Abstract
Bacterial community composition was monitored in four shallow eutrophic lakes during one year using denaturing gradient gel electrophoresis (DGGE) of PCR-amplified prokaryotic rDNA genes. Of the four lakes investigated, two were of the clearwater type and had dense stands of submerged macrophytes while two others were of the turbid type characterized by the occurrence of phytoplankton blooms. One turbid and one clearwater lake had high nutrient levels (total phosphorus, >100 μg liter−1) while the other lakes had relatively low nutrient levels (total phosphorus, <100 μg liter−1). For each lake, seasonal changes in the bacterial community were related to bottom-up (resources) and top-down (grazers) variables by using canonical correspondence analysis (CCA). Using an artificial model dataset to which potential sources of error associated with the use of relative band intensities in DGGE analysis were added, we found that preferential amplification of certain rDNA genes over others does not obscure the relationship between bacterial community composition and explanatory variables. Besides, using this artificial dataset as well as our own data, we found a better correlation between bacterial community composition and explanatory variables by using relative band intensities compared to using presence/absence data. While bacterial community composition was related to phytoplankton biomass in the high-nutrient lakes no such relation was found in the low-nutrient lakes, where the bacterial community is probably dependent on other organic matter sources. We used variation partitioning to evaluate top-down regulation of bacterial community composition after bottom-up regulation has been accounted for. Using this approach, we found no evidence for top-down regulation of bacterial community composition in the turbid lakes, while grazing by ciliates and daphnids (Daphnia and Ceriodaphnia) was significantly related to changes in the bacterial community in the clearwater lakes. Our results suggest that in eutrophic shallow lakes, seasonality of bacterial community structure is dependent on the dominant substrate source as well as on the food web structure.
In aquatic ecosystems, bacteria play a key role in the breakdown of organic matter and the remineralization of nutrients. They are grazed upon by protozoa and some metazoans and, as such, form the base of a heterotrophic aquatic food chain. Since the development of the microbial loop concept in the early 1980s (2), substantial research efforts have been invested in evaluating the factors regulating bacteria in aquatic ecosystems. Bottom-up (resources) as well as top-down (predation) factors have been shown to regulate bacterial populations. Temperature or organic substrate concentrations may regulate bacterial growth (38). Exudates produced by phytoplankton are an important organic substrate for bacteria in many aquatic ecosystems (3), although in some lakes allochthonous humic substances may be equally important (4). Under oligotrophic conditions, inorganic nutrients may limit bacterial growth (10). Heterotrophic nanoflagellates are often the dominant grazers on bacteria in aquatic ecosystems (45), but, especially in eutrophic environments, ciliates can be important grazers too (22). In lakes, the metazoan Daphnia is an important grazer on bacteria (18).
While several studies have been carried out to evaluate the role of bottom-up or top-down regulation on biomass or production of the aggregate bacterial community, little information is available on the influence of the same regulating mechanisms on bacterial community composition. This has been due mainly to the lack of suitable techniques for studying bacterial community composition. About a decade ago, genetic methods were developed to obtain a fingerprint of the bacterial community in field samples (34). These new techniques have resulted in field studies on the distribution of bacterial communities in a variety of aquatic ecosystems (several references are given below). Recently, experimental studies have been carried out as well, whose results suggest that bottom-up as well as top-down factors may influence bacterial community composition. The bacterial community composition was found to respond to the development or degradation of phytoplankton blooms (see, e.g., references 43 and 55), suggesting that the composition and/or concentration of organic matter regulates bacterial community composition. Grazing by protozoan as well as metazoan organisms has been identified as a force driving changes in bacterial community composition (see, e.g., references 24 and 56).
Multivariate analyses techniques provide the appropriate statistical tools for describing variation in ecological communities (indirect multivariate analysis) and describing ways to relate this variation to changes in the environment (direct multivariate analysis). While these techniques have in the past been used extensively in a variety of disciplines dealing with ecological communities (e.g., terrestrial vegetation and phytoplankton), they have rarely been used to study bacterial community composition. Indirect multivariate analyses methods are purely descriptive in nature and lack statistical testing for relations between explanatory variables and community composition. They are used principally to reduce the multidimensionality typical of ecological community data and to extract common patterns from distributions of many species. Multidimensional scaling is an indirect multivariate analysis that was used by Van Hannen et al. (55, 56) and Schafer et al. (46) to describe changes in the bacterial community in mesocosms. Direct multivariate analyses are used to relate changes in ecological communities to changes in the environment. These techniques yield measures of correlation between explanatory variables and community composition and provide statistical tests for this correlation. In field studies of bacterial communities, direct multivariate analyses were used by Lindström (28) and Van der Gucht et al. (54) to demonstrate a relation between bacterial community composition and measured environmental variables. Interpretative problems may arise when many different explanatory variables are intercorrelated. Multicollinearity between several explanatory variables makes it difficult to distinguish between the separate effects of these different variables on the community composition. Variation partitioning, an extension of direct multivariate analysis used to separate the effects of different explanatory variables on community composition in a way analogous to partial correlation analysis, provides a solution for this problem (7).
The aim of this study was to evaluate the impact of bottom-up and top-down factors on seasonal changes in planktonic bacterial communities in eutrophic shallow lakes. At a given nutrient concentration, eutrophic shallow lakes can occur at two alternative stable states that are characterized by very different food web structures. Shallow lakes can be of the clearwater type characterized by a high zooplankton grazing pressure and low phytoplankton biomass, or they can be of the turbid type characterized by the incidence of dense phytoplankton blooms and an inability of the zooplankton to regulate these blooms (49). Differences between clearwater and turbid lakes are not restricted to the classic food chain (phytoplankton > zooplankton > fish) but also involve the microbial food web: all components of the microbial food web tend to attain higher biomass in turbid lakes than in clearwater shallow lakes (29). For this study, four lakes were selected which differed not only in food web structure but also in trophic status. Two clearwater and two turbid shallow lakes were selected, one turbid and one clearwater lake characterized by high nutrient concentrations (>100 μg of total P liter−1) and the other two lakes characterized by relatively low nutrient concentrations (<100 μg of total P liter−1). The bacterial community composition in each lake was monitored for 1 year by using denaturing gradient gel electrophoresis (DGGE) analysis of PCR-amplified prokaryotic rRNA genes. Seasonal changes in bacterial community composition in each of the lakes were related to bottom-up and top-down variables by using canonical correspondence analysis, a direct multivariate analysis method. The effects of bottom-up and top-down variation were subsequently separated using variation partitioning. Here we relate seasonal changes in bacterial community composition directly to explanatory variables and suggest that different control mechanisms on bacterial community composition may operate in clearwater and turbid shallow lakes.
MATERIALS AND METHODS
Sampling
Being situated in a low-lying and densely populated region of Europe, most lakes in Belgium are shallow and have, during the last century, shifted from the clearwater type characterized by dense stands of submerged macrophytes to the turbid type characterized by high phytoplankton biomass. For this study, four shallow lakes from two nature reserves situated in two different regions in Belgium were selected: a clearwater lake (Lake Visvijver) and a turbid lake (Lake Blankaart) situated in the Blankaart nature reserve and a clearwater lake (Lake Maten 13) and a turbid lake (Lake Maten 12) situated in the De Maten nature reserve. The lakes were sampled monthly in winter and biweekly in summer throughout 1999. Subsurface samples were collected at a fixed site within each lake. pH and temperature were measured in the field. For determination of bacterioplankton community composition and measurement of nutrient and suspended particulate matter (SPM) concentrations, a 2-liter water sample was transported to the laboratory in a cooler box to be processed within 5 h. Samples for determination of bacterioplankton abundance were fixed with formalin (2% final concentration). Samples for enumeration of microzooplankton (ciliates and heterotrophic nanoflagellates [HNF]) were fixed with Lugol-formalin-thiosulfate (50). For collecting macrozooplankton, the entire water column (usually <1 m) was sampled using a Shindler-Patalas trap and samples were fixed with sucrose-saturated formalin (13). In the laboratory, nutrient samples were filtered over a GF/C filter and stored frozen until used for analysis by the method of Strickland and Parsons (52). SPM concentrations were determined gravimetrically after filtration onto a GF/C filter. In each lake, cover by submerged macrophytes was estimated twice during the summer period by a transect survey (W. Rommens, personal communication).
DGGE analyses.
For analysis of bacterial diversity using DGGE, a 200-ml subsample was filtered over a membrane filter (pore size, 0.22 μm; filter diameter, 45 mm [Millipore]). The filter was cut into quarters using a sterile scalpel, and each quarter was stored at −80°C until used for further processing. DNA was extracted directly using the bead-beating method concomitant with phenol extraction and ethanol precipitation (57). Following extraction, the DNA was purified on a Wizard column (Promega, Madison, Wis.) as recommended by the manufacturer. For DGGE analysis, a small rDNA fragment was amplified with primers 357F-GC-clamp (5′-CGCCCGCCGCGCCCCGCGCCCGGCCCGCCGCCCCCGCCCCCCTACGGGAGGCAGCAG-3′) and 518R (5′-ATTACCGCGGCTGCTGG-3′). Using these specific primers, our analysis is restricted to the domain Bacteria.
Due to the use of this universal prokaryote primer, our community profiles may include sequences not only from heterotrophic bacteria but also from autotrophic cyanobacteria and from chloroplast DNA. In summer 1998, in a separate study (Van der Gucht, unpublished results), a clone library was constructed from each of the lakes studied. Of all sequences analyzed, 11, 7, 3, and 2% were attributable to cyanobacterial or chloroplast DNA in lake Maten 12, Lake Blankaart, Lake Visvijver, and Lake Maten 13, respectively. When we investigated the relationship between cyanobacterial biomass and bacterial community composition in a CCA ordination (see below), we never found a significant relation. Therefore, we think that these sequences contributed in only a minor way to our bacterial community profiles and therefore do not severely influence our conclusions.
PCR amplification procedures were performed with a Genius temperature cycler. Each mixture, containing 5 μl of template DNA, each primer at 0.5 μM, each deoxynucleoside triphosphate at 200 μM, 1.5 mM MgCl 2, 20 ng of bovine serum albumin, 5 μl of 10× PCR buffer (100 mM Tris-HCl [pH 9], 500 mM KCl), and 2.5 U of Taq DNA polymerase (Ampli-Taq; Perkin Elmer), was adjusted to a final volume of 50 μl with sterile water (Sigma). Following incubation for 5 min at 94°C, a touchdown PCR was performed using 20 cycles consisting of denaturation at 94°C for 1 min, annealing at 65°C (the temperature was decreased by 0.5°C every cycle until the touchdown temperature of 56°C was reached) for 1 min, and primer extension at 72°C for 1 min. Five additional cycles were carried out at an annealing temperature of 55°C. The tubes were then incubated for 10 min at 72°C. The presence of PCR products and their concentrations were determined by analyzing 5 μl of product on 2% agarose gels, staining with ethidium bromide, and comparing them to a molecular weight marker (Smartladder; Eurogentec).
DGGE was performed with the D-Code System from Bio-Rad Laboratories. PCR samples were loaded onto 1-mm-thick 8% (wt/vol) polyacrylamide gels in 1× TAE (20 mM Tris-acetate [pH 7.4], 10 mM acetate, 0.5 mM disodium EDTA). The denaturing gradient contained 35 to 70% denaturant (100% denaturant corresponded to 7 M urea and 40% [vol/vol] formamide). Equal amounts of PCR product were applied to the DGGE gel. Electrophoresis was performed for 16 h at 75 V. The temperature was set at 60°C. DGGE gels were stained with ethidium bromide and photographed on a UV transillumination table with a charge-coupled device camera.
As standards, we used a mixture of DNA from nine clones, obtained from a clone library of the 16S rRNA genes from one of the lakes studied. On every gel, three standard lanes were analyzed in parallel with the samples. All samples from one lake were analyzed on two parallel DGGE gels. Since these bands always should be formed at the same denaturant concentration in the gel, their position was used to compare the patterns formed in different gels. This procedure was semiautomated by using the Bionumerics 5.1 software package (Applied Maths BVBA, Kortrijk, Belgium). Since previous studies have shown that the DGGE band intensity is stable and reproducible, we decided to use band presence as well as relative intensity in our analyses because it provides extra information (47). The Bionumerics software measures an optical density profile through each lane (corresponding to a single sample), identifies band positions, and calculates the percent contribution of the intensity of each band to the total intensity of the lane. This procedure yielded a matrix with the relative intensity of each band in all samples.
Plankton biomass.
Samples for enumeration of bacteria and HNF were processed in the laboratory within 24 h of collection. Bacteria and HNF were stained with 4′,6-diamidino-2-phenylindole (DAPI) and counted on membrane filters using epifluorescence microscopy (51); 0.2-μm-pore-size filters were used for bacteria, and 0.8-μm-pore-size filters were used for HNF. Low vacuum pressure (<10 kPa) was used for preparing filters for enumeration of bacteria and HNF. Bacterial abundance was converted to biomass by assuming a constant conversion factor of 0.2 pg of C cell −1 (26). HNF were enumerated in three size classes (<4 μm, 4 to 10 μm, and >10 μm) and converted to biomass using a conversion factor of 0.15 pg of C μm−3 (12). Ciliates and phytoplankton were enumerated using inverted light microscopy. Ciliates and phytoplankton were usually identified to the genus level. For each taxon considered, about 30 cells were measured and the biovolume was calculated assuming ideal spherical shapes. Ciliate biovolume was converted to biomass by assuming a conversion factor of 0.22 pg of C μm−3 (39). For calculating phytoplankton biomass, the formulations given by Menden-Deuer and Lessard (30) were used. For determination of macrozooplankton biomass, all individuals in the sample were identified to the species level and enumerated using a dissection microscope. All individuals were measured, and abundance was converted to biomass using published length-weight regressions (8).
Data analyses.
We used multivariate statistics to investigate the relation between bacterial community composition and explanatory variables. The software package CANOCO 4.0 for Windows (53) was used for all analyses. Variation in community composition and the relationship of this variation to explanatory variables were analyzed for each lake separately. Analyses were done on presence-absence data as well as relative band intensity data. The data matrices containing relative band intensities were log(x + 1)-transformed before analysis. Explanatory variables were log(x + 1)-transformed where necessary to approximate normal distribution. We used ordination techniques based on weighted averaging, correspondence analysis (CA), and canonical correspondence analysis (CCA), which assume a unimodal response of species to the environment. The suitability of weighted averaging techniques as opposed to linear methods was tested for by performing a detrended correspondence analysis (DCA) with detrending by segments (17). Exploratory DCA analyses of the datasets of the four lakes showed that the gradient length of the first axes in standard deviation units always exceeded 2 units, confirming the suitability of weighted-averaging-based techniques for analyzing our data. In our study, CA was used to determine the total amount of variation in the data while CCA was used to quantify the amount of variation in community composition explained by a single variable or sets of explanatory variables. The significance of the relation between explanatory variables (or sets thereof) and community composition was tested using Monte Carlo permutation tests (999 unrestricted permutations, P < 0.05).
The goal of this study was to investigate the relative contribution of bottom-up (resource) and top-down (predators) variables to explaining seasonal changes in bacterial community composition in the four lakes studied. However, this may be problematic if bottom-up and top-down variables that are both related to variation in the bacterial community are themselves correlated. A significant relation between community composition and biomass of potential predators may arise not only from a direct trophic interaction but also through indirect effects. This may be the case when bacteria and potential predators of bacteria are dependent on the same resource. For instance, phytoplankton biomass may provide a resource to the bacterial community via the production of exudates. At the same time, phytoplankton is a resource for cladocerans, which graze on most phytoplankton groups. If, in this particular example, a significant relationship is observed between bacterial community composition and biomass of phytoplankton as well as biomass of cladocerans, the significant relationship between the bacterial community and biomass of cladocerans is not necessarily a direct relationship. Bacterial community composition and cladoceran biomass may both be influenced by phytoplankton biomass in the absence of a direct effect of cladocera on the bacterial community. This results in a significant correlation between grazers and bacterial community composition. Multivariate analysis with variation partitioning as described by Borcard et al. (7) provides a solution to this problem and allows us to avoid these indirect relationships in the interpretation of multivariate analyses. This technique, which can be viewed as a multivariate analogue of partial correlation analysis, reevaluates the relationship between community composition and a given explanatory variable, A, after the effect of another variable, B, to which variable A may be correlated, has been accounted for. We used this technique to test for relations between potential bacterial grazers and the bacterial community after possible relations between bottom-up variables (e.g., phytoplankton biomass and nutrients), which may also influence grazers, and the bacterial community have been accounted for. If the significant relationship between bacterial community composition and grazer biomass persists after removal of possible sources of bottom-up variation from the data, we can be sure that the relation between grazers and the bacterial community is not influenced by indirect effects of bottom-up variables on grazers. This method allowed us to distinguish between true and apparent top-down effects on bacterial community composition in the lakes studied.
For the variation partitioning analysis, all explanatory variables were divided into two groups: variables related to bottom-up regulation (temperature, phytoplankton biomass, nitrogen and phosphorus concentrations, pH, and SPM) and variables related to top-down regulation (biomass of HNF, oligotrich and other ciliates, Daphnia, and Ceriodaphnia). HNF, ciliates, and large cladocerans like Daphnia and Ceriodaphnia are all capable of grazing on bacteria. Bacteria may also be influenced by temperature, phytoplankton biomass, nutrient concentrations, pH, and SPM concentration. These bottom-up variables may also directly or indirectly influence all or some of the potential predators of the bacteria. Variation partitioning was used to evaluate whether predators affected the bacterial community independently of the effect of bottom-up variables. First, for each lake, we selected only variables that independently explained a significant amount of the variation in bacterial community composition. Then, for the sets of bottom-up and top-down variables separately, we generated a minimal set of explanatory variables that explained variation in the community composition just as well as the full set by using the forward selection procedure in CANOCO. Finally, we removed bottom-up variation from the community data by introducing the bottom-up-related variation selected in the forward selection procedure in the CCA analyses as a covariable. After that, the variation explained by the top-down factors was quantified again. This variation, which is always smaller than or equal to the total top-down-related variation in the community data, is pure top-down-related variation and represents the variation explained by top-down factors in which bottom-up variation has been accounted for.
Data simulation.
Although the intensity of DGGE bands provides a relative measure of the abundance of the corresponding bacterial taxa in a sample and although band intensity has in the past been used as a proxy for biomass (15, 47), there are strong indications that band intensity is not linearly related to the biomass of the bacterial taxa present in a sample (11, 56). On the other hand, a lot of information is lost when only presence-absence data are used. To evaluate how bias associated with the use of band intensities might influence the results of multivariate analyses, we constructed an artificial dataset, added potential sources of error to it, and evaluated how these possible sources of error influence the relationship between the community and an imaginary explanatory variable.
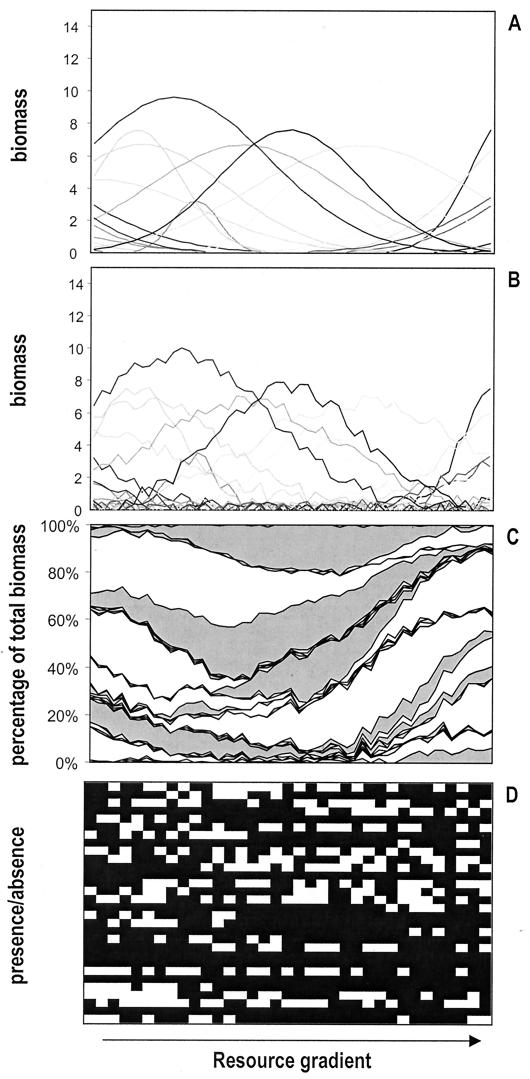
We created an artificial dataset containing information on biomass (B) of 30 imaginary taxa that display a unimodal response along a resource gradient with concentration R. This unimodal response is defined by a maximal biomass, Bmax, attained at the optimum resource concentration μ and characterized by spreading, σ, around this optimum:
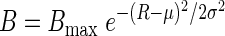 |
Our artificial dataset contained 30 taxa and 30 random samples from a resource gradient varying between 50 and 150 arbitrary units. For these taxa, the optimum μ was allowed to vary randomly between 0 and 200 units along the resource gradient while the spreading σ varied randomly between 5 and 25 units along the resource gradient and Bmax varied randomly between 0 and 10 biomass units. Dataset 1 comprised absolute biomass data of the 30 taxa in the 30 samples and represented the ideal situation where the biomass of all taxa is determined with absolute precision (Fig. 1A). In community studies, datasets often contain substantial errors and the biomass of the most abundant taxa is generally quantified more precisely than that of rare taxa. To simulate these errors, we added a random error of 0 to 10 biomass units, corresponding to 0 to 10% of the maximal biomass, to our data. If the addition of this error term resulted in negative biomass, the corresponding taxon was not observed in that sample and the biomass was set to 0. This dataset 2 corresponds to “the best we could hope for” in bacterial community studies (Fig. 1B). In dataset 3, we added an error typical for DGGE analysis of PCR-amplified genes in addition to the same error terms as those used in dataset 2. In DGGE analysis of bacterial communities, as it is currently applied, not all DNA is extracted with equal efficiency (32, 44). Moreover, during the PCR amplification step, sequences from some taxa may be preferentially amplified over others (see, e.g., references 33 and 42). This may result in the overestimation of some taxa and underestimation of others. We assume that this over- or underestimation is constant for a given taxon in different samples. We included this source of error in dataset 3 by multiplying the biomass of all taxa by a factor that was constant for each taxon and which varied randomly between a twofold overestimation and a twofold underestimation among the different taxa. Finally, after adding all error terms to dataset 3, we transformed the biomass of each taxon to obtain relative biomass, as is done during analysis of DGGE gels in Bionumerics (= biomass of a taxon divided by total biomass) (Fig. 1C). Finally, dataset 4 (based on dataset 3) was constructed, in which only presence-absence data were included (Fig. 1D). In all artificial datasets, total community biomass or number of taxa per sample did not vary systematically along the resource gradient. Given the modeled unimodal response of species biomass to resource concentration, we used CCA to quantify the variation in resource concentration explained by community composition. All datasets except dataset 4 were log(x + 1) transformed prior to analysis. The analysis was done for the four datasets separately and was repeated five times for different sets of taxa (with randomly chosen values for Bmax, σ, and μ) and samples (chosen randomly from the artificial resource gradient). Results from the analyses of datasets 1 versus 2, 2 versus 3, and 3 versus 4 were compared using paired t tests.
FIG. 1.
Examples of the four different artificial datasets used in the data simulation exercise. (A) Dataset 1; (B), dataset 2; (C), dataset 3; (D) dataset 4. For a detailed description of the construction of the four datasets, see the text.
RESULTS
Comparison of the lakes studied.
A comparison of the four lakes studied is presented in Table 1. Compared to the De Maten lakes, the Blankaart lakes were characterized by much higher nutrient concentrations. There was also a marked difference in pH: both De Maten lakes had a lower pH than the Blankaart lakes. Also, zooplankton biomass tended to be higher in the Blankaart lakes than in the De Maten lakes. Apart from zooplankton biomass, nutrient levels, and pH, the differences between clearwater and turbid lakes from the two regions were more pronounced than the regional differences. Submerged macrophyte cover was higher and SPM concentrations were lower in the clearwater lakes than in the turbid lakes. Despite a fivefold difference in nutrient concentrations between the lakes from the two regions, phytoplankton biomass was found to be more closely related to water clarity than to nutrient levels: phytoplankton biomass was more than twice as high in the turbid lakes as in the clearwater lakes. Green algae were the dominant component of the phytoplankton community in the turbid lakes, whereas cryptophytes were the most important phytoplankton group in the clearwater lakes. In 1999, cyanobacteria were never dominant in any of the lakes. Euglenophytes were common in the De Maten lakes and rare in the Blankaart lakes. Like phytoplankton biomass, the biomass of the components of the microbial loop (bacteria, HNF, and ciliates) tended to be higher in the turbid lakes than in the clearwater lakes, although the differences were less pronounced than for phytoplankton biomass. In all lakes studied, the ciliate biomass was higher than the HNF biomass. Oligotrich ciliates were a relatively more important component of the ciliate community in the Blankaart lakes than in the De Maten lakes. Regarding the community composition of the zooplankton, we observed a larger contribution of daphnids (Daphnia and Ceriodaphnia) to total biomass in the clearwater lakes than in the turbid lakes.
TABLE 1.
Environmental variables and biomasses of the most important groups in the plankton of the four lakes studieda
| Variable | Blankaart reserve
|
De Maten reserve
|
||
|---|---|---|---|---|
| Lake Blankaart | Lake Visvijver | Lake Maten 12 | Lake Maten 13 | |
| Nitrogen (μg liter −1) | 5170 ± 5430 | 457 ± 632 | 245 ± 604 | 70 ± 95 |
| Phosphorus (μg liter −1) | 427 ± 970 | 506 ± 332 | 53 ± 30 | 10 ± 12 |
| SPM (mg liter −1) | 49 ± 39 | 5.0 ± 2.1 | 23 ± 11 | 11.0 ± 14.6 |
| pH | 8.52 ± 0.70 | 8.15 ± 0.43 | 7.30 ± 0.24 | 6.94 ± 0.25 |
| Phytoplankton (μg of C liter −1) | 1587 ± 1490 | 140 ± 165 | 459 ± 340 | 228 ± 175 |
| % chlorophytes | 51 | 11 | 29 | 26 |
| % euglenophytes | 6 | 1 | 27 | 26 |
| % cyanobacteria | 7 | 6 | 4 | 1 |
| % cryptophytes | 15 | 53 | 17 | 31 |
| Bacteria (μg of C liter −1) | 187 ± 221 | 111 ± 95 | 134 ± 118 | 79 ± 49 |
| HNF (μg of C liter −1) | 21 | 1 | 7 | 2 |
| Ciliates (μg of C liter −1) | 180 | 37 | 301 | 84 |
| % oligotrich ciliates | 64 | 88 | 42 | 52 |
| Macrozooplankton (μg of C liter −1) | 967 ± 1100 | 547 ± 834 | 254 ± 259 | 134 ± 176 |
| % daphnids | 16 | 22 | 6 | 19 |
| Submerged macrophytes (% cover) | 0 | 55 | 0 | 43 |
Results are given as mean ± standard deviation for concentration data; no standard deviation is given for the percentage data. Since the percent cover by submerged macrophytes was determined only twice during the summer period, no standard deviation is given for these data.
Data simulation.
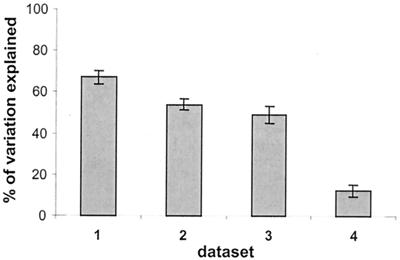
The results of the data simulation are presented in Fig. 2. Inclusion of a random error in biomass determination (dataset 2) resulted in a significant (P < 0.001) decrease in the variation explained by resource concentration compared to the “ideal” dataset 1. The errors typical of bacterial community analysis as assessed by DGGE, together with the use of relative band intensity (dataset 3), however, resulted only in a slight and nonsignificant (P = 0.09) decrease in the variation explained by resource concentration compared to dataset 2. The decrease in variation explained was quite variable among the five replicate datasets tested (range, 0.5 to 10.4%). When only presence-absence data were used (dataset 4), a significantly (P < 0.001) smaller fraction of community variation was explained by resource concentration.
FIG. 2.
Results of the data simulation exercise. Variation in community composition is explained by resource concentration in the four artificial datasets. Bars represent averages of five replicate datasets; error bars represent ±95% confidence intervals.
Multivariate analyses.
Seasonal succession in bacterial community composition was studied using an indirect CA analysis; only the results for the relative band intensity data are presented (Fig. 3). In Lake Blankaart and the two lakes from the De Maten reserve, a relatively clear seasonal pattern was reproduced in the first two CA axes, indicating a relatively gradual seasonal succession of bacterial communities in these lakes. In Lake Visvijver, however, no such pattern was detectable, indicating that rapid changes in bacterial community composition often occurred over short timescales.
FIG. 3.
CA ordination plots (axes 1 and 2) for the four lakes studied. Circles represent samples. The first sample taken in the season is represented by a solid circle, while the broken line indicates the sampling sequence. Arrows represent correlation coefficients between explanatory variables and the first two ordination axes. Correlation coefficients were multiplied by 2 to give a better fit in the ordination plot. Only explanatory variables significantly explaining variation in the data are displayed.
The relation between bacterial community composition and explanatory variables was investigated for the presence-absence data as well as for the relative band intensity data (Table 2). All variables that significantly explained variation in the presence-absence data also significantly explained variation in the relative band intensity data. On four occasions, a significant relation was found for the analysis of relative band intensity data that was not found for the analysis of the presence-absence data. When a variable significantly explained variation in both datasets, the amount of variation explained was not always higher for relative band intensity data than for presence-absence data. On eight occasions, more variation was explained in the presence-absence data, but this difference amounted to an average of only 0.9% (0.3 to 1.4%). On seven occasions, more variation was explained in the relative band intensity data, with a larger difference, amounting to an average of 2.4% (0.3 to 5.6%). The total contribution of bottom-up and top-down variables to explaining bacterial community composition was determined by following the forward selection procedure in CANOCO. Subsequently, to determine pure top-down-related variation, bottom-up-related variation was removed from the data by introducing this variation as a covariable in the CCA analyses. Differences between the two types of datasets in the variation explained by these groups of explanatory variables were similar to differences in the variation explained by separate variables except when a different number of variables was selected. This was the case for the bottom-up variation in Lake Maten 13, where pH and phosphorus concentration were included by forward selection during analysis of the presence-absence dataset while only phosphorus concentration was included in the analysis of relative band intensity data. This was also the case for the pure top-down variation in Lake Visvijver, where both Daphnia and nonoligotrich ciliates were included in the analysis of the relative band intensity data while only nonoligotrich ciliates were included in the analysis of the presence-absence data. In the following discussion, we address only the results of the analyses of the relative band intensity data.
TABLE 2.
Percentage of variation in bacterial community composition explained by the different environmental variables and sets of bottom-up and top-down variables in the four lakesa
| % Variation | Blankaart reserve
|
De Maten reserve
|
||||||
|---|---|---|---|---|---|---|---|---|
| Lake Blankaart
|
Lake Visvijver
|
Lake Maten 12
|
Lake Maten 13
|
|||||
| P-A | RBI | P-A | RBI | P-A | RBI | P-A | RBI | |
| Bottom-up variation | ||||||||
| Temperature | 16.5 | 15.6 | 15.6 | 16.8 | 00.0 | 13.1 | ||
| Phytoplankton | 11.8 | 11.5 | 12.7 | 13.4 | ||||
| Nitrogen | 00.0 | 11.2 | ||||||
| Phosphorus | 14.6 | 16.3 | ||||||
| pH | 15.1 | 13.7 | ||||||
| SPM | ||||||||
| Total | 16.5 | 15.6 | 12.7 | 13.4 | 15.6 | 16.8 | 27.6 | 16.3 |
| Top-down variation | ||||||||
| HNF | ||||||||
| Oligotrich ciliates | 13.8 | 14.9 | ||||||
| Nonoligotrich ciliates | 15.0 | 18.8 | ||||||
| Daphnia | 15.4 | 15.0 | 13.0 | 15.7 | 14.9 | 13.9 | ||
| Ceriodaphnia | 16.5 | 16.2 | 17.3 | 16.0 | ||||
| Total | 15.4 | 15.0 | 40.2 | 42.6 | 0.0 | 0.0 | 30.1 | 28.6 |
| Pure top-down variation | ||||||||
| HNF | ||||||||
| Oligotrich ciliates | ||||||||
| Nonoligotrich ciliates | 13.2 | 18.8 | ||||||
| Daphnia | 00.0 | 14.0 | 00.0 | 12.1 | ||||
| Ceriodaphnia | 17.3 | 16.0 | ||||||
| Total | 0.0 | 0.0 | 13.2 | 26.1 | 0.0 | 0.0 | 17.3 | 16.0 |
Two percentages are presented corresponding to the analyses of the matrices containing only presence-absence data (P-A) and the matrices containing relative band intensities (RBI). The underlined values denote variables which were included in the set of bottom-up, top-down, or pure top-down-related variables by means of forward selection. Pure top-down variation denotes the variation explained by top-down variables after bottom-up variation had been accounted for. Only significant (Monte Carlo permutation test, 999 unrestricted permutations, P < 0.05) relationships are shown.
In Lake Blankaart, temperature, nitrogen concentration, and phytoplankton and Daphnia biomass significantly explained the variation in bacterial community composition. In Lake Visvijver, the biomass of phytoplankton, Daphnia, Ceriodaphnia, and oligotrich as well as nonoligotrich ciliates significantly explained the variation in the dataset. In Lake Maten 12, a significant relation was found only for temperature, while in Lake Maten 13, temperature, pH, phosphorus concentration, and biomass of Daphnia and Ceriodaphnia were significantly related to bacterial community composition.
Bottom-up variables explained about the same percentage of the total variation (13.4 to 16.8%) in the four lakes studied. In all lakes, only one variable was included in the set of bottom-up variables selected by the forward selection procedure: temperature in both turbid lakes, phytoplankton biomass in Lake Visvijver, and phosphorus concentration in Lake Maten 13. Top-down variables explained more variation in the clearwater lakes than in the turbid lakes. None of the top-down variables significantly explained the variation in Lake Maten 12, while Daphnia explained 15.0% of the variation in Lake Blankaart. In Lake Maten 13, 28.6% (nonoligotrich ciliates, Ceriodaphnia, and Daphnia selected) of the variation and in Lake Visvijver 42.6% (both Ceriodaphnia and Daphnia selected) of the variation was related to top-down variables.
To evaluate whether the variables measured in this study are able to explain the observed patterns in bacterial community composition, we compared the output of the CA analyses with the output of a CCA analysis including all variables that independently significantly explain variation in the datasets. We calculated correlation coefficients between the sample scores on the first and (if applicable) second CCA axes and the sample scores on the corresponding CA axes. For the analyses based on the presence-absence data as well as the relative band intensity data, correlation coefficients were always higher than 0.87, corresponding to a significance level of >0.00025. This indicates that the explanatory variables measured in this study explain the main variation in the data.
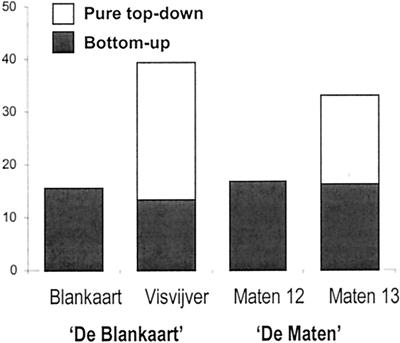
The results of the variation partitioning analysis are summarized in Fig. 4. When bottom-up-related variation was removed in the data from Lake Blankaart, the only top-down variable significantly related to bacterial community composition (Daphnia) no longer explained any variation in the data. In Lake Maten 12, no top-down variation was related to bacterial community composition before or after removal of bottom-up variation. After removal of bottom-up-related variation, only the biomass of nonoligotrich ciliates and Daphnia continued to explain the variation in the data for Lake Visvijver. Both were included by forward selection, and together they explained 26.1% of the pure top-down-related variation. In Lake Maten 13, both Daphnia and Ceriodaphnia significantly explained variation in the data after removal of bottom-up-related variation. Only Ceriodaphnia was included in the set of pure top-down-related variation by forward selection and explained 16.8% of the total variation.
FIG. 4.
Results of the variation partitioning analysis for the four lakes studied. For each lake, the total variation in bacterial community composition explained is partitioned among bottom-up variation and pure top-down variation. The results presented are based on the analyses of the relative band intensity data.
DISCUSSION
In previous studies of bacterioplankton communities, different multivariate approaches were used to describe variation in community composition in space or time. To our knowledge, these studies only used presence-absence data in the analyses. Techniques used in these studies have included cluster analysis (see, e.g., reference 47) or other analysis methods based on similarity indices such as multidimensional scaling (46, 55). In a study of bacterial diversity in five Swedish lakes, Lindström (28) used CCA to relate the presence-absence data of DGGE bands directly to variation in the environment. In our study, we applied the same technique (CCA) but used band intensity in addition to presence-absence data.
We used an artificial dataset to evaluate whether sources of error associated with the use of relative band intensity in DGGE profiles of PCR-amplified genes could affect the relation between bacterial community composition and explanatory variables. Compared to presence-absence data, the use of relative band intensities resulted in a significantly better relationship of the community data to the explanatory variable. This relationship was not influenced by preferential amplification of certain genotypes over others and or by the use of relative biomass over absolute biomass. This is not surprising, since multivariate analyses take into account changes in the relative composition of the community. These changes are not influenced by incorrect estimates of individual biomass of the different taxa. For our own datasets, we compared analyses based on presence-absence data with analyses based on relative band intensity data. As in our model analysis, the use of relative band intensity data resulted in more variables significantly explaining more variation in the data. Therefore, we used relative band intensities instead of presence-absence data to investigate the relationship between bacterial community composition and explanatory variables. Although problems associated with DGGE analysis of PCR-amplified genes cannot be used to infer information on biomass of different bacterial taxa, these problems do not obscure the relation between bacterial community composition and the environment. Admittedly, this conclusion assumes that the amplification efficiency of a given bacterial genotype is not influenced by the presence of other genotypes in the sample.
The direct multivariate analyses revealed several significant relationships between explanatory variables and bacterial community composition. Bottom-up factors (temperature, phytoplankton biomass, nitrogen and phosphorus concentrations, and pH) as well as top-down factors (biomass of oligotrich and non-oligotrich ciliates, Daphnia, and Ceriodaphnia) were found to be significantly related to changes in the bacterial communities of the four lakes studied. The relationship between seasonality in bacterial community structure and explanatory variables differed according to the region as well as to the food web structure of the lake, which differed in the turbid and the clearwater lakes.
The most important regional differences were total nutrient concentrations and pH. Whereas the De Maten lakes are situated in a watershed that is only moderately impacted by human activities, the Blankaart lakes are situated in an area of intensive agriculture. This explains the higher nutrient concentrations observed in the Blankaart lakes than in the De Maten lakes. Nutrient concentration may directly influence bacterial biomass (10) as well as community composition (37) through effects on growth. However, a significant relationship between bacterioplankton community structure and nutrients may also arise from covariation of nutrient concentrations with phytoplankton or submerged macrophyte biomass, since these aquatic plants can take up a large fraction of the dissolved nutrients during summer and may at the same time influence bacterial dynamics via release of carbon in the water (see reference 3 for phytoplankton and reference 41 for macrophytes). Only in the clearwater Lake Maten 13 did dissolved-phosphorus concentrations frequently drop below 10 μM, a level low enough to limit bacterial growth and therefore to structure the bacterial community. In Lake Blankaart, total inorganic nitrogen levels were never below 10 μM. The relationship with nitrogen observed in Lake Blankaart is therefore probably indirect and caused by covariation with phytoplankton biomass.
In both Blankaart lakes, the bacterial community was significantly related to phytoplankton biomass, while no such relation was found in the De Maten lakes. Probably, the bacterioplankton in the Blankaart lakes relies mainly on phytoplankton exudates as a carbon source while the bacterioplankton in the De Maten lakes depends mainly on other carbon sources, possibly allochthonous sources of carbon like humic acids. In a survey of six Adirondack lakes, a relationship was observed between dissolved organic carbon concentration and bacterial community composition, suggesting that the dominant organic matter sources may influence bacterial community composition (31). Covariation between bacterioplankton community composition and phytoplankton biomass has been observed in other lakes (14, 35) as well as in marine ecosystems (1, 40). Bacterial community composition in other lakes was shown to be dependent on humic acids (27). Further research is needed to determine which factors regulate bacterial communities in the De Maten lakes.
Variation partitioning can be used to detect covariation between different explanatory variables or sets of explanatory variables. In this study, this approach was adopted to separate top-down from bottom-up effects on bacterioplankton dynamics. It allowed us to quantify pure top-down effects on the seasonality of bacterioplankton community composition. The results of these analyses suggest strong differences between turbid and clearwater lakes. Pure top-down-related variation was important (16.8 and 26.1% of seasonal variation) in the clearwater lakes but was not observed in the turbid lakes. Although Daphnia biomass was related to bacterioplankton dynamics in Lake Blankaart, this relationship was no longer significant when bottom-up effects were accounted for, probably because Daphnia biomass covaried with phytoplankton biomass. In the turbid Lake Maten 12, no relation was found to the biomass of any grazer organism. In the clearwater lakes, Daphnia, Ceriodaphnia and nonoligotrich ciliates explained a large fraction of the bacterial seasonality after bottom-up effects were accounted for. Many ciliates are potentially important grazers on bacteria, and it has been shown that ciliates select certain size classes of bacteria over others (23). Daphnids such as Daphnia and Ceriodaphnia are large filter feeders that play a key role in shallow lakes (16). They not only regulate phytoplankton biomass but also have a strong impact on the microbial food web, including the bacteria (20). Daphnids are able to feed on a broad particle size range but their filter apparatus is unable to retain the smallest bacteria, and selective grazing by daphnids is probably related to size selection (9). Given the relationship observed by Bernard et al. (5) between bacterial size and taxonomic composition, size-selective grazing by ciliates or daphnids may explain the impact of these grazers on bacterial community composition. In a field study of a eutrophic lake, Höfle et al. (14) also observed a relationship between bacterial diversity and Daphnia biomass.
The results of our study do not imply that top-down control is of no importance in determining bacterial community composition in turbid lakes. From this study, we can only conclude that top-down factors are not related to seasonal changes in the structure of the bacterial community. We found no relationship between HNF and bacterial community structure in any of the lakes studied, although it is well known that HNF are important grazers on bacteria in most aquatic systems including lakes. During prey capture, HNF select certain bacteria over others (6, 19), and a strong influence of HNF grazing on bacterial community composition has also been demonstrated in experiments (21, 24, 25, 56). In the lakes studied, however, HNF biomass was very low compared to ciliate biomass, and ciliates were probably more important with respect to bacterivory. A relatively small grazing impact of HNF on bacteria in the lakes studied may explain the absence of a relationship to bacterial community composition.
Our results suggest that bacterial succession may differ according to the dominant substrate source in the lake (phytoplankton versus other sources). In clearwater shallow lakes, as opposed to turbid lakes, the effect of top-down control by grazers is superimposed on regulation by substrates. Large metazoan filter feeders such as Daphnia and Ceriodaphnia, as well as protozoan ciliates, may play an important role in the structuring of the bacterial community. Organic matter degradation is related to the composition of the bacterial community (36, 43). Therefore, by removing taxa responsible for the degradation of certain organic matter fractions or by selecting for species with lower growth rates, grazers may influence the rate of degradation of organic matter and remineralization of nutrients in clearwater shallow lakes. Next to grazing of phytoplankton and many other feedback mechanisms that stabilize the clearwater state in shallow eutrophic lakes (see reference 48), grazer control of bacterial community composition may represent another process contributing to the maintainance of water clarity in clearwater shallow lakes.
Acknowledgments
The research presented in this paper was carried out in the framework of the Flemish VLINA project 97/03 “Restoration of nature values in shallow lakes: investigating the structure and functioning of the microbial loop and the trophic cascade in a series of model systems.” We thank participants in this project for providing data on the four lakes studied: Jeroen Van Wichelen, Vanessa Geenens, Steven Declerck, Jochen Vandekerckhove, Hanne Degans, Danny Rejas and Wouter Rommens. K.M. is a postdoctoral fellow of the Belgian Fund for Scientific Research.
Erik Van Hannen is acknowledged for providing comments on a previous version of the manuscript.
REFERENCES
- 1.Acinas, S. G., F. Rodriguez-Valera, and C. Pedrosalio. 1997. Spatial and temporal variation in marine bacterioplankton diversity as shown by RFLP fingerprinting of PCR amplified 16s rDNA. FEMS Microbiol. Ecol. 24:27-40. [Google Scholar]
- 2.Azam, F., T. Fenchel, J. G. Field, J. S. Gray, L. A. Meyer-Reil, and F. Thingstadt. 1983. The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 10:257-263. [Google Scholar]
- 3.Baines, S. B., and M. L. Pace. 1991. The production of dissolved organic matter by phytoplankton and its importance to bacteria—patterns across marine and fresh-water systems. Limnol. Oceanogr. 36:1078-1090. [Google Scholar]
- 4.Bano, N., M. A. Moran, and R. E. Hodson. 1997. Bacterial utilization of dissolved humic substances from a freshwater swamp. Aquat. Microb. Ecol. 12:233-238. [Google Scholar]
- 5.Bernard, L., C. Courties, P. Servais, M. Troussellier, M. Petit, and P. Lebaron. 2000. Relationships among bacterial cell size, productivity, and genetic diversity in aquatic environments using cell sorting and flow cytometry. Microb. Ecol. 40:148-158. [DOI] [PubMed] [Google Scholar]
- 6.Boenigk, J., and H. Arndt. 2000. Comparative studies on the feeding behavior of two heterotrophic nanoflagellates: the filter-feeding choanoflagellate Monosiga ovata and the raptorial-feeding kinetoplastid Rhynchomonas nasuta. Aquat. Microb. Ecol. 22:243-249. [Google Scholar]
- 7.Borcard, D., P. Legendre, and P. Drapeau. 1992. Partialling out the spatial component of ecological variation. Ecology 73:1045-1055. [Google Scholar]
- 8.Bottrell, H. H., A. Duncan, Z. M. Gliwicz, E. Grygierek, A. Herzig, A. Hillbrichtilkowska, H. Kurasawa, P. Larsson, and T. Weglenska. 1976. Review of some problems in zooplankton production studies. Norw. J. Zool. 24:419-456. [Google Scholar]
- 9.Brendelberger, H. 1991. Filter mesh size of cladocerans predicts retention efficiency for bacterial. Limnol. Oceanogr. 36:884-894. [Google Scholar]
- 10.Chrzanowski, T. H., R. W. Sterner, and J. J. Elser. 1995. Nutrient enrichment and nutrient regeneration stimulate bacterioplankton growth. Microb. Ecol. 29:221-230. [DOI] [PubMed] [Google Scholar]
- 11.Farrelly, V., F. A. Rainey, and E. Stackebrandt. 1995. Effect of genome size and rRNA gene copy number on PCR amplification of 16SrRNA genes from a mixture of bacterial species. Appl. Environ. Microbiol. 61: 2798-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fenchel, T. 1982. Ecology of heterotrophic microflagellates. 2. Bioenergetics and growth. Mar. Ecol. Progr. Ser. 8:225-231. [Google Scholar]
- 13.Haney, J. F., and D. J. Hall. 1973. Sugar-coated Daphnia—preservation technique for Cladocera. Limnol. Oceanogr. 18:331-333. [Google Scholar]
- 14.Höfle, M. G., H. Haas, and K. Dominik. 1999. Seasonal dynamics of bacterioplankton community structure in a eutrophic lake as determined by 5S rRNA analysis. Appl. Environ. Microbiol. 65:3164-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson, C. R., P. F. Churchill, and E. E. Roden. 2001. Successional changes in bacterial assemblage structure during epilithic biofilm development. Ecology 82:555-566. [Google Scholar]
- 16.Jeppesen, E., J. P. Jensen, M. Sondergaard, T. Lauridsen, L. J. Pedersen, and L. Jensen. 1997. Top-down control in freshwater lakes: the role of nutrient state, submerged macrophytes and water depth. Hydrobiologia 342:151-164. [Google Scholar]
- 17.Jongman, R. H. G., C. J. F. ter Braak, and O. F. R. van Tongeren. 1987. Data analysis in community and landscape ecology. Pudoc, Wageningen, The Netherlands.
- 18.Jürgens, K. 1994. Impact of Daphnia on planktonic microbial food webs: a review. Mar. Microb. Food Webs 8:295-324. [Google Scholar]
- 19.Jürgens, K., and W. R. DeMott. 1995. Behavioral flexibility in prey selection by bacterivorous nanoflagellates. Limnol. Oceanogr. 40:1503-1507. [Google Scholar]
- 20.Jürgens, K., and E. Jeppesen. 2000. The impact of metazooplankton on the structure of the microbial food web in a shallow, hypertrophic lake. J. Plankton Res. 22:1047-1070. [Google Scholar]
- 21.Jürgens, K., J. Pernthaler, S. Schalla, and R. Amann. 1999. Morphological and compositional changes in a planktonic bacterial community in response to enhanced protozoan grazing. Appl. Environ. Microbiol. 65:1241-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kisand, V., and P. Zingel. 2000. Dominance of ciliate grazing on bacteria during spring in a shallow eutrophic lake. Aquat. Microb. Ecol. 22:135-142. [Google Scholar]
- 23.Kivi, K., and O. Setala. 1995. Simultaneous measurement of food particle selection and clearance rates of planktonic oligotrich ciliates (Ciliophora: Oligotrichina). Mar. Ecol. Prog. Ser. 119:125-137. [Google Scholar]
- 24.Langenheder, S., and K. Jürgens. 2001. Regulation of bacterial biomass and community structure by metazoan and protozoan predation. Limnol. Oceanogr. 46:121-134. [Google Scholar]
- 25.Lebaron, P., P. Servais, M. Troussellier, C. Courties, G. Muyzer, L. Bernard, H. Schafer, R. Pukall, E. Stackebrandt, T. Guindulain, and J. Vives-Rego. 2001. Microbial community dynamics in mediterranean nutrient-enriched seawater mesocosms: changes in abundances, activity and composition. FEMS Microbiol. Ecol. 34:255-266. [DOI] [PubMed] [Google Scholar]
- 26.Lee, S., and J. A. Fuhrman. 1987. Relationships between biovolume and biomass of naturally derived marine bacterioplankton. Appl. Environ. Microbiol. 53:1298-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindström, E. S. 1998. Bacterioplankton community composition in a boreal forest lake. FEMS Microbiol. Ecol. 27:163-174. [Google Scholar]
- 28.Lindström, E. S. 2000. Bacterioplankton community composition in five lakes differing in trophic status and humic content. Microb. Ecol. 40:104-113. [DOI] [PubMed] [Google Scholar]
- 29.Mathes, J., and H. Arndt. 1994. Biomass and composition of protozooplankton in relation to lake trophy in north German lakes. Mar. Microb. Food Webs 8:357-375. [Google Scholar]
- 30.Menden-Deuer, S., and E. J. Lessard. 2000. Carbon to volume relationships for dinoflagellates, diatoms, and other protist plankton. Limnol. Oceanogr. 45:569-579. [Google Scholar]
- 31.Methé, B. A., and J. P. Zehr. 1999. Diversity of bacterial communities in Adirondack lakes: do species assemblages reflect lake water chemistry? Hydrobiologia 401:77-96. [Google Scholar]
- 32.More, M. I., J. B. Herrick, M. C. Silva, W. C. Ghiorse, and E. L. Madsen. 1994. Quantitative cell-lysis of indigenous microorganisms and rapid extraction of microbial DNA from sediment. Appl. Environ. Microbiol. 60:1572-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mutter, G. L., and K. A. Boynton. 1995. PCR bias in amplification of androgen receptor alleles, a trinucleotide repeat marker used in clonality studies. Nucleic Acids Res. 23:1411-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muyzer, G., E. C. Dewaal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S ribosomal-RNA. Appl. Environm. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pernthaler, J., F. O. Glockner, S. Unterholzner, A. Alfreider, R. Psenner, and R. Amann. 1998. Seasonal community and population dynamics of pelagic Bacteria and Archaea in a high mountain lake. Appl. Environ. Microbiol. 64:4299-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pinhassi, J., F. Azam, J. Hemphala, R. A. Long, J. Martinez, U. L. Zweifel, and A. Hagstrom. 1999. Coupling between bacterioplankton species composition, population dynamics, and organic matter degradation. Aquat. Microb. Ecol. 17:13-26. [Google Scholar]
- 37.Pinhassi, J., and A. Hagstrom. 2000. Seasonal succession in marine bacterioplankton. Aquat. Microb. Ecol. 21:245-256. [Google Scholar]
- 38.Pomeroy, L. R., and W. J. Wiebe. 2001. Temperature and substrates as interactive limiting factors for marine heterotrophic bacteria. Aquat. Microb. Ecol. 23:187-204. [Google Scholar]
- 39.Putt, M., and D. K. Stoecker. 1989. An experimentally determined carbon:volume ratio for marine “oligotrichous” ciliates from estuarine and coastal waters. Limnol. Oceanogr. 34:1097-1103. [Google Scholar]
- 40.Rehnstam, A. S., S. Backman, D. C. Smith, and F. Azam, and A. Hagstrom. 1993. Blooms of sequence-specific culturable bacteria in the sea. FEMS Microbiol. Ecol. 102:161-166. [Google Scholar]
- 41.Reitner, B., A. Herzig, and G. J. Herndl. 1999. Dynamics in bacterioplankton production in a shallow, temperate lake (Lake Neusiedl, Austria): evidence for dependence on macrophyte production rather than on phytoplankton. Aquat. Microb. Ecol. 19:245-254. [Google Scholar]
- 42.Reysenbach, A. L., L. J. Giver, G. S. Wickham, and N. R. Pace. 1992. Differential amplification of rRNA genes by polymerase chain-reaction. Appl. Environ. Microbiol. 58:3417-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Riemann, L., G. F. Steward, and F. Azam. 2000. Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl. Environ. Microbiol. 66:578-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rochelle, P. A., J. C. Fry, R. J. Parkes, and A. J. Weightman. 1992. DNA extraction for 16s ribosomal-RNA gene analysis to determine genetic diversity in deep sediment communities. FEMS Microbiol. Lett. 100:59-65. [DOI] [PubMed] [Google Scholar]
- 45.Sanders, R. W., K. G. Porter, S. J. Bennett, and A. E. DeBiase. 1989. Seasonal patterns of bacterivory by flagellates, ciliates, rotifers, and cladocerans in a fresh-water planktonic community. Limnol. Oceanogr. 34:673-687. [Google Scholar]
- 46.Schafer, H., L. Bernard, C. Courties, P. Lebaron, P. Servais, R. Pukall, E. Stackebrandt, M. Troussellier, T. Guindulain, J. Vives-Rego, and G. Muyzer. 2001. Microbial community dynamics in mediterranean nutrient-enriched seawater mesocosms: changes in the genetic diversity of bacterial populations. FEMS Microbiol. Ecol. 34:243-253. [DOI] [PubMed] [Google Scholar]
- 47.Schauer, M., R. Massana, and C. Pedros-Allo. 2000. Spatial differences in bacterioplankton composition along the Catalan coast (NW Mediterranean) assessed by molecular fingerprinting. FEMS Microbiol. Ecol. 33:51-59. [DOI] [PubMed] [Google Scholar]
- 48.Scheffer, M. 1998. Ecology of shallow lakes. Chapman & Hall, London, United Kingdom.
- 49.Scheffer, M., S. H. Hosper, M. L. Meijer, B. Moss, and E. Jeppesen. 1993. Alternative equilibria in shallow lakes. Trends Ecol. Evol. 8:275-279. [DOI] [PubMed] [Google Scholar]
- 50.Sherr, B. F., E. B. Sherr, and C. Pedrós-Alió. 1989. Simultaneous measurements of bacterioplankton production and protozoan herbivory. Mar. Ecol. Progr. Ser. 54: 209-219. [Google Scholar]
- 51.Sherr, E. B., D. A. Caron, and B. F. Sherr. 1993. Staining of heterotrophic protists for visualisation via epifluorescence microscopy, p. 213-228. In P. F. Kemp, B. F. Sherr, E. B. Sherr, and J. J. Cole (ed.), Aquatic microbial ecology. Lewis Publishers, Boca Raton, Fla.
- 52.Strickland, J. D. H., and T. R. Parsons. 1975. A practical handbook of seawater analysis. Fish. Res. Board Can. Bull. 167:1-310. [Google Scholar]
- 53.ter Braak, C. J. F. 1998. CANOCO for Windows. Centre for Biometry, Wageningen, The Netherlands.
- 54.Van der Gucht, K., K. Sabbe, L. De Meester, N. Vloemans, G. Zwart, M. Gillis, and W. Vyverman. 2001. Contrasting bacterioplankton community composition and seasonal dynamics in two neighbouring hypertrophic freshwater lakes. Environ. Microbiol. 3:680-690. [DOI] [PubMed] [Google Scholar]
- 55.van Hannen, E. J., G. Zwart, M. P. Van Agterveld, H. J. Gons, J. Ebert, and H. J. Laanbroek. 1999a. Changes in bacterial and eukaryotic community structure after mass lysis of filamentous cyanobacteria associated with viruses. Appl. Environ. Microbiol. 65:795-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Hannen, E. J., M. Veninga M., J. Bloem, H. J. Gons, and H. J. Laanbroek. 1999b. Genetic changes in the bacterial community structure associated with protistan grazers. Arch. Hydrobiol. 145:25-38. [Google Scholar]
- 57.Zwart, G., W. D. Hiorns, B. A. Methe, M. P. Van Agterveld, R. Huismans, S. C. Nold, J. P. Zehr, and H. J. Laanbroek. 1998. Nearly identical 16s rRNA sequences recovered from lakes in North America and Europe indicate the existence of clades of globally distributed freshwater bacteria. Syst. Appl. Microbiol. 21:546-556. [DOI] [PubMed] [Google Scholar]