Abstract
rRNA transcription in Saccharomyces cerevisiae is performed by RNA polymerase I and regulated by changes in growth conditions. During log phase, ∼50% of the ribosomal DNA (rDNA) genes in each cell are transcribed and maintained in an open, psoralen-accessible conformation. During stationary phase, the percentage of open rDNA genes is greatly reduced. In this study we found that the Rpd3 histone deacetylase was required to inactivate (close) individual rDNA genes as cells entered stationary phase. Even though ∼50% of the rDNA genes remained open during stationary phase in rpd3Δ mutants, overall rRNA synthesis was still reduced. Using electron microscopy of Miller chromatin spreads, we found that the number of RNA polymerases transcribing each open gene in the rpd3Δ mutant was significantly reduced when cells grew past log phase. Bulk levels of histone H3 and H4 acetylation were reduced during stationary phase in an RPD3-dependent manner. However, histone H3 and H4 acetylation was not significantly altered at the rDNA locus in an rpd3Δ mutant. Rpd3 therefore regulates the number of open rDNA repeats.
Keywords: deacetylation/repression/RNA polymerase I/RPD3/yeast
Introduction
Ribosome biogenesis is highly regulated by cellular growth conditions and the demand for protein synthesis: fast growth demands larger numbers of ribosomes and cessation of growth correlates with decreased ribosome synthesis. Ribosomes in all cells are made up of ribosomal proteins and several non-coding rRNA molecules. As a result, ribosome biogenesis is controlled primarily through the regulation of rRNA transcription by RNA polymerase I (Pol I), and ribosomal protein gene transcription by RNA polymerase II (Pol II). RNA Pol I is a multiprotein complex that is conserved in all eukaryotes. Therefore, the powerful genetic methods in the budding yeast, Saccharomyces cerevisiae, have led to its use as a model system in the study of rRNA transcriptional regulation. The ∼150 ribosomal DNA (rDNA) gene copies of S.cerevisiae are organized in the nucleolus as a single tandem array on chromosome XII (for a review, see Planta, 1997). Each 9.1 kb rDNA repeat contains the Pol I-transcribed 35S pre-rRNA gene and the Pol III-transcribed 5S rRNA gene (see Figure 1A). The 5S gene is located in the middle of a non-transcribed spacer (NTS), which separates each 35S transcriptional unit.
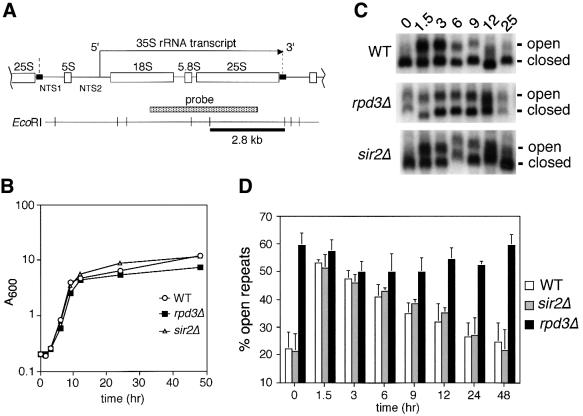
Fig. 1. Deletion of RPD3 prevents inactivation of rDNA repeats as cells exit log phase. (A) Schematic representation of the rDNA in S.cerevisiae, showing the initiation site and direction of 35S transcription by Pol I. The Pol III-transcribed 5S rRNA gene separates the non-transcribed spacer regions (NTS1 and NTS2). A Pol I enhancer element is located at the 3′ end of each 35S repeat (small black rectangles). The 2.8 kb EcoRI restriction fragment and probe are indicated. The boundaries of one rDNA repeat are indicated by vertical dashed lines. (B) Representative growth curve of yeast cultures used for the psoralen cross-linking assay. The strains used were JS311 (WT), JS490 (rpd3Δ) and JS218 (sir2Δ). (C) Gel retardation of actively transcribed rDNA genes through psoralen cross-linking. Aliquots harvested at the indicated time points were photoreacted with psoralen. Genomic DNA was digested with EcoRI and rDNA-specific fragments were detected using the 35S probe. For simplicity, only the 2.8 kb fragment is shown. The actively transcribed repeats (open) and inactive repeats (closed) are indicated on the right. (D) Quantitation of actively transcribed rDNA repeats. By PhosphorImager analysis, the percentage of the total rDNA repeats that were in the open chromatin conformation (slow migrating band) was calculated for each strain and time point. The average percentages from three independent experiments are shown in a bar graph along with standard deviation error bars.
Previous studies have determined that the rate of rRNA transcription is high in exponentially growing yeast cells and is repressed as cells enter stationary phase (Ju and Warner, 1994). Sogo and co-workers have developed an in vivo psoralen cross-linking assay in yeast that can distinguish between Pol I-transcribed and non-transcribed rDNA genes based on their differential accessibility to the cross-linker (Dammann et al., 1993). They determined that only a subset of rDNA repeats in each cell are transcribed at a given time. Approximately 50% of the genes are transcribed in log phase cells, but this percentage is reduced as cells enter stationary phase (Dammann et al., 1993). These results led to the proposal that the rate of rRNA transcription in yeast is determined in part by the number of transcribed rDNA genes in each cell (Dammann et al., 1993). This hypothesis fits well with electron microscope (EM) images of ‘Miller’ rDNA chromatin spreads from Xenopus oocytes showing that rDNA genes are either highly transcribed by RNA Pol I or not transcribed at all (Osheim et al., 1996).
The initiation of yeast Pol I transcription is dependent on four different transcription factors: TATA binding protein (TBP), Rrn3, upstream activating factor (UAF) and core factor (CF). CF comprises three subunits, Rrn6, Rrn7 and Rrn11, and binds to a core promoter element surrounding the transcriptional start site (Lalo et al., 1996; Lin et al., 1996). CF functions analogously to the TIF-IB (SL1) factor of mammalian cells in that it binds to the core promoter region. UAF contains six subunits, Rrn5, Rrn9, Rrn10, Uaf30 and histones H3 and H4 (Keys et al., 1996; Keener et al., 1997; Siddiqi et al., 2001). UAF binds tightly to an element upstream of the start site called the upstream control element (UCE) (Keys et al., 1996). Mammalian upstream binding factor (UBF) is an HMG-box protein that binds to an upstream 35S gene promoter element (Jantzen et al., 1990), and could therefore potentially have some functional similarity to yeast UAF. However, UAF and UBF are structurally very different and many of their functional characteristics are non-overlapping. TBP binds to both UAF and CF and is required for the UAF-dependent recruitment of CF to the promoter (Steffan et al., 1996). Rrn3 is the functional homolog of mammalian TIF-IA (Bodem et al., 2000; Moorefield et al., 2000). It associates with Pol I to form an initiation-competent Pol I complex (Yamamoto et al., 1996). Biochemical studies have demonstrated that the downregulation of rRNA transcription during stationary phase is caused by disappearance of the initiation- competent Pol I–Rrn3 complex (Milkereit and Tschochner, 1998). rRNA transcription therefore has the potential to be regulated at two levels: (i) activation (opening) and inactivation (closing) of individual rDNA genes; and (ii) transcriptional initiation frequency and/or elongation from each open rDNA gene. Such a model has been proposed previously by Reeder and colleagues (Reeder, 1989; Reeder, 1992; Aprikian et al., 2001).
While a great deal of progress has been made in elucidating the basic mechanism of rRNA transcriptional initiation, the influence of chromatin structure and the post-translational modifications of histones and non-histone proteins on Pol I transcription have not been thoroughly investigated. Histone acetylation and deacetylation play important roles in the regulation of Pol II-mediated gene expression. The reversible acetylation of conserved N-terminal lysine residues on histones H3 and/or H4 modulates the accessibility of various transcription factors to enhancer and promoter elements in chromatin (reviewed in Roth et al., 2001). In general, histone acetylation is correlated with transcriptional activity. Indeed, several transcriptional activators and coactivators contain intrinsic histone acetyltransferase activities. For example, the yeast Gcn5 histone acetyltransferase is the catalytic subunit of the Spt-Ada-Gcn5-acetyltransferase (SAGA) transcriptional coactivator complex (Grant et al., 1997). Recently, histone methylation has also been shown to be involved in regulating Pol II transcription (Kouzarides, 2002), although its role (if any) in the regulation of Pol I transcription is unknown.
One of the most well studied histone deacetylases (HDACs) is the Rpd3 protein, which is homologous to mammalian HDAC1 (Taunton et al., 1996). Rpd3 contributes to the repression of multiple genes in yeast (Vidal and Gaber, 1991), and is the catalytic subunit of a multiprotein deacetylase complex that contains among other proteins, Sin3 and Sap30 (Rundlett et al., 1996; Kasten et al., 1997; Zhang et al., 1998). Sin3 is a corepressor which, through interactions with specific DNA binding repressors such as Ume6, targets the RPD3 complex to specific promoters, resulting in transcriptional repression (Kadosh and Struhl, 1997). Repression is the result of localized deacetylation of histones H3 and H4 in the vicinity of the promoter (Kadosh and Struhl, 1998; Rundlett et al., 1998). However, Ume6 does not target Rpd3 to all RPD3-regulated genes (Robyr et al., 2002).
In this paper we report a novel role for Rpd3 in repressing rRNA transcription in stationary phase. In the absence of RPD3, cells were unable to reduce the number of transcriptionally active (open) rDNA repeats as they entered stationary phase. However, the rpd3Δ mutants were still able to repress rRNA transcription by reducing the number of Pol I molecules transcribing each open gene, indicating that rRNA transcriptional repression in yeast occurs by two independent mechanisms.
Results
RPD3 is required for the inactivation of rDNA repeats as cells approach stationary phase
In previous work, rpd3 mutations caused the NTS to become highly accessible to cross-linking by psoralen, suggesting that Rpd3 could potentially regulate Pol I-mediated rRNA transcription (Smith et al., 1999). To test this hypothesis, we first determined the percentage of rDNA genes that were actively transcribed (open) during log and stationary phases. Wild-type (RPD3+), rpd3Δ and sir2Δ strains were first grown to stationary phase (time = 0), and then reinoculated into fresh YPD and allowed to grow back into stationary phase (Figure 1B). SIR2 was included because it was previously reported that deletion of SIR2 increased the percentage of rDNA genes that were open (Smith and Boeke, 1997). Aliquots of cells were harvested at the indicated time points and photoreacted with psoralen. Genomic DNA from these cells was digested with EcoRI, separated on a native agarose gel and detected by Southern blotting with a probe specific for the Pol I-transcribed region of the rDNA (Figure 1A). A representative Southern blot is shown in Figure 1C and average quantitative data from three independent experiments are shown in Figure 1D. As expected, the 2.8 kb rDNA fragment was separated into two different bands representing the open (slower mobility) and closed (faster mobility) rRNA genes (Figure 1C). In the stationary phase cultures (time = 0), the percentage of open genes was very low in the wild-type and sir2Δ strains. However, ∼50% of the genes were open in the rpd3Δ mutant. Within 1.5 h following reinoculation, the wild-type and sir2Δ strains had reactivated ∼50% of the rDNA genes even though the cells had not started to double at this time point. After the 1.5 h time point, the open percentage steadily decreased in the wild-type and sir2Δ strains as they passed through log phase and re-entered stationary phase. In the rpd3Δ mutant, the proportion of active repeats remained at ≥45% throughout the entire time course. Therefore, RPD3, but not SIR2, was required to inactivate (close) rDNA repeats as cells approached stationary phase.
Repression of rRNA transcription still occurs in rpd3Δ mutants approaching stationary phase
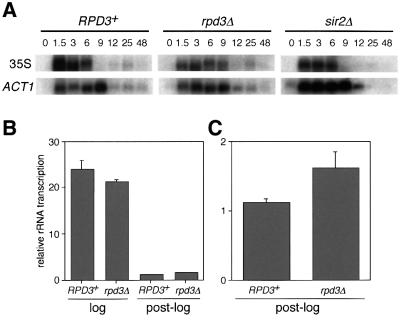
It was possible that loss of Rpd3 function completely eliminated the ability of cells to regulate rRNA transcription in response to changes in growth conditions. The psoralen cross-linking results in Figure 1 suggested that rpd3Δ mutant cells in stationary phase would have the same level of rRNA transcription as they did in log phase. To test this hypothesis, the relative amount of rRNA transcription in RPD3+, rpd3Δ and sir2Δ cells was determined after various growth times using RNA blot analysis (Figure 2). The time course was the same as described in Figure 1B. 35S precursor rRNA was detected using an oligonucleotide probe specific for the 5′ externally transcribed spacer (ETS) region, which is a good measurement for the amount of nascent rRNA transcript. The peak of rRNA transcription occurred at time = 1.5–3 h for each strain (Figure 2A), which was consistent with the psoralen cross-linking results. As expected, rRNA transcription was repressed in the wild-type and sir2Δ strains at the diauxic shift (9 h) and beyond. Surprisingly, rRNA transcription was also repressed in the rpd3Δ mutant as cells entered stationary phase (Figure 2A), even though ∼50% of the rDNA genes were in the open chromatin conformation. As expected, the ACT1 control mRNA was also repressed as the cells entered stationary phase (Wenzel et al., 1995).
Fig. 2. rRNA transcription from log phase and post-log phase yeast cultures. (A) Northern blot analysis of total RNA isolated from JS311 (RPD3+), JS490 (rpd3Δ) or JS566 (sir2Δ) cells that were harvested at the indicated time points after culture inoculation. Oligonucleotide probes were specific for the 35S rRNA precursor and the ACT1 mRNA. (B) In vivo transcriptional run-on assay for rRNA. Relative rRNA transcription amounts were plotted for log and post-log cells. (C) Enlarged graph of the post-log phase run-on data. Error bars are equal to the standard deviation from three independent experiments.
To confirm the repression of Pol I transcription in stationary phase cells observed in the rpd3Δ mutant by northern blotting, we performed an in vivo transcriptional run-on assay. RPD3+ and rpd3Δ strains were grown into log phase or past the diauxic shift (post-log phase), permeabilized, labeled with [α-32P]UTP for 10 min and then chased with unlabeled UTP. The reactions were performed in the presence of α-amanitin to inhibit Pol II-mediated transcription. The relative levels of labeled transcript are plotted in Figure 2B and C. In log phase, there was very little difference in rRNA production in the rpd3Δ mutant compared with wild type. If anything, there was a slight reduction in the mutant. In the post-log phase cells, there was a large reduction in rRNA transcription in both the wild-type and rpd3Δ mutant (Figure 2B), with the mutant having slightly more rRNA transcription than the wild type (Figure 2C). These data were fully consistent with what we observed by northern blotting.
These results suggested that, in stationary phase, the rpd3Δ mutant contained many transcriptionally active (open) rDNA genes, but that Pol I transcription from each of those open genes was repressed by an Rpd3-independent mechanism. Even though SIR2 is critical for rDNA silencing of Pol II-transcribed reporter genes and suppressing rDNA recombination (Gottlieb and Esposito, 1989; Bryk et al., 1997; Smith and Boeke, 1997), it appears that SIR2 does not play a major role in the regulation of Pol I-mediated rRNA transcription under the conditions tested.
Pol I occupancy on each active rDNA gene decreases as rpd3Δ mutants enter stationary phase
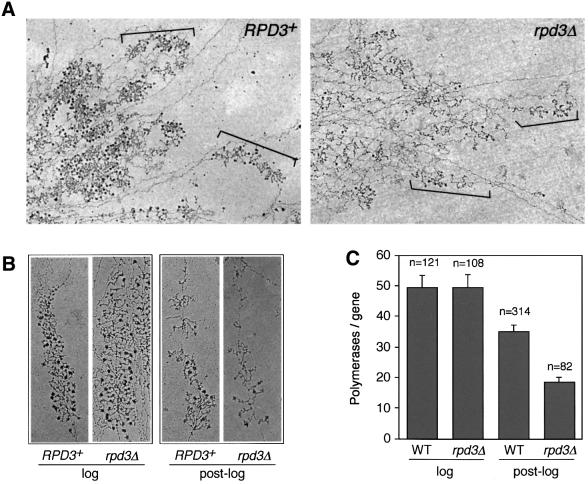
rRNA transcription in the rpd3Δ mutant decreased as cells entered stationary phase (Figure 2), even though the number of open genes did not change (Figure 1). One prediction from these results was that, in stationary phase, the rpd3Δ mutant should have fewer Pol I complexes loaded onto each open rDNA gene. We tested this hypothesis using Miller chromatin spreads. Hypo tonically lysed yeast cells were spread onto carbon-coated EM grids, fixed and the dispersed chromatin was visualized using an electron microscope (Figure 3). Actively transcribed rDNA genes were easily identified by their characteristic Christmas tree-like appearance (Figure 3). Miller spreads were performed on RPD3+ and rpd3Δ strains that were grown into log phase or into the diauxic shift (post-log phase). Since the rDNA is organized into a large tandem array, the active genes were usually clustered close to each other on the grids. An example of this clustering is shown for the post-log phase cultures in Figure 3A. The number of polymerases per active gene was then manually counted from high magnification images. Representative rDNA genes for each condition are shown in Figure 3B. In exponentially growing RPD3+ and rpd3Δ cells, the average number of polymerases loaded was ∼50 (Figure 3C). Rpd3p is therefore not required for the normal loading of Pol I onto actively transcribing rDNA genes during exponential growth. However, after the diauxic shift, the RPD3+ strain had ∼2-fold more polymerases loaded per gene compared with the rpd3Δ mutant (35 compared with 18; Figure 3C). Even the RPD3+ strain partially reduced the polymerase loading in stationary phase. We conclude that the regulation of rRNA transcription in response to changes in growth conditions occurs by two different processes. The first is control of the number of transcriptionally active (open) rDNA genes by Rpd3. The second process is the regulation of Pol I initiation frequency from each open gene.
Fig. 3. Miller chromatin spread analysis of yeast rRNA transcription. (A) Typical clusters of rDNA genes from RPD3+ and rpd3Δ strains grown beyond log phase, approximating the diauxic shift. Examples of individual 35S genes are bracketed. Dark knobs at the end of transcripts represent rRNA processing machinery. (B) High magnification views of representative 35S genes from RPD3+ and rpd3Δ strains growing in log phase or post-log phase. (C) Graphical representation of the average number of polymerases that transcribe each active 35S gene in each strain and growth condition. Error bars represent the 99% confidence limits for the mean. The number (n) of genes analyzed for each condition is indicated.
Bulk histone acetylation analysis
Since Rpd3 is a known HDAC, we hypothesized that it reduced the number of open rDNA genes in stationary phase through either global deacetylation of histones H3 and/or H4 at the rDNA array or localized deacetylation of histones H3 and/or H4 at the Pol I promoter. To test this hypothesis, we first analyzed bulk histone H3 and H4 acetylation status as the cells progressed from log into stationary phase using whole-cell extracts (WCEs). Histone H3-acetyl- and H4-acetyl-specific antibodies were used to examine their acetylation status by western blotting (Figure 4). There was little difference in H3 or H4 acetylation between RPD3+ and rpd3Δ strains in log phase (time = 6 h). However, the level of H3 acetylation (H3 K9,14-ac antibody) and H4 acetylation (penta antibody) decreased as RPD3+ cells entered stationary phase. This H3 and H4 deacetylation was largely Rpd3 dependent because as cultures entered stationary phase the level of acetylation failed to decrease as much in the rpd3Δ mutant (Figure 4). It is currently unclear why the rpd3Δ effect on H4 acetylation was not as dramatic at the 96 h time point.
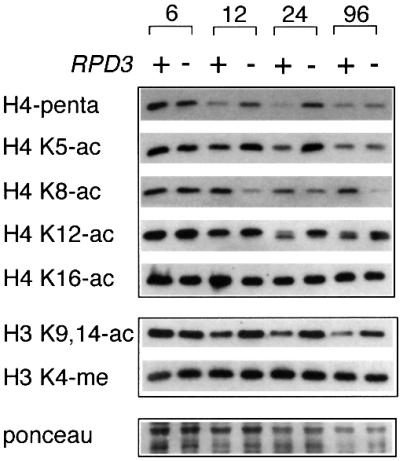
Fig. 4. Bulk histone H3 and H4 acetylation status in stationary phase cells. JS311 (RPD3+) and JS490 (rpd3Δ) strains were grown from log into stationary phase. WHEs were analyzed by western blotting with a panel of histone H3- or H4-specific antibodies indicated on the left side of the panel. Hours of growth in YPD medium are indicated at the top. H3 K4-me antibody is specific for H3 methylated on lysine 4. This row represents a loading control, along with the Ponceau staining at the bottom.
The H4 penta antibody was presumably specific for fully acetylated histone H4. To determine which specific lysine residues of H4 were being deacetylated by Rpd3p, we probed the western blots with antibodies specific for acetylated lysines 5, 8, 12 or 16 on the N-terminal tail. Lysines 5 and 12 displayed patterns of acetylation that were very similar to the penta antibody pattern, specifically RPD3-dependent deacetylation in stationary phase (Figure 4). For the K12-ac antibody, histone H4 unexpectedly ran as a doublet in stationary phase. The nature of the lower band is currently unclear. K16 acetylation did not change at all during stationary phase and was not affected by the rpd3Δ mutation. Surprisingly, K8-ac appeared to become deacetylated in stationary phase rpd3Δ mutants (Figure 4). However, further ELISA analysis of the K8-ac antibody revealed that it only recognized an H4 tail that was monoacetylated on K8 (W.Cheung, unpublished data). Therefore, when K5 and K12 acetylation decreased in the wild-type strain compared with rpd3Δ during stationary phase, the K8 antibody recognized the deacetylated H4 better. The penta H4 antibody result therefore probably reflects the K5 and K12 acetylation status of histone H4. If K8 acetylation changed, we were unable to detect it at this time. Each lane was loaded equally according to Ponceau staining of the membranes and the abundance of K4-methylated histone H3, which was not affected by growth conditions or rpd3Δ. We conclude from this experiment that Rpd3 is required for global histone H3 and H4 deacetylation during the transition to stationary phase.
Histone H3 and H4 acetylation at the rDNA locus is unchanged in an rpd3Δ mutant
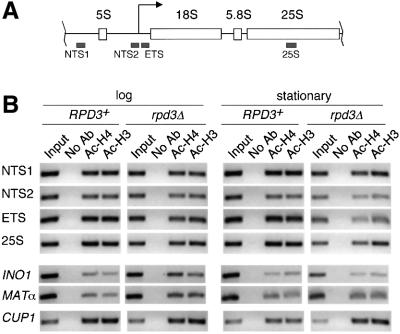
To determine whether the rpd3Δ mutation also altered histone H4 or H3 acetylation at the rDNA, we performed chromatin immunoprecipitation (ChIP) assays with the α-acetyl H4 (penta) and α-acetyl H3 (9,14) antibodies. We predicted that the Pol I promoter region may be hyperacetylated in the rpd3Δ mutant. JS311 (RPD3+) and JS490 (rpd3Δ) were grown to mid-log phase (A600 = 1.0) or early stationary phase (A600 = ∼7.0) and then subjected to ChIP with the histone H4 and H3 antibodies. Associated rDNA sequences were amplified by PCR and detected by ethidium bromide staining (Figure 5). Four different locations in the rDNA repeat were examined: NTS1, NTS2, the 5′ ETS and the 25S region (Figure 5A). The NTS2 primer pair was specific for the Pol I promoter region upstream of the transcriptional start site.
Fig. 5. Histone H3 and H4 acetylation status of rDNA-associated chromatin in log and stationary phases. Specific DNA sequences that were associated with acetylated histone H3 or H4 were immunoprecipitated from RPD3+ (JS311) and rpd3Δ (JS490) WCEs. (A) The rDNA-specific sequences included NTS1, NTS2 (the Pol I promoter region), the 5′ ETS and the 25S transcribed region. The non-rDNA control sequences were the CUP1, MATα and INO1 genes. (B) PCR amplifications were performed with each primer pair. For the input lanes, 1/20 the amount of DNA was amplified for each rDNA location and CUP1 to remain in the linear range for the reaction. Ac-H4 represents the Penta H4 antibody. Ac-H3 represents the H3 K9/K14 acetyl antibody. The data shown are representative of four independent experiments. The fold differences between RPD3+ and rpd3Δ strains are shown in Table I.
In log phase, there were no significant increases in H3 or H4 acetylation at any of the rDNA locations in the rpd3Δ mutant compared with the RPD3+ strain (Figure 5B). Similar results were observed for the CUP1 control locus. As a positive control, we examined the INO1 promoter, which was already known to be deacetylated by Rpd3. As expected, H3 and H4 acetylation at the INO1 promoter increased ∼3.7- and 3.3-fold, respectively, in the rpd3Δ mutant (Figure 5B; Table I). Another single copy locus, MATα, also showed a slight increase in H3 and H4 acetylation, but not as much as at the INO1 promoter. We know that large changes in rDNA histone acetylation are detectable by this type of assay because log phase sir2Δ mutants show a large increase in H3 acetylation at the rDNA (J.Sandmeier, unpublished data; Bryk et al., 2002).
Table I. Fold change in histone acetylation (rpd3Δ/WT)a.
| |
Log |
Post-log |
||
|---|---|---|---|---|
| Acetyl-H4 | Acetyl-H3 | Acetyl-H4 | Acetyl-H3 | |
| NTS1 | 1.1 | 1.0 | 0.9 | 1.3 |
| NTS2 | 1.1 | 1.4 | 1.0 | 2.0 |
| ETS | 0.8 | 1.1 | 0.4 | 1.1 |
| 25S | 0.9 | 1.3 | 0.8 | 1.8 |
| INO1 | 3.3 | 3.7 | 2.7 | 2.1 |
| MATα | 1.9 | 2.6 | 1.5 | 2.0 |
| CUP1 | 1.1 | 1.6 | 3.0 | 4.2 |
aData are compiled from Figure 5. Values are the IP/input ratio for the rpd3Δ strain divided by the IP/input ratio for the wild-type strain.
Stationary phase was where we expected to see a significant difference in acetylation based on the bulk histone analysis. Surprisingly, the rpd3Δ mutation still did not induce consistent and dramatic hyperacetylation of H3 or H4 at any rDNA positions during stationary phase compared with RPD3+ (Figure 5B; Table I). There was actually slight H4 hypoacetylation of the ETS and 25S regions in the mutant. In contrast, the INO1 and CUP1 regions showed consistent H4 and H3 hyperacetylation in the stationary phase rpd3Δ mutant when quantitated (Table I). The CUP1 hyperacetylation was unexpected, but was particularly strong on histone H3 (∼4.2-fold increase, Table I). Further chromatin IPs with the mono-acetyl H4 antibodies confirmed that there was no increase in H4 acetylation in the stationary phase rpd3Δ mutant (data not shown). Therefore, even though bulk histones H3 and H4 remain acetylated in stationary phase rpd3Δ cells compared with wild type (Figure 4), the rDNA largely remains in a normal acetylation state. We conclude that if Rpd3p directly deacetylates histone H3 or H4 at the rDNA, then it must occur at a very localized region not covered in this study, or it only occurs in a subset of the ∼150 rDNA genes in each cell, which would make its detection very difficult.
Discussion
In this paper we demonstrate that the RPD3 HDAC complex functions in the transcriptional inactivation of rDNA genes as yeast cells enter stationary phase. There is now a wealth of information about the roles of histone acetyltransferases and HDACs in the regulation of Pol II-mediated transcription. In contrast, the function of HATs and HDACs in Pol I transcriptional regulation is only beginning to be investigated. Studies on nucleolar dominance in plants were among the first to implicate histone deacetylation in Pol I regulation. This epigenetic silencing of Pol I transcription in plants can be weakened by treatment with inhibitors of DNA methylation and histone deacetylation (Chen and Pikaard, 1997).
Yeast rRNA transcription is regulated by two mechanistically distinct processes
In this study we observed that rRNA transcription in rpd3Δ mutants was repressed during stationary phase compared with log phase, even though the proportion of open rDNA repeats was the same in each growth condition. This result in effect separates rRNA transcriptional regulation into two modes. The first mode of Pol I transcriptional regulation is control of the number of transcriptionally active rDNA repeats by Rpd3, perhaps through promoter accessibility. Rpd3 does not appear to directly deacetylate histones H3 or H4 at the rDNA during stationary phase. Therefore, the mechanism of how Rpd3 regulates rDNA promoter accessibility is still unclear. A possible model is discussed in the next section.
The second mode of Pol I transcriptional regulation is the control of Pol I initiation frequency on each active repeat. The predominant view of rRNA transcription is that when an rDNA gene is activated for transcription, it is transcribed at a high level, with Pol I molecules lined up one after another. Such genes were seen in logarithmically growing cells (Figure 4), as observed in many previous EM studies from vertebrate (e.g. Miller and Beatty, 1969) and yeast cells (Rattner et al., 1982). In addition, our results show for the first time that polymerase density, and thus initiation frequency, decreases on rDNA genes as wild-type cells enter stationary phase. Furthermore, as rpd3Δ mutant cells enter stationary phase, the number of polymerases transcribing each active gene is further reduced compared with the wild-type reduction (∼18 versus ∼35 pols/gene). Perhaps the simplest interpretation of this latter result is that the initiation-competent Pol I molecules are able to distribute on more open genes, which have not been inactivated by Rpd3, resulting in fewer polymerases per active gene. This mode of regulation probably involves Rrn3. Pol I is only competent for transcription when it is complexed with Rrn3, and stationary phase cells actually lack the active Rrn3–Pol I complex (Yamamoto et al., 1996; Milkereit and Tschochner, 1998). A similar model for two mechanistically distinct levels of rRNA transcriptional regulation in yeast was proposed previously (Reeder, 1989, 1992; Aprikian et al., 2001).
Our data suggest that both modes of rRNA transcriptional regulation are biologically relevant in wild-type yeast cells. The number of active repeats is clearly reduced in stationary phase, as measured by psoralen cross-linking. In addition, fewer Pol I molecules are loaded on the repeats that remain active. In the absence of Rpd3, cells may rely completely on Rrn3p-mediated regulation. By combining both modes of regulation, yeast cells are then better equipped for a rapid response to changing environmental conditions.
The relationship between RPD3 and rDNA silencing of Pol II-transcribed reporter genes
RPD3 paradoxically counteracts SIR2-mediated transcriptional silencing. Mutations in components of the RPD3 complex result in strengthened repression at the silent mating type loci (HML and HMR), telomeres and the rDNA (De Rubertis et al., 1996; Rundlett et al., 1996; Vannier et al., 1996; Smith et al., 1999; Sun and Hampsey, 1999). The conventional view of repressive chromatin predicts that loss of a deacetylase activity would weaken silencing, not enhance it. The enhanced silencing caused by rpd3 mutations could be due to an indirect effect, such as increased expression of a dosage-sensitive silencing factor. If this hypothesis were true, then the same indirect effect would probably occur in Drosophila, because mutations in Drosophila RPD3 enhance position effect variegation (De Rubertis et al., 1996). A more direct model has been proposed by Michael Hampsey’s laboratory (Sun and Hampsey, 1999). Their model is based on the earlier observation that histone H4 is specifically acetylated on lysine 12 in metazoan and yeast heterochromatin (Turner et al., 1992; Braunstein et al., 1996). In the model, rpd3 mutations result in a greater proportion of histone H4 that is acetylated on lysines 5 and 12, which they referred to as the ‘inactive’ or heterochromatin form of H4. The incorporation of this inactive H4 into silent chromatin is then carried out by chromatin assembly factor, which binds this form of H4 in vivo (Verreault et al., 1996) and helps assemble it into silent chromatin where K12 remains acetylated (Braunstein et al., 1996; Kaufman et al., 1997). More available inactive H4 in the rpd3Δ mutant would translate into more efficient assembly of the silent chromatin. We did not observe preferential acetyl ation of lysine 12 at the rDNA compared with non-rDNA loci (data not shown), which is consistent with ChIP data from other groups that observed hypoacetylation of all four N-terminal lysine residues of H4 at HMR and telomeres (de Bruin et al., 2000; Suka et al., 2001). The above model is not as attractive if K12 is not preferentially acetylated in yeast heterochromatin.
How can RPD3 be favorable for Pol I transcriptional repression and antagonistic for Pol II transcriptional repression (rDNA silencing) at the same time? It turns out that Pol I-mediated rRNA transcription from the tandem array is absolutely required for the silencing of Pol II-transcribed reporter genes integrated into the same tandem array (S.Buck and J.Smith, unpublished data). Based on this new finding, we now propose that the effect of an rpd3Δ mutation on rDNA silencing is due to its effect on Pol I transcription. For example, many cells within a yeast colony are likely to be in a growth state similar to stationary phase. Since 50% of the rDNA genes in rpd3Δ mutant cells remain open, silencing would remain strong in the entire colony population. rDNA silencing has not been directly measured in stationary phase cells, but it is known that passage through stationary phase advances the replicative aging of yeast cells (Ashrafi et al., 1999), which is consistent with a loss of rDNA silencing. Future experiments will test this new idea. The downside of this model is that it does not explain why rpd3Δ enhances TPE or HM silencing. However, silencing at these loci is significantly different than silencing at the rDNA. For example, rDNA silencing does not require SIR3 and SIR4 (Bryk et al., 1997; Smith and Boeke, 1997).
Model for the regulation of rRNA transcription by RPD3
We found that bulk histone H4 from WCEs was deacetylated on lysines 5 and 12 in an RPD3-dependent manner when cells were grown into stationary phase. We therefore predicted that rDNA repeats remained open in stationary phase rpd3Δ cells because the nucleosomes surrounding the rDNA promoter were presumably still acetylated in the mutant. However, according to ChIPs (Figure 5), histone acetylation at the rDNA was not enhanced in the mutant. If elevated histone acetylation did remain in the mutant, it would have to occur in an rDNA gene location not covered in this study. Furthermore, if the change in acetylation was minor or only occurred in a fraction of the repeats, then the difference would be difficult to observe by ChIPs because the rDNA consists of a mixture of ∼150 repeats.
A common developing theme is that rRNA transcription by Pol I is regulated by the acetylation and deacetylation of Pol I-specific transcription factors. Based on the available information, we propose that Rpd3 controls the number of open rDNA genes by deacetylating a Pol I-specific transcription factor, not nucleosomal histones. Such a scenario is not unprecedented. In mammalian cells, UBF is acetylated by CBP histone acetyltransferase and deacetylated by a complex of Rb and HDAC1 (Pelletier et al., 2000). Acetylation by CBP activates Pol I transcription and deacetylation by Rb–HDAC1 represses Pol I transcription (Pelletier et al., 2000). In addition, the TAFI68 subunit of TIF-IB (SL1) is acetylated by the PCAF acetyltransferase and deacetylated by the NAD+-dependent deacetylase mSir2α (Muth et al., 2001). SL1 is the mammalian counterpart of yeast CF. The acetylation of TAFI68 activates Pol I transcription in vitro (Muth et al., 2001). It is, therefore, surprising that yeast Sir2 does not appear to play a role in rRNA transcriptional regulation (Figures 1 and 2). In a previous study, there was a consistent increase in the number of rDNA genes that were transcriptionally active (open) in a sir2Δ mutant. However, based on statistics from the psoralen time course in this study, we now believe that the small difference observed is not significant.
The most obvious Pol I transcription factor candidate for potential post-translational modification in yeast is UAF. Histones H3 and H4 are actually subunits of UAF (Keener et al., 1997). Therefore, Rpd3p may deacetylate the H3 or H4 subunits of UAF to inactivate individual rDNA genes. UAF is tightly bound to the UCE and does not readily exchange once it is bound (Keys et al., 1996). UAF has even been proposed to assemble on the rDNA during DNA replication and remain there in the absence of active rRNA transcription (Keener et al., 1997), perhaps in a nucleosomal-like structure. The histone H3/H4 tetramer is able to form a nucleosome-like structure with DNA. Similarly, the TAF subunits of TFIID have been suggested to form a nucleosome-like structure that functions in promoter binding (Hoffmann et al., 1996; Xie et al., 1996). Therefore, if UAF were acetylated on the H4 and/or H3 subunits, the promoter would presumably become more accessible to the rest of the Pol I initiation machinery. Deacetylation by Rpd3 would close the genes. If H3 or H4 acetylation in the UAF particle was changed in the rpd3Δ mutant, then it might be possible to detect the change by ChIP at the rDNA upstream promoter. Our NTS2 primer pair was designed to detect such a change, but we were still unable to see one (Figure 5). The same reasons why we do not see nucleosomal acetylation changes in the rpd3Δ mutant at the rDNA also apply to UAF, if not more so. Rpd3 could also potentially target one of the non-histone UAF subunits or CF subunits. In a related version of the model, the higher concentration of H3 or H4 that is acetylated on K5 and K12 in stationary phase rpd3Δ mutants (Figure 4) could cause preferential incorporation of acetylated H3 and H4 into the UAF complex that is assembled at the rDNA promoter. Future work will be aimed at testing these models.
Materials and methods
Strains and media
To generate JS490, JS493 and JS566, the RPD3, SIN3 or SIR2 open reading frames (ORFs), respectively, were deleted from JS311 and replaced with the kanMX4 marker by a one-step gene replacement method (Smith et al., 1999). Miller spreads were performed on the S288C derivatives, JS772 and JS777. The genotype for each strain is described in Table II.
Table II. Yeast strains.
| Strain | Genotype |
|---|---|
| JS311a | MATα his3Δ200 leu2Δ1 met15Δ0 trp1Δ63 ura3-167 RDN1::Ty1-MET15, mURA3-HIS3 |
| JS490a | JS311 made rpd3Δ::kanMX4 |
| JS566 | JS311 made sir2Δ::kanMX4 |
| JS218b | MATα his3Δ200 leu2Δ1 ura3-167 sir2Δ::HIS3 RDN1::Ty1- mURA3 |
| JS772c | MATa his3Δ0 leu2Δ0 ura3Δ0 |
| JS777 | JS772 made rpd3Δ::kanMX4 |
Psoralen cross-linking
Psoralen cross-linking assays were performed as described previously (Dammann et al., 1993; Smith and Boeke, 1997). Stationary phase cultures were used to inoculate fresh 250 ml YPD cultures to an A600 of ∼0.2. Aliquots of ∼1 × 108 cells were harvested at the indicated time points. Washed cells were resuspended in 1.4 ml of ice-cold TE and 0.7 ml of this mixture was aliquoted into a 24-well tissue culture plate; 40 µl of a 200 µg/ml solution of 4,5′,8-trimethylpsoralen (Sigma) were added to each well. The cells were then UV irradiated (UV lamp model B-100A; Ultraviolet Products, Inc.) on ice for five doses of 5 min at a distance of 6 cm. Cells were spheroplasted with zymolyase, lysed, proteinase K treated, phenol–chloroform extracted and the resulting nucleic acid was ethanol precipitated. The pellet was resuspended in TE and normalized to a nucleic acid concentration of 2 mg/ml, and 4 µl were digested with EcoRI in a 30 µl reaction. Cleaved DNA was separated on a 1.3% agarose gel and transferred to a positively charged nylon membrane. rDNA-specific bands were detected by hybridization with a 3.6 kb XbaI–XbaI fragment containing most of the 35S region of yeast rDNA (Figure 1A).
RNA blot analysis
Yeast cultures were grown as described above for psoralen cross-linking. Cells were harvested (∼4 × 108) at the indicated time points and total RNA was isolated using the acid–phenol method (Ausubel et al., 2000). Total RNA (15 µg) was separated on a 1.2% agarose–2% formaldehyde gel and transferred to a nylon filter. The filter was hybridized for 90 min with the 32P-labeled probe JS45 (5′-TCGGGTCTCTCTGCTGCC GGAAATGCTCTCTGTTCA-3′), which hybridizes to the 5′ ETS of the 35S rRNA. Hybridization was performed in Quikhybe solution (Stratagene) at 65°C. Filters were washed twice (5 min) at room temperature with 2× SSC/0.1% SDS and then twice more at 60°C with 0.1× SSC/0.1% SDS. 35S rRNA bands were visualized by autoradiography. The ACT1 oligonucleotide probe was JB2351 (5′-GTC ACCGGCAAAACCGGCTTTACACATACCAGAACCGTTATCAAT AACCAAAGCAGCAAC-3′). Hybridization with this probe was the same as with JS45 except that the hybridization temperature was 60°C and the high temperature wash was at 55°C.
Transcriptional run-on assay
In vivo run-on assays were performed as described previously (Elion and Warner, 1986; Cormack and Struhl, 1992), with several modifications. Cells were harvested (∼8 × 107) from log phase cultures (OD600 = 0.5) or cultures that were approaching stationary phase (OD600 = 5.6). Cells were washed twice with ice-cold TMN buffer (10 mM Tris–HCl pH 7.4, 100 mM NaCl, 5 mM MgCl2), resuspended in 1 ml of 0.5% sodium N-lauroyl sarcosine and incubated for 15 min on ice. Cells were then pelleted and resuspended in a 100 µl reaction mixture containing 50 mM Tris–HCl pH 7.9, 100 mM KCl, 5 mM MgCl2, 1 mM MnCl2, 2 mM DTT, 0.5 mM ATP, 0.25 mM CTP, 0.25 mM GTP, 10 mM phosphocreatine, 10 mg/ml creatine phosphokinase, 100 µCi [α-32P]UTP (800 Ci/mmol) and 25 µg/ml α-amanitin. This reaction was incubated at 25°C for 10 min, and then stopped by the addition of 1 ml of ice-cold TMN supplemented with 1 mM UTP. Total RNA was isolated and separated by RNA blotting as described above. Labeled RNA was detected by autoradiography and quantitated using a Molecular Dynamics Storm PhosphorImager.
Electron microscopy
Yeast was grown in YPD plus 1 M sorbitol to mid- (A600 = 0.4) or post- (A600 = 2.5) log phase. Miller chromatin spreads of yeast strains were prepared after brief digestion of the yeast cell wall with zymolyase followed by hypotonic lysis in 0.025% Triton X-100 pH 9 (Hamkalo and Miller, 1973; Rattner et al., 1982). Volumes of 1 and 0.2 ml were harvested, and zymolyase concentrations of 5 and 25 mg/ml were used for mid- and post-log phase cells, respectively. Cell wall digestion was carried out at 30°C with shaking in the presence of actinomycin D (0.1 mg/ml). After 4 min, the digests were spun at maximum speed in an Eppendorf microcentrifuge for 6 s and the yeast pellet was resuspended in 1 ml of the lysis solution, which was then further diluted with an additional 3 ml of lysis solution. The samples were swirled gently for 20 min to allow for dispersal of cellular contents before being fixed with 0.4 ml of 0.1 M sucrose, 3.7% formalin and centrifuged onto carbon-coated EM grids. The grids were then stained with phosphotungstic acid and uranyl acetate and examined in a JEOL 100 CX microscope.
Western blotting
WCEs were separated on a 12% SDS–polyacrylamide gel and then transferred to PVDF membranes. Antibodies specific for acetylated H4 (penta H4), acetylated H3 (K9,14-acetyl) and H3 methylated on K4 were kindly provided by David Allis, and were incubated with the membrane at 1:5000 dilution in PBS-T (1× PBS, 0.05% Tween-20) containing 2% milk for 1 h at room temperature. All other antibodies were incubated at 1:10 000 dilutions for 2 h at room temperature. Membranes were washed with PBS-T (2% milk). Antibodies specific for histone H4 acetylated on lysine 5, 8, 12 or 16 were purchased from Serotec. The secondary antibody was a goat α-rabbit–HRP conjugate used at a dilution of 1:5000 (Amersham Biosciences). Post-translationally modified histones H3 and H4 were detected using ECL reagents from Amersham Biosciences.
ChIPs
YPD cultures (50 ml) of JS311 and JS490 were grown into log phase (A600 = 1) or early stationary phase (A600 = 7), then fixed for 1 h with 1% formaldehyde. Chromatin extraction and immunoprecipitations were performed as described previously (Kuo and Allis, 1999). Cell pellets were resuspended in 400 µl of FA-lysis buffer (50 mM HEPES–KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% deoxycholic acid, 1 mM PMSF, 1 µg/ml leupeptin, 1 µg/ml pepstatin). Cells were disrupted with glass beads in a BioSpec Products Mini Beadbeater. The extract was sonicated to fragment the chromatin and then centrifuged at 14 000 r.p.m. for 20 min to clarify the WCE.
For the immunoprecipitations, 200 µl of FA-lysis buffer were combined with 40 µl of WCE and 1 µl of the histone antibodies. This mixture was incubated at 4°C overnight on a rotator. Bound chromatin was precipitated with protein A–Sepharose beads (Amersham Biosciences) for 2 h at 4°C. The beads were washed extensively and the immune complexes were eluted twice with 200 µl of 1% SDS/0.1 M NaHCO3 at room temperature. Following reversal of cross-linking, the recovered material was treated with RNase A and then proteinase K. The remaining DNA was phenol extracted, ethanol precipitated and resuspended in a final volume of 150 µl of TE. Each sample (3 µl) was used in 50 µl PCRs specific for the NTS1, NTS2, 5′-ETS or 25S regions of the rDNA, or the INO1, MATα and CUP1 genes. PCR products were separated on a 2% agarose–1× TBE gel, stained with ethidium bromide and images captured using an AlphaImager 2000 documentation and analysis system from Alpha Innotech. Antibodies were the same as those used for western blotting.
Acknowledgments
Acknowledgements
We thank Jef Boeke for early support of this work and for helpful discussions. We also thank David Allis for providing multiple histone H3 and H4 antibodies and financial support to W.L.C. Technical assistance with EM work was provided by Martha Sikes. This work was supported by NIH grants GM63952 to A.L.B. and GM61692 to J.S.S.
References
- Aprikian P., Moorefield,B. and Reeder,R.H. (2001) New model for the yeast RNA polymerase I transcription cycle. Mol. Cell. Biol., 21, 4847–4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi K., Sinclair,D., Gordon,J.I. and Guarente,L. (1999) Passage through stationary phase advances replicative aging in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 96, 9100–9105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (2000) Current Protocols in Molecular Biology, Vol. 2. John Wiley & Sons, New York, NY.
- Bodem J., Dobreva,G., Hoffmann-Rohrer,U., Iben,S., Zentgraf,H., Delius,H., Vingron,M. and Grummt,I. (2000) TIF-IA, the factor mediating growth-dependent control of ribosomal RNA synthesis, is the mammalian homolog of yeast Rrn3p. EMBO Rep., 1, 171–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunstein M., Sobel,R.E., Allis,C.D., Turner,B.M. and Broach,J.R. (1996) Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol. Cell. Biol., 16, 4349–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryk M., Banerjee,M., Murphy,M., Knudsen,K.E., Garfinkel,D.J. and Curcio,M.J. (1997) Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev., 11, 255–269. [DOI] [PubMed] [Google Scholar]
- Bryk M., Briggs,S.D., Strahl,B.D., Curcio,M.J., Allis,C.D. and Winston,F. (2002) Evidence that Set1, a factor required for methylation of histone H3, regulates rDNA silencing in S.cerevisiae by a Sir2-independent mechanism. Curr. Biol., 12, 165–170. [DOI] [PubMed] [Google Scholar]
- Chen Z.J. and Pikaard,C.S. (1997) Epigenetic silencing of RNA polymerase I transcription: a role for DNA methylation and histone modification in nucleolar dominance. Genes Dev., 11, 2124–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack B.P. and Struhl,K. (1992) The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell, 69, 685–696. [DOI] [PubMed] [Google Scholar]
- Dammann R., Lucchini,R., Koller,T. and Sogo,J.M. (1993) Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res., 21, 2331–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin D., Kantrow,S.M., Liberatore,R.A. and Zakian,V.A. (2000) Telomere folding is required for the stable maintenance of telomere position effects in yeast. Mol. Cell. Biol., 20, 7991–8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubertis F., Kadosh,D., Henchoz,S., Pauli,D., Reuter,G., Struhl,K. and Spierer,P. (1996) The histone deacetylase RPD3 counteracts genomic silencing in Drosophila and yeast. Nature, 384, 589–591. [DOI] [PubMed] [Google Scholar]
- Elion E.A. and Warner,J.R. (1986) An RNA polymerase I enhancer in Saccharomyces cerevisiae. Mol. Cell. Biol., 6, 2089–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb S. and Esposito,R.E. (1989) A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell, 56, 771–776. [DOI] [PubMed] [Google Scholar]
- Grant P.A. et al. (1997) Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev., 11, 1640–1650. [DOI] [PubMed] [Google Scholar]
- Hamkalo B.A. and Miller,O.L.,Jr (1973) Electron microscopy of genetic activity. Annu. Rev. Biochem., 42, 379–396. [DOI] [PubMed] [Google Scholar]
- Hoffmann A., Chiang,C.M., Oelgeschlager,T., Xie,X., Burley,S.K., Nakatani,Y. and Roeder,R.G. (1996) A histone octamer-like structure within TFIID. Nature, 380, 356–359. [DOI] [PubMed] [Google Scholar]
- Jantzen H.M., Admon,A., Bell,S.P. and Tjian,R. (1990) Nucleolar transcription factor hUBF contains a DNA-binding motif with homology to HMG proteins. Nature, 344, 830–836. [DOI] [PubMed] [Google Scholar]
- Ju Q. and Warner,J.R. (1994) Ribosome synthesis during the growth cycle of Saccharomyces cerevisiae. Yeast, 10, 151–157. [DOI] [PubMed] [Google Scholar]
- Kadosh D. and Struhl,K. (1997) Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell, 89, 365–371. [DOI] [PubMed] [Google Scholar]
- Kadosh D. and Struhl,K. (1998) Targeted recruitment of the Sin3–Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol. Cell. Biol., 18, 5121–5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasten M.M., Dorland,S. and Stillman,D.J. (1997) A large protein complex containing the yeast Sin3p and Rpd3 transcriptional regulators. Mol. Cell. Biol., 17, 4852–4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman P.D., Kobayashi,R. and Stillman,B. (1997) Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev., 11, 345–357. [DOI] [PubMed] [Google Scholar]
- Keener J., Dodd,J.A., Lalo,D. and Nomura,M. (1997) Histones H3 and H4 are components of upstream activation factor required for the high-level transcription of yeast rDNA by RNA polymerase I. Proc. Natl Acad. Sci. USA, 94, 13458–13462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keys D.A., Lee,B.-S., Dodd,J.A., Nguyen,T.T., Vu,L., Fantino,E., Burson,L.M., Nogi,Y. and Nomura,M. (1996) Multiprotein transcription factor UAF interacts with the upstream element of the yeast RNA polymerase I promoter and forms a stable preinitiation complex. Genes Dev., 10, 887–903. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. (2002) Histone methylation in transcriptional control. Curr. Opin. Genet. Dev., 12, 198–209. [DOI] [PubMed] [Google Scholar]
- Kuo M.H. and Allis,C.D. (1999) In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods, 19, 425–433. [DOI] [PubMed] [Google Scholar]
- Lalo D., Steffan,J.S., Dodd,J.A. and Nomura,M. (1996) RRN11 encodes the third subunit of the complex containing Rrn6p and Rrn7p that is essential for the initiation of rDNA transcription by yeast RNA polymerase I. J. Biol. Chem., 271, 21062–21067. [DOI] [PubMed] [Google Scholar]
- Lin C.W., Moorefield,B., Payne,J., Aprikian,P., Mitomo,K. and Reeder,R.H. (1996) A novel 66-kilodalton protein complexes with Rrn6, Rrn7, and TATA-binding protein to promote polymerase I transcription initiation in Saccharomyces cerevisiae. Mol. Cell. Biol., 16, 6436–6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkereit P. and Tschochner,H. (1998) A specialized form of RNA polymerase I, essential for initiation and growth-dependent regulation of rRNA synthesis, is disrupted during transcription. EMBO J., 17, 3692–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller O.L. Jr, and Beatty,B.R. (1969) Visualization of nucleolar genes. Science, 164, 955–957. [DOI] [PubMed] [Google Scholar]
- Moorefield B., Greene,E.A. and Reeder,R.H. (2000) RNA polymerase I transcription factor Rrn3 is functionally conserved between yeast and human. Proc. Natl Acad. Sci. USA, 97, 4724–4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muth V., Nadaud,S., Grummt,I. and Voit,R. (2001) Acetylation of TAFI68, a subunit of TIF-IB/SL1, activates RNA polymerase I transcription. EMBO J., 20, 1353–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osheim Y.N., Mougey,E.B., Windle,J., Anderson,M., O’Reilly,M., Miller,O.L.,Jr, Beyer,A. and Sollner-Webb,B. (1996) Metazoan rDNA enhancer acts by making more genes transcriptionally active. J. Cell Biol., 133, 943–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier G., Stefanovsky,V.Y., Faubladier,M., Hirschler-Laszkiewicz,I.I., Savard,J., Rothblum,L.I., Cote,J. and Moss,T. (2000) Competitive recruitment of CBP and Rb-HDAC regulates UBF acetylation and ribosomal transcription. Mol. Cell, 6, 1059–1066. [DOI] [PubMed] [Google Scholar]
- Planta R.J. (1997) Regulation of ribosome synthesis in yeast. Yeast, 13, 1505–1518. [DOI] [PubMed] [Google Scholar]
- Rattner J.B., Saunders,C., Davie,J.R. and Hamkalo,B.A. (1982) Ultrastructural organization of yeast chromatin. J. Cell Biol., 93, 217–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder R.H. (1989) Regulatory elements of the generic ribosomal gene. Curr. Opin. Cell Biol., 1, 466–474. [DOI] [PubMed] [Google Scholar]
- Reeder R.H. (1992) The regulation of transcription by RNA polymerase I. In Yakamoto,K. and McKnight,S. (eds), Transcriptional Regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Robyr D., Suka,Y., Xenarios,I., Kurdistani,S.K., Wang,A., Suka,N. and Grunstein,M. (2002) Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell, 109, 437–446. [DOI] [PubMed] [Google Scholar]
- Roth S.Y., Denu,J.M. and Allis,C.D. (2001) Histone acetyltransferases. Annu. Rev. Biochem., 70, 81–120. [DOI] [PubMed] [Google Scholar]
- Rundlett S.E., Carmen,A.A., Kobayashi,R., Bavykin,S., Turner,B.M. and Grunstein,M. (1996) HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl Acad. Sci. USA, 93, 14503–14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundlett S.E., Carmen,A.A., Suka,N., Turner,B.M. and Grunstein,M. (1998) Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature, 392, 831–835. [DOI] [PubMed] [Google Scholar]
- Siddiqi I.N., Dodd,J.A., Vu,L., Eliason,K., Oakes,M.L., Keener,J., Moore,R., Young,M.K. and Nomura,M. (2001) Transcription of chromosomal rRNA genes by both RNA polymerase I and II in yeast uaf30 mutants lacking the 30 kDa subunit of transcription factor UAF. EMBO J., 20, 4512–4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J.S. and Boeke,J.D. (1997) An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev., 11, 241–254. [DOI] [PubMed] [Google Scholar]
- Smith J.S., Caputo,E. and Boeke,J.D. (1999) A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol. Cell. Biol., 19, 3184–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan J.S., Keys,D.A., Dodd,J.A. and Nomura,M. (1996) The role of TBP in rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae: TBP is required for upstream activation factor-dependent recruitment of core factor. Genes Dev., 10, 2551–2563. [DOI] [PubMed] [Google Scholar]
- Suka N., Suka,Y., Carmen,A.A., Wu,J. and Grunstein,M. (2001) Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol. Cell, 8, 473–479. [DOI] [PubMed] [Google Scholar]
- Sun Z.-W. and Hampsey,M. (1999) A general requirement for the Sin3–Rpd3 histone deacetylase complex in regulating silencing in Saccharomyces cerevisiae. Genetics, 152, 921–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taunton J., Hassig,C.A. and Schreiber,S.L. (1996) A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science, 272, 408–411. [DOI] [PubMed] [Google Scholar]
- Turner B.M., Birley,A.J. and Lavender,J. (1992) Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell, 69, 375–384. [DOI] [PubMed] [Google Scholar]
- Vannier D., Balderes,D. and Shore,D. (1996) Evidence that the transcriptional regulators SIN3 and RPD3, and a novel gene (SDS3) with similar functions, are involved in transcriptional silencing in S.cerevisiae. Genetics, 144, 1343–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verreault A., Kaufman,P.D., Kobayashi,R. and Stillman,B. (1996) Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell, 87, 95–104. [DOI] [PubMed] [Google Scholar]
- Vidal M. and Gaber,R.F. (1991) RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol. Cell. Biol., 11, 6317–6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel T.J., Teunissen,A.W.R.H. and Steensma,H.Y. (1995) PDA1 mRNA: a standard for quantitation of mRNA in Saccharomyces cerevisiae superior to ACT1 mRNA. Nucleic Acids Res., 23, 883–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler E.A. et al. (1999) Functional characterization of the S.cerevisiae genome by gene deletion and parallel analysis. Science, 285, 901–906. [DOI] [PubMed] [Google Scholar]
- Xie X., Kokubo,T., Cohen,S.L., Mirza,U.A., Hoffmann,A., Chait,B.T., Roeder,R.G., Nakatani,Y. and Burley,S.K. (1996) Structural similarity between TAFs and the heterotetrameric core of the histone octamer. Nature, 380, 316–322. [DOI] [PubMed] [Google Scholar]
- Yamamoto R.T., Nogi,Y., Dodd,J.A. and Nomura,M. (1996) RRN3 gene of Saccharomyces cerevisiae encodes an essential RNA polymerase I transcription factor which interacts with the polymerase independently of DNA template. EMBO J., 15, 3964–3973. [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Sun,Z.W., Iratni,R., Erdjument-Bromage,H., Tempst,P., Hampsey,M. and Reinberg,D. (1998) SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol. Cell, 1, 1021–1031. [DOI] [PubMed] [Google Scholar]