Abstract
The extent of hyperthermophilic microbial diversity associated with siliceous sinter (geyserite) was characterized in seven near-boiling silica-depositing springs throughout Yellowstone National Park using environmental PCR amplification of small-subunit rRNA genes (SSU rDNA), large-subunit rDNA, and the internal transcribed spacer (ITS). We found that Thermocrinis ruber, a member of the order Aquificales, is ubiquitous, an indication that primary production in these springs is driven by hydrogen oxidation. Several other lineages with no known close relatives were identified that branch among the hyperthermophilic bacteria. Although they all branch deep in the bacterial tree, the precise phylogenetic placement of many of these lineages is unresolved at this time. While some springs contained a fair amount of phylogenetic diversity, others did not. Within the same spring, communities in the subaqueous environment were not appreciably different than those in the splash zone at the edge of the pool, although a greater number of phylotypes was found along the pool's edge. Also, microbial community composition appeared to have little correlation with the type of sinter morphology. The number of cell morphotypes identified by fluorescence in situ hybridization and scanning electron microscopy was greater than the number of phylotypes in SSU clone libraries. Despite little variation in Thermocrinis ruber SSU sequences, abundant variation was found in the hypervariable ITS region. The distribution of ITS sequence types appeared to be correlated with distinct morphotypes of Thermocrinis ruber in different pools. Therefore, species- or subspecies-level divergences are present but not detectable in highly conserved SSU sequences.
A picture of the kinds of hyperthermophilic microorganisms inhabiting slightly alkaline (pH 7.8 to 8.9) near-boiling hot springs around the world is beginning to emerge. Molecular and cultivation studies show that many of these ecosystems, including high- and low-sulfide springs in Japan, Iceland, Kamchatka, and Yellowstone National Park, are dominated by organisms belonging to the order Aquificales (11, 12, 13, 25, 29, 34). Recent cultivation of Thermocrinis ruber, the pink filaments isolated from Yellowstone's Octopus Spring, indicates that primary production in these ecosystems is by chemoautotrophic hydrogen oxidation (12). Other organisms in these ecosystems belong to known bacterial divisions, including the Thermotogales, the Thermus clade, and the Thermodesulfobacterium clade. Some organisms are unrelated to any known cultured divisions; they include the Korarchaeota (found in Yellowstone's Calcite Springs and Obsidian Pool) (3, 4), the lineage clone 8 cluster III associated with silica scale in a geothermal power plant (14), EM19 from Octopus Spring (25), and several other new candidate divisions from Obsidian Pool (4, 13).
The microbial communities in these near-boiling springs are closely associated with siliceous sinter (SiO2 · nH2O), commonly known as geyserite. Geyserite, by definition, precipitates from near-boiling fluids (temperatures greater than 80°C) along the bottoms and edges of hot springs and geysers (33). The morphology of the sinter deposits varies in different regions of the pools. Spicular geyserite forms at the air-water interface where vent water splashes along the inner rim of springs (Fig. 1). Spicules are elongated columnar or domal sinter structures, typically a few millimeters in diameter and several millimeters in length. Subaqueous siliceous sinter in the permanently submerged environment has a much different appearance and forms a flat, laminated geyserite deposit. In some springs, loose sediment accumulates in the bottom of the pool.
FIG. 1.
Edge of the source pool of Queen's Laundry. Spicular geyserite is shown in the splash zone at the air-water interface. Stratiform geyserite is formed beneath the water in the subaqueous environment.
Cady and Farmer (6; S. L. Cady, J. D. Farmer, D. J. Des Marais, and D. F. Blake, abstract from the 1995 annual meeting of the Geological Society of America [27:A304]) showed, using scanning electron microscopy (SEM), that these high-temperature geyserite surfaces are colonized by thin biofilms that consist primarily of filamentous microorganisms. The filaments are commonly silicified and gradually become entombed in silica, becoming part of the sinter fabric (17, 18, 19, 23, 27). Some workers have proposed that the filaments serve an active role in silica precipitation by providing favorable sites for nucleation; however, direct evidence of mineral templating has yet to be demonstrated (9, 16, 19, 20, 27). Cady et al. (Cady et al., Geol. Soc. Am. 27:A304, 1995) proposed that these organisms direct silica accretion and that the distribution of the microbes affects the morphological development of the geyserite. Though microorganisms have a direct role in the formation of sinter fabric, the factors controlling the formation of various sinter morphologies are often not known.
The aim of this study was to characterize the extent of thermophilic microbial diversity in a variety of microenvironments associated with subaqueous and spicular geyserite both within the same pool and among several different pools throughout Yellowstone. Analyses were done using environmental PCR amplification of the small-subunit rRNA gene (SSU rDNA), the large-subunit (LSU) rDNA, and the internal transcribed spacer (ITS) region, combined with in situ fluorescence microscopy and SEM.
(This work is part of a Ph.D. dissertation under the supervision of Brent Mishler, Department of Integrative Biology, University of California, Berkeley.)
MATERIALS AND METHODS
Locations sampled and DNA extraction.
Seven near-boiling silica-depositing springs were studied in three different locations of Yellowstone National Park (Table 1). Subaqueous sinter samples of loose sediment or stratiform geyserite were collected by scooping the sinter or sediment into a polypropylene tube attached to the end of a pole. Spicular geyserite at the air-water interface was collected using a chisel and tweezers. Growth surfaces (glass microscope slides) were incubated in a slide tray in the springs for 3 to 5 days as close as possible to the vent (5). Sinter samples were either preserved in 70% ethanol or were collected on ice and then extracted immediately in the laboratory. Microscope slides were collected in polypropylene tubes filled with 70% ethanol and stored at 4°C. Small pieces of sinter (approximately 0.5 g) were used to extract DNA from organisms present on sinter surfaces. Samples that were on ice were extracted directly, while those preserved in ethanol were first spun in an Eppendorf tube and then the ethanol was removed with a pipette. Cells on microscope slides were harvested with a razor blade and transferred to an Eppendorf tube by rinsing the slide with ethanol. The ethanol was removed by spinning the cells for 20 min and pipetting off the liquid. DNA was extracted by treating cells with lysozyme and proteinase K (13), followed by a freeze-thaw step (instead of bead beating). The samples were then extracted twice with equal volumes of phenol and chloroform and then twice with chloroform, purified on a Microcon (Millipore Corporation, Bedford, Mass.) YM-100 spin column, and resuspended in a small volume of Tris-EDTA (TE) (10 to 25 μl).
TABLE 1.
SSU rDNA clone libraries used in this study
| Springa | Temp (°C) | pHh | Type of samplec | Library name | Frequency of clonesg | Repre- sentative clones | Affiliation | % Identity to nearest hit by BLASTf | Concn (mg/liter) of:h
|
|
|---|---|---|---|---|---|---|---|---|---|---|
| SO4 | H2S | |||||||||
| Octopus Spring | 89-93 | 8.9 | LS near vent | OctSp | 56/78 | OctSp9 | Thermotogales | 97% to EM3 | 19 (31) | <0.016 (2) |
| 13/78 | OctSp106 | OctSpA1-106 | 100% to OctSpA1-106 | |||||||
| 8/78 | OctSp78 | Aquificales | 98% to Thermocrinis ruber | |||||||
| 1/78 | OctSp200 | EM19 | 96% to EM19 | |||||||
| Octopus Spring | 89-93b | 8.9 | SG | OctSpSin | 58/85 | OctSpSin2 | Aquificales | 97% to OPS165 | 19 (31) | <0.016 (2) |
| 23/85 | OctSpSin6 | Thermotogales | 98% to EM3 | |||||||
| 2/85 | OctSpSin69 | OctSpA1-106 | 100% to OctSpA1-106 | |||||||
| 1/85 | OctSpSin76 | Thermus | 98% to Thermus OPS15 | |||||||
| 1/85 | OctSpSin54 | OS Type L | 90% to OS-type L | |||||||
| Octopus Spring | 84 | 8.9 | LS, main pool | CS | 35/64 | CS1 | Aquificales | 98% to Thermocrinis ruber | NA | NA |
| 18/64 | CS5 | Thermus | 97% to Thermus aquaticus | |||||||
| 8/64 | CS2 | OS Type L | 95% to OS-type L | |||||||
| 3/64 | CS19 | CS19 | Unaffiliated | |||||||
| Queen's Laundry | 94-98 | 7.8 | SSG | QL3 | 76/80 | QL3-1 | Aquificales | 98% to Thermocrinis ruber | 21-22 (31) | 0.07-0.08 (30) |
| 4/80 | QL3-47 | Thermotogales | 96% to EM3 | 12-25 (30) | ||||||
| Queen's Laundry | 85-88b | 7.8 | SG | QL4d | 39/50 | QL4-1 | Aquificales | 98% to Thermocrinis ruber | 12-25 (30) | 0.07-0.08 (30) |
| 5/50 | QL4-12 | Bacillus | 96% to Bacillus sporother- modurans | |||||||
| 3/50 | QL4-19 | Thermotogales | 99% to EM3 | |||||||
| 2/50 | QL4-43 | EM19 | 98% to EM19 | |||||||
| 1/50 | QL4-2 | Clone 8 cluster III | 97% to clone 8 of cluster III | |||||||
| Queen's Laundry | 85-88b | 7.8 | SG | QL14 | 48/53 | QL14-1 | Bacillus | 97% to Bacillus sporosarcina | 12-25 (30) | 0.07-0.08 (30) |
| 4/53 | QL14-3 | Aquificales | 98% to OPS165 | |||||||
| 1/53 | QL14-64 | Thermotogales | 97% to EM3 | |||||||
| Queen's Laundry | 70.9 | 8.1 | SSG | QL5 | 65/66 | QL5-1 | Aquificales | 98% to Thermocrinis ruber | 12-25 (30) | 0.07-0.08 (30) |
| 1/66 | QL5-73 | Thermodesulfo- bacterium | 97% to OPT4 | |||||||
| Eclipse Geyser | 89-93 | NA | SSG | EGN | 63/63 | EGN2 | Aquificales | 97% to Thermocrinis ruber | NA | NA |
| Spindle Spring | 92-94 | 8.4 | GMS | SpinN | 72/72 | SpinN1 | Aquificales | 98% to Thermocrinis ruber | NA | NA |
| Boulder Spring | 92-95 | 8.5 | SSG | BSpN | 46/76 | BSpN2 | Aquificales | 98% to Thermocrinis ruber | 13-22 (30) | 0.29-0.49 (30) |
| 30/76 | BSpN50 | Thermodesulfo- bacterium | 97% to OPT4 | |||||||
| Abyss Pool | 85 | NA | LS | AbyssN | 52/69 | AbyssN1 | Thermotogales | 98% to EM3 | 55 (28) | NA |
| 12/69 | AbyssN9 | EM19 | 98% to EM19 | |||||||
| 5/69 | AbyssN61 | Aquificales | 97% to Thermocrinis ruber | |||||||
| Black Pool | 85 | 7.9 | LS | BH | 67/85 | BH1 | BH1 | Unaffiliated | NA | NA |
| 9/85 | BH9 | Thermotogales | 98% to EM3 | |||||||
| 4/85 | BH81, 92 | EM19 | 97% to EM19 | |||||||
| 3/85 | BH60 | BH60 | Unaffiliated | |||||||
| 2/85 | BH3 | Clone 8 cluster III | 96% to clone 8 cluster III | |||||||
| Black Pool | 85 | 7.9 | LS | BH4e | 16/27 | BH4-1 | EM19 | 97% to EM19 | NA | NA |
| 3/27 | BH4-13 | Thermus | 98% to Thermus aquaticus | |||||||
| 3/27 | BH4-26 | Aquificales | 98% to Thermocrinis ruber | |||||||
| 2/27 | BH4-63 | BH4-63 | Unaffiliated | |||||||
| 1/27 | BH4-59 | OS-Type L | 94% to OS-type L | |||||||
| 1/27 | BH4-10 | BH1 | Unaffiliated | |||||||
| 1/27 | BH4-50 | Clone 8 cluster III | 96% to clone 8 cluster III | |||||||
Locations of the springs: Octopus Spring, White Creek Area, Lower Geyser Basin; Queen's Laundry, Sentinel Meadows, Lower Geyser Basin; Eclipse Geyser, White Creek Area; Spindle Spring, White Creek Area; Boulder Spring, Sentinel Meadows; Abyss Pool, West Thumb Geyser Basin; Black Pool, West Thumb Geyser Basin.
Temperature refers to that of the vent water and may not be the temperature on the surface of the spicule.
Abbreviations: LS, loose sediment; SG, spicular geyserite; SSG, subaqueous stratiform geyserite; GMS, glass microscope slide (growth surface).
515F-1391R primer set.
8F-907R primer set.
Unaffiliated, nearest BLAST hit showed < 87% sequence identity to any known cultivated or uncultivated bacterial sequence.
Number of occurrences/total number analyzed.
NA, not available.
PCR amplification and cloning.
Typically, 1 μl of the prepared DNA was used in the PCR amplification of SSU and LSU rDNAs. Reaction mixtures contained 30 mM Tricine (pH 8.4), 50 mM KCl, 1.5 mM MgCl2, a 100 μM concentration of each deoxynucleoside triphosphate, 0.05% NP-40, 3 μl of Taq polymerase, and 2 ng of primer in a 50-μl volume. SSU reaction mixtures were incubated in an MJ Research thermal cycler at 94°C for 3 min, followed by 30 cycles at 92°C for 30 s, 37°C for 30s, and 72°C for 45 s, with a 72°C extension for 10 min. Most SSU libraries (Table 1) were amplified with the universal forward primer 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and reverse primer 1492R (5′ GGTTACCTTGTTACGACTT-3′) (21). Clone library QL4 was amplified with 515F and reverse primer 1391R (5′-GACGGGCGGTGWGTRCA-3′) (21). Clone library BH4 was amplified with bacterial primer 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 907R (5′-CCGTCAATTCCTTTRAGTTT-3′) (21).
DNA was extracted from Chloroflexus aurantiacus type strain J-10-f cells (provided by Beverly Pierson, University of Puget Sound) as described above, except a bead-beating step followed the freeze-thaw, and the DNA was ethanol precipitated instead of passing it over a spin column. Approximately 1.7 kb of the LSU rDNA was amplified using the LSU forward primer 208F (5′-TGAAACATCTNAGTA-3′) and the LSU bacterium-specific reverse primer 1939R (5′-ATTTCGCTACCTTAGGA-3′) by denaturing at 94°C for 3 min and then cycling 35 times at 92°C for 30 s, 45°C for 1 min, and 72°C for 2 min, with a 10 min 72°C extension. Similarly, the LSU rDNA was amplified from Deinococcus radiodurans (source: N. Pace's laboratory's genomic DNA archive). Approximately 1.9 kb of the LSU rDNA from the unidentified bacterium OctSpA1-106 was amplified from DNA extracted from Octopus Spring sediment. The primary amplification was performed with a 37°C annealing temperature using the SSU forward primer OctSpF (5′-GGAAGCCGCTGCACCTGA-3′) and the LSU universal reverse primer 2744R (5′-CTTAGATGCNTTCAGC-3′). A secondary amplification was performed with a 50°C annealing temperature using 1492F (reverse complement of 1492R) and 1939R. LSU primer sequences were similar to those in reference 21. The OctSpF primer sequence was chosen from an alignment of thermophilic bacterial SSU sequences.
SSU-ITS-LSU clone libraries (Table 2) spanned approximately 100 bp of the SSU, the entire 325 bp of the ITS region, and 1.9 kb of the LSU. The primary amplification was performed with a 50°C annealing temperature using the SSU primer AqalesF (5′-CGTCAGGTCAGTATGC-3′) and LSU primer 1939R. A secondary amplification was performed with a 53°C annealing temperature using 1391F (reverse complement of 1391R) and 1939R or 1059R (5′-GGCTGCYTCTAAGCMAA-3′). All PCR products were cloned, screened by restriction fragment length polymorphism analysis (except for the BH4 library), and sequenced as previously described (13). Most clones were sequenced with vector primers; however, internal reads were also done when ambiguities were present. Chimeras were identified with distance matrices using an internal database of rDNAs from bacterial thermophiles.
TABLE 2.
LSU and SSU-ITS-LSU rDNA clone libraries used in this study
| DNA source | Temp (°C) | Location of sample | Type of sample | Library name | Region amplified | Amplification primersa | Annealing temp (°C)a |
|---|---|---|---|---|---|---|---|
| Chloroflexus aurantiacus | NAb | NA | J-10-f cells | Chlaurant | LSU | 208F-1939R | 45 |
| Deinococcus radiodurans | NA | NA | Genomic DNA | Deinrad | LSU | 208F-1939R | 45 |
| Octopus Spring | 89-93 | Source pool | Subaqueous sediment | OctSp23Sa | SSU-ITS-LSU | OctSpF-2744R/1492F-1939R | 37/50 |
| Black Pool | 85 | Main pool | Subaqueous sediment | BHla | SSU-ITS-LSU | 515F-1939R | 50 |
| Queen's Laundry | 85-88 | Source pool | Growth surface | QLAqLSU | SSU-ITS-LSU | AqalesF-1939R/1492F-1059R | 50/55 |
| Boulder Spring | 92-95 | Source pool | Growth surface | BSpLa | SSU-ITS-LSU | AqalesF-1939R/1391F-1059R | 50/53 |
| Octopus Spring | 89-93 | Source pool | Subaqueous sediment | 202 | SSU-ITS-LSU | AqalesF-1939R/1391F-1939R | 50/53 |
| Octopus Spring | 75 | Main pool below pho- tosynthetic mat | Subaqueous sediment | 206 | SSU-ITS-LSU | AqalesF-1939R/1391F-1939R | 50/53 |
| Octopus Spring | 75 | Main pool below pho- tosynthetic mat | Subaqueous sediment | 306 | SSU-ITS-LSU | AqalesF-1939R/1391F-1059R | 50/53 |
| Octopus Spring | 85-87 | Outflow channel of source pool | Pink filaments | 310 | SSU-ITS-LSU | AqalesF-1939R/1391F-1059R | 50/53 |
| Queen's Laundry | 85-88 | Source pool | Subaqueous stratiform geyserite | 312 | SSU-ITS-LSU | AqalesF-1939R/1391F-1059R | 50/53 |
| Octopus Spring | 85 | Main pool near source pool | Subaqueous sediment | 308 | SSU-ITS-LSU | AqalesF-1939R/1391F-1059R | 50/53 |
| Queen's Laundry | 70.9 | Main pool near out- flow channel | Subaqueous stratiform geyserite | 316 | SSU-ITS-LSU | AqalesF-1939R/1391F-1059R | 50/53 |
Conditions indicated for primary amplification (left side of slash) and secondary amplification (right side of slash).
NA, not available.
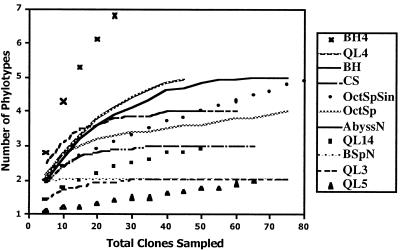
Rarefaction curves.
Rarefaction curves were generated using the program Resampling Rarefaction 1.0, made available by Steve Holland, University of Georgia Statigraphy Lab (http://www.uga.edu/∼strata/software/). Small subsets of each clone library were resampled with increasing sample sizes, with 100 replicates performed for each sample size (Fig. 2).
FIG. 2.
Rarefaction curves for SSU rDNA libraries. The number of phylotypes sampled was plotted against the number of individual clones subsampled from the original clone library. Plots for libraries that did not plateau (BH4, OctSpSin, QL14, and QL5) are indicated by tick marks, and libraries that did plateau are indicated by smooth lines. Libraries EGN and SpinN are not shown.
Phylogenetic analyses.
Sequences were manually aligned with other known rRNA sequences according to their predicted secondary structure using Genetic Data Environment software (version 2.2; http://bimas.dcrt.nih.gov/gde_sw.html), and trees were inferred using maximum-parsimony (MP), minimum-evolution (ME), and maximum-likelihood (ML) methods provided in PAUP* (version 4; Sinauer Associates, Sutherland, Mass.). Alignment A contained SSU sequences with a character set of 1,148 nucleotides (hypervariable characters were excluded from the analysis). Alignment B contained both SSU and LSU sequences (with 1,119 and 1,079 nucleotides in the character set, respectively). Bootstrap values were calculated using MP (with 20 random taxon additions performed per bootstrap replicate), ME (using the Hasegawa-Kishino-Yano [HKY] model and a gamma shape parameter estimated by ML), ML (where the gamma shape parameter, proportion of invariant nucleotides, and the transition/transversion ratio were estimated by ML), and fastDNAml (22). The number of changes per character per branch (λ) was estimated by dividing the number of character changes along a branch by the total number of characters in the analysis (1).
SEM.
Specimens of sinter and growth surfaces were prepared for SEM either by air drying or by dehydration in a graded ethanol series followed by critical-point drying. The specimens were then coated with a thin conductive layer (∼50 Å) of ion-sputtered tungsten and viewed with a Hitachi 4000 SEM operating between 2 and 5 keV (6).
In situ hybridizations.
Fluorescein-labeled RNA probes complementary to the SSU rRNA were transcribed using Qiagen-purified plasmid DNA. Two micrograms of plasmid DNA was cut with PvuII, phenol-chloroform extracted, ethanol precipitated, and then incubated in a solution containing 40 mM Tris (pH 8.0); 2 mM spermidine; 5 mM dithiothreitol; bovine serum albumin (50 μg/ml); 0.01% NP-40; 9 mM MgCl2; a 0.6 mM concentration (each) of GTP, ATP, and CTP; 10 mM fluorescein-12-UTP (Boehringer Mannheim); 20 U of RNase inhibitor; 2.5 U of pyrophosphatase; and 40 U of T7 RNA polymerase at 37°C for at least 6 h. Transcripts were ethanol precipitated and separated on a 4% denaturing acrylamide gel, excised with a razor blade, and eluted overnight in 200 to 400 μl of solution containing TE and 0.1% sodium dodecyl sulfate. RNAs were ethanol precipitated and resuspended in 20 to 50 μl of TE. Growth surfaces were fixed with 3.7% formaldehyde for 20 min at room temperature and then rinsed several times with 70% ethanol and air dried. The slides were broken into small pieces and hybridized with 40 ng of probe in 12 μl of hybridization buffer [373 mM NaCl, 2.5 mM EDTA, 50 mM Tris (pH 8.0), poly(A) (1 mg/ml), 0.2% bovine serum albumin, 5% formamide] in a heat-sealable plastic bag for 4 to 6 h at temperatures ranging from 50 to 60°C. Slides were washed in 1.5 ml of wash buffer (20 mM Tris [pH 8.0], 5 mM EDTA, 450 mM NaCl) for 5 min at room temperature three times, rinsed briefly in water, and air dried. Slide pieces were mounted with an antifade medium (Vectashield; Vector Laboratories, Inc.), and imaged with a Zeiss E600 fluorescent microscope and Optronix charge-coupled device camera. Except in the case of the Thermocrinis ruber probe (see below), hybridization temperatures were increased by 2 or 3°C increments until only one morphotype remained. Growth surfaces from Eclipse Geyser served as a negative stringency control for the BSpN50/Thermodesulfobacterium, EM19, QL4-2, and OctSpA1-106 probes (not shown), since these phylotypes were not found in the Eclipse Geyser SSU clone library. Thus, conditions were chosen so that the probes hybridized with one morphology and not with cells from Eclipse Geyser. Because Thermocrinis ruber was found in all springs, conditions were optimized, using growth surfaces from Boulder Spring, so that the BSpN50/Thermodesulfobacterium probe labeled one morphotype and the Thermocrinis ruber probe labeled another.
Cluster analysis.
A multivariate cluster analysis was performed on the microbial composition data of each spring using Jaccard's coefficients. Phylotypes (treated as character states) were scored for presence or absence in each clone library (treated as taxa), and a cluster diagram of clone libraries was constructed by using the unweighted pair group method using arithmetic averages in PAUP*.
Nucleotide sequence accession numbers.
Nucleotide sequences have been submitted to GenBank under the accession numbers AF015925 and AF352532 through AF352554.
RESULTS
SEM.
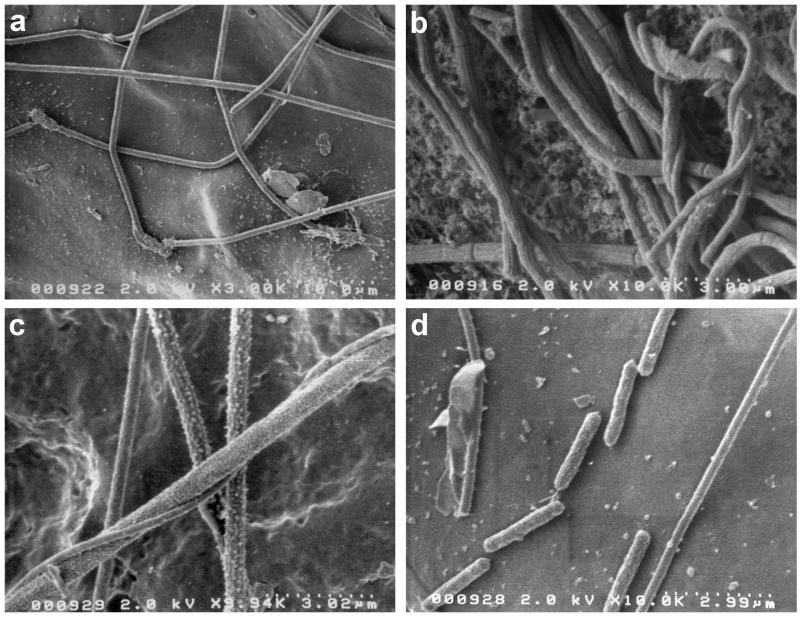
Growth surfaces (glass microscope slides) incubated in the subaqueous region of the Octopus Spring source pool (89 to 92°C) for several days were quickly colonized by microbes. SEM photomicrographs revealed at least five different cell morphotypes, including four types of filaments and short rods seen individually or in chains (Fig. 3). In contrast, the cells of the pink filamentous streamers from the Octopus Spring outflow channel (84 to 88°C), a meter or so downstream of the source pool, had a different morphotype than cells in the source pool.
FIG. 3.
Scanning electron micrographs of bacterial morphotypes growing on a glass microscope slide incubated in the Octopus Spring source pool for several days (a, c, and d) and of the pink filamentous streamers (b) from the outflow channel of Octopus Spring. The scale bar is at the lower right of each panel.
Community analyses.
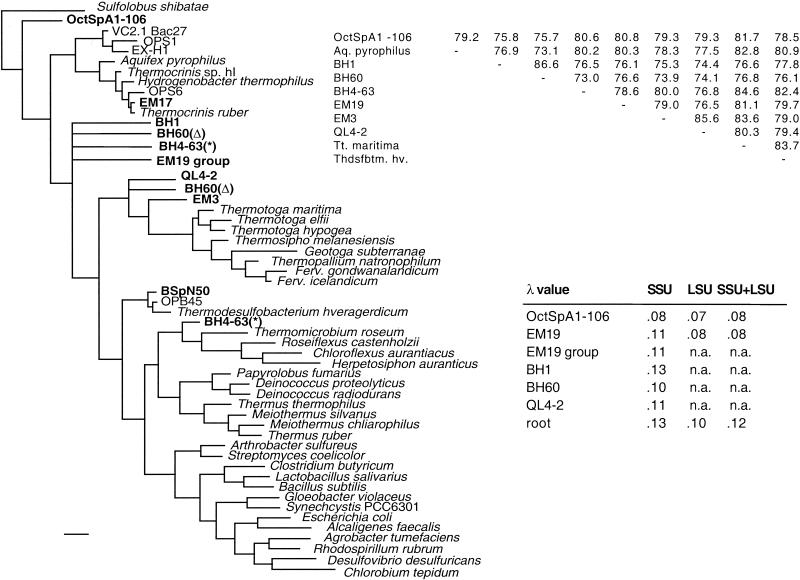
The results of the SSU rDNA analyses for seven springs are shown in Table 1. Some springs contained only one phylotype (e.g., Eclipse Geyser and Spindle Spring), and one spring contained only two phylotypes (Boulder Spring), while most springs contained three to five phylotypes. Black Pool, which is at a lower temperature, contained at least nine phylotypes. Lineages EM3, EM19, and OS-type L are all uncultivated lineages previously found in Octopus Spring that have no known close relatives (25, 32). Lineage QL4-2 is a novel lineage closely related to a very short sequence obtained from silica scale in a Japanese geothermal power plant (14). Lineages OctSpA1-106, CS19, BH1, BH60, and BH4-63 are novel lineages found in this study that also have no known close relatives.
Black Pool, a large quiescent pool near Abyss Pool, contained a community that was unique. Library BH, constructed from subaqueous sediment, contained five phylotypes: BH1, EM3, EM19, BH60, and QL4-2. An SSU-ITS-LSU clone library (Table 2) made from the same DNA sample also amplified rDNAs corresponding to OctSpA1-106 and Thermocrinis ruber, showing that these lineages were present even though they were not detected in the SSU clone library. In addition, the BH4 library (Table 1), constructed with bacterium-specific SSU primers, amplified other lineages (BH4-63, a close relative of Thermus spp. and OS-type L) that were not found in either the universal SSU or the SSU-ITS-LSU libraries.
For most of the phylotypes found in this study (OctSpA1-106, Thermocrinis ruber, EM19, EM3, QL4-2, and Thermodesulfobacterium), sequences from geographically separated pools were often nearly identical. To compare sequence diversity in the Thermocrinis ruber phylotype, multiple clones from the same pool and from many other pools were sequenced completely (25,375 bp analyzed). On average, all sequences were 99.6% identical, while the two most divergent sequences in this clade were 98.4% identical. For comparison, EM19 sequences among different springs were on average 98.9% identical (out of 11,325 bp analyzed), EM3 sequences showed 98.6% identity (out of 5,220 bp analyzed), QL4-2 sequences were 99.5% identical (out of 2,310 bp), and Thermodesulfobacterium sequences were 99.5% identical (out of 4,645 bp analyzed). Population-level SSU trees were not constructed from these sequences, because the lack of sequence diversity resulted in poorly supported topologies.
Though rarefaction analyses (Fig. 2) of the clone libraries showed that most of the libraries were adequately sampled, they also indicated that four libraries (BH4, OctSpSin, QL14, and QL5) were not (libraries EGN and SpinN were not plotted since they only contained one phylotype). It is possible that additional sampling of clones may result in a larger number of phylotypes detected, although if the trend lines for the OctSpSin, QL14, and QL5 libraries are extended (assuming they will not flatten out), only 0.5 to 1.1 more clones would be expected by sampling 200 clones. The BH4 library contained a larger number of phylotypes than the BH library, despite inadequate sampling of the BH4 library, indicating primer specificity or selective amplification.
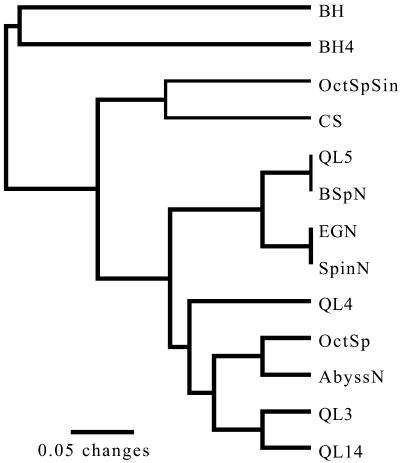
A multivariate cluster analysis of the community composition in each spring (Fig. 4) showed that the subaqueous community in the Octopus Spring source pool was most similar to the subaqueous community of Abyss Pool. In contrast, the community colonizing the spicular geyserite of the Octopus Spring source pool (OctSpSin) was most similar to the subaqueous community (CS) in the cooler (84°C) main pool of Octopus Spring (this pool lacks its own source and is filled by fluids from the hotter neighboring source pool). Communities from spicular geyserite and subaqueous geyserite clustered randomly through the tree and with each other, exhibiting polyphyletic relationships. The community from Black Pool was the most unique of those sampled.
FIG. 4.
Cluster (unweighted pair group method using arithmetic averages) diagram using Jaccard's coefficient. Clone libraries are represented here as taxa. Clone libraries with the greatest number of shared phylotypes cluster together.
In situ hybridizations.
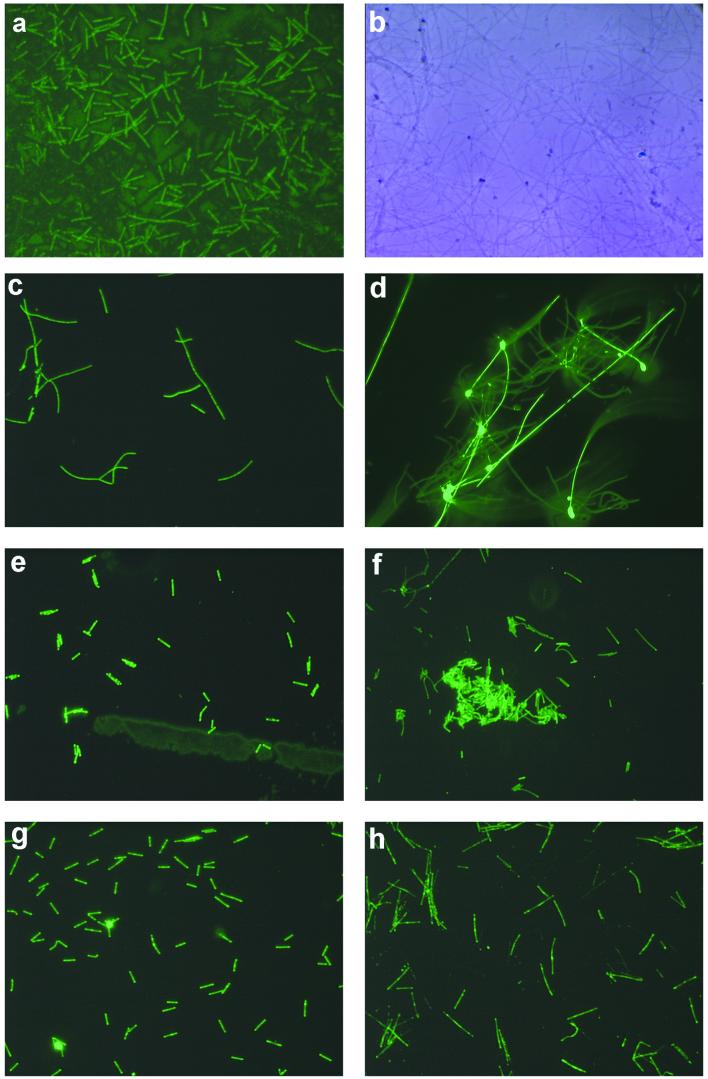
Hybridizations with the QL4-2 probe against slides incubated in the Queen's Laundry source pool showed that the QL4-2 phylotype corresponds to short filaments that were frequently found to be attached to each other at one end, forming rosettes (Fig. 5a ). The corresponding phase-contrast image (Fig. 5b) shows a large diversity of morphotypes that did not hybridize with the probe.
FIG. 5.
In situ hybridizations using glass microscope slides. Digital camera integration times and the hybridization temperature are indicated in parentheses. (a) QL4-2 probe against a slide from Queen's Laundry (0.5 s, 53°C). (b) Phase-contrast image of the same field of view showing specificity of the QL4-2 probe and variety of morphotypes on the slide. (c and d) Thermocrinis ruber probe against slides from the Octopus Spring source pool (2 and 1 s, both 53°C). (e) Thermocrinis ruber probe against a slide from Boulder Spring (2 s, 53°C). (f) BSpN50/Thermodesulfobacterium probe against a slide from Boulder Spring (2 s, 55°C). (g) OctSpA1-106 probe against a contact slide from the Octopus Spring source pool (1 s, 50°C). (h) EM19 probe against a slide from the Octopus Spring source pool (1 s, 58°C). All images are taken at a magnification of ×400. For comparison of scale, the long filaments with holdfasts (d) are the same as those in Fig. 3a.
Figures 5c, d, and e show hybridizations carried out with the Thermocrinis ruber probe against slides incubated in the Octopus Spring source pool. Two distinct morphotypes of filaments hybridized with the probe. One had a variable length with regular septa along the filament and appeared to be branched (Fig. 5c). Another morphotype had a very long filament with a brightly fluorescing holdfast and only occasional septa (Fig. 5d). This probe also hybridized to short rods (not shown), but this morphotype was not very abundant. Hybridizations to contact slides from Spindle Spring and Eclipse Geyser (not shown) showed dense populations of long filaments with holdfasts. On Spindle Spring samples, a small, solitary patch of short rods was seen to hybridize; however, this morphotype was rare. Hybridizations against cells from Boulder Spring (Fig. 5e) revealed another distinct morphotype, short rods that were often paired. They fluoresced unevenly, having bright regions inside the cells and at the ends. In contrast, the Thermodesulfobacterium probe (Fig. 5f) hybridized to thinner rods of various length which often formed microcolonies.
Cells from the Octopus Spring source pool that hybridized with the OctSpA1-106 probe (Fig. 5g) were rods, also with uneven fluorescence (Fig. 5e). The EM19 probe (Fig. 5h), in contrast, hybridized to longer rods that were rare in abundance and occurred unevenly in patches on the growth surfaces.
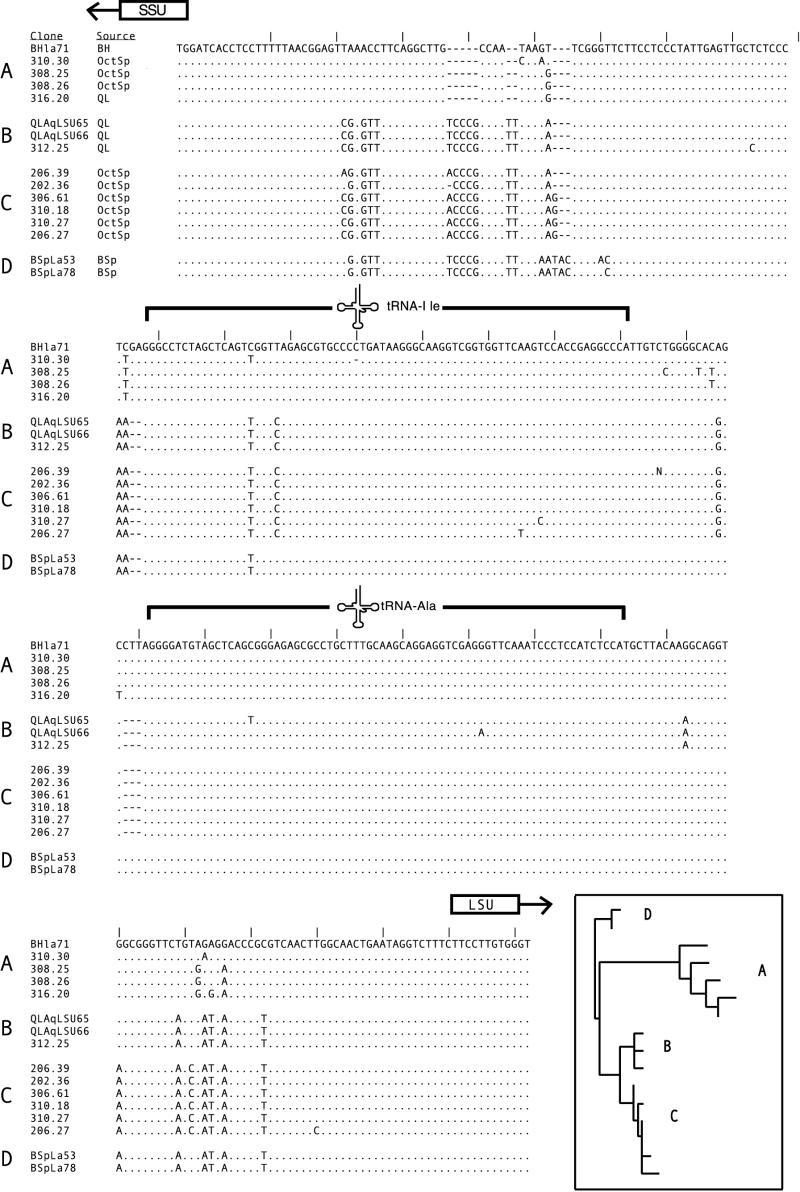
Thermocrinis ruber ITS diversity among different springs.
In order to investigate the sequence diversity within the Thermocrinis ruber clade, SSU-ITS-LSU clone libraries were constructed from DNA isolated from several locations in Queen's Laundry pool, Octopus Spring pool, Black Pool sediment, and Boulder Spring subaqueous sinter. Sequencing of several random clones from each of these libraries (Fig. 6) showed that despite having nearly identical SSU and LSU sequences, a great deal more sequence diversity occurred on the level of the ITS region. The sequences from Black Pool, Octopus Spring, and Queen's Laundry fell into three distinct monophyletic groups, while those from Boulder Spring were unique. ITS sequences from cells growing on a growth surface in Queen's Laundry pool were nearly identical and belonged to the same cluster as those found in the subaqueous stratiform geyserite from the same pool, suggesting that the sample source (i.e., glass slide versus natural sinter surface) did not bias the community composition.
FIG. 6.
Sequence alignment of the ITS region of Thermocrinis ruber from various springs. The locations of the two tRNA genes, SSU, and LSU rDNAs are indicated by brackets and arrows. Four main ITS clusters were found (the inset shows an ME tree). The clone library and clone number are designated in the clone name (e.g., BHla71 corresponds to clone 71 in the BHla library). Abbreviations for the sources are as follows: BH, Black Pool; OctSp, Octopus Spring; QL, Queen's Laundry; and BSp, Boulder Spring. Sequences identical to the BHla71 sequence are indicated with a period; gaps with respect to the BHla71 sequence are indicated with a dash.
Phylogenetic analyses of the OctSpA1-106 SSU and LSU sequences.
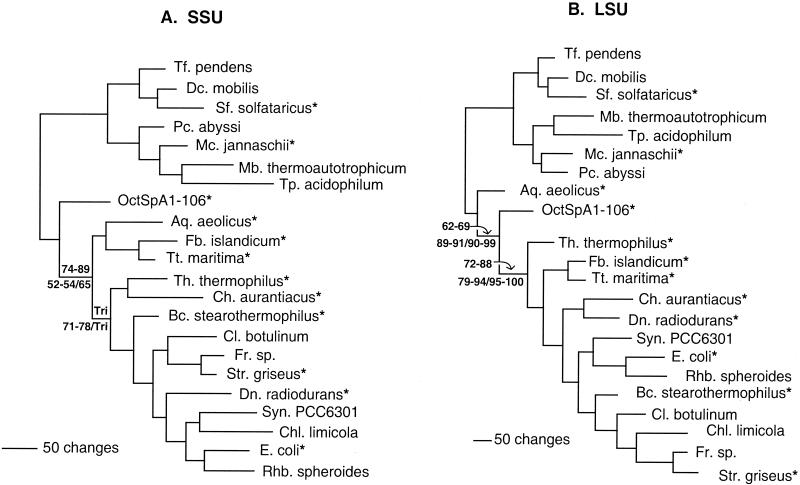
OctSpA1-106, found in the Octopus Spring source pool subaqueous sediment and spicular geyserite clone libraries, was found to be a novel SSU sequence. A nearly identical sequence was also found in an SSU-ITS-LSU library constructed from Black Pool sediment. It was studied in detail because phylogenetic analyses showed that it was the deepest diverging branch in the bacterial line of descent (Fig. 7A). Bootstrap support for this placement was moderate to good (ranging from 52 to 95%) and varied according to the taxa, character set, and phylogenetic method used. MP and ME tended to show the lowest bootstrap support (75 to 93%), while ML (both ML and fastDNAml) showed higher bootstrap values (88 to 92%). Bootstrap support was sensitive to inclusion of EM19, whose branching position is poorly resolved within the thermophilic bacteria. Removal of EM19 from analyses increased bootstrap support (by 4 to 25%) for OctSpA1-106 being the first branch to diverge from the bacterial line of descent. The number of changes per character (λ) along the branch leading to OctSpA1-106 is 0.80, while that leading to EM19 is 0.11 (Fig. 8).
FIG. 7.
MP trees constructed using SSU (A) and LSU (B) rRNA with 10,000 random addition replicates. Bootstrap calculations using MP were performed with five random addition replicates per bootstrap replicate, and bootstrap values are indicated for the OctSpA1-106 and Aquifex aeolicus clades. ME bootstrap values (below the branch to the left of the slash) were calculated with HKY as a nucleotide substitution model with a gamma shape parameter (0.4677947158 for the SSU data set and 0.5141387521 for LSU) and proportion of invariable characters (0.06518549129 for SSU and 0.02032941966 for LSU) estimated by ML. ML bootstrap values (below the branch to the right of the slash) were obtained with PAUP* using a restricted number of taxa (indicated by asterisks). “Tri” indicates that the bootstrap tree showed a trifurcation at this node. Abbreviations: Tf., Thermofilum; Dc., Desulfurococcus; Sf., Sulfolobus; Pc., Pyrococcus; Mc., Methanococcus; Mb., Methanobacterium; Tp., Thermoplasma; Aq., Aquifex; Fb., Fervidobacterium; Tt., Thermotoga; Th., Thermus; Ch., Chloroflexus; Bc., Bacillus; Cl., Clostridium; Fr., Frankia; Str., Streptomyces; Dn., Deinococcus; Syn., Synechocystis; Chl., Chlorobium; E., Escherichia; Rhb., Rhodobacter.
FIG. 8.
Consensus phylogenetic tree of novel bacterial SSU rDNA sequences found in this study calculated using MP. A distance matrix of the sequences (percent identity) is shown in the upper right corner. λ values for the SSU and LSU of various sequences are tabulated in the lower right corner (n.a., sequence not available).
To test the sensitivity of this topology to the stem G+C content of outgroup taxa, analyses were done with three different sets of archaeal taxa. A high stem G+C content set (averaging 84.1%) resulted in the highest bootstrap support for the deeply branching nature of OctSpA1-106 (95%, using fastDNA-ml). A low stem G+C content set (averaging 72.4%) showed a lower bootstrap support for the OctSpA1-106 lineage (80%), while a large outgroup set containing both high and low stem G+C content archaeal sequences resulted in an intermediate bootstrap support (88%).
The LSU rDNA from the OctSpA1-106 organism was isolated by constructing a clone library of the SSU-ITS-LSU region with DNA from the Octopus Spring source pool sediment (Table 2). Trees made with the OctSpA1-106 LSU sequence had a different topology than those made with the SSU gene: LSU trees had moderate (62 to 69%) to high (89 to 99%) bootstrap support for OctSpA1-106 branching between Aquifex pyrophilus (or A. aeolicus) and Thermotoga maritima (Fig. 6B). Again, bootstrap support for this branching topology varied according to the character set, taxa, and method used. A combined analysis with both LSU and SSU had moderate to high bootstrap support for OctSpA1-106 branching between the Aquificales and Thermotoga maritima (not shown). The value for λ for the OctSpA1-106 branch was 0.07 for the LSU analysis and 0.08 for the combined LSU and SSU analysis.
Phylogenetic analysis of other SSU sequences.
Bootstrap support for the EM19 sequence was previously reported to be poor (25). It was hoped that by expanding the sequence diversity in this clade the phylogenetic resolution of this group would improve. However, this was not the case. Two relatives of EM19 were found in the Black Pool and Queen's Laundry pool SSU libraries: BH92 and QL4-43 were 97.7 and 96.3% identical to EM19, respectively. The EM19 sequence alone had a branch length of 0.11, and the EM19 clade (with BH92 and QL4-43) was also 0.11 (Fig. 8). Bootstrap support for the branching location of this EM19 clade remained poor yet showed two alternative branching places: between the Thermotogales and the Thermodesulfobacterium clade and between the Thermotogales and the Aquificales.
The QL4-2 lineage branched with EM3 (deep in the Thermotogales) and in some cases brought EM3 out of the Thermotogales into a new clade. Branching of QL4-2 with EM3 was likely a long branch attraction artifact, since the two sequences shared little sequence similarity. Indeed, when EM3 was removed from the analysis, QL4-2 occasionally could be found to branch deep in the Thermodesulfobacterium clade or between the Thermotogales and the Thermodesulfobacterium clade.
Similarly, the BH1, BH60, and BH4-63 lineages had poor bootstrap support. BH1 and BH60 appeared to branch somewhere deep in the bacterial tree, while BH4-63 branched slightly higher, sometimes with the green nonsulfur bacteria or between the Thermodesulfobacterium clade and the Thermus-Deinococcus clade in others.
DISCUSSION
Organisms belonging to the Aquificales (specifically the Thermocrinis ruber clade) appear to be ubiquitous in the microbial communities of slightly alkaline, silica-depositing springs in Yellowstone National Park. Other studies suggest that this is also the case for similar springs throughout the world (12, 13, 25, 29, 34). Because many of the members of the order Aquificales are hydrogen oxidizers, it is likely that primary production in these ecosystems is driven by chemoautotrophic hydrogen oxidation. The results of a recent comparative study of the carbon isotopic signature of the Octopus Spring pink filamentous streamer community and cultured cells of Thermocrinis ruber suggest that these organisms may also be using formate as a carbon and energy source (15).
In general, these communities appear to be composed entirely of thermophilic bacteria. PCRs using archaeal primers failed to amplify, despite several attempts with various archaeon-specific primers (not shown), suggesting that Archaea are rare in these communities. Analyses of lipids in the subaqueous community of the Octopus Spring source pool show that although archaeol is present, it is in very small quantities relative to bacterial lipids (15). The abundance of bacteria in these environments is supported by DNA hybridization analyses of various springs in Yellowstone (13), and by an SSU rDNA study of Japanese alkaline springs (34). In contrast, a large diversity of archaeal sequences has been found in two non-silica-depositing, near-neutral-pH springs in Yellowstone: Obsidian Pool (4) and Calcite Springs (24).
EM3, a divergent member of the Thermotogales, is common in many of these springs, being found in Abyss Pool, Black Pool, and on both the subaqueous sinter and spicular geyserite in Queen's Laundry pool and Octopus Spring. EM19 is also present in these four springs; however, it was not consistently found in all microenvironments in each spring. For example, it was observed only in the spicular geyserite library from Queen's Laundry and the loose sediment communities of Octopus Spring, Abyss Pool, and Black Pool. In situ hybridizations suggest that EM19 is rare in abundance and formed patchy microcolonies on growth substrates. This observation may explain why EM19 is found in some clone libraries but not others. A close relative of Thermodesulfobacterium was abundant in Boulder Spring (as evidenced by the clone library and by in situ hybridization) yet was a minor component of the subaqueous sinter library of Queen's Laundry pool. This phylotype is found in springs with measurable H2S and is consistent with Thermodesulfobacterium being a thermophilic sulfate reducer (Table 1). The OctSpA1-106 phylotype has a rather limited distribution and is largely restricted to the Octopus Spring source pool. PCR amplifications with primers specific for this lineage using DNA from Queen's Laundry, Spindle Spring, Eclipse Geyser, and Boulder Spring all failed (not shown), although this phylotype was found in an SSU-ITS-LSU library constructed from Black Pool sediment.
The QL4-2 lineage was present in the spicular geyserite community in Queen's Laundry pool and in Black Pool sediment. A 275-bp DNA fragment closely related to this sequence was reported from an 85°C community associated with silica scale in a Japanese geothermal power plant (14). However, it was mistakenly identified as a member of the anaerobic gram-positive bacterial clade containing Thermoanaerobacter, Caldocellum, and Anaerocellum spp. Phylogenetic analysis of the entire SSU gene shows that this lineage clearly branches much deeper in the bacterial domain, although the precise branching location is unresolved due to long branch attraction artifacts. The distribution of QL4-2 suggests that this organism may be common in springs with temperatures around 85°C. CS19 was another novel sequence found in sediment from the cooler (84°C) main pool of Octopus Spring, and may also be common in springs in this temperature range (although neither QL4-2 nor CS19 was found in Abyss Pool sediment).
Several novel lineages were found in the sediment of Black Pool (BH1, BH60, BH4-63) that were not found in any other spring. Black Pool is unusual in that it has been undergoing a systematic change in chemistry over the past several years. Unfortunately, detailed documentation of this change in chemistry is lacking, although a log of temperatures and visual observations is kept by the park rangers at West Thumb Geyser Basin (N. Wilson [Grant Visitor Center, Yellowstone National Park], personal communication). The samples for this study were taken in July 1998; however, the pool had decreased in temperature by almost 10°C by June 2000. It is not known if we sampled a transitory population resulting from a shift in temperature and/or chemistry or if stable communities containing these lineages will be found in other Yellowstone springs.
Although all SSU rDNA sequences found in this study branch among the hyperthermophilic bacteria at the base of the bacterial domain, resolution of the precise branching order of many of the lineages is problematic. This is because most lineages have no close relatives and there is little to no sequence diversity within clades, making long branch attraction a serious issue. One strategy to deal with this problem is to expand the diversity within each clade in order to break up the long branches (10). Finding two close relatives of the EM19 sequence in this study, however, was not enough to break up the long branch leading to this clade. Branch lengths of 0.05 are easily resolved in phylogenetic analyses using nucleotide data (1). Saitou (26) showed that branch lengths with λ values approaching 0.10 are long enough that they become hard to estimate accurately, and branch length is often underestimated. In addition, the Felsenstein zone, a zone where phylogenetic methods are inconsistent (8), becomes larger as λ approaches 0.10 (1, 26). Thus, it appears that placement of the EM19 clade (with a λ of 0.11) will remain unresolved until more divergent related taxa are found that can break up the long branch.
Another strategy for resolving the position of long branching taxa is to obtain more sequence data (7). This, also, did not improve phylogenetic resolution in the case of EM19, probably because the branch length to EM19 was still long. It also did not improve the situation with the OctSpA1-106 clade, because the LSU and SSU analyses produced two different topologies. These differences were essentially due to a rooting problem: does the root of the bacterial domain attach to the OctSpA1-106 branch or to the base of the Aquifex clade? More data are needed to resolve this issue.
The deep placement of the OctSpA1-106 branch in SSU analyses may be due to “attraction” of the stem G+C-rich OctSpA1-106 sequence to stem G+C-rich archaeal sequences. Analyses with higher-stem G+C-content archaeal sequences had a slightly higher bootstrap support than analyses with lower-stem G+C-content archaeal sequences. This result could be interpreted in two ways: either there is a small attraction of the OctSpA1-106 sequence to high-stem G+C outgroup taxa, or the lower-stem G+C outgroup taxa have longer branches that slightly affect the ability to resolve branching relationships deep in the bacterial domain. However, analyses with only bacterial sequences produce largely the same topology as trees with archaeal outgroups, suggesting that the topology of the bacteria is unaffected by the presence of distantly related outgroups.
Several lines of evidence suggest that Thermocrinis ruber exhibits morphological variability in these springs. First, the morphotypes in Boulder Spring were different from those found in Octopus Spring, Eclipse Geyser, and Spindle Spring. The clone library of Boulder Spring contained two phylotypes (Thermocrinis ruber and a Thermodesulfobacterium relative), while those from Eclipse Geyser and Spindle Spring contained one (Thermocrinis ruber). No long filaments (only rods) were observed on growth surfaces from Boulder Spring. One morphology of rods hybridized with the Thermocrinis ruber probe, while another hybridized to the BSpN50/Thermodesulfobacterium probe (Fig. 5e and f). In contrast, growth surfaces from Eclipse Geyser and Spindle Spring were densely colonized by very long filaments with holdfasts, which also hybridized with the Thermocrinis ruber probe (and did not hybridize with any other probe [not shown]). Similarly, morphologies in the pink filamentous community in the outflow channel of Octopus Spring were different from those in found the source pool, even though both clone libraries contained the same phylotypes (25) (Fig. 3; Table 1). Collectively, these observations cannot be easily explained by nonspecificity of the probe (where one would expect multiple morphologies to hybridize in all springs) or insufficient sampling of the clone libraries (because the Thermocrinis ruber sequence was found in all samples).
Second, the number of cell morphotypes identified in the Octopus Spring source pool using fluorescence in situ hybridization and SEM was larger (five or more) than the number of phylotypes in the corresponding SSU clone library (four). While this could be explained by incomplete sampling of the clone libraries, it could also be explained by morphological variation within phylotypes. In situ hybridizations on springs with simpler communities (e.g., Spindle Spring, Eclipse Geyser, and Boulder Spring) suggest that the community analyses are picking up all, or at the very least most, of the lineages present. Indeed, Thermocrinis ruber has been shown to form single rods in liquid batch culture and long filaments in a flowthrough culture, showing that the cells can exhibit some degree of phenotypic plasticity (12).
Third, a large amount of sequence diversity was found in the hypervariable ITS sequences of Thermocrinis ruber, which correlated with the observed distribution of morphotypes. The ITS sequence from Boulder Spring was unique from that in any other spring, correlating with the unique morphotypes found in this spring by fluorescence in situ hybridization. In Octopus Spring and Queen's Laundry Pool, different subsets of ITS sequence types were found in different microenvironments within the same pool. These results suggest that there is a high degree of population level diversity within the Thermocrinis ruber clade, both on the morphological and ITS levels. This interpretation is supported by comparative lipid analyses of the Octopus Spring source pool and pink filamentous streamer populations, which also indicate population level diversity within the Thermocrinis ruber clade (15).
While some springs exhibited a fair amount of sequence diversity in terms of number of phylotypes, others did not. These results are supported by microscopic observations of growth surfaces; springs with limited phylotypic diversity (e.g., Eclipse Geyser and Spindle Spring) contained only one morphotype of Thermocrinis ruber (or, in the case of Spindle Spring, almost entirely one morphotype), while springs with more phylotypic diversity (e.g., Octopus Spring and Queen's Laundry) had more than one morphotype of Thermocrinis ruber. It is not clear what the controlling factors are for determining the amount of phylotypic diversity in these springs. Simple correlations between the size of the spring, type of spring or bubbler, temperature, and variety of siliceous sinter, etc., could not be readily identified. The controlling factors are most likely to be geochemical differences (either in gas or fluid chemistry), which were not measured in this study.
Communities in the subaqueous environment were not found to be significantly different from communities associated with spicular geyserite at the air-water interface within the same spring. There were, however, a slightly larger number of phylotypes associated with the spicular geyserite than with the subaqueous geyserite. The increasing amount of diversity at the air-water interface can be correlated with a slight decrease in temperature. For example, the subaqueous sinter in the Octopus Spring source pool contained four phylotypes (OctSpA1-106, EM3, Thermocrinis ruber, and EM19), while the nearby spicular geyserite lacked EM19 but also contained Thermus sp. and OS-type L. A cluster analysis of community composition showed that the spicular geyserite community (OctSpSin) was most similar to the CS in a slightly lower temperature region of the pool. This similarity can be attributed to a slight decrease in temperature due to evaporative cooling, because the spicular geyserite forms in the splash zone.
Three arguments suggest that the particular microbial composition of these communities has little influence over the morphology of sinters in these springs: (i) the communities associated with spicular geyserite in Queen's Laundry and Octopus Spring were different (although they both contained Thermocrinis ruber and EM3), (ii) the communities associated with spicular geyserite and subaqueous sediment or stratiform geyserite within the same spring are very similar, and (iii) springs precipitating many of the same sinter morphologies exhibit large differences in the amount of phylotypic diversity (some springs contain only one phylotype and others contain more phylotypes). This study shows that much more needs to be done in order to determine whether or how biofilm communities influence sinter morphologies. In addition, more work needs to be done to examine the population level diversity within these clades, for variation in morphological and metabolic properties could be important contributors to sinter fabric development. An important future direction will be to correlate community diversity on several levels with the geochemical variation in these springs.
Acknowledgments
This work was supported by a NASA Graduate Student Researcher's Program fellowship (NGT 2-52233) to C.E.B. This work was also supported by two grants to S.L.C. (NASA-NAG5-9579 and NSF-EAR-9809471). This research was funded in part by a LExEn grant from the NSF and the NASA Astrobiology Institute to N.R.P.
We acknowledge the laboratory of N.R.P., where the cloning, sequencing, in situ hybridizations, and some phylogenetic analyses were carried out. Many thanks go to Bob Lindstrom, Smokey Sturtevant, Ann Deutch, and Mary Wilson of Yellowstone National Park for their kind assistance. We thank Brent Mishler, John Taylor, and Jere Lipps, whose detailed comments and suggestions improved the manuscript.
REFERENCES
- 1.Albert, V. A., M. W. Chase, and B. D. Mishler. 1993. Character-state weighting for cladistic analysis of protein-coding DNA sequences. Ann. Missouri Bot. Gard. 80:752-766. [Google Scholar]
- 2.Ball, J. W., D. K. Nordstrom, E. A. Jenne, and D. V. Vivit. 1998. Chemical analyses of hot springs, pools, geysers, and surface waters from Yellowstone National Park, Wyoming, and vicinity, 1974-1975. U.S. Geological Survey open-file report 98-182. U.S. Geological Survey, Reston, Va.
- 3.Barns, S. M., C. F. Delwiche, J. D. Palmer, and N. R. Pace. 1996. Perspectives on archaeal diversity, thermophily and monophyly from environmental rDNA sequences. Proc. Natl. Acad. Sci. USA 93:9188-9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barns, S. M., R. E. Fundyga, M. W. Jeffries, and N. R. Pace. 1994. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc. Natl. Acad. Sci. USA 91:1609-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brock, T. D. 1978. Thermophilic microorganisms and life at high temperatures. Springer-Verlag, New York, N.Y.
- 6.Cady, S. L., and J. D. Farmer. 1996. Fossilization processes in siliceous thermal springs: trends in preservation along thermal gradients in evolution of hydrothermal ecosystems on Earth (and Mars?). Ciba Found. Symp. 202:150-173. [DOI] [PubMed] [Google Scholar]
- 7.Charleston, M. A., M. D. Hendy, and D. Penny. 1994. The effects of sequence length, tree topology, and number of taxa on the performance of phylogenetic methods. J. Comput. Biol. 1:133-151. [DOI] [PubMed] [Google Scholar]
- 8.Felsenstein, J. 1978. Cases in which parsimony or compatibility methods will be positively misleading. Syst. Zool. 27:401-410. [Google Scholar]
- 9.Ferris, F. G., T. J. Beveridge, and W. S. Fyfe. 1986. Iron-silica crystallite nucleation by bacteria in a geothermal sediment. Nature 320:609-611. [Google Scholar]
- 10.Hendy, M. D., and D. Penny. 1989. A framework for the quantitative study of evolutionary trees. Syst. Zool. 38:297-309. [Google Scholar]
- 11.Hjorleifsdottir, S., S. Skirnisdottir, G. O. Hreggvidsoon, O. Holst, and J. K. Kristjansson. 2001. Species composition of cultivated and noncultivated bacteria from short filaments in an Icelandic hot spring. Microb. Ecol. 42:117-125. [DOI] [PubMed] [Google Scholar]
- 12.Huber, R., W. Eder, S. Heldwein, G. Wanner, H. Huber, R. Rachel, and K. O. Stetter. 1998. Thermocrinis ruber gen. nov., sp. nov., a pink-filament-forming hyperthermophilic bacterium isolated from Yellowstone National Park. Appl. Environ. Microbiol. 64:3576-3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inagaki, F., S. Hayashi, K. Doi, Y. Motomura, E. Izawa, and S. Ogata. 1997. Microbial participation in the formation of siliceous deposits from geothermal water and analysis of the extremely thermophilic bacterial community. FEMS Microbiol. Ecol. 24:41-48. [Google Scholar]
- 15.Jahnke, L. L., W. Eder, R. Huber, J. M. Hope, K.-I. Hinrichs, J. M. Hayes, D. J. Des Marais, S. L. Cady, and R. E. Summons. 2001. Signature lipids and stable carbon isotope analyses of Octopus Spring hyperthermophilic communities compared with those of Aquificales representatives. Appl. Environ. Microbiol. 67:5179-5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones, B., and R. W. Renaut. 1996. Influence of thermophilic bacteria on calcite and silica precipitation in hot springs with water temperatures above 90°C: evidence from Kenya and New Zealand. Can. J. Earth Sci. 33:72-83. [Google Scholar]
- 17.Jones, B., and R. W. Renaut. 1997. Formation of silica oncoids around geysers and hot springs at El Tatio, northern Chile. Sedimentology 44:287-304. [Google Scholar]
- 18.Jones, B., R. W. Renaut, and M. R. Rosen. 1997. Vertical zonation of biota in microstromatolites associated with Hot Springs, North Island, New Zealand. Palaios 12:220-236. [Google Scholar]
- 19.Jones, B., R. W. Renaut, and M. R. Rosen. 1997. Biogenicity of silica precipitation around geysers and hot-spring vents, North Island, New Zealand. J. Sediment. Res. 67:88-104. [Google Scholar]
- 20.Konhauser, K. O., and F. G. Ferris. 1996. Diversity of iron and silica precipitation by microbial mats in hydrothermal waters, Iceland: implications for Precambrian iron formations. Geology 24:323-326. [Google Scholar]
- 21.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, N.Y.
- 22.Olsen, G. J., H. Matsuda, R. Hagstrom, and R. Overbeek. 1994. FastDNAml: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput. Appl. Biosci. 10:41-48. [DOI] [PubMed] [Google Scholar]
- 23.Renaut, R. W., B. Jones, and M. R. Rosen. 1996. Primary silica oncoids from Orakeikorako Hot Springs, North Island, New Zealand. Palaios 11:446-458. [Google Scholar]
- 24.Reysenbach, A. L., M. Ehringer, and K. Hershberger. 2000. Microbial diversity at 83°C in Calcite Springs, Yellowstone National Park: Another environment where the Aquificales and “Korarchaeota” coexist. Extremophiles 4:61-67. [DOI] [PubMed] [Google Scholar]
- 25.Reysenbach, A. L., G. S. Whickham, and N. R. Pace. 1994. Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring, Yellowstone National Park. Appl. Environ. Microbiol 60:2113-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saitou, N. 1989. A theoretical study of the underestimation of branch lengths by the maximum parsimony principle. Syst. Zool. 38:1-6. [Google Scholar]
- 27.Schultze-Lam, S., F. G. Ferris, K. O. Konhauser, and R. G. Wiese. 1995. In situ silicification of an Icelandic hot spring microbial mat: implications for microfossil formation. Can. J. Earth Sci. 32:2021-2026. [Google Scholar]
- 28.Scott, R. C. 1964. Records of post earthquake chemical quality of groundwater in Hegben Lake, Montana, earthquake of August 17, 1959, p. 179-184. U.S. Geological Survey professional paper no. 435. U.S. Geological Survey, Reston, Va.
- 29.Skirnisdottir, S., G. O. Hreggvidsson, S. Hjorleifsdottir, V. T. Marteinsson, S. P. Petursdottir, O. Holst, and J. K. Kristjansson. 2000. The influence of sulfide and temperature on the species composition and community structure of hot spring microbial mats. Appl. Environ. Microbiol. 66:2835-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson, J. M., and J. M. DeMonge. 1996. Chemical analyses of hot springs, pools and geysers from Yellowstone National Park, Wyoming, and vicinity, 1980-1993. U.S. Geological Survey open-file report 96-68. U.S. Geological Survey, Reston, Va.
- 31.Thompson, J. M., and S. Yadav. 1979. Chemical Analyses of waters from geysers, hot springs and pools in Yellowstone National Park, Wyoming, from 1974 to 1978. U.S. Geological Survey open-file report 79-704. U.S. Geological Survey, Reston, Va
- 32.Ward, D. M., M. M. Bateson, R. Weller, and A. L. Ruff-Roberts. 1992. Ribosomal RNA analysis of microorganisms as they occur in nature. Adv. Microb. Ecol. 12:219-286. [Google Scholar]
- 33.White, D. E., Thompson, G. A., Sandberg, G. H. 1964. Rocks, structure, and geologic history of Steamboat Springs thermal area, Washoe County, Nevada. U.S. Geological Survey professional paper 458B. U.S. Geological Survey, Reston, Va.
- 34.Yamamoto, H., A. Hiraishi, K. Kato, H. X. Chiura, Y. Maki, and A. Shimizu. 1998. Phylogenetic evidence for the existence of novel thermophilic bacteria in hot spring sulfur-turf microbial mats in Japan. Appl. Environ. Microbiol. 64:1680-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]