Abstract
The initiation of DNA replication is a key event in the cell cycle of all organisms. In bacteria, replication initiation occurs at specific origin sequences that are recognized and processed by an oligomeric complex of the initiator protein DnaA. We have determined the structure of the conserved core of the Aquifex aeolicus DnaA protein to 2.7 Å resolution. The protein comprises an AAA+ nucleotide-binding fold linked through a long, helical connector to an all-helical DNA-binding domain. The structure serves as a template for understanding the physical consequences of a variety of DnaA mutations, and conserved motifs in the protein suggest how two critical aspects of origin processing, DNA binding and homo-oligomerization, are mediated. The spatial arrangement of these motifs in DnaA is similar to that of the eukaryotic-like archaeal replication initiation factor Cdc6/Orc1, demonstrating that mechanistic elements of origin processing may be conserved across bacterial, archaeal and eukaryotic domains of life.
Keywords: bacteria/DnaA/DNA replication initiation/structure
Introduction
Replication origins are specific DNA elements that define the initiation point of bi-directional DNA replication (Kornberg and Baker, 1992). In all cellular organisms and many viruses, origin-associated proteins recognize cis DNA elements and initiate replication by processing the origin DNA to allow for the assembly of the replication machinery. Genetic and biochemical studies have highlighted general properties of DNA replication initiation in a variety of model systems. The initiation process has been particularly well characterized in the bacterium Escherichia coli.
The E. coli chromosome has a discrete replication origin site (oriC) in their chromosomes that mediates the formation of a large, oligomeric assembly of the conserved initiator protein, DnaA, a member of the AAA+ family of ATPases (Neuwald et al., 1999; Messer et al., 2001). In E.coli, the nucleoprotein complex at oriC spans ∼200 bp of the origin and is thought to contain 5–20 DnaA molecules (Kornberg and Baker, 1992; Carr and Kaguni, 2001; Messer et al., 2001). Once formed on negatively supercoiled DNA, the nucleoprotein origin complex can be remodeled in the presence of ATP and the architectural factors HU and IHF (Grimwade et al., 2000), melting an AT-rich DNA unwinding element (DUE) within oriC (Hwang and Kornberg, 1992). While ATP-DnaA and ADP-DnaA are both able to form nucleoprotein complexes, only ATP-DnaA induces DUE remodeling, possibly through an allosteric mechanism that increases the affinity of DnaA for single-stranded DNA, stabilizing the DUE open complex (Speck and Messer, 2001). Origin processing is completed with the recruitment of two hexamers of the replicative helicase, DnaB, to exposed single-stranded DNA regions with the help of an additional factor, the helicase loader DnaC (Kornberg and Baker, 1992). Once DnaB is loaded to form the prepriming complex (Funnell et al., 1987), the open DUE expands and replisome assembly is initiated by the recruitment of the DnaG primase (Fang et al., 1999). The assembled replisome then stimulates ATP hydrolysis by DnaA through interactions with the β-clamp subunit of the polymerase III holoenzyme (Kurokawa et al., 1999), preventing multiple origin firings (Mizushima et al., 1997; Katayama et al., 1998). Subsequent initiation cycles require the reactivation of ADP-DnaA through nucleotide exchange. This process is slow in solution and appears to be stimulated through direct interactions with acidic membrane lipids in vitro (Sekimizu and Kornberg, 1988) and in vivo (Crooke, 2001; Zheng et al., 2001).
While no in vitro origin-processing assay has been developed fully for eukaryotic DNA replication initiation, many aspects of this complex process have nonetheless been elucidated. In Saccharomyces cerevisiae, replication initiation sites have been mapped to specific cis elements known as autonomous replication sequences (ARSs) (Bell, 1995). ARS elements are recognized by an ensemble of proteins, conserved among all eukaryotes, which includes the multicomponent origin recognition complex (ORC) and other initiation factors such as Cdc6p, Cdt1p, Cdc48p and the MCM replicative helicases (Bell, 2002). The six-subunit ORC assembly (Bell and Stillman, 1992) contains three subunits (Orc1p, Orc4p and Orc5p) that, like DnaA, each possess a single AAA+-type ATP-binding module. ORC binds specific origin sites and, in conjunction with another AAA+ protein (Cdc6p), processes replication origins and recruits the MCM replicative helicase complex to initiation sites (Bell et al., 1995; Bell, 2002). The underlying events leading to origin processing appear to be conserved in other eukaryotes such as Schizosaccharomyces pombe, Drosophila melanogaster and Xenopus laevis, even though these organisms’ origins of replication are not as compact and defined as that of S.cerevisiae (Bell, 2002).
The decoding of archaeal genomes has revealed that these organisms likewise possess AAA+ replication initiation proteins. Archaeal initiators, however, are homologous to eukaryotic factors and related only weakly to their bacterial equivalents at the amino acid sequence level (Edgell and Doolittle, 1997). In addition, rather than possessing a full complement of all MCM, ORC and Cdc6p subunits, archaea instead retain a pared down ensemble of such factors; for example, most archaeal organisms possess only a single copy of a protein that is approximately equally related to both Orc1p and Cdc6p (Edgell and Doolittle, 1997; Bernander, 2000; Liu et al., 2000). Despite these similarities to their eukaryotic counterparts, recent studies reveal that the origin of replication in at least one member of the euryarchaeotal kingdom is bacteria-like in its overall organization (Myllykallio et al., 2000). These findings have, therefore, hinted that commonalities exist between the origin-processing functions among the three domains of life (Bernander, 2000; Kelman, 2000), although the extent of mechanistic conservation between them at the structural level has not been defined.
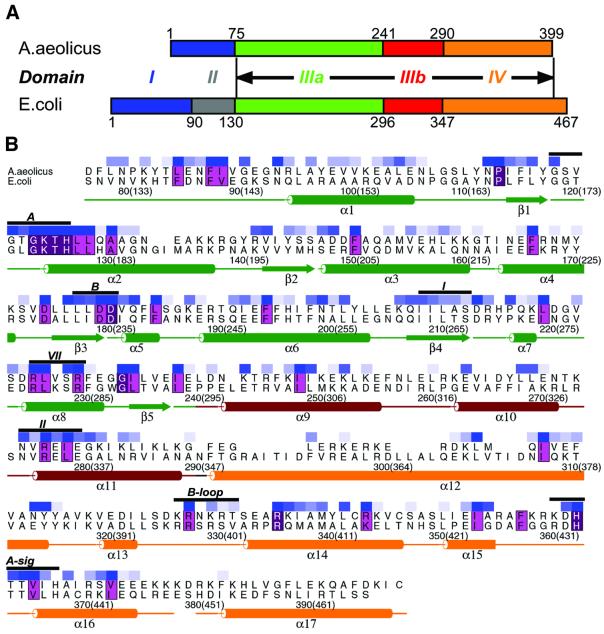
To elucidate DnaA function further and to help compare and contrast the origin-processing proteins of different cellular organisms, we have initiated structural studies of the DnaA protein. Genetic and biochemical data have already allowed for the diverse functions of E.coli DnaA to be assigned to four distinct regions of the protein (Sutton and Kaguni, 1997a; Messer et al., 1999) (Figure 1A): (I) an N-terminal module that is involved in the recruitment of DnaB; (II) an extended linker segment; (III) an ATP-binding cassette of the AAA+ family of macromolecular remodeling factors; and (IV) a C-terminal DNA-binding domain that recognizes a conserved 9 bp DNA recognition sequence (TTA/TTNCACC) within oriC known as the DnaA box (Speck et al., 1997). Comparative sequence alignments of 67 DnaA proteins found in the non-redundant protein database define the AAA+ and DNA-binding domains as a conserved core present in all bacterial DnaA proteins. Moreover, in E.coli, while equivalent DnaA domain III/IV constructs are unable to recruit DnaB to the origin, they are both necessary and sufficient to generate the nucleoprotein complex and unwind the DUE (Sutton et al., 1998; Speck and Messer, 2001). Additional studies have also indicated that domains I and II are dispensable for the multimerization of DnaA in the presence of ADP or ATP (Messer et al., 2001). Together, these findings indicate that DnaA oligomerization and DNA binding/remodeling functions are contained within domains III and IV. Here, we present the three-dimensional structure of conserved core domains III/IV from DnaA of the thermophilic bacterium Aquifex aeolicus (Figure 1A), and discuss its implications for DnaA function and for replication origin processing in archaea and eukaryotes.
Fig. 1. DnaA domain organization and secondary structure. (A) Comparison of the domain organization of E.coli and A.aeolicus DnaA proteins. Domains are numbered according to earlier boundary predictions (Schaper and Messer, 1997). The boundary between domains III and IV has been adjusted using the DnaA structure. Black arrows define the extent of the conserved DnaA core region described in this study that spans the C-terminal 324 residues of A.aeolicus DnaA. There is 35% identity and 65% similarity between A.aeolicus DnaA and the E.coli ortholog across this region. Full-length A.aeolicus DnaA lacks domain II and has a poorly conserved domain I (15% identity compared with E.coli). (B) Sequence alignment and secondary structure elements of the conserved core of A.aeolicus and E.coli DnaA. Aquifex aeolicus residue numbers and their E.coli equivalents (in parentheses) are indicated. Secondary structure elements are in excellent agreement with previously published predictions (Messer et al., 1999). The coloring of the boxes above the residues reflects the degree of conservation at each position among all 67 currently available DnaA sequences, as determined by Clustal X Q scores, with dark blue representing high conservation and lighter blue shadings indicating progressively less conservation. Residues with a Q score <0.5 remain uncolored (see Materials and methods). In addition, invariant residues are highlighted in magenta and residues with strong chemical conservation in pink. Black bars indicate conserved sequence motifs: A, Walker A; B, Walker B; I, Sensor I; II, Sensor II; B-loop, DnaA basic loop; A-sig, DnaA signature sequence.
Results and discussion
Overall structure of DnaA
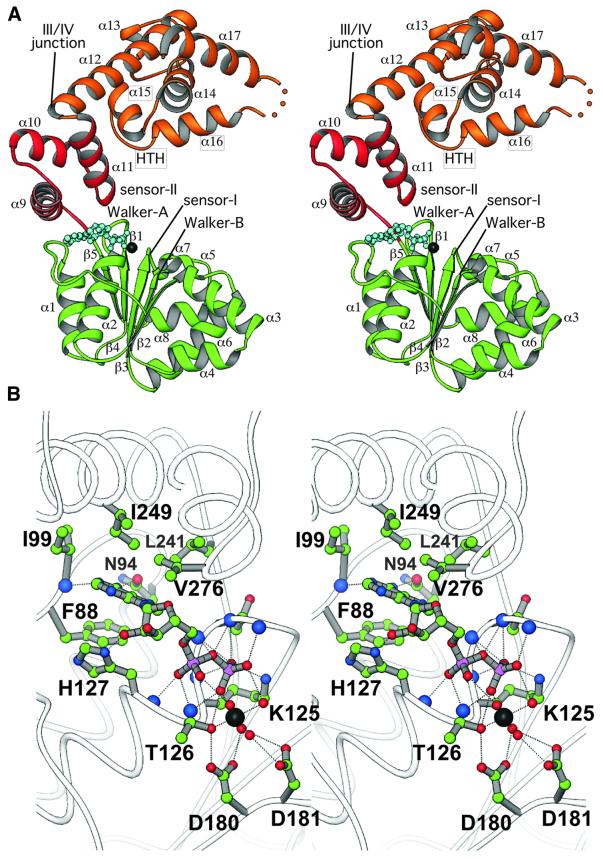
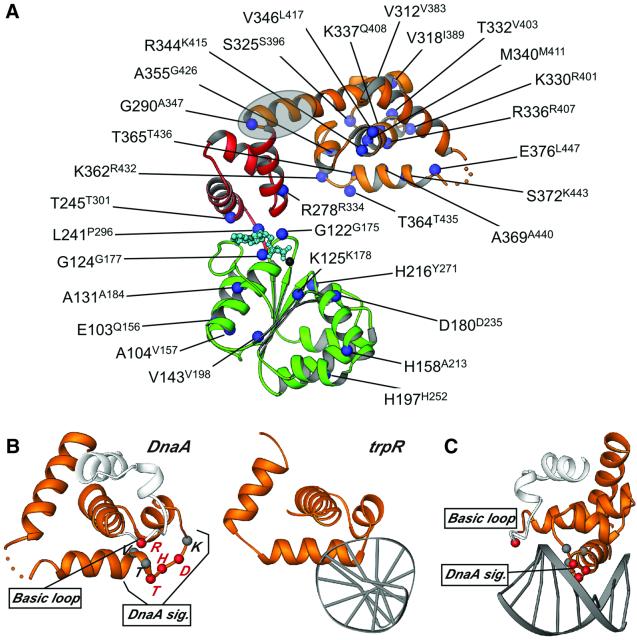
Aquifex aeolicus DnaA was solved to 2.7 Å resolution in the presence of ADP by multiwavelength anomalous dispersion (MAD) with selenomethionine-labeled protein (Table I). The DnaA core is an elongated protein of dimensions 70 × 55 × 25 Å with three distinct structural regions (Figures 1B and 2A). In keeping with the original nomenclature proposed for E.coli DnaA, we will refer to these regions as domains IIIa, IIIb and IV. Domain IIIa is a five-stranded parallel β-sheet (β1–β5) sandwiched on one side by helices α1 and α2 and on the other by helices α3–α8. The organization of domain IIIa is an abbreviated RecA-type fold common to many nucleotide-binding proteins. Domain IIIb is an antiparallel three-helix bundle connected to domain IIIa through a short linker segment. The Walker A, Walker B and sensor I sequence motifs of domain IIIa, together with the sensor II region of domain IIIb, coordinate ADP and a Mg2+ ion within the nucleotide-binding site (Figure 2). This organization, together with additional conserved motifs, is typical of the AAA+ family of ATPases (Neuwald et al., 1999). Extensive mutagenesis analysis of E.coli DnaA confirms the importance of ATP binding and hydrolysis in the function of this protein, as many temperature-sensitive mutations, including some of the ‘classic’ DnaA mutants, map to the domain IIIa/b region (Figure 3A). A summary of available mutagenesis data for E.coli DnaA, correlated with A.aeolicus amino acid positions, is shown in Table II.
Table I. Structure determination statistics.
| Data collection | |||
| Crystal I (remote) | Crystal II (peak) | ||
| Radiation (Å) | 1.1272 | 0.9797 | |
| Resolution (Å) | 30–2.7 | 30–3.0 | |
| Completeness % (last shell) | 98.6 (98.9) | 99.8 (99.0) | |
|
Rsyma % (last shell) |
6.1 (41.5) |
9.0 (40.3) |
|
| Structure refinement (crystal I) | |||
| Resolution range (Å) | 20–2.7 | No. of reflections (overall) | 15 831 |
| No. of non-hydrogen atoms | 2718 | No. of reflections (test set) | 1458 |
| No. of protein atoms | 2638 | Rwork/Rfreeb (%) | 22.9/25.5 |
| No. of ligand atoms | 28 | R.m.s.d.bonds/angles | 0.011 Å/1.17° |
| No. of water atoms | 52 | Ramachandranmost favored/allowed (%) | 88.2/11.8 |
aRsym = ΣΣj|Ij – <I>|/ΣIj, where Ij is the intensity measurement for reflection j and <I> is the mean intensity for multiple recorded reflections.
bRwork, free = Σ||Fobs| – |Fcalc||/Σ|Fobs|, where the working and free R factors are calculated using the working and free reflection sets, respectively. The free reflections (8.4% of the total) were held aside throughout refinement.
Fig. 2. Structure of DnaA. (A) Stereo RIBBONS diagram of the DnaA model. Domains are colored as follows: IIIa, green; IIIb, red; IV, gold. The bound nucleotide is shown as a ball-and-stick model. Key nucleotide-binding motifs and the HTH motif of domain IV are labeled. (B) Stereo RIBBONS diagram of the nucleotide binding cleft of DnaA. Residues located within 4 Å of the bound nucleotide and the coordinated Mg2+ ion are labeled and shown as ball-and-stick models.
Fig. 3. (A) RIBBONS diagram highlighting the position of E.coli DnaA mutations mapped to the A.aeolicus DnaA model (blue spheres). Identification and phenotype of mutations can be found in Table II. The shaded oval (gray) marks the location of the α12 hinge region reported to play a role in cardiolipin-mediated effects. (B) RIBBONS diagrams of DnaA domain IV and trp repressor DNA-binding domain/DNA complex (Otwinowski et al., 1988) highlighting the closely related HTH motif in gold. Highly conserved residues in the basic loop and the DnaA signature sequences are indicated by spheres. Residues determined by mutagenesis to be critical for DNA binding in E.coli that map to the HTH and the basic loop motifs are highlighted in red. (C) RIBBONS model of the DnaA–DNA complex based on the trp repressor/DNA complex. The positions of the signature sequence motif and the basic loop are indicated.
Table II. List of E.coli DnaA mutants and their equivalent A.aeolicus residues.
| Escherichia coli mutation (allele) | Aquifex aeolicus equivalent | In vivo defect | In vitro defect | Reference |
|---|---|---|---|---|
| Classic mutations | ||||
| A184V, H252Y (dnaA46) | A131, H197 | TS alleles | N/A | Hansen et al. (1992) |
| A184V, G426S (dnaA5) | A131, A355 | TS alleles | N/A | Hansen et al. (1992) |
| A184V, P296Q (dnaA601/602) | A131, L241 | TS alleles | N/A | Hansen et al. (1992) |
| A184V, A347V (dnaA604/606) | A131, G290 | TS alleles | N/A | Hansen et al. (1992) |
| V157E (dnaA167) | A104 | TS alleles | N/A | Hansen et al. (1992) |
| I389N (dnaA203/204) | V318 | TS alleles | N/A | Hansen et al. (1992) |
| V383 (dnaA205) | V312 | TS alleles | N/A | Hansen et al. (1992) |
| M411 (dnaA211) | M340 | TS alleles | N/A | Hansen et al. (1992) |
| Q156L, A184V, H252Y, Y271H (dnaAcos) | E103, A131, H197, H216 | Replication overinitiation | N/A | Braun et al. (1987) |
| A213D (dnaA73) | H158 | Cold sensitive. Suppressors of dnaX(Ts) strain | N/A | Gines-Candelaria et al. (1995) |
| R432L (dnaA721) | K362 | Cold sensitive. Suppressors of dnaX(Ts) strain | DNA-binding defect | Blaesing et al. (2000); Gines-Candelaria et al. (1995) |
| T435K (dnaA71) | T364 | Cold sensitive. Suppressors of dnaX(Ts) strain | DNA-binding defect | Blaesing et al. (2000); Gines-Candelaria et al. (1995); Sutton and Kaguni (1997b) |
| G426S | A355 | N/A | DNA-binding defect | Carr and Kaguni (1996) |
| A184V |
A131 |
Asynchronous replication initiation |
ATP-binding defect |
Carr and Kaguni (1996); Skarstad et al. (1988) |
| Domain III | ||||
| G175D | G122 | TS alleles | N/A | Sutton and Kaguni (1997b) |
| G177D | G124 | TS alleles | N/A | Sutton and Kaguni (1997b) |
| A184T | A131 | TS alleles | N/A | Sutton and Kaguni (1997b) |
| K171I | K125 | Non-complementing | ATP binding and oriC unwinding defect | Mizushima et al. (1998) |
| D235N | D180 | Non-complementing | ATP binding and oriC unwinding defect | Mizushima et al. (1998) |
| V198M | V143 | TS allele | Thermolabile | Sutton and Kaguni (1995) |
| T301I | T245 | TS allele | Thermolabile | Sutton and Kaguni (1995) |
| R334A |
R278 |
Overinitiation |
Defective ATP hydrolysis |
Nishida et al. (2002) |
| Domain IV | ||||
| S396F | S325 | N/A | DNA-binding defect | Blaesing et al. (2000) |
| R401A | K330 | N/A | DNA-binding defect | Blaesing et al. (2000) |
| V403A | T332 | N/A | DNA-binding defect | Blaesing et al. (2000) |
| R407A | R336 | N/A | DNA-binding defect | Blaesing et al. (2000) |
| Q408R | K337 | N/A | DNA-binding defect | Blaesing et al. (2000) |
| K415E | R344 | N/A | DNA-binding defect | Blaesing et al. (2000) |
| L417P | V346 | N/A | DNA-binding defect | Blaesing et al. (2000) |
| T436A | T365 | N/A | DNA-binding defect | Blaesing et al. (2000) |
| A440V | A369 | N/A | DNA-binding defect | Blaesing et al. (2000) |
| K443E | S372 | N/A | DNA-binding defect | Blaesing et al. (2000) |
| L447W | E376 | TS allele | DNA-binding defect | Sutton and Kaguni (1995) |
Phenotypic defects are indicated for both in vivo and in vitro studies.
TS, temperature sensitive; N/A, data not available.
DNA-binding domain
Domain IV, the DNA-binding domain, comprises a long connector helix (α12) linked to a helix–turn–helix (HTH) motif (α15 and α16) that is buttressed by two additional helices (α14 and α17). There are no significant intraprotomer contacts between the globular regions of domains III and IV, and the position of domain IV appears to be solely stabilized by interactions with adjacent monomers in the crystal lattice. In addition, two and a half helical turns of the domain IV connector helix are solvent exposed (residues E295V359–K304K372, where corresponding E.coli residue numbers are indicated in superscript italics next to A.aeolicus amino acid positions), and the overall B factors of domain IV are higher than those of domains IIIa and IIIb. Together, these observations suggest that a significant degree of flexibility is present in helix α12 of the protein and that a hinge point probably exists at the domain III/IV boundary.
A DALI search (Holm and Sander, 1996) reveals that the closest structural homolog of domain IV is the DNA-binding domain of the trp operon repressor (r.m.s.d. of 2.8 Å over 61 residues), a member of the NarL/FixJ family of DNA-binding proteins (CATH code 1.10.10.60) (Orengo and Taylor, 1996) (Figure 3B). To gain insight into how DnaA might interact with target DNA sequences, we modeled DNA on the domain IV HTH using the trp repressor/DNA structure (Otwinowski et al., 1988) as a guide (Figure 3C). Direct modeling of duplex DNA onto domain IV produces a modest steric clash with the AAA+ region; however, this overlap can be relieved by a small rigid body swivel motion of domain IV about the domain III/IV connector hinge. The domain IV/DNA model is consistent with biochemical and genetic studies that define two key functional elements of DnaA required for efficient DNA binding. The first element, considered the DnaA signature sequence motif (KDHTTVI in A.aeolicus), has been shown by extensive mutational analyses to define both the affinity and specificity of DnaA box binding (Sutton and Kaguni, 1997b; Blaesing et al., 2000). This sequence maps to the α15–α16 junction and falls within the turn of the HTH motif that is positioned in the major groove of the modeled DNA (Figure 3B and C). The second element is a conserved region known as the ‘basic loop’, which contains an arginine (R328R399) essential for DNA binding (Blaesing et al., 2000). This element is proximal to the HTH motif, and our model suggests that it may be important in mediating DNA backbone or minor groove interactions (Figure 3C), perhaps helping to induce DNA conformational changes thought to occur upon binding (Schaper and Messer, 1995).
Oligomerization determinants
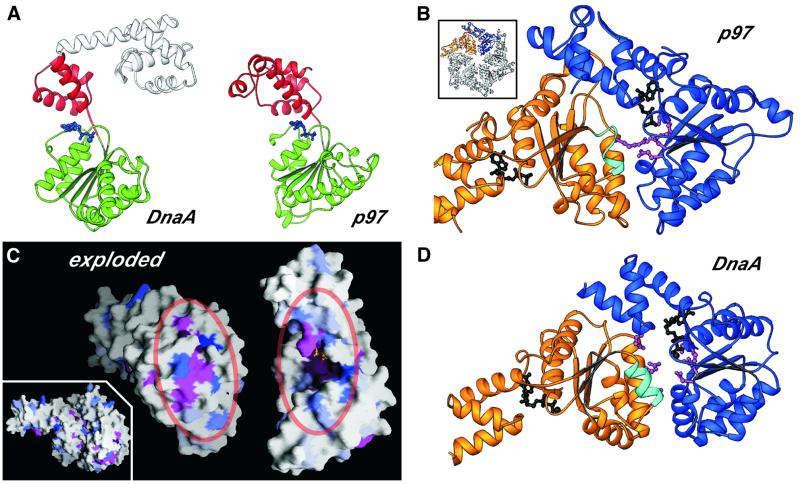
In addition to its ability to bind DNA, self-oligomerization is a second critical feature of the DnaA origin-processing function. An inspection of the crystal packing contacts between A.aeolicus DnaA protomers did not reveal potential DnaA oligomerization interfaces. Nevertheless, the inclusion of DnaA in the AAA+ family of molecular remodeling factors (Neuwald et al., 1999) allows us to infer how DnaA oligomerization may be mediated. A search of the structural database (Holm and Sander, 1996), using domain III of DnaA as a probe, identified the closest structural homologs of DnaA among known oligomeric AAA+ proteins to be p97 and NSF (Figure 4A), which are hexameric proteins involved in the disassembly of SNARE complexes during membrane fusion (Lenzen et al., 1998; Yu et al., 1998; Zhang et al., 2000). The oligomeric organization of p97 and NSF is related, and is also seen in other oligomeric AAA+ proteins such as the HslU protease and the bacterial and archaeal clamp loader complexes (Bochtler et al., 2000; Jeruzalmi et al., 2001; Oyama et al., 2001).
Fig. 4. (A) RIBBONS diagram of DnaA and p97 (residues 191–458) showing the high degree of structural conservation across the AAA+ domain of the two proteins (the r.m.s.d. between the two proteins is 3.2 Å over 192 Cα positions). (B) RIBBONS diagram of the AAA+ region of a p97 dimer excised from the hexameric structure (inset) (Zhang et al., 2000) depicting the typical oligomeric arrangement of AAA+ protomers. The ADP bound at the interface is shown in black. The helix containing the Box VII motif is shown in cyan and the key arginine residue present at the dimerization interface is depicted as a magenta ball-and-stick model. (C) Inset: surface depiction (Nicholls et al., 1991) of the DnaA dimer modeled on p97, showing the high degree of complementarity between the monomers. The structural alignment of the AAA+ regions was generated using least-squares fitting of the DnaA AAA+ domain on each of two neighboring p97 AAA+ domains from the p97 hexamer. The exploded view reveals the clustering of conserved residues at the dimerization interface to form the bipartite nucleotide-interaction site. The degree of conservation among all known DnaA sequences is indicated by the degree of blue shading (Figure 1B); invariant (magenta) and chemically conserved residues (pink) are also highlighted. (D) RIBBONS diagram of the DnaA AAA+ domain (residues D77S130–G290N348) model dimer shown in blue and gold. Critical elements present at the predicted dimer interface are highlighted as in Figure 4B.
All oligomeric AAA+ proteins structurally characterized to date appear to share a common mode of assembly, in which the protomer of one AAA+ protein projects residues from a conserved motif known as Box VII (Neuwald et al., 1999) into the nucleotide binding cleft of a neighboring protomer, forming a bipartite ATP-interaction site (Figure 4B). A particularly important feature of this interface is the interaction of a conserved arginine residue in the Box VII motif with the γ-phosphate of the bound nucleotide in the adjacent monomer (Figure 4B) (Lenzen et al., 1998; Yu et al., 1998; Bochtler et al., 2000). Nucleotide hydrolysis in this site is thought to mediate allosteric effects within AAA+ oligomers, although the oligomerization interfaces are not radically altered by the presence of the γ-phosphate, as demonstrated by the fact that the hexameric structures of NSF and p97 are quite similar, even though NSF is bound to AMP-PNP or ATP, and p97 to ADP (Lenzen et al., 1998; Yu et al., 1998; Zhang et al., 2000).
To determine whether the DnaA structure could accommodate an AAA+-like oligomerization state, we modeled DnaA onto the p97 hexamer (Figure 4C and D). Such an approach has been used previously to identify the oligomerization surface of RuvB, an AAA+ protein involved in DNA branch migration (Putnam et al., 2001; Yamada et al., 2001), as well as to interpret mutational data obtained for the AAA+ protease FtsH (Karata et al., 1999). Our resulting DnaA model displays no significant clashes between adjacent AAA+ modules (Figure 4A and B) and only two minor overlaps between domains IIIb and IV of adjacent protomers, which can be readily relieved by the same swiveling motion of domain IV about the domain III–IV connector hinge used to accommodate our DNA-binding model. Significantly, a high degree of general conservation (Figure 4C) and charge/hydrophobicity complementarity (data not shown) is observed along the predicted DnA interaction surface. Moreover, our oligomeric model places two Box VII residues of DnaA, R226R281 and R230R285, within 4 Å of the nucleotide-binding pocket of an adjacent monomer, in an arrangement that resembles closely the interaction of analogous regions of p97 (Figure 4B and D). These arginines are invariant in 66 out of 67 DnaA proteins and are substituted by histidines only in Caulobacter crescentus. Our DnaA oligomerization model thus offers a framework to explain the ability of domains III and IV of DnaA to support self-association.
Origin DNA binding and nucleoprotein complex formation by DnaA
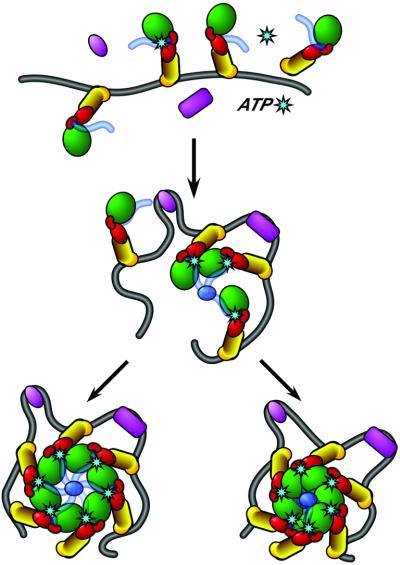
Bacterial replication origin processing begins with the assembly of a number of DnaA molecules on specific DNA-binding elements within oriC (Figure 5). The monomer structure of DnaA suggests how the processes of DNA binding and oligomerization may be carried out during the initiation event. Binding of DnaA to oriC is mediated initially by associations of the HTH and basic loop motifs in domain IV with appropriate DnaA boxes. The presence of multiple DnaA boxes within oriC localizes several DnaA molecules in close proximity to one another, fostering the association of both domain I and domains IIIa/IIIb. Based on the strong conservation of sequence motifs, domain III interactions are expected to form bipartite nucleotide binding sites similar to those seen in other AAA+ oligomers.
Fig. 5. Model for DnaA assembly on oriC. (Top) oriC is recognized by DnaA (red, green and yellow) and architectural factors such as IHF and HU (purple). (Middle) Concomitant with oriC binding, the AAA+ domains oligomerize, stabilizing the nucleoprotein complex through intermonomer contacts around the ATP-binding site. Additional stability may be provided by domain I self-oligomerization (light blue). (Bottom) Self-assembly of DnaA molecules eventually leads to formation of the complete nucleoprotein complex. Note that the DnaA oligomer could conceivably accommodate either a closed ring (left) or a helical filament (right) arrangement of monomers. DUE opening may occur spontaneously through local strain induced by assembly of the nucleoprotein complex in the presence of ATP.
We envisage that during this process, domain IV is only loosely tethered through α12 to the rest of DnaA, so as to allow the protein to bind DnaA boxes with different relative spacings and orientations within oriC, while also accommodating the spatial configurations necessary for the AAA+ domains to associate. Indeed, the mobile properties of domain IV may be more pronounced in DnaA proteins from organisms other than A.aeolicus, as the connector helix is significantly longer in all other DnaA homologs (Figures 1B and 2A; data not shown). The connector helix may also play a role in regulating the conformational states induced by nucleotide binding and hydrolysis. Studies have shown that the E.coli connector is less susceptible to tryptic degradation in the presence of ATP (Carr and Kaguni, 1996) and that this region of the protein is important for lipid-mediated nucleotide exchange in E.coli DnaA (Garner et al., 1998; Makise et al., 2000; Zheng et al., 2001), suggesting that DnaA reactivation may be mediated through a regulatory link between the connector helix region and the nucleotide-binding site. The concept of the connector helix as an effector element that mediates the assembly and disassembly of the nucleoprotein complex may also explain why mutations in this region of the protein have been particularly difficult to interpret (Makise et al., 2000; Zheng et al., 2001).
Comparison of bacterial and archaeal initiation factors
Genomic sequence mining has shown that, within the eukaryotic and archaeal kingdoms, several replication initiation factors share the AAA+ ATP-binding motif also found in DnaA. Beyond those sequence motifs, however, the degree to which architectural conservation exists between bacterial and eukaryotic/archaeal replication initiatiors has been unclear. While no high-resolution models of ORC are currently available, structural studies of an archaeal Cdc6/Orc1 protein have shown that the AAA+ region of this molecule is linked to a winged-helix domain (WHD) at its C-terminus (Liu et al., 2000). The presence of the WHD suggested that the protein may bind origin DNA directly, an activity known to be possessed by its eukaryotic Orc1 ortholog. Moreover, in at least one kingdom of archaea, the euryarchaeota, Cdc6/Orc1 has been shown to associate with a bacterial-like replication origin (Myllykallio et al., 2000; Matsunaga et al., 2001). These findings have led to speculation that the archaeal Cdc6/Orc1 protein may possess cellular functions similar to those of DnaA (Bernander, 2000; Lee and Bell, 2000; Liu et al., 2000; Myllykallio et al., 2000).
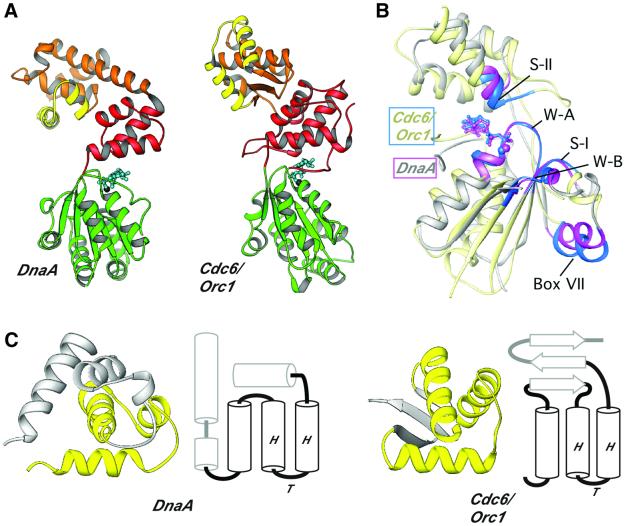
This hypothesis is supported by the striking degree of similarity between the structures and general domain organization of DnaA and Cdc6/Orc1 (Figure 6A). Both proteins possess helical domains (DnaA domain IV and Cdc6/Orc1 WHD) that are fused C-terminally to AAA+ modules. Interestingly, although these C-terminal elements belong to the same HTH topological superfamily (Figure 6C), differences in their folds argue against a strict evolutionary relationship. Rather, it seems that a conversion or rearrangement of HTH modules may have occurred during evolution. In this view, we would propose that the C-terminal HTH domains of these initiator proteins serve as origin-localization elements (OLEs) for a common AAA+ remodeling machinery.
Fig. 6. Structural comparison of DnaA and Cdc6/Orc1. (A) RIBBONS diagrams of DnaA and its closest structural homolog, Cdc6/Orc1. Domains are labeled as in Figure 2A, except that the HTH motifs are highlighted in yellow. (B) RIBBONS diagram superimposing the AAA+ regions of DnaA (light gray) and Cdc6/Orc1 (pale yellow). Elements critical for the spatial conservation of the bipartite ATP-interaction cleft are highlighted in magenta (DnaA) and blue (Cdc6/Orc1). The two proteins exhibit an r.m.s.d. of 2.3 Å across 139 of the 213 Cα residues that are present in core secondary structure elements. (C) Comparisons of DnaA and Cdc6/Orc1 OLEs. The RIBBONS diagram shows the related HTH topology (in yellow) of the C-terminal modules next to secondary structure topology diagrams that indicate how the HTH motif is anchored within each C-terminal fold. Secondary structure comparisons were performed using the sequential structure alignment program SSAP (Orengo and Taylor, 1996).
The ability to ‘reprogram’ OLEs could provide a mechanism by which similar origin-processing AAA+ modules have been adapted to novel origin sites. The idea that the ATP-dependent remodeling cores have been conserved in DnaA and Cdc6/Orc1 is bolstered by the close structural similarity of the proteins’ AAA+ folds to each other (Figure 6A). Indeed, the closest structural relative of DnaA domain III among all AAA+ proteins found in the RCSB database is archaeal Cdc6/Orc1 (Figure 6B), even though the two proteins share only 15% sequence identity over this region. The spatial arrangement of DnaA and Cdc6/Orc1 AAA+ motifs, including the Box VII segment (Figure 6B), is correlated particularly strongly. Taken together, the structural data suggest that DnaA and Cdc6/Orc1 may form comparable higher-order oligomeric structures on origin sequences, and that nucleotide-dependent effects within a given nucleoprotein complex may manifest themselves similarly at the structural level. Given the degree of homology between archaeal and eukaryotic processing factors, these considerations also seem applicable to eukaryotic replication initiation.
Biochemical and genomic studies have hinted at the existence of universal mechanistic commonalities in the process of DNA replication initiation, but structural evidence of such similarities between components of the initiation process has been lacking. Although many bacterial replication proteins do not share high sequence similarity to their archaeal or eukaryotic counterparts, structural conservation nevertheless exists between many replication proteins of similar function, as seen, for example, in the processivity clamps and single-stranded DNA-binding proteins (Kong et al., 1992; Krishna et al., 1994; Shamoo et al., 1995; Bochkarev et al., 1997). The structure of the bacterial DnaA and its architectural relationship to archaeal Cdc6/Orc1 extend these evolutionary relationships further, and suggest that common functional themes are likely to continue to emerge among the origin-processing proteins of all domains of life.
Materials and methods
Protein purification
The sequence encoding residues 76–399 of A.aeolicus DnaA was PCR amplified from genomic DNA and cloned into vector pSV272, generating a His6-MBP fusion construct with a linker encoding the TEV-protease cleavage sequence. Seleno-methionine (Se-Met) His6-MBP–DnaA fusion protein was expressed in BL21 RIL cells (Novagen). Cells were grown at 37°C in minimal media supplemented with Se-Met and induced at an A600 of 0.3 with 0.1 mM IPTG for 4–5 h. After centrifugation, the cells were resuspended in lysis buffer [50 mM HEPES pH 7.5, 500 mM KCl, 10% glycerol, 1 µM pepstatin-A, 1 µM leupeptin, 1 mM PMSF, 5 mM β-mercaptoethanol (β-ME) and 50 µg/ml of lysozyme] and lysed by sonication. Se-Met His6-MBP–DnaA was purified using Ni-affinity chromatography (POROS MC) and ion-exchange chromatography (HT Heparin). Purified Se-Met His6-MBP–DnaA was then cleaved with His6-tagged TEV protease (Kapust and Waugh, 1999) for 20 h at 4°C in cleavage buffer (50 mM HEPES 7.5, 1 M KCl, 15% glycerol, 5 mM β-ME, 5 mM MgCl2 and 1 mM ADP). After orthogonal purification through Ni-affinity (Qiagen) and amylose columns (NEBiolabs) to remove uncleaved product, Se-Met DnaA was concentrated and applied to an S200 gel filtration column in cleavage buffer plus 1 mM Tris (2-carboxyethyl) phosphine (TCEP). Similar to its E.coli ortholog (Schaper and Messer, 1997), purified A.aeolicus DnaA is a monomer as judged by gel-filtration chromatography. The presence of Se-Met substitutions was verified by mass spectrometry.
Crystallization
Following gel filtration, Se-Met DnaA was concentrated to 10 mg/ml and dialyzed overnight against 10 mM HEPES, 400 mM KCl, 25 mM MgCl2, 5 mM ADP and 1 mM TCEP. Se-Met DnaA-ADP was crystallized by hanging-drop vapor diffusion by mixing 1 µl of protein and 1 µl of well solution (100 mM NaOAc pH 5.0, 14% hexanediol, 15% PEG MME 2000 and 1 mM TCEP). Before sealing the well, 100 µl of 4 M KCl were added to the well solution (1 ml). Crystals (∼200 × 200 × 75 µm) grew overnight and reached maximal size after 2–3 days. Crystals were transferred to harvesting solution (well solution plus 11% PEG MME 550) for 2–5 min and flash frozen in liquid nitrogen.
Data collection
Data were collected at Beamline 8.3.1 at the Advanced Light Source (ALS) and processed using Elves (J.M.Holton and T.A.Alber, manuscript in preparation) and Denzo/Scalepack (Otwinowski and Minor, 1997). The crystals belong to the I23 space group with unit cell dimensions a = b = c = 155.75 Å and a solvent content of ∼70%. There is one DnaA monomer per asymmetric unit.
Structure determination and refinement
Positions of selenium sites were determined using Elves and Solve (Terwilliger and Berendzen, 1999) and initial maps generated using DM (Cowtan, 1994). Additional maps for model building were obtained using Sharp (de La Fortelle and Bricogne, 1997) and the model was built using o (Jones et al., 1991). Refinement was performed using REFMAC (Lamzin and Wilson, 1993). The refined model includes all residues except for K76, K379 and D380, and has no residues in either the generously allowed or disallowed regions of the Ramachandran plot as determined by PROCHECK (Laskowski et al., 1993). Sequence alignments of DnaA protein sequences identified by PSI-BLAST (Altschul et al., 1997) searches against the non-redundant protein database were generated with Clustal X (Thompson et al., 1997) using a Gonnet 350 matrix (Benner et al., 1994). Figures were generated using RIBBONS (Carson, 1991) and ALSCRIPT (Barton, 1993).
Coordinates and structure factors
Coordinates and structure factors have been deposited with the RCSB database under accession code 1L8Q.
Acknowledgments
Acknowledgements
We would like to thank James Holton and Emmanuel Skordalakes for data collection assistance at ALS Beamline 8.3.1, Dave King for mass spectrometry analysis, Navin Pokala for the pSV272 vector, Jon Kaguni for helpful comments, and James Keck, Mike Botchan, Nick Cozzarelli and Berger laboratory members for critical reading of the manuscript. This work was generously supported by the G.Harold and Leila Y.Mathers Charitable Foundation and the NCI (CA77373).
References
- Altschul S.F., Madden,T.L., Schäffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton G.J. (1993) ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng., 6, 37–40. [DOI] [PubMed] [Google Scholar]
- Bell S.P. (1995) Eukaryotic replicators and associated protein complexes. Curr. Opin. Genet. Dev., 5, 162–167. [DOI] [PubMed] [Google Scholar]
- Bell S.P. (2002) The origin recognition complex: from simple origins to complex functions. Genes Dev., 16, 659–672. [DOI] [PubMed] [Google Scholar]
- Bell S.P. and Stillman,B. (1992) ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature, 357, 128–134. [DOI] [PubMed] [Google Scholar]
- Bell S.P., Mitchell,J., Leber,J., Kobayashi,R. and Stillman,B. (1995) The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell, 83, 563–568. [DOI] [PubMed] [Google Scholar]
- Benner S.A., Cohen,M.A. and Gonnet,G.H. (1994) Amino acid substitution during functionally constrained divergent evolution of protein sequences. Protein Eng., 7, 1323–1332. [DOI] [PubMed] [Google Scholar]
- Bernander R. (2000) Chromosome replication, nucleoid segregation and cell division in archaea. Trends Microbiol., 8, 278–283. [DOI] [PubMed] [Google Scholar]
- Blaesing F., Weigel,C., Welzeck,M. and Messer,W. (2000) Analysis of the DNA-binding domain of Escherichia coli DnaA protein. Mol. Microbiol., 36, 557–569. [DOI] [PubMed] [Google Scholar]
- Bochkarev A., Pfuetzner,R.A., Edwards,A.M. and Frappier,L. (1997) Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature, 385, 176–181. [DOI] [PubMed] [Google Scholar]
- Bochtler M., Hartmann,C., Song,H.K., Bourenkov,G.P., Bartunik,H.D. and Huber,R. (2000) The structures of HsIU and the ATP-dependent protease HsIU-HsIV. Nature, 403, 800–805. [DOI] [PubMed] [Google Scholar]
- Braun R.E., O’Day,K. and Wright,A. (1987) Cloning and characterization of dnaA(Cs), a mutation which leads to over initiation of DNA replication in Escherichia coli K-12. J. Bacteriol., 169, 3898–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr K.M. and Kaguni,J.M. (1996) The A184V missense mutation of the dnaA5 and dnaA46 alleles confers a defect in ATP binding and thermolability in initiation of Escherichia coli DNA replication. Mol. Microbiol., 20, 1307–1318. [DOI] [PubMed] [Google Scholar]
- Carr K.M. and Kaguni,J.M. (2001) Stoichiometry of DnaA and DnaB protein in initiation at the Escherichia coli chromosomal origin. J. Biol. Chem., 276, 44919–44925. [DOI] [PubMed] [Google Scholar]
- Carson M. (1991) RIBBONS 2.0. J. Appl. Crystallogr., 24, 958–961. [Google Scholar]
- Cowtan K. (1994) ‘DM’: an automated procedure for phase improvement by density modification. Joint CCP4 ESF-EACBM Newslett. Protein Crystallogr., 31, 34–38. [Google Scholar]
- Crooke E. (2001) Escherichia coli DnaA protein–phospholipid interactions: in vitro and in vivo. Biochimie, 83, 19–23. [DOI] [PubMed] [Google Scholar]
- de La Fortelle E. and Bricogne,G. (1997) Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol., 276, 472–494. [DOI] [PubMed] [Google Scholar]
- Edgell D.R. and Doolittle,W.F. (1997) Archaea and the origin(s) of DNA replication initiation. Cell, 87, 995–998. [DOI] [PubMed] [Google Scholar]
- Fang L., Davey,M.J. and O’Donnell,M. (1999) Replisome assembly at oriC, the replication origin of E.coli, reveals an explanation for initiation sites outside an origin. Mol. Cell, 4, 541–553. [DOI] [PubMed] [Google Scholar]
- Funnell B.E., Baker,T.A. and Kornberg,A. (1987) In vitro assembly of a prepriming complex at the origin of the Escherichia coli chromosome. J. Biol. Chem., 262, 10327–10334. [PubMed] [Google Scholar]
- Garner J., Durrer,P., Kitchen,J., Brunner,J. and Crooke,E. (1998) Membrane-mediated release of nucleotide from an initiator of chromosomal replication, Escherichia coli DnaA, occurs with insertion of a distinct region of the protein into the lipid bilayer. J. Biol. Chem., 273, 5167–5173. [DOI] [PubMed] [Google Scholar]
- Gines-Candelaria E., Blinkova,A. and Walker,J.R. (1995) Mutations in Escherichia coli dnaA which suppress a dnaX(Ts) polymerization mutation and are dominant when located in the chromosomal allele and recessive on plasmids. J. Bacteriol., 177, 705–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimwade J.E., Ryan,V.T. and Leonard,A.C. (2000) IHF redistributes bound initiator protein, DnaA, on supercoiled oriC of Escherichia coli. Mol. Microbiol., 35, 835–844. [DOI] [PubMed] [Google Scholar]
- Hansen F.G., Koefoed,S. and Atlung,T. (1992) Cloning and nucleotide sequence determination of twelve mutant dnaA genes of Escherichia coli. Mol. Gen. Genet., 234, 14–21. [DOI] [PubMed] [Google Scholar]
- Holm L.L. and Sander,C. (1996) Mapping the protein universe. Science, 273, 595–602. [DOI] [PubMed] [Google Scholar]
- Hwang D.S. and Kornberg,A. (1992) Opening of the replication origin of Escherichia coli by DnaA protein with protein HU or IHF. J. Biol. Chem., 267, 23083–23086. [PubMed] [Google Scholar]
- Jeruzalmi D., O’Donnell,M. and Kuriyan,J. (2001) Crystal structure of the processivity clamp loader γ (γ) complex of E.coli DNA polymerase III. Cell, 106, 429–441. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kapust R.B. and Waugh,D.S. (1999) Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci., 8, 1668–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karata K., Inagawa,T., Wilkinson,A.J., Tatsuta,T. and Ogura,T. (1999) Dissecting the role of a conserved motif (the second region of homology) in the AAA family of ATPases. Site-directed mutagenesis of the ATP-dependent protease FtsH. J. Biol. Chem., 274, 26225–26232. [DOI] [PubMed] [Google Scholar]
- Katayama T., Kubota,T., Kurokawa,K., Crooke,E. and Sekimizu,K. (1998) The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E.coli chromosomal replicase. Cell, 94, 61–71. [DOI] [PubMed] [Google Scholar]
- Kelman Z. (2000) The replication origin of archaea is finally revealed. Trends Biochem. Sci., 25, 521–523. [DOI] [PubMed] [Google Scholar]
- Kong X.P., Onrust,R., O’Donnell,M. and Kuriyan,J. (1992) Three-dimensional structure of the β subunit of E.coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell, 69, 425–437. [DOI] [PubMed] [Google Scholar]
- Kornberg A. and Baker,T.A. (1992) DNA Replication. W.H. Freeman and Co., New York, NY.
- Krishna T.S., Kong,X.P., Gary,S., Burgers,P.M. and Kuriyan,J. (1994) Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell, 79, 1233–1243. [DOI] [PubMed] [Google Scholar]
- Kurokawa K., Nishida,S., Emoto,A., Sekimizu,K. and Katayama,T. (1999) Replication cycle-coordinated change of the adenine nucleotide-bound forms of DnaA protein in Escherichia coli. EMBO J., 18, 6642–6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamzin V.S. and Wilson,K.S. (1993) Automated refinement of protein models. Acta Crystallogr. D, 49, 129–147. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Lee D.G. and Bell,S.P. (2000) ATPase switches controlling DNA replication initiation. Curr. Opin. Cell Biol., 12, 280–285. [DOI] [PubMed] [Google Scholar]
- Lenzen C.U., Steinmann,D., Whiteheart,S.W. and Weis,W.I. (1998) Crystal structure of the hexamerization domain of N-ethylmaleimide-sensitive fusion protein. Cell, 94, 525–536. [DOI] [PubMed] [Google Scholar]
- Liu J., Smith,C.L., DeRyckere,D., DeAngelis,K., Martin,G.S. and Berger,J.M. (2000) Structure and function of Cdc6/Cdc18: implications for origin recognition and checkpoint control. Mol. Cell, 6, 637–648. [DOI] [PubMed] [Google Scholar]
- Makise M., Mima,S., Tsuchiya,T. and Mizushima,T. (2000) Identification of amino acids involved in the functional interaction between DnaA protein and acidic phospholipids. J. Biol. Chem., 275, 4513–4518. [DOI] [PubMed] [Google Scholar]
- Matsunaga F., Forterre,P., Ishino,Y. and Myllykallio,H. (2001) In vivo interactions of archaeal Cdc6/Orc1 and minichromosome maintenance proteins with the replication origin. Proc. Natl Acad. Sci. USA, 98, 11152–11157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer W. et al. (1999) Functional domains of DnaA proteins. Biochimie, 81, 819–825. [DOI] [PubMed] [Google Scholar]
- Messer W. et al. (2001) Bacterial replication initiator DnaA. Rules for DnaA binding and roles of DnaA in origin unwinding and helicase loading. Biochimie, 83, 5–12. [DOI] [PubMed] [Google Scholar]
- Mizushima T., Nishida,S., Kurokawa,K., Katayama,T., Miki,T. and Sekimizu,K. (1997) Negative control of DNA replication by hydrolysis of ATP bound to DnaA protein, the initiator of chromosomal DNA replication in Escherichia coli. EMBO J., 16, 3724–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima T., Takaki,T., Kubota,T., Tsuchiya,T., Miki,T., Katayama,T. and Sekimizu,K. (1998) Site-directed mutational analysis for the ATP binding of DnaA protein. Functions of two conserved amino acids (Lys-178 and Asp-235) located in the ATP-binding domain of DnaA protein in vitro and in vivo. J. Biol. Chem., 273, 20847–20851. [DOI] [PubMed] [Google Scholar]
- Myllykallio H., Lopez,P., Lopez-Garcia,P., Heilig,R., Saurin,W., Zivanovic,Y., Philippe,H. and Forterre,P. (2000) Bacterial mode of replication with eukaryotic-like machinery in a hyperthermophilic archaeon. Science, 288, 2212–2215. [DOI] [PubMed] [Google Scholar]
- Neuwald A.F., Aravind,L., Spouge,J.L. and Koonin,E.V. (1999) AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res., 9, 27–43. [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Nishida S., Fujimitsu,K., Sekimizu,K., Ohmura,T., Ueda,T. and Katayama,T. (2002) A nucleotide switch in the Escherichia coli DnaA protein initiates chromosomal replication: evidence from a mutant DnaA protein defective in regulatory ATP hydrolysis in vitro and in vivo. J. Biol. Chem., 277, 14986–14995. [DOI] [PubMed] [Google Scholar]
- Orengo C.A. and Taylor,W.R. (1996) SSAP: sequential structure alignment program for protein structure comparison. Methods Enzymol., 266, 617–635. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 472–494. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Schevitz,R.W., Zhang,R.G., Lawson,C.L., Joachimiak,A., Marmorstein,R.Q., Luisi,B.F. and Sigler,P.B. (1988) Crystal structure of trp repressor/operator complex at atomic resolution. Nature, 335, 321–329. [DOI] [PubMed] [Google Scholar]
- Oyama T., Ishino,Y., Cann,I.K., Ishino,S. and Morikawa,K. (2001) Atomic structure of the clamp loader small subunit from Pyrococcus furiosus. Mol. Cell, 8, 455–463. [DOI] [PubMed] [Google Scholar]
- Putnam C.D., Clancy,S.B., Tsuruta,H., Gonzalez,S., Wetmur,J.G. and Tainer,J.A. (2001) Structure and mechanism of the RuvB Holliday junction branch migration motor. J. Mol. Biol., 311, 297–310. [DOI] [PubMed] [Google Scholar]
- Schaper S. and Messer,W. (1995) Interaction of the initiator protein DnaA of Escherichia coli with its DNA target. J. Biol. Chem., 270, 17622–17626. [DOI] [PubMed] [Google Scholar]
- Schaper S. and Messer,W. (1997) Prediction of the structure of the replication initiator protein DnaA. Proteins, 28, 1–9. [PubMed] [Google Scholar]
- Sekimizu K. and Kornberg,A. (1988) Cardiolipin activation of dnaA protein, the initiation protein of replication in Escherichia coli. J. Biol. Chem., 263, 7131–7135. [PubMed] [Google Scholar]
- Shamoo Y., Friedman,A.M., Parsons,M.R., Konigsberg,W.H. and Steitz,T.A. (1995) Crystal structure of a replication fork single-stranded DNA binding protein (T4 gp32) complexed to DNA. Nature, 376, 362–366. [DOI] [PubMed] [Google Scholar]
- Skarstad K., von Meyenburg,K., Hansen,F.G. and Boye,E. (1988) Coordination of chromosome replication initiation in Escherichia coli: effects of different dnaA alleles. J. Bacteriol., 170, 852–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck C. and Messer,W. (2001) Mechanism of origin unwinding: sequential binding of DnaA to double- and single-stranded DNA. EMBO J., 20, 1469–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck C., Weigel,C. and Messer,W. (1997) From footprint to toeprint: a close-up of the DnaA box, the binding site for the bacterial initiator protein DnaA. Nucleic Acids Res., 25, 3242–3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton M.D. and Kaguni,J.M. (1995) Novel alleles of the Escherichia coli dnaA gene are defective in replication of pSC101 but not of oriC. J. Bacteriol., 177, 6657–6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton M.D. and Kaguni,J.M. (1997a) The Escherichia coli dnaA gene: four functional domains. J. Mol. Biol., 274, 546–561. [DOI] [PubMed] [Google Scholar]
- Sutton M.D. and Kaguni,J.M. (1997b) Threonine 435 of Escherichia coli DnaA protein confers sequence-specific DNA binding activity. J. Biol. Chem., 272, 23017–23024. [DOI] [PubMed] [Google Scholar]
- Sutton M.D., Carr,K.M., Vicente,M. and Kaguni,J.M. (1998) Escherichia coli DnaA protein. The N-terminal domain and loading of DnaB helicase at the E.coli chromosomal origin. J. Biol. Chem., 273, 34255–34262. [DOI] [PubMed] [Google Scholar]
- Terwilliger T. and Berendzen,J. (1999) Automated structure solution for MIR and MAD. Acta Crystallogr. D, 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and Higgins,D.G. (1997) The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res., 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Kunishima,N., Mayanagi,K., Ohnishi,T., Nishino,T., Iwasaki,H., Shinagawa,H. and Morikawa,K. (2001) Crystal structure of the Holliday junction migration motor protein RuvB from Thermus thermophilus HB8. Proc. Natl Acad. Sci. USA, 98, 1442–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu R.C., Hanson,P.I., Jahn,R. and Brunger,A.T. (1998) Structure of the ATP-dependent oligomerization domain of N-ethylmaleimide sensitive factor complexed with ATP. Nat. Struct. Biol., 5, 803–811. [DOI] [PubMed] [Google Scholar]
- Zhang X. et al. (2000) Structure of the AAA ATPase p97. Mol. Cell, 6, 1473–1484. [DOI] [PubMed] [Google Scholar]
- Zheng W., Li,Z., Skarstad,K. and Crooke,E. (2001) Mutations in DnaA protein suppress the growth arrest of acidic phospholipid-deficient Escherichia coli cells. EMBO J., 20, 1164–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]