFP alone; lane 3, λO162–GFP plus ClpXP. Lanes 4 and 5, separate incubations. Lane 4, λO162–GFP plus ClpXP at 0 time; lane 5, λO1620–GFP plus ClpXP after 60 min incubation. The identity of the degradation product was confirmed in separate experiments (H.S.Lee and M.R.Maurizi, manuscript in preparation).
To this end, we mixed λO162–GFP with 2:1 ClpXPin complexes at room temperature in the presence of 1 mM ATPγS and removed samples for negative staining and EM analysis (Figure 2B, upper panel). Averaged images of λO162–GFP-bound complexes at zero time showed empty digestion chambers and extra density at either one or both ends (Figure 2A, bottom left). Compared with GFP–SsrA or λO bound to ClpXPin, the λO162–GFP density was considerably more dispersed. We conclude that the λO162 moiety interacts with the distal surface of ClpX, and the GFP portion protrudes distally from the complex. Averaging of the different view angles generates a ring of λO162 density near the surface of ClpX and a diffuse volume of GFP density extending further out.
After 20 min incubation at room temperature with ATPγS, the complexes had undergone a dramatic change. The predominant species had its digestion chamber at least partly occupied, and the density remaining bound was concentrated on the axis and closely apposed to the surface of ClpX (Figure 2B, lower left). In averaged images (Figure 2A, bottom center), the surface-bound density differed from the diffuse density initially seen upon λO162–GFP binding, but closely resembled the density seen when GFP–SsrA was bound to ClpXPin (cf. Figure 1A). We interpret these images as showing the translocated λO162 moiety inside ClpP and the GFP portion in a folded globular state, stalled at the entrance of the translocation channel. Virtually identical partially translocated complexes were produced when ClpXPin– λO162–GFP complexes were assembled in ATPγS and excess ATP was added. Density appeared at the center of ClpPin within 2 min of ATP addition, many of which had a compact globular density closely apposed to ClpX at one end of the complex (Figure 2A, bottom right and B, lower right). The compact globular density allowed us to identify the end of the complex from which translocation had taken place. This density was seldom seen at both ends, suggesting that translocation took place primarily from one end. However, because relatively few initial complexes (20%) had two λO162–GFP molecules originally bound, these data did not definitively show whether λO162–GFP was translocated from one or both ends.
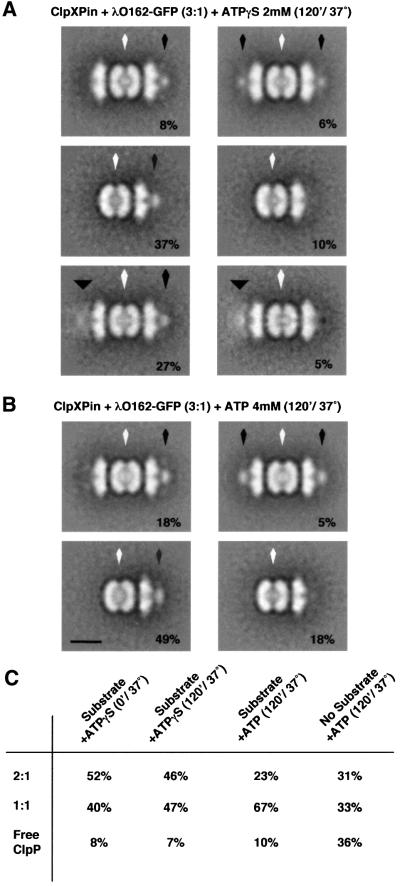
To increase the number of complexes doubly loaded with substrate, we assembled ClpXPin in the presence of either ATP or ATPγS and added a 3-fold excess of λO162–GFP over ClpX. Incubations were continued at 37°C for 120 min to maximize translocation. Immediately after assembly of ClpXPin, the distribution of complexes was 52% 2:1 complexes, 40% 1:1 complexes, and 8% ClpPin rings. ClpXPin complexes remain stable in ATPγS, which is hydrolyzed very slowly, but they tend to fall apart when ATP is hydrolyzed. After 120 min in ATP, only 31% of the particles were 2:1 complexes, while 33% were 1:1 complexes, and 36% were free ClpPin (Figure 4C). ClpX rings or other sub-species of ClpX were not clearly identifiable. ClpXPin was incubated with λO162–GFP in the presence of ATP for 2 h, after which all the complexes had undergone translocation. During this time, the proportion of 2:1 complexes had decreased to 23%, while that of 1:1 complexes increased to 67%; free ClpPin made up the remaining 10% of the particles (Figure 4B and C). In addition to internalized density, the majority of 1:1 complexes also had a compact density at the surface of ClpX, indicating that translocation had occurred from that end. In 78% of the 2:1 complexes (18% of all particles), a similar compact density was found at only one end. A few 2:1 complexes (5% of the total) had a compact density at both ends, suggesting that translocation of a second substrate might have occurred, but was inefficient, even after 2 h. Translocation reactions carried out in the presence of ATPγS gave similar results, except that most 2:1 complexes remained assembled and only 7% of the complexes dissociated to give ClpPin. In 77% of the particles (1:1 and 2:1 complexes combined), translocation took place from only one end. Again, very few complexes (6%) had a compact density at both ends, indicating translocation from both ends.
Fig. 4. Translocation of λO162–GFP occurs from one end of ClpXPin complexes. We mixed ClpXPin complexes with a 3-fold molar excess of λO162–GFP with respect to ClpX in the presence of either ATPγS or ATP, and samples were incubated at 37°C for 120 min. As a control, ClpXPin complexes without any substrate added were assembled in the presence of ATP and incubated in the same conditions. The percentages of 2:1, 1:1 complexes and a free ClpP were determined for each sample (C), and ClpXPin complexes were classified according to the density associated with the outer surface of ClpX. (A) and (B) are displayed average images of the different complexes observed after incubation in the presence of either ATPγS or ATP, respectively. The number in the lower right corner of each average is the percentage that each class represented in the total population of complexes. Each average image combined between 200 and 300 particles for resolutions of between 30 and 32 Å. The scale bar represents 100 Å.
After 2 h, >70% of the remaining ClpX–ClpP interfaces showed evidence of partially translocated substrate. That observation, together with the greater recovery of ClpXPin complexes when substrate was present, suggests that having partially translocated substrate bridging the ClpX surface and the ClpP chamber may help hold the complex together. In this case, dissociation of ClpX would mean that no translocation occurred from that end of the complex, further supporting the interpretation of unilateral translocation. The results obtained in the presence of ATPγS are consistent with this interpretation. Further more, in 32% of the stable complexes obtained in ATPγS, the non-translocated substrate remained bound to the other end of the 2:1 complexes. Since translocation is fast (<2 min in ATP and <20 min in ATPγS), the low frequency of translocation from both ends suggests that there is an inhibitory effect on further translocation once a substrate has been translocated. We conclude that the presence of protein in the translocation channel or within the ClpP chamber sends an inhibitory signal to the other end of the ClpXP complex, preventing further translocation.
Between 15 and 20% of the complexes had no detect able end-associated density after incubation, although they had stain-occluded cores indicative of translocation (Figure 4A and B). Such complexes were unexpected because no degradation of the GFP moieties has been observed with active ClpXP. Possibly, some of the GFP might not have been well folded, in which case it would have been translocated along with the λO162. In addition, poorly folded GFP would be less visible in the negative stain. It is also possible that a few ClpXPin complexes retained a minor amount of active ClpP that cleaved between the λO162 and the GFP allowing release of the GFP moiety. Further analysis will be needed to test these alternatives.
Translocation of λO–GFP–SsrA
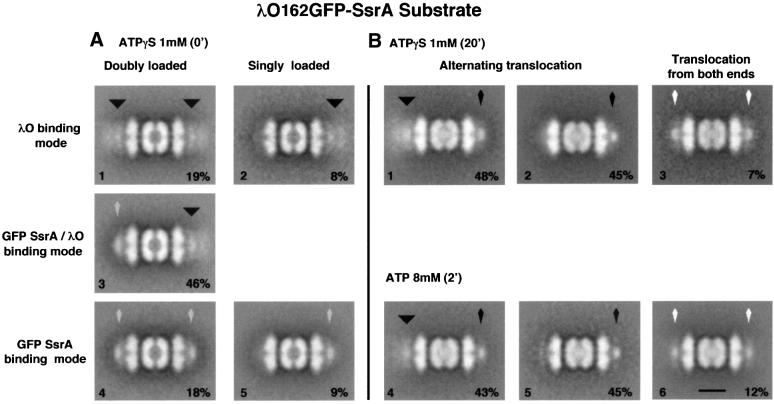
We constructed a fusion, λO162–GFP–SsrA, which we found provided a high proportion of doubly loaded complexes and could be translocated from the N- or C-terminus. With active ClpXP, λO162–GFP–SsrA is completely degraded when translocation is initiated from the C-terminal SsrA tag, and a fraction of the fusions are degraded from the λO side, resulting in transient accumulation of the GFP portion (H.S.Lee and M.R.Maurizi, manuscript in preparation). We assembled complexes of λO162–GFP–SsrA and ClpXPin in the presence of ATPγS and examined them by EM. More than 80% of the complexes had extra density at both ends. λO162– GFP–SsrA bound in two modes: the ‘λO mode’, with a broad, diffuse density extending out from the surface of ClpX (Figure 5A, top); and the ‘SsrA mode’, with a compact density apposed to the surface of ClpX (Figure 5A, bottom). More than half of the doubly loaded complexes (46% of the total) had one λO162–GFP–SsrA bound in the SsrA mode and one in the λO mode (Figure 5A, middle); the remainder had both molecules bound in one or the other mode (Figure 5A, top and bottom left). A minority of 2:1 complexes had a single λO162– GFP–SsrA bound, and they were about evenly split between the two modes (Figure 5A, right, top and bottom). This mixture of complexes was a good starting point for translocation assays.
Fig. 5. Binding and translocation of a substrate with N-terminal and C-terminal recognition tags. (A) λO162–GFP–SsrA bound to ClpXPin in the presence of 1 mM ATPγS. Particles were subjected to classification according to the density associated with the outer surface of ClpX, and averages were obtained of five different classes. λO162–GFP–SsrA can interact with ClpXPin either by its N-terminal λO recognition motif (λO mode) or by its C-terminal SsrA motif (SsrA mode). The latter class was identifiable because of the intense, axially aligned density observed when SsrA-tagged proteins bind to ClpX (see Figure 1). Complexes with λO162–GFP–SsrA bound to one or both faces of ClpXPin in the λO mode (A1 and A2) and in the SsrA mode (A4 and A5) were found. About half of the complexes had substrate molecules in the λO mode and the SsrA mode (A3) on either side. (B) After translocation of λO162–GFP–SsrA in 8 mM ATP or in ATPγS for 20 min electron micrographs were recorded and scanned. Particles with translocated density in ClpP were further classified according to the appearance of the residual density bound to the surface of ClpX, and average images of each class were generated. The majority of particles (B1, B2, B4 and B5) had one side with a condensed axially aligned density similar to the non-translocated GFP domain seen previously with λO162–GFP, indicative of translocation from the λO end. With either ATP or ATPγS, about half of these particles retained unaltered substrate bound to ClpX in the λO mode (B1 and B4), and in half, the substrate had dissociated from the other surface (B2 and B5). A minority of particles (12% with ATP, and 7% with ATPγS) appeared to have a reduced amount of substrate associated with ClpX on both ends of the complex (B3 and B6), implying that translocation occurred from both sides. Each average image combined between 150 and 350 particles for resolutions of between 27 and 32 Å. The scale bar represents 100 Å.
After incubation for 20 min with ATPγS or 2 min with ATP, the centers of the complexes filled with stain-occluding protein and there were changes in the bound substrate. With either nucleotide, a small, axially aligned density, similar to the compact GFP domain remaining after translocation of the λO moiety of λO162–GFP, remained on one end of ∼90% of the particles (Figure 5B, left and middle). In about half of these, the other end of the post-translocation complexes had density very similar to the original substrate density (Figure 5B, left), suggesting little, if any, translocation from that side. In the other particles, clearly evident in the ATPγS-treated samples, there was little or no density visible on the other end (Figure 5B, middle). A faint density was visible on the left side of the images of ATP-treated complexes (Figure 5B, bottom middle), but examination of individual particles showed no discrete densities that we could identify. We believe this density may result from the N-terminal domains of ClpX that are occasionally visible in side views or may be a result of misclassification of a small number of particles. The absence of significant density on the non-translocating side in half of the complexes reflects a greater tendency of substrates to dissociate from filled complexes, as was also seen with the λO162–GFP complexes (Figure 4).
Another 7% of post-translocation complexes with ATPγS and 12% with ATP appeared to have translocated substrate from both ends (Figure 5B, right). The density on both ends of these particles assumed a somewhat compact, axially aligned form, suggestive of at least partial translocation from both ends. These data do not indicate whether the two translocation events occurred simultaneously or sequentially.
We could not count significant numbers of λO162–GFP–SsrA bound in the ‘SsrA mode’ after translocation with either ATPγS or ATP. This was unexpected, because in separate experiments, we showed that the presence of partially translocated λO162–GFP on one side of 2:1 ClpXPin did not prevent binding of GFP–SsrA to the other side (see below), although we cannot rule out decreased affinity for SsrA binding to translocation complexes. It is also possible that our classification program was unable to distinguish between λO162–GFP– SsrA bound in the ‘SsrA mode’ and the GFP density remaining after partial translocation of the λO162 moiety. In this case, our estimates of the amount of substrate translocated from both ends would be slightly overestimated, which only lends further support to the model of one-sided translocation.
2:1 ClpXP complexes are more active than 1:1 complexes
When ClpP is in excess over ClpX, 1:1 complexes predominate, whereas when ClpX is in excess, 2:1 complexes predominate. We wanted to know whether 1:1 complexes were as active as 2:1 complexes. In this context, earlier studies with ClpAP indicated that 2:1 and 1:1 complexes had the same activity (Maurizi et al., 1998). However, we found that with a protein substrate, λO, or a peptide substrate, FAPHMALVPV, the specific activity of 2:1 ClpXP complexes was twice that of 1:1 complexes (Table I). Assays were conducted with fixed amounts of either ClpP or ClpX, and the other component was varied until saturation was reached. Substrate was in excess and the assay time was kept short to ensure that only ClpX or ClpP was limiting. The specific activities suggest that substrates are translocated to ClpP more rapidly when two ClpX hexamers are bound. This result is consistent with our model of alternating translocation by 2:1 complexes if initiation of translocation or a step preceding translocation is rate limiting (see Discussion).
Table I. Activity of 1:1 and 2:1 ClpXP complexesa.
| Limiting component | Active complex | Activity |
|
|---|---|---|---|
| µg λO/µg protein | µmol λO/µmol oligomer | ||
| ClpP14 | 2:1 ClpXP | 0.73 ± 0.06 | 3.3 ± 0.3 (2.0) |
| ClpX6 | 1:1 ClpXP | 0.35 ± 0.03 | 1.5 ± 0.1 (0.9) |
aActivity was determined by the amount of [3H]λO acid solubilized per min at saturating substrate concentration. The activity per oligomer was calculated from the molecular weights of the ClpP tetradecamer (300 kDa), ClpX hexamer (275 kDa) and λO dimers (66 kDa). The data suggest that at limiting ClpP, when two hexamers of ClpX are bound, the contribution of both ClpX hexamers is required for maximum activity.
Translocation of native λO protein
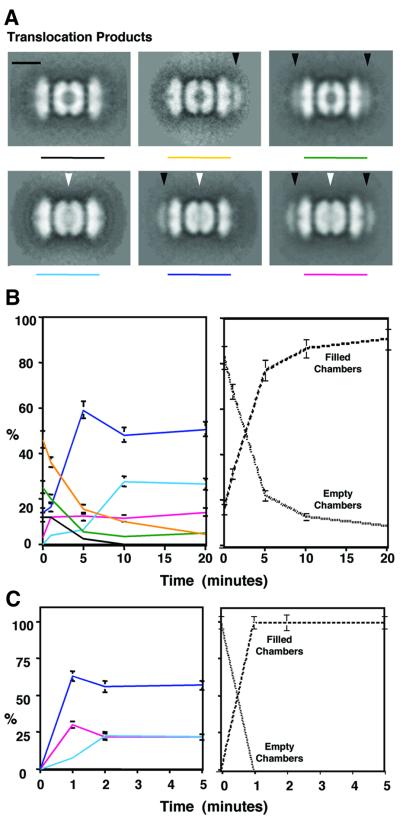
To confirm that the native substrate, λO, is translocated primarily from one end of ClpXPin, we analyzed the complexes present at different times after initiating translocation. To slow the rate of translocation, λO was added to complexes of ClpXPin in ATPγS, and the reaction mix was sampled for negative staining after 1, 5, 10 and 20 min. The images from each time point were classified by an automated procedure that distinguished six principal reaction products: complexes with λO bound at two, one or no ends; and in each case, with the internal chamber of ClpXPin empty or full (Figure 6A). The filling of the internal chamber took place within 10 min, as shown by plotting the percentage of all complexes with stain-occluded centers as a function of time (Figure 6B, right). The major trends of these data are as follows. (i) Accumulation of complexes with stain-occluded ClpPin chambers, and λO either at one end (59%) or at both ends (28%). Both types of complexes can be interpreted as having translocated from one end only. (ii) A marked decrease in the number of complexes with stain-penetrable (i.e. empty) centers from an initial 83% (46% with λO at one end, 25% with λO at both ends, and 12% with no λO) to 9%; and a concomitant increase in the number of complexes with stain-occluded ClpXPin (i.e. with internalized λO) to 91% (27% with no end-bound λO, 50% with one end-bound λO, and 14% with λO at both ends; Figure 6B). (iii) Only 4% or less of the stain-occluded complexes with a significantly smaller λO-associated density at both ends was indicative of having translocated λO from both ends.
Fig. 6. Binding and translocation of native λO. The slower translocation observed with ATPγS allowed us to identify (A) and quantitate (B) complexes with zero (A, left), one (A, middle, black arrow) or two (A, right, black arrows) λO dimers bound on the ends of complexes or within the proteolytically inactive ClpP chamber (A, bottom row, white arrows) along an ATPγS-driven time course reaction. Average images combined between 300 and 500 particles for resolutions between 27 and 30 Å. The scale bar represents 100 Å. (B) Evolution of the intermediates along the time course translocation reaction (left plot). The plot on the right displays the percentage of all complexes with either stain-occluded or non-stain-occluded centers as a function of time. (C) Species of ClpXPin complexes isolated in an ATP-driven time course reaction were very similar to the three translocated products observed in the ATPγS-driven translocation (A, bottom row). Evolution of the ClpXPin complexes with zero, one or two λO dimers along the time course ATP translocation reaction (left plot). On the right, the percentage of all complexes with either stain- occluded or non-stain-occluded centers as a function of time was plotted. Error bars were calculated by (√N/N) × %, where N is the number of particles that represent the intermediate at that time point, and % is the percentage that these particles represent with respect to the total population of complexes at that time point.
To determine whether rapid ATP hydrolysis influenced the ability of ClpXPin to translocate λO from one or both ends, ATP was added to ClpXPin–λO complexes pre-assembled in the presence of ATPγS. Aliquots were removed after 1, 2 or 5 min, and analyzed by EM (data not shown). At each time point after ATP addition, the internal chambers of all ClpXPin complexes were occupied (Figure 6C, right) and the averaged images of the translocation complexes were indistinguishable from those observed in the ATPγS-driven reaction (data not shown). Most complexes had either no end-bound density or had a density that resembled that of the original pre-translocation complex, whereas <4% had reduced amounts of density at both ends. Thus, partial translocation from both ends was very uncommon. At the start, 63% of complexes had λO bound at one end, which decreased to ∼56% 1 min later, and subsequently remained around this value (Figure 6C, left). ClpXPin with λO bound at both ends formed 30% of the initial complexes, decreasing to 22% after 1 min (Figure 6C, left). Finally, ClpXPin with no λO bound represented ∼7% of the starting population, increased to ∼23% after 1 min, and remained at that value (Figure 6C, left).
In the complexes seen with ATPγS (Figure 6A) or ATP (data not shown), the end-bound density after translocation was similar to the initially bound density attributable to λO dimers. The kinetics of translocation, with density appearing in the center of ClpPin while density disappears from one side and is retained on the other, strongly suggest that translocation takes place from one end at a time. One complication with this interpretation is that reversibly bound λO dimers can redistribute as the reaction proceeds, as shown by the appearance of complexes with filled chambers and with a λO dimer at both ends. However, the redistribution of λO argues that protein bound on the ends of complexes is not partially translocated or unfolded but is bound in its initial mode.
Translocated ClpXP complexes can bind additional substrate
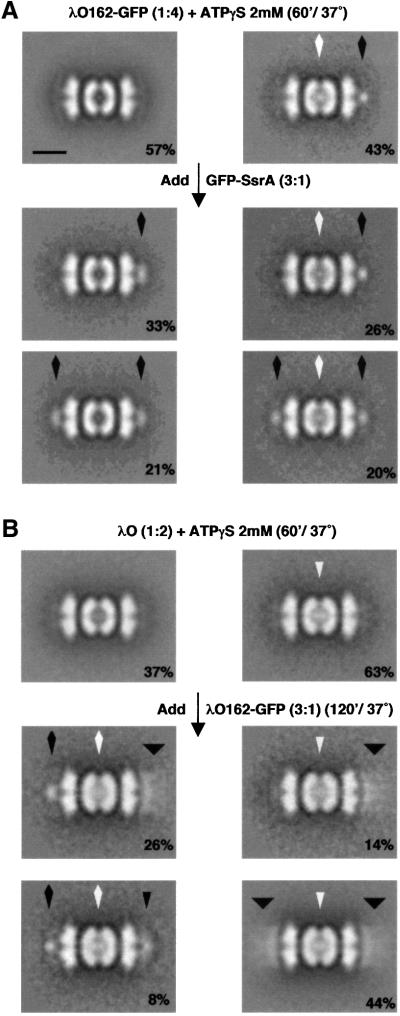
The previous experiments showed that complexes whose digestion chambers are filled with λO can bind additional λO dimers. We wanted to compare the binding of sub strates with translocated and non-translocated ClpXPin complexes. A mixed population of complexes was formed by incubation with a sub-stoichiometric amount of λO162–GFP in ATPγS. EM showed that 43% of the complexes had partially translocated λO162–GFP on one end, and 57% had no substrate bound or translocated (Figure 7A, upper row). A 3-fold molar excess of GFP–SsrA with respect to ClpX was added and the complexes were imaged and classified. Empty complexes were able to bind GFP–SsrA at one or both ends (Figure 7A, left panels). About 70% of the available sites bound GFP–SsrA. Filled complexes were also able to bind GFP–SsrA. About 43% displayed an additional compact globular density at the opposite end (Figure 7A, bottom row, right panel), although we could not distinguish between the bound GFP–SsrA and the non-translocated portion of the substrate, λO162–GFP. The slightly lower binding to the filled complexes might indicate an allosteric effect whereby translocated substrate lowers the substrate-binding affinity of the other end of the complex. In a separate experiment, we found that ClpXP complexes filled with translocated GFP–SsrA were able to bind additional GFP–SsrA at the end (data not shown). Thus, binding of substrates to partially filled complexes is not prevented by pre-internalized substrates.
Fig. 7. Substrate binding to ClpXPin complexes in which the digestion chamber is occupied. (A) We obtained a mixed population of ClpXPin complexes that had undergone translocation of λO162–GFP from one end (upper row, right), and of unloaded and stained-penetrated ClpXPin complexes (upper row, left). A 3-fold molar excess of GFP–SsrA with respect to ClpX was added and complexes observed under negative stained EM were classified. GFP–SsrA was able to bind to initially empty ClpXPin complexes either in one (middle, left) or both ends (bottom, left). Twenty percent of the initially translocated complexes displayed a compact globular density attached to both ends (bottom, right), and 26% of the initially translocated complexes remained unaltered (middle, right). (B) λO and ClpXPin complexes were mixed in 1:2 molar ratio, respectively. A mixed population of translocated and non-translocated complexes was obtained. The upper row shows averages built with non-translocated (left) and translocated (right) complexes. λO162–GFP was added in 3-fold molar excess with respect to ClpX, and further incubated for 120 min at 37°C. Particles were subjected to image classification according to the density associated with the outer surface of ClpX, and averages for each class are displayed. Figures written on the averages indicate the percentage that each kind of complex represent with respect to the total population. Average images combined between 150 and 450 particles for resolutions between 25 and 32 Å. The scale bar represents 100 Å.
When the ClpXP digestion chamber is occupied by λO, additional substrate cannot be translocated into it
To test for translocation of additional substrate into complexes already containing λO protein, we prepared such complexes by incubating ClpXPin with λO in the presence of ATPγS for 1 h. A mixed population of 63% translocated complexes and 37% empty complexes was obtained (Figure 7B, upper row). Most of the complexes did not have any end-bound density, while a few (12%) had densities corresponding to λO dimers at one or both ends (data not shown). A 3-fold molar excess of λO162–GFP was added and the samples were again incubated until all complexes were filled. Only 33% of them had translocated the λO162 moiety, as indicated by the appearance of a compact density apposed to the axial channel in one end of the complex (Figure 7B, bottom and middle row). The number of complexes with partially translocated λO162–GFP correlated well with the number of empty complexes, suggesting that only empty complexes translocated substrate during the second incubation. These complexes also had either λO162–GFP (26%) or λO (8%) attached to the opposite end. Of the remaining filled complexes, 58% had λO162–GFP-like density at one or both ends, and 8% had λO bound to one end. The binding of λO162–GFP to >90% of λO-translocated complexes without signs of additional translocation suggests that the presence of translocated substrate prevents further translocation. Interestingly, these data also indicate that substrate within the digestion chamber is trapped and cannot be displaced by additional translocation of a different substrate.
Discussion
This study extends earlier observations on the structural basis of substrate binding and internalization by the ClpXP (Ortega et al., 2000) and ClpAP (Ishikawa et al., 2001) proteases. In particular, it provides insight into the dynamics of substrate translocation in 2:1 ClpXP complexes. Our studies were aided by the use of a substrate with a domain that was resistant to unfolding by ClpX, which allowed us to discriminate between the end products of translocation and the initial substrate complex, and thereby identify the end from which translocation had occurred. We found that, in 2:1 ClpXP complexes, translocation occurs primarily from one end at a time. In addition, after translocation occurs from one end, translocation of substrate from the other end is very inefficient. These conclusions stem from two observations. First, in experiments in which incubation times were limited, translocation from one end appeared complete and, in ∼90% of the complexes, the substrate at the other end was not translocated (Figures 2, 5 and 6). Secondly, when excess substrate was used and translocation reactions were continued for 120 min, only ∼5% of the complexes appeared to have translocation from both ends (Figure 4).
Two factors could contribute to translocation being favored from one end of ClpXP complexes at a time. First, if initiation of translocation is rate-limiting and occurs in a stochastic manner, the probability of initiating translocation from both sides ‘simultaneously’ would be expected to be low. Secondly, there could be negative cooperativity between the two ClpX rings that affects translocation, but not the initial binding of substrates. The latter model implies a mechanism of signaling between the respective ends of translocating complexes presumably occasioned by the presence of substrate and mediated through ClpP. In general terms, if not specific detail, such cooperativity would be reminiscent of the communication between the cis and trans rings of another bipolar complex, GroEL, during protein folding (Burston et al., 1995; Llorca et al., 1996). Physiologically, a mechanism for delaying translocation from the opposite end of ClpXP may be needed to ensure that substrates are fully unfolded and internalized once the process is initiated to avoid release of unfolded or partially degraded intermediates.
Alternatively, the presence of non-degraded substrate within ClpPin might sterically block translocation from the other side, and we would observe one-sided translocation. However, the ClpP chamber might accommodate as much as 50 kDa of protein, much more than the 16 kDa portion of λO translocated in λO162–GFP, so steric interference seems unlikely as the entire explanation. Even after long incubations, there was little evidence for translocation from the second side. A second possibility is that there is negative feedback from undigested protein (or under degradative conditions, large peptides) within the ClpP chamber. Conformational effects on ClpP produced by the undigested protein could affect interactions with ClpX at the other end of the complex, inhibiting its protein unfolding or translocating activity. This interpretation is somewhat debatable because Singh et al. (2000) showed earlier that filled ClpXPin complexes can unfold GFP– SsrA, and Kim et al. (2000) reported that filled complexes can translocate additional protein that displaces the internalized protein. However, our experiments failed to see displacement of the internalized protein when trying to translocate additional substrate (Figure 7B).
If translocation is favored from one end at a time, why is the proteolytic activity of 2:1 ClpXP complexes higher (∼2-fold) than 1:1 complexes? Since there is only one degradation chamber and degradation itself is 1–2 orders of magnitude faster than unfolding and translocation, the implication is that one of these latter rate-limiting steps occur more efficiently in 2:1 complexes. One explanation derives from a simple stochastic model for initiation of substrate unfolding/translocation. If initiation is very slow and subsequent steps are fast so that ClpX recycles rapidly to the pre-initiation phase, initiation would occur twice as frequently in 2:1 complexes. Our data support this model, because whether translocation was complete (as with λO) or was arrested by a stable domain (as with λO162–GFP), the process was largely all or none; in ≥85% of the complexes, substrate was observed either in its initial binding mode or in its final translocated state or had dissociated. The infrequent occurrence of translocation intermediates indicates that, once initiated, translocation proceeds rapidly. As discussed above, failure to observe translocation from both ends even after long periods also suggests that translocation from one end results in a signal that blocks initiation of translocation from the opposite end. This signal may normally be removed when degradation is complete, resetting both ends of the complex to the same state. Alternatively, translocation could alternate between the two ends, in which case the unfolding/translocation cycle would have to be more efficient in 2:1 complexes compared with 1:1 complexes. For example, the non-translocating end could begin to unfold substrate while translocation from the other end is proceeding and would thus be primed to initiate a new round of translocation immediately after completion of the previous cycle. Future studies will be addressed to these issues.
Negative cooperativity in translocation may facilitate release of oligopeptides in processive degradation
In the degradation of proteins by ClpXP, the proteolytic step is much faster than unfolding and translocation, and peptide bond cleavage is 10–50 times faster for peptides compared with proteins (Thompson and Maurizi, 1994; Singh et al., 1999). Thus, the rate of peptide bond hydrolysis is not an impediment to simultaneous translocation of substrate from both ends. Nevertheless, our results indicate that substrate is internalized primarily from one end of doubly loaded 2:1 ClpXP complexes, pointing to negative cooperativity between the two sides of the complex.
An advantage of this alternating mode of translocation is that the pore in the trans ClpP and ClpX rings is not blocked and a possible exit pathway is available for the oligopeptide products of proteolysis. It has been reported that the binding of ClpY to one end of ClpQ elicits asymmetric changes in pore size (Wang et al., 2001b). These authors suggested that opening the trans ClpQ pore might facilitate the release of oligopeptide products. Also, a gating mechanism involving the axial pore of the 20 S particle has been shown (Groll et al., 2000), and it has been proposed that release of reaction products from the 20S peptidase is facilitated by the binding of a regulatory Reg complex to the trans ring (Whitby et al., 2000). The utility of this exit pathway through the trans ring may be contingent on whether this ring has protein substrate bound, which might be expected to impede the passage of peptide products.
Binding sites for N- and C-terminal recognition motifs
Our images show protein substrates bound to the distal surface of ClpX in ClpXP complexes. Do λO with its N-terminal tag and GFP–SsrA with its C-terminal tag bind to the same site? The top view difference images of bound substrates (Figure 1B, bottom) imply that the respective binding sites are close, if not necessarily the same. All the extra density is confined within the hexameric ring, with no evidence for substrate binding on the edges or sides of ClpX rings. The close juxtaposition of GFP–SsrA to the ClpX channel suggests the possibility that the SsrA tag may bind near or within the channel. From structural studies and modeling of the ClpX structure (Kim et al., 2000; Ortega et al., 2000; Singh et al., 2000), the ClpX surface sites that contact λO and SsrA are far from a proposed sensor and substrate discrimination (SSD) domain (Smith et al., 1999). Although the recombinant SSD domain expressed independently was shown to bind to proteins carrying C-terminal recognition motifs, our data suggest that this domain does not participate in the initial binding of SsrA-tagged proteins to ClpX to enter the degradation pathway.
In side views of ClpXP, both substrates extend a similar distance axially from ClpX, but λO extends further in the dimension perpendicular to the axis. We suggest that the λO dimer may bind via one of its subunits with the other extending further from the axis; in top views, this second subunit may be poorly represented as a result of disorder, exacerbating the low occupancy in the averaged image. Although the binding sites for N- and C-terminal motifs appear to be close, we note that they differ functionally in that GFP is unfolded and translocated when targeted via SsrA but not when targeted via λO.
Slowed translocation in the presence of ATPγS
The key factor that enabled us to observe functional polarity in translocating complexes was the greatly reduced rate of λO translocation in the presence of ATPγS. With ATPγS, complete translocation requires 5–10 min (Figure 6B, right plot). In contrast, the ATP-driven reaction was too rapid and was usually complete in ∼1 min. The reactions catalyzed by ATPγS and ATP both favor the alternating translocation mechanism. However, unfolding and degradation of GFP–SsrA in the presence of ATPγS is inefficient at best, with the result that translocation of λO–GFP–SsrA in ATPγS proceeded from the λO end, whereas with ATP translocation proceeded from either end of the substrate. The products of the two reactions could not be distinguished in our micrographs, suggesting that the pathway followed and the disposition of the non-translocated portion of the substrate were the same, regardless of which end went in first.
Only one λO subunit at a time passes through an axial channel
Many ClpXP substrates associate with themselves or other proteins to form multimeric complexes. One question regarding homo-oligomers bearing multiple degradation signals is whether one or more than one subunit is translocated simultaneously into ClpP. It was reported recently that up to three polypeptide chains may be able to pass through the axial pore of ClpP at the same time (Burton et al., 2001), but whether translocation of independent subunits through a single channel can occur simultaneously has not been shown. Our data with λO162–GFP suggest that, at least with this substrate, only one subunit at a time is translocated through the axial channel. With λO162–GFP, in which only the λO portion is translocated, the resulting complex exhibited a single GFP-like on-axis density attached to the distal surface of ClpX, indicating that only a single fusion protein had undergone translocation. A smaller density was also found adjacent to the intact GFP domain in these complexes (Figures 2A, 4A and B), but it had none of the characteristics of GFP. Most likely, this faint density represents a non-translocated portion of λO in some of the complexes, which would be consistent with biochemical assays showing accumulation of products in which the λO domain is only partly degraded (H.S.Lee and M.R.Maurizi, unpublished data). We conclude that only one λO162 domain is translocated at a time, while associated subunits are released. This model is consistent with studies showing that unfolding of a single subunit of tetrameric MuA is sufficient to disrupt the strand transfer complex during Mu transposition (Burton et al., 2001).
Materials and methods
Protein preparation
ClpX (Grimaud et al., 1998) and ClpP (Maurizi et al., 1994) were purified as described previously. ClpXPin was prepared by treating ClpP with carbobenzoxy-Leu-Tyr-chloromethyl ketone (Singh et al., 1999). λO was purified from overexpressing cells by chromatography on SP-Sepharose, followed by gel filtration on Superdex 200 and a polishing step on MonoS (all columns from Pharmacia Biotech). Preparation of λO162–GFP and λO162–GFP–SsrA fusions is described elsewhere (H.S.Lee and M.R.Maurizi, in preparation). Proteins were quantitated by calibrated dye binding assays using standard solutions of the purified proteins. Concentrations of the standard solutions were determined from the absorbance at 280 nm (or 495 nm for GFP derivatives) using the proteins’ absorption coefficients.
Degradation assays
λO162–GFP degradation was assayed at 37°C in 50 mM Tris–HCl pH 8.0, 0.1 M KCl, 10 mM MgCl2 and 4 mM ATP. Samples were removed at zero time and after 60 min, mixed with hot SDS sample buffer, run on 12% acrylamide–SDS gels and stained with Coomassie Blue. The specific activity of ClpXP was determined under conditions of limiting ClpX or limiting ClpP. When ClpP was limiting (0.2 µM tetradecamer), ClpX was varied from 1 to 10 µM hexamer; when ClpX was limiting (0.2 µM hexamer), ClpP was varied from 1 to 20 µM tetradecamer. Degradation of λO was measured at 37°C in the above buffer with either 4 mM ATP or ATPγS and 8 µM [3H]λO (8000 c.p.m./µg). Reactions were terminated after 4–10 min by adding 10% (w/v) trichloroacetic acid, and the amount of radioactivity in the supernatant solution was measured by scintillation counting.
Substrate translocation assays
To favor 2:1 ClpXPin complexes, ClpX hexamers were assembled by incubating ClpX (final concentration, 50 µg/ml) for 10 min at room temperature in 50 mM Tris–HCl pH 7.5, 0.2 M KCl, 20 mM MgCl2, 1 mM EDTA, 10% glycerol and 1 mM ATPγS. Then ClpPin (final concentration, 30 µg/ml) was added. After addition of the appropriate molar ratio of substrate to assembled ClpXPin, an aliquot was taken immediately and applied to a grid prepared as described below. After 2 min to allow time for adsorption, the excess solution was removed by blotting, the grid was treated with uranyl acetate and processed as described below. Translocation reactions were continued in ATPγS by incubation at room temperature, and samples were taken after 1, 5, 10 and 20 min. In other cases, complexes were diluted into 8 mM ATP (final concentration of ATPγS was 50–100 µM) immediately after assembly in ATPγS, a sample was removed immediately, and additional samples were taken after 1, 5 and 10 min at room temperature.
To measure translocation of λO162–GFP into partially filled ClpXPin, λO was added (in 1:2 molar ratio with respect to ClpX) to ClpXPin complexes assembled in 2 mM ATPγS, and samples were incubated for 60 min at 37°C. Samples were taken for negative staining, and λO162–GFP (3-fold molar excess with respect to ClpX) was added to the remainder. After 120 min at 37°C, samples were taken for negative staining. In another experiment, λO162–GFP (in 1:4 molar ratio with respect to ClpX) was translocated into assembled ClpXPin complexes by incubation for 60 min at 37°C in 2 mM ATPγS. Samples were removed for negative staining, and 0.5 µg of GFP–SsrA (3-fold molar excess of substrate with respect to ClpX) were added to the remainder. Incubation at 37°C was continued for 120 min, and samples were taken for negative staining.
Electron microscopy
Grids bearing carbon-coated nitrocellulose films were glow discharged and floated on a 0.1% aqueous solution of polylysine hydrobromide (mol. wt 60–120 K; Polysciences, Inc.) for 2 min. After blotting, the grids were floated for 2 min on 5 µl drops of sample at ∼50 µg/ml; 1% uranyl acetate was used for negative staining. Specimens were observed on a Philips CM120 or an EM400T electron microscope.
Image analysis
Micrographs were digitized at 0.30 nm/pixel, using a SCAI scanner (Z/I Imaging Huntsville, AL). Image processing was carried out using the PIC-III program (Trus et al., 1996). Particles were extracted using X3D (Conway et al., 1993) and translationally aligned and subjected to correlation averaging according to Kocsis et al. (1995). Averages were normalized by standardizing the background density and the peak density of ClpP. Resolution was assessed according to the spectral signal-to-noise ratio criterion (Unser et al., 1987).
Image classification
Side-view images of ClpXPin complexes were centered and aligned before classifying them with the self-organizing feature maps algorithm implemented in the Xmipp image processing package (Marabini and Carazo, 1994; Marabini et al., 1996). Typically, we combined two steps of classification to sort all the different ClpXPin complexes out in each experiment. First, a mask was applied to the images covering everything except the digestion chamber of ClpPin. The set of images was classified and an array of five-by-five node images was obtained. The two expected classes, corresponding to particles containing translocated protein and those without, appeared on opposite edges of the array. Particles from the two classes were selected and counted. Images in each one of these two classes were masked by covering everything except the distal surface of ClpX and classified in a second round of classification to sort out ClpXPin complexes according to the density attached to ClpX. An array of five-by-five node images was obtained displaying the different classes. Average images were calculated for each class.
Acknowledgments
Acknowledgements
We thank Drs N.Cheng, T.Ishikawa and D.Winkler for help with EM, Drs D.Belnap, B.Trus and B.Heymann for guidance in computational matters, Drs J.M.Carazo and A.Pascual for kindly providing Xmipp, and Drs T.Ishikawa and O.Llorca for helpful discussions. J.O. is a recipient of fellowships from National Institutes of Health (USA) and Ministerio de Educacion y Cultura (Spain).
References
- Bochkareva E.S., Lissin,N.M. and Girshovich,A.S. (1992) Positive cooperativity in the functioning of molecular chaperone GroEL. J. Biol. Chem., 267, 6796–6800. [PubMed] [Google Scholar]
- Bochtler M., Ditzel,L., Groll,M. and Huber,R. (1997) Crystal structure of heat shock locus V (HslV) from Escherichia coli. Proc. Natl Acad. Sci. USA, 94, 6070–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochtler M., Hartmann,C., Song,H.K., Bourenkov,G.P., Bartunik,H.D. and Huber,R. (2000) The structures of HsIU and the ATP-dependent protease HsIU-HsIV. Nature, 403, 800–805. [DOI] [PubMed] [Google Scholar]
- Burston S.G., Ranson,N.A. and Clarke,A.R. (1995) The origins and consequences of asymmetry in the chaperonin reaction cycle. J. Mol. Biol., 249, 138–152. [DOI] [PubMed] [Google Scholar]
- Burton B.M., Williams,T.L. and Baker,T.A. (2001) ClpX-mediated remodeling of mu transpososomes: selective unfolding of subunits destabilizes the entire complex. Mol. Cell, 8, 449–454. [DOI] [PubMed] [Google Scholar]
- Conway J.F., Trus,B.L., Booy,F.P., Newcomb,W.W., Brown,J.C. and Steven,A.C. (1993) The effects of radiation damage on the structure of frozen hydrated HSV-1 capsids. J. Struct. Biol., 111, 222–233. [DOI] [PubMed] [Google Scholar]
- Fenton W.A. and Horwich,A.L. (1997) GroEL-mediated protein folding. Protein Sci., 6, 743–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonciarz-Swiatek M., Wawrzynow,A., Um,S.J., Learn,B.A., McMacken,R., Kelley,W.L., Georgopoulos,C., Sliekers,O. and Zylicz,M. (1999) Recognition, targeting and hydrolysis of the λO replication protein by the ClpP/ClpX protease. J. Biol. Chem., 274, 13999–14005. [DOI] [PubMed] [Google Scholar]
- Gottesman S. and Maurizi,M.R. (1992) Regulation by proteolysis: energy-dependent proteases and their targets. Microbiol. Rev., 56, 592–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S., Roche,E., Zhou,Y. and Sauer,R.T. (1998) The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev., 12, 1338–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantcharova V., Alm,E.J., Baker,D. and Horwich,A.L. (2001) Mechanisms of protein folding. Curr. Opin. Struct. Biol., 11, 70–82. [DOI] [PubMed] [Google Scholar]
- Grimaud R., Kessel,M., Beuron,F., Steven,A.C. and Maurizi,M.R. (1998) Enzymatic and structural similarities between the Escherichia coli ATP-dependent proteases, ClpXP and ClpAP. J. Biol. Chem., 273, 12476–12481. [DOI] [PubMed] [Google Scholar]
- Groll M., Bajorek,M., Kohler,A., Moroder,L., Rubin,D.M., Huber,R., Glickman,M.H. and Finley,D. (2000) A gated channel into the proteasome core particle. Nat. Struct. Biol. 7, 1062–1067. [DOI] [PubMed] [Google Scholar]
- Ishikawa T., Belnap,D., Maurizi,M.R. and Steven,A.C. (2000) Docking configuration of the proteolytic and ATPase components of the energy-dependent protease HslVU (ClpYQ) in active complexes. Nature, 408, 667–668. [DOI] [PubMed] [Google Scholar]
- Ishikawa T., Beuron,F., Kessel,M., Wickner,S., Maurizi,M.R. and Steven,A.C. (2001) Translocation pathway of protein substrates in ClpAP protease. Proc. Natl Acad. Sci. USA, 98, 4328–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler K.C., Waller,P.R. and Sauer,R.T. (1996) Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science, 271, 990–993. [DOI] [PubMed] [Google Scholar]
- Kessel M., Maurizi,M.R., Kim,B., Kocsis,E., Trus,B.L., Singh,S.K. and Steven,A.C. (1995) Homology in structural organization between E.coli ClpAP protease and the eukaryotic 26 S proteasome. J. Mol. Biol., 250, 587–594. [DOI] [PubMed] [Google Scholar]
- Kessel M., Wu,W., Gottesman,S., Kocsis,E., Steven,A.C. and Maurizi,M.R. (1996) Six-fold rotational symmetry of ClpQ, the E.coli homolog of the 20S proteasome and its ATP-dependent activator, ClpY. FEBS Lett., 398, 274–278. [DOI] [PubMed] [Google Scholar]
- Kim Y.I., Burton,R.E., Burton,B.M., Sauer,R.T. and Baker,T.A. (2000) Dynamics of substrate denaturation and translocation by the ClpXP degradation machine. Mol. Cell, 5, 639–648. [DOI] [PubMed] [Google Scholar]
- Klumpp M. and Baumeister,W. (1998) The thermosome: archetype of group II chaperonins. FEBS Lett., 430, 73–77. [DOI] [PubMed] [Google Scholar]
- Kocsis E., Trus,B.L., Cerritelli,M., Cheng,N. and Steven,A.C. (1995) Improved methods for determination of rotational symmetries in macromolecules. Ultramicroscopy, 60, 219–228. [DOI] [PubMed] [Google Scholar]
- Llorca O., Carrascosa,J.L. and Valpuesta,J.M. (1996) Biochemical characterization of symmetric GroEL–GroES complexes. Evidence for a role in protein folding. J. Biol. Chem., 271, 68–76. [DOI] [PubMed] [Google Scholar]
- Lupas A., Flanagan,J.M., Tamura,T. and Baumeister,W. (1997) Self-compartmentalizing proteases. Trends Biochem. Sci., 22, 399–404. [DOI] [PubMed] [Google Scholar]
- Marabini R. and Carazo,J.M. (1994) Pattern recognition and classification of images of biological macromolecules using artificial neural networks. Biophys. J., 66, 1804–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marabini R., Masegosa,I.M., San Martín,M.C., Marco,S., Fernández,J.J., de la Fraga,L.G., Vaquerizo,C. and Carazo,J.M. (1996) Xmipp: an image processing package for electron microscopy. J. Struct. Biol., 116, 237–240. [DOI] [PubMed] [Google Scholar]
- Maurizi M.R., Thompson,M.W., Singh,S.K. and Kim,S.H. (1994) Endopeptidase Clp: ATP-dependent Clp protease from Escherichia coli. Methods Enzymol., 244, 314–331. [DOI] [PubMed] [Google Scholar]
- Maurizi M.R., Singh,S.K., Thompson,M.W., Kessel,M. and Ginsburg,A. (1998) Molecular properties of ClpAP protease of Escherichia coli: ATP-dependent association of ClpA and clpP. Biochemistry, 37, 7778–7786. [DOI] [PubMed] [Google Scholar]
- Ortega J., Singh,S.K., Maurizi,M.R. and Steven,A.C. (2000) Visualization of substrate binding and translocation by the ATP-dependent protease. Mol. Cell, 6, 1515–1521. [DOI] [PubMed] [Google Scholar]
- Rohrwild M., Pfeifer,G., Santarius,U., Muller,S.A., Huang,H.C., Engel,A., Baumeister,W. and Goldberg,A.L. (1997) The ATP-dependent HslVU protease from Escherichia coli is a four-ring structure resembling the proteasome. Nat. Struct. Biol., 4, 133–139. [DOI] [PubMed] [Google Scholar]
- Singh S.K., Guo,F. and Maurizi,M.R. (1999) ClpA and ClpP remain associated during multiple rounds of ATP-dependent protein degradation by ClpAP protease. Biochemistry, 38, 14906–14915. [DOI] [PubMed] [Google Scholar]
- Singh S.K., Grimaud,R., Hoskins,J.R., Wickner,S. and Maurizi,M.R. (2000) Unfolding and internalization of proteins by ClpXP and ClpAP. Proc. Natl Acad. Sci. USA, 97, 8898–8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S.K., Rozycki,J., Ortega,J., Ishikawa,T., Lo,J., Steven,A.C. and Maurizi,M.R. (2001) Functional domains of the ClpA and ClpX molecular chaperones identified by limited proteolysis and deletion analysis. J. Biol. Chem., 276, 29420–29429. [DOI] [PubMed] [Google Scholar]
- Smith C.K., Baker,T.A. and Sauer,R.T. (1999) Lon and Clp family proteases and chaperones share homologous substrate-recognition domains. Proc. Natl Acad. Sci. USA, 96, 6678–6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa M.C., Trame,C.B., Tsuruta,H., Wilbanks,S.M., Reddy,V.S. and McKay,D. (2000) Crystal and solution structures of an HslUV protease-chaperone complex. Cell, 103, 633–643. [DOI] [PubMed] [Google Scholar]
- Thompson M.W. and Maurizi,M.R. (1994) Processive degradation of proteins by the ATP-dependent Clp protease from Escherichia coli. J. Biol. Chem., 269, 18209–18215. [PubMed] [Google Scholar]
- Trus B.L., Kocsis,E., Conway,J.F. and Steven,A.C. (1996) Digital image processing of electron micrographs: the PIC system-III. J. Struct. Biol., 116, 61–67. [DOI] [PubMed] [Google Scholar]
- Unser M., Trus,B.L. and Steven,A.C. (1987) A new resolution criterion based on spectral signal-to-noise ratios. Ultramicroscopy, 23, 39–51. [DOI] [PubMed] [Google Scholar]
- Wang J., Hartling,J.A. and Flanagan,J.M. (1997) The structure of ClpP at 2.3 Å resolution suggests a model for ATP-dependent proteolysis. Cell, 91, 447–456. [DOI] [PubMed] [Google Scholar]
- Wang J. et al. (2001a) Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism. Structure, 9, 177–184. [DOI] [PubMed] [Google Scholar]
- Wang J., Song,J.J., Seong,I.S., Franklin,M.C., Kamtekar,S., Eom,S.H. and Chung,C.H. (2001b) Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU. Structure, 9, 1107–1116. [DOI] [PubMed] [Google Scholar]
- Weber-Ban E.U., Reid,B.G., Miranker,A.D. and Horwich,A.L. (1999) Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature, 401, 90–93. [DOI] [PubMed] [Google Scholar]
- Whitby F.G., Masters,E.I., Kramer,L., Knowlton,J.R., Yao,Y., Wang,C.C. and Hill,C.P. (2000) Structural basis for the activation of 20S proteasomes by 11S regulators. Nature, 408, 115–120. [DOI] [PubMed] [Google Scholar]
- Wickner S., Maurizi,M.R. and Gottesman,S. (1999) Post-translational quality control: folding, refolding and degradation of proteins. Science, 286, 1888–1893. [DOI] [PubMed] [Google Scholar]
- Zwickl P., Baumeister,W. and Steven,A. (2000) Dis-assembly lines: the proteasome and related ATPase-assisted proteases. Curr. Opin. Struct. Biol., 10, 242–250. [DOI] [PubMed] [Google Scholar]