Abstract
Water quality assessment involves the specific, sensitive, and rapid detection of bacterial indicators and pathogens in water samples, including viable but nonculturable (VBNC) cells. This work evaluates the specificity and sensitivity of a new method which combines a fluorescent in situ hybridization (FISH) approach with a physiological assay (direct viable count [DVC]) for the direct enumeration, at the single-cell level, of highly diluted viable cells of members of the family Enterobacteriaceae in freshwater and drinking water after membrane filtration. The approach (DVC-FISH) uses a new direct detection device, the laser scanning cytometer (Scan RDI). Combining the DVC-FISH method on a membrane with Scan RDI detection makes it possible to detect as few as one targeted cell in approximately 108 nontargeted cells spread over the membrane. The ability of this new approach to detect and enumerate VBNC enterobacterial cells in freshwater and drinking water distribution systems was investigated and is discussed.
Freshwater and drinking water quality assessment involves the specific and rapid detection of viable enteric indicators and pathogens in collected samples. Since the presence of these indicators or pathogens is often the main cause of noncompliance and subsequent boiling events, sensitive detection of bacterial contaminants in distributed water is a major goal for utilities.
Currently, water quality assessment requires time-consuming, classical culture-based methods involving sample membrane filtration, incubation, and biochemical confirmation tests (24). However, depending on environmental pressures (starvation, oxidative stress, and so forth), variable proportions of enteric bacteria can be disseminated in water in viable but nonculturable (VBNC) states (for a review, see reference 9). Since drinking water is generally characterized as being highly oligotrophic and having oxidative stress, it can be assumed that distribution systems contain bacterial cells in VBNC states which cannot be detected through routine culture-based methods. Moreover, pathogens in a VBNC state could remain virulent or produce enterotoxins (20, 21). As a consequence, the occurrence of VBNC enteric pathogens or indicators in aquatic systems may change the approach to measuring health-related risks from classical culture-based methods.
A large number of probes and methods enabling the physiological characterization of bacteria at the single-cell level were recently developed (for a review, see reference 11). Most of these involve fluorescence-based methods and include the direct viable count (DVC) method combined with nucleic acid staining (10, 14, 29), the measurement of respiratory activity with the fluorogenic dye 5-cyano-2,3-ditoyl tetrazolium chloride (13, 23, 27, 28), the measurement of esterase activity with the ChemChrome fluorogenic substrate (7, 19, 22), and the measurement of membrane integrity (4, 16). However, even though most physiological probes make it possible to characterize metabolic activities or functions at the single-cell level, these do not provide information on the identity of the nonculturable cells. Rare studies in which methods combining physiological and taxonomic probes to detect specific VBNC cells in water have been described. Brayton et al. (6) combined the fluorescent-antibody method with the DVC procedure to detect viable Vibrio cholerae O1 in water. Nishimura et al. (18) and Kalmbach et al. (12) combined the DVC procedure and fluorescent in situ hybridization (FISH) to analyze the phylogenetic affiliation and the metabolic potential of single cells in seawater samples and in drinking water biofilm, respectively. FISH applied to the identification of a specific 16S rRNA bacterial sequence is a powerful tool for obtaining taxonomic information at the whole-cell level with a high degree of specificity but without the need for a complex cultivation step (for reviews, see references 1 and 3). These methods permit the direct identification of bacteria within hours, instead of the days required with classical culture-based methods. Combining the DVC assay, which increases intracellular rRNA levels, with FISH performed on rRNA-targeted sequences could prove useful in detecting and identifying viable whole cells in mixed microbial communities.
However, single-cell detection methods are severely limited when applied to the enumeration of cells present at very low concentrations (such as pathogens, Escherichia coli, and enterobacteria disseminated in surface water and drinking water). Indeed, the quantitative limitations of the direct analytical devices currently in use, such as the epifluorescence microscope and the flux cytometer, do not permit the enumeration of highly diluted cells. Recently, Joux and Lebaron (11) reported that detecting fewer than 103 targeted cells per square centimeter of membrane following sample filtration with a microscope is a very tedious process. Similarly, flow cytometric detection may range from difficult to impossible when targeted cells represent fewer than 1 cell in 1,000 or 1 cell in 1,000,000 nontargeted cells (11). Recently, the Scan RDI, a laser scanning cytometer (Chemunex, Ivry-sur-Seine, France), was developed as a rapid and sensitive device for the enumeration of highly diluted fluorescence-labeled cells in water samples. It is characterized by its ability to detect and enumerate 1 to 300 targeted cells spread over a membrane (5).
The purpose of the present study was to assess the specificity and sensitivity of a new FISH method involving a conjugated DNA probe and a fluorescence amplification technique based on tyramide signal amplification (TSA) in combination with a DVC test. This method was applied to the direct detection of cells of members of the family Enterobacteriaceae by using the Scan RDI described above for the rapid enumeration of highly diluted viable enterobacteria cells in freshwater and drinking water.
The specificity of the new DVC-FISH approach was evaluated with a wide diversity of enterobacteria strains and its quantitative sensitivity limitations were assessed based on the density of the associated microflora and the concentrations of fluorescence-labeled cells. Its application to freshwater and drinking water samples was also investigated.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains used in this study and their origins are listed in Table 1. Reference strains and environmental isolates were grown on tryptic soy agar (Difco, Detroit, Mich.) at 35 ± 2°C for 24 h. To obtain cells in the late stationary growth phase (low rRNA content), 1% of an overnight culture (growth in tryptic soy broth) at 35 ± 2°C was inoculated into fresh tryptic soy broth and cultured at 35 ± 2°C for 18 to 24 h.
TABLE 1.
Origins and phylogenetic affiliations of bacterial strains used in this study
| Strain | Origin or strain no. | Phylogenetic affiliation |
|---|---|---|
| Gram negative | ||
| Escherichia colia | ATCC 25922 | Enterobacteriaceae |
| E. coli 1b | Drinking water | Enterobacteriaceae |
| E. coli 2b | Freshwater | Enterobacteriaceae |
| Klebsiella pneumoniaec | ATCC 13883 | Enterobacteriaceae |
| Enterobacter cloacaec | Drinking water | Enterobacteriaceae |
| Enterobacter aerogenesb | ATCC 13048 | Enterobacteriaceae |
| Citrobacter freundiic | ATCC 60902 | Enterobacteriaceae |
| C. freundii 1b | Drinking water | Enterobacteriaceae |
| C. freundii 2b | Freshwater | Enterobacteriaceae |
| Salmonella enterica serovar Typhimuriumb | ATCC 14028 | Enterobacteriaceae |
| Proteus vulgarisc | ATCC 13313 | Enterobacteriaceae |
| Aeromonas spp.c | Freshwater | Aeromonadaceae |
| Pseudomonas aeruginosac | ATCC 27853 | Pseudomonadaceae |
| Alcaligenes faecalisc | ATCC 337 | Alcaligenaceae |
| Gram positive | ||
| Staphylococcus aureusc | ATCC 25923 | Bacillus-Staphylococcus group |
| Streptococcus faecalisc | ATCC 19433 | Streptococcaceae |
| Streptococcus pyogenesc | ATCC 19615 | Streptococcaceae |
Strain used for specificity testing of the HRP-ENT1 probe and the TSA system and for the assessment of the DVC-FISH-Scan RDI assay.
Strain used for the assessment of the DVC-FISH-Scan RDI assay.
Strain used for specificity testing of the HRP-ENT1 probe and the TSA system.
Probe.
The 16S rRNA probe for members of the family Enterobacteriaceae used in this study and previously described by Loge et al. (15) has the sequence 5′-CCGCTTGCTCTCGCGAG-3′; the sequence of this probe (ENT1 probe) complements positions 1273 to 1289 of the E. coli 16S rRNA sequence (15). To increase the fluorescence intensity signals of hybridized cells, hybridizations were performed with the ENT1 probe conjugated with horseradish peroxidase (HRP). DNA-RNA hybrids were revealed with a fluorescein isothiocyanate (FITC)-tyramide substrate by using the direct TSA system (Perkin-Elmer, Life Science, Woodbridge, Ontario, Canada) as described by Schönhuber et al. (26). The oligonucleotides were purchased from Interactiva (Ulm, Germany). Our protocol is a modification of the hybridization protocol of Loge et al. (15), since in our study hybridization was performed with an HRP-labeled probe.
Specificity testing of the HRP-ENT1 probe.
Specificity tests were performed with reference and environmental strains (six strains of Enterobacteriaceae and six strains of non-Enterobacteriaceae) (Table 1). In situ hybridizations were performed with glass slides. Following the cultivation procedure, cells were fixed as reported by Amann et al. (2). Ten microliters of cellular suspension was deposited into each well and air dried. Fixed cells were treated by an ethanol dehydration step with 50, 80, and 94% ethanol baths, each of a 3-min duration. Cells were treated with lysozyme prior to hybridization, covered with 10 μl of lysozyme-TE solution (100 μg of lysozyme [Sigma; 50,000 U/mg] per ml in Tris-EDTA [TE] buffer, which contained 100 mM Tris-HCl [pH 8.2] and 50 mM EDTA), and incubated at room temperature for 10 min. The enzyme reaction was stopped by rinsing the slides with TE solution, and cells were covered with 10 μl of TE solution and incubated at room temperature for 3 min. Hybridizations with the HRP-ENT1 probe were performed by covering cells with 10 μl of hybridization solution (0.9 M NaCl, 20 mM Tris-HCl [pH 7.2], 0.01% sodium dodecyl sulfate, 20% formamide, 0.2% diethyl pyrocarbonate) prewarmed to 45°C and containing the HRP-ENT1 probe (final concentration, 5 ng · μl−1). Glass slides were incubated in hybridization chambers at 45°C for 2.5 h. Following hybridization, the slides were immersed in washing buffer (180 mM NaCl, 20 mM Tris-HCl [pH 7.2], 5 mM EDTA, 0.01% sodium dodecyl sulfate) and incubated at 48°C for 30 min. Cells were covered with 10 μl of TNT buffer (0.1 M Tris-HCl [pH 7.5], 0.15 M NaCl, 0.05% Tween 20) and incubated at room temperature for 15 min. Excess buffer was removed, and cells were covered with 10 μl of FITC-tyramide (1/50 in diluent provided by the distributor) (TSA system). After 10 min at room temperature, the slides were covered with 10 μl of TNT buffer and incubated at room temperature for 15 min. The slides were air dried and mounted in Citifluor AF1 (Citifluor Ltd., Houdon, United Kingdom). Microscopic observations were made by using a BX60 epifluorescence microscope with a WIBA block filter (Olympus, Hamburg, Germany).
Sample characteristics.
The DVC-FISH procedure combined with the Scan RDI was applied to the enumeration of diluted cells of members of the Enterobacteriaceae among cultured cells and in spiked and unspiked drinking water and freshwater samples in order to test the specificity and sensitivity of the procedure.
(i) Cultured cells.
Strains of members of the Enterobacteriaceae (Table 1) were prepared as described above, and the cells were diluted in sterile MilliQ water for analysis of approximately 102 cells.
(ii) Spiked drinking water.
The aim of the study was to determine the sensitivity of the DVC-FISH-Scan RDI method for enumerating low numbers of cells of members of the Enterobacteriaceae mixed within high-density endogenous microflora. Analyses were performed with three types of water spiked with E. coli ATCC 25922 cells in stationary growth phase at different concentrations (ranging between approximately 1 and 1,000 culturable cells per 100 ml). The water samples analyzed were as follows: (i) water without endogenous microflora and consisting of prefiltered sterile MilliQ water (water sample A), (ii) water with endogenous microflora and consisting of 10% tap water mixed with 90% prefiltered sterile MilliQ water (water sample B), and (iii) water with endogenous microflora and consisting of 100% tap water (water sample C). The free chlorine in the tap water was first neutralized by the addition of sodium thiosulfate (final concentration, 0.01%). Analyses were performed with 100 ml of each type of water in duplicate.
(iii) Freshwater and drinking water.
Water samples were provided by a North American water treatment plant and distribution system. Three types of water were analyzed at two times (14 April 2001 and 17 April 2001): raw water (RW, inlet water), sand-filtered water (SFW), and chlorinated water (CW, finished water). Free chlorine was measured after sampling and was neutralized by the addition of sodium thiosulfate (final concentration, 0.01%).
Solid-phase DVC-FISH procedure. (i) DVC procedure.
Cellular viability, represented by metabolic synthesis activity, was measured by the modified DVC procedure of Kogure et al. (14). The assay included a bacterial cell metabolism revivification step in the presence of a DNA gyrase inhibitor (nalidixic acid), which stops cell division and increases the intracellular rRNA content of sensitive cells. An optimized DVC protocol applied to cells of members of the Enterobacteriaceae spread over membranes after filtration was developed. Samples were filtered through 25-mm, 0.45-μm-pore-size black polyester membrane filters (type CB04; Chemunex). Individual membranes were placed on cellulose pads (25 mm; Millipore, Bedford, Mass.) soaked in 650 μl of nonselective nutritive broth (70% phosphate-buffered saline [PBS], which contained 130 mM NaCl, 2.5 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4 [pH 7.2]; 0.036% yeast extract; 0.36% Casamino Acids) (10) containing nalidixic acid (Sigma; final concentration, 10 μg · ml−1) in petri dishes. The samples containing the cultured cells and the spiked drinking water samples were incubated at 35 ± 2°C for 2 h, and the freshwater and drinking water samples were incubated for 4 h. An untreated DVC control was made up for each sample analyzed.
(ii) Cell fixation procedure.
Both untreated cells and those treated by the DVC procedure were fixed on cellulose pads soaked in 650 μl of 4% paraformaldehyde prepared in PBS. The membranes were incubated in petri dishes at 4°C for between 4 and 16 h. The membranes were washed in 10 ml of PBS under low vacuum and then stored at −20°C until analyzed.
(iii) FISH.
The DVC procedure was combined with FISH to allow the specific detection of enterobacteria with metabolic synthesis activity [DVC-FISH(+)]. An optimized in situ hybridization protocol applied to cells spread over CB04 membranes was developed. Individual membranes were placed on cellulose pads soaked in 650 μl of ethanol solution at 50, 80, and 94% in petri dishes and incubated at room temperature for 4 min in each bath. Membranes were placed on 100 μl of lysozyme-TE solution in petri dishes and incubated at room temperature for 10 min. The enzyme reaction was stopped by placing the membranes on cellulose pads soaked in 650 μl of TE buffer for 5 min. Fifteen microliters of hybridization solution warmed to 45°C and containing the HRP-ENT1 probe (final concentration, 5 ng · μl−1) was poured onto the membranes. Cover slides were added to limit evaporation during the hybridization period (45°C for 2.5 h). Membranes were washed on cellulose pads soaked in 650 μl of washing buffer and incubated at 48°C for 30 min. They were placed on cellulose pads soaked in TNT buffer and incubated at room temperature for 15 min. To reveal the HRP, the membranes were placed on 100 μl of FITC-tyramide (1/50) and incubated in the dark at room temperature for 10 min. They were washed on cellulose pads soaked in TNT buffer at room temperature until they were analyzed.
(iv) Cell enumeration with the Scan RDI.
The direct detection and enumeration of DVC-FISH-processed cells were performed with the Scan RDI. A membrane was placed on a stainless support on top of a 25-mm, 0.45-μm-pore-size black cellulose membrane (type HABP; Millipore) saturated in 100 μl of washing buffer. Membranes were scanned with an argon laser (488-nm emission wavelength), and fluorescent events emitting at 515 nm were detected. These optical characteristics are compatible with those of fluorescein dye. Typical targeted fluorescent signals were discriminated from raw data by use of a set of internal discriminants (17). Results were plotted on a schematic membrane on which fluorescent events were positioned (x and y coordinates). At the end of the analysis procedure, all events positively selected after the discrimination process were validated by using a BX60 epifluorescence microscope with a WIBA filter fitted on a motorized stage driven by the computer of the cytometer (7, 17). With this validation step, it is possible to confirm fluorescent events as targeted fluorescent bacterial signals or as false-positive signals (autofluorescent particles). In this study, two discriminant parameters, determined with the Scan RDI software, were used to characterize hybridized cells: peak intensity of the dominant peak expressed in arbitrary fluorescence units (FU) and number of scan lines (SL), which were linked to the fluorescence intensity and to the cellular length at the detected event, respectively. These parameters were considered to be an expression of the intracellular content of 16S rRNA targets and of the rate of cell elongation, respectively. Cells elongated and showing fluorescence intensity higher than that of control cells were considered to be viable cells with potential synthesis activity [DVC-FISH(+)].
Colony counts.
Culturable cells were enumerated by means of the plate count procedure after sample filtration through 47-mm, 0.45-μm-pore-size membranes (type HVLP; Millipore). CFU were enumerated on m-Endo agar for cultured cell samples and on both m-Endo and MacConkey media for water samples. m-Endo medium is recommended for the standard enumeration of coliform groups in drinking water samples (8). MacConkey medium is specifically designed for the isolation of Enterobacteriaceae (8). For cultured cell samples, serial dilutions were performed until approximately 102 cultured enterobacteria were spread over membranes. For spiked drinking water samples, analyses were performed after filtration of a 100-ml water sample. For environmental water samples, analyses were performed after filtration of 50- and 10-ml RW samples obtained on 14 April 2001 and 17 April 2001, respectively, and 100-ml SFW and CW samples obtained on those dates. Membranes were placed on agar media and incubated at 35 ± 2°C for 24 h for both media. Typical colonies of total coliforms and Enterobacteriaceae were enumerated and averaged for each sample and each medium in duplicate.
Total cell counts.
Samples were fixed with formaldehyde (final concentration, 3.7%). Total cells were enumerated following nucleic acid staining with 4′,6′-diamidino-2-phenylindole (DAPI; Sigma; final concentration, 10 μg · ml−1) for 15 min in the dark. Stained samples were filtered through 25-mm, 0.2-μm-pore-size black polycarbonate membrane filters under low vacuum. Membranes were mounted in oil for microscopic observation (nd = 1.516; Olympus). Total cells were counted by using a BX60 epifluorescence microscope with a WU block filter (Olympus) and the ×100 fluorescent oil immersion objective lens. Total cells were enumerated and averaged from 20 microscopic fields, which represented 300 to 500 cells per membrane. Analyses were performed in duplicate.
Statistical analysis.
Statistical comparisons between different concentrations were made by using the Mann-Whitney test, the t test on transformed log data, and regression analysis (25). All analyses were carried out with the STATISTICA software package (Statsoft, Tulsa, Okla.).
RESULTS AND DISCUSSION
Specificity of the HRP-ENT1 probe revealed by the TSA system.
The specificity of the HRP-ENT1 probe revealed by the TSA system was evaluated with various strains from different sources, whether they belonged to the family Enterobacteriaceae or not (Table 1). This part of the study focused on pure cultured cells in stationary growth phase fixed on glass slides. All enterobacteria strains showed homogeneous and highly fluorescent hybridization signals. No positive hybridization signal was observed for strains not belonging to the family Enterobacteriaceae. The specificity of the ENT1 probe was already demonstrated by Loge et al. (15) for a wide variety of enterobacteria strains collected by them, as well as for isolated strains taken from wastewater. Hybridization of the strains listed in Table 1 has made it possible to confirm the specificity of the ENT1 probe for a wide variety of strains of members of the Enterobacteriaceae, some of which were isolated from drinking water.
Unlike Loge et al. (15), who used an ENT1 probe in direct combination with a fluorochrome, we have developed a hybridization protocol in which the ENT1 probe is used in combination with HRP and is revealed by the TSA system, with the aim of increasing the fluorescence intensity of the hybridized cells. The use of a probe in combination with HRP requires a permeabilization treatment prior to the hybridization reaction in order to facilitate penetration of the probe-HRP complex through the membranes, which are approximately 100 times larger than the fluorescein molecule (2). The results of this initial study have made it possible to validate the permeabilization and hybridization protocol applied to a probe marked with HRP for a large number of enterobacteria strains from different origins. No false-positive results were generated by the permeabilization and hybridization protocol for strains that were not members of the Enterobacteriaceae.
Solid-phase DVC-FISH assay combined with Scan RDI enumeration.
The specificity and sensitivity of the DVC-FISH method combined with Scan RDI enumeration of highly diluted enterobacteria cells were first studied with cultured cells in stationary growth phase (Table 1). This initial study was aimed at verifying the sensitivity of the combined DVC-FISH-Scan RDI protocol, which had been optimized on membranes, for various enterobacteria strains from a variety of environments. The DVC procedure was applied prior to the hybridization step in order to (i) increase the fluorescence intensity of the cells hybridized by increasing the intracellular content of rRNA targets and (ii) generate cellular elongation significantly greater than that measured for hybridized cells not treated by the DVC procedure. The nalidixic acid concentration used in this study was 10 μg · ml−1, and the conditions under which the cells were incubated in the presence of nonselective nutritive broth containing the antibiotic were 2 h at 35 ± 2°C for all the strains tested.
The numbers of cells measured by the DVC-FISH-Scan RDI method were compared to the numbers of colonies enumerated on m-Endo medium after 24 h of incubation at 35 ± 2°C. Under these conditions, cell numbers determined by the two methods (Table 2) were not considered statistically significantly different for all strains tested (Mann-Whitney test; P = 0.15).
TABLE 2.
DVC-FISH-Scan RDI and colony counts for various strains of members of the Enterobacteriaceae
| Strain | Mean (SD) counts · ml−1 determined bya:
|
|
|---|---|---|
| DVC-FISH-Scan RDI approach | Colony countingb | |
| E. coli ATCC 25922 | 152.0 (19.8) | 166.0 (22.6) |
| E. coli 1 | 119.5 (30.4) | 165.0 (7.1) |
| E. coli 2 | 112.0 (19.8) | 141.0 (4.2) |
| E. aerogenes ATCC 13048 | 129.0 (7.1) | 128.0 (14.1) |
| C. freundii 1 | 172.0 (19.8) | 231.0 (21.2) |
| C. freundii 2 | 611.4 (46.9) | 499.0 (106.1) |
| S. enterica serovar Typhimurium ATCC 14028 | 175.0 (74.9) | 217.0 (15.5) |
Standard deviations were calculated for the means of duplicate membrane counts.
CFU were enumerated on m-Endo medium after 24 h of incubation at 35 ± 2°C and after the membrane filtration procedure.
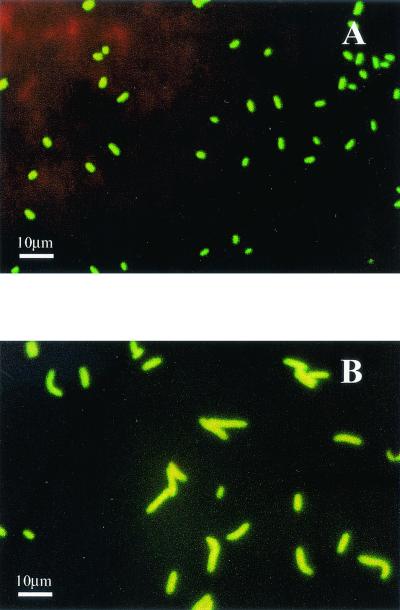
Detection assays performed with fluorescent cells showed that untreated DVC control cells exhibited fluorescence visible by epifluorescence microscopy, but no cells were detected by the Scan RDI (under the detection threshold of the Scan RDI). In contrast, DVC-sensitive hybridized cells (elongated fluorescent cells) were easily detected by the Scan RDI and were considered DVC-FISH-Scan RDI(+) counts. For all enterobacteria strains tested, fluorescence intensities (mean and standard deviation) of DVC-FISH-Scan RDI(+)counts were between 269.7 ± 55.2 FU (S. typhimurium ATCC 14028) and 441.5 ± 22.1 FU (E. coli 1), and cellular lengths (mean and standard deviation) were between 3.0 ± 0.1 SL (E. coli 2) and 4.2 ± 0.4 SL (E. coli 1) (Table 3). Figure 1 shows E. coli ATCC 25922 cells in stationary growth phase hybridized with the HRP-ENT1 probe revealed with an FITC-tyramide substrate (TSA system) prior to and following the DVC procedure.
TABLE 3.
Fluorescence intensities and cellular lengths for cells detected by the DVC-FISH-Scan RDI approach for various strains of members of the Enterobacteriaceae
| Strain | Mean (SD)a
|
|
|---|---|---|
| FU | SL | |
| E. coli ATCC 25922 | 399.8 (10.7) | 4.1 (0.5) |
| E. coli 1 | 441.5 (22.1) | 4.2 (0.4) |
| E. coli 2 | 281.9 (20.5) | 3.0 (0.1) |
| E. aerogenes ATCC 13048 | 416.3 (91.8) | 3.9 (0.2) |
| C. freundii 1 | 406.5 (93.8) | 3.7 (0.2) |
| C. freundii 2 | 393.7 (98.6) | 3.8 (0.2) |
| S. enterica serovar Typhimurium ATCC 14028 | 269.7 (55.2) | 3.5 (0.3) |
Standard deviations were calculated for the means of positive counts enumerated from duplicate membranes.
FIG. 1.
In situ hybridization performed with E. coli ATCC 25922 cells in stationary growth phase. Hybridization was performed with the HRP-ENT1 probe (A) and with the HRP-ENT1 probe after the DVC-FISH assay (B) (nalidixic acid, 10 μg · ml−1; 35 ± 2°C; 2 h of incubation in nutritive broth). Hybridized cells were revealed with FITC-TSA system.
The DVC-FISH procedure with the HRP-ENT1-TSA system probe, combined with the detection of hybridized cells by the Scan RDI, has the potential to specifically detect members of the Enterobacteriaceae from different origins, at the cellular level, following a short treatment of about 1 day. For pure cultured cells, it offers the same detection sensitivity for viable cells as do culture-based methods.
Quantitative sensitivity of the DVC-FISH-Scan RDI method for E. coli-spiked drinking water samples.
The quantitative detection and enumeration limits of the DVC-FISH- Scan RDI method were evaluated with E. coli ATCC 25922 cells in stationary growth phase added at different concentrations to three types of water. The concentrations (mean and standard deviation) were 0, (1.8 ± 0.2) × 107, and (1.8 ± 0.2) × 108 total cells per 100 ml for water samples A, B, and C, respectively. The level of free chlorine measured in the tap water (water sample C) was lower than the detection limit of the method used. As was done previously, the DVC-FISH-Scan RDI counts were compared to the numbers of colonies enumerated following filtration of the sample and 24 h of incubation at 35 ± 2°C on m-Endo medium. The counts were obtained from analyses carried out with a 100-ml spiked sample.
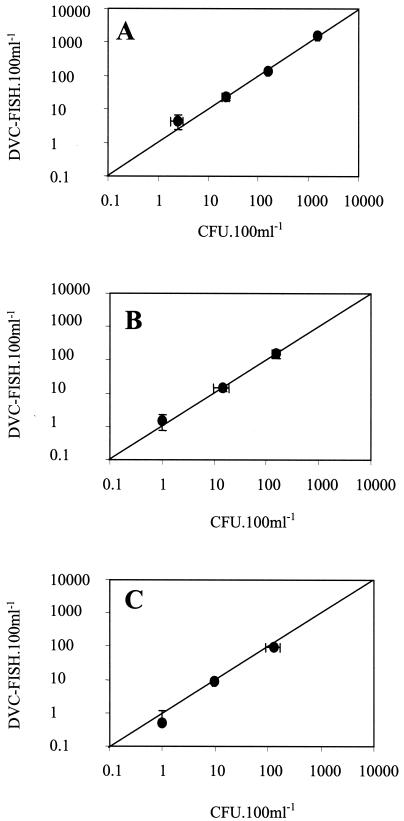
For the three types of water samples, the DVC-FISH-Scan RDI counts correlated well with the colony counts in a log-log plot (P ≤ 0.01) (Fig. 2). The linear regression slopes indicated correlations of close to 100% (water samples A and B) and 71.2% (water sample C) between the E. coli cells enumerated by the DVC-FISH-Scan RDI method and the numbers of colonies enumerated on m-Endo medium. The DVC-FISH- Scan RDI(+) counts were easily confirmed as fluorescent bacterial cells by epifluorescence microscopy. Fluorescence intensities (mean and standard deviation) were between 326.2 ± 30.5 FU (water sample A) and 422.8 ± 110.6 FU (water sample C), and cellular lengths (mean and standard deviation) were between 3.6 ± 0.26 SL (water sample A) and 3.8 ± 0.7 SL (water sample B).
FIG. 2.
Relationship between E. coli cell numbers enumerated by the Scan RDI following application of the DVC-FISH method and colony counts determined on m-Endo medium after membrane filtration of 100 ml of water sample A (A), water sample B (B), and water sample C (C) (see Materials and Methods for descriptions of water samples). Horizontal and vertical bars are standard deviations calculated for the means of duplicate plate and DVC-FISH-Scan RDI counts, respectively. Colony counts above 100 CFU were measured after extrapolation of the CFU enumerated after analysis with the appropriate dilution.
These results showed that the DVC-FISH approach combined with the enumeration of fluorescent cells by the Scan RDI provides a very low detection limit and permits the enumeration of a single viable E. coli cell in a sample with a high total cell density (a total of approximately 108 cells spread over the membrane). However, under our conditions, viable E. coli cell numbers were underestimated by an order of 30% compared with colony counts when 100 ml of tap water was analyzed (water sample C). Because of the large numbers of autofluorescent particles spread over the membrane, microscopic confirmation after Scan RDI detection would be a tedious process. Consequently, only a small portion of the membrane can be analyzed for the purposes of this confirmation step. Total enumeration of targeted cells is thus achieved by extrapolation of the cell numbers detected, weighted by the proportion of confirmed positive events. This extrapolation, which we performed, could account for the underestimation. A solution would be to perform the analysis on a smaller volume of sample to decrease the number of false-positive events detected on the membrane.
Direct enumeration of viable cells of members of the Enterobacteriaceae in freshwater and drinking water samples.
The DVC-FISH-Scan RDI approach was applied to the analysis of naturally contaminated water samples for direct enumeration of viable enterobacteria cells at the single-cell level. Colony counts were determined following incubation on m-Endo and MacConkey media and were compared to DVC-FISH-Scan RDI counts to measure the proportion of VBNC enterobacteria cells. Three types of water (RW, SFW, and CW) were analyzed. Free chlorine levels measured on 14 April 2001 and 17 April 2001 in CW samples were 0.84 and 0.86 mg · liter−1, respectively.
Counts of cells enumerated by the DVC-FISH-Scan RDI approach and determined by culture-based methods involving m-Endo and MacConkey media, as well as total cell counts, are reported in Table 4 for each sample analyzed. For the water samples investigated, total cell counts ranged from approximately 108 total cells per 100 ml (RW) to 107 total cells per 100 ml (SFW and CW).
TABLE 4.
Characteristics of cells of members of the Enterobacteriaceae detected by the DVC-FISH-Scan RDI method in freshwater and drinking water samples
| Type of water | Sampling date (yr/mo/day) | Characteristics of cells of members of the Enterobacteriaceae detected by the DVC-FISH-Scan RDI method, mean (SD)
|
Mean (SD) counts
|
||||
|---|---|---|---|---|---|---|---|
| Counts · 100 ml−1a | FUb | SLb | Colonies · 100 ml−1 on the following mediuma:
|
Total cells · 100 ml−1 (107) | |||
| m-Endo | MacConkey | ||||||
| RW | 01/04/14 | 135.2 (47.3) | 343.3 (35.8) | 3.4 (0.05) | 20 (3) | 20 (3) | 6.9 (1.3) |
| SFW | 01/04/14 | 42.5 (0.7) | 279.7 (17.5) | 3.1 (0.1) | 7 (3) | 6 (1) | 7.5 (1.4) |
| CW | 01/04/14 | 0 | None detected | None detected | 0 | 0 | 6.0 (1.3) |
| RW | 01/04/17 | 1,727 (184) | 284.5 (16.7) | 3.2 (0.1) | 205 (149) | 55 (21) | 15.0 (2.9) |
| SFW | 01/04/17 | 330 (14.1) | 257.2 (13.8) | 2.7 (0) | 144 (23) | 20 (1) | 9.7 (1.8) |
| CW | 01/04/17 | 20 (7.1) | 222.0 (25.2) | 2.5 (0.2) | 0 | 0 | 7.2 (1.2) |
Standard deviations were calculated for the means of duplicate membrane counts.
Standard deviations were calculated for the means of positive counts enumerated from duplicate membranes.
A comparison between counts measured by the DVC-FISH- Scan RDI approach and by culture-based methods following 24 h of incubation showed that the counts were higher when cells of members of the Enterobacteriaceae were enumerated by the DVC-FISH-Scan RDI approach (t test on transformed log data; the P values were 0.0002 for a comparison with counts on m-Endo medium and 0.00006 for a comparison with counts on MacConkey medium), except for the 14 April 2001 CW sample, for which neither DVC-FISH-Scan RDI counts nor colony counts were measured. Consequently, for the DVC-FISH-Scan RDI(+) samples, the proportions of VBNC cells of members of the Enterobacteriaceae (counts on MacConkey medium) ranged from 85.2% (14 April 2001 RW sample) to 100% (17 April 2001 CW sample).
This work involving the new DVC-FISH-Scan RDI approach is original in its capacity to detect and enumerate as few as 20 ± 7.1 VBNC cells of members of the Enterobacteriaceae per 100 ml of CW. Moreover, enterobacteria cells detected by the DVC-FISH-Scan RDI approach were easily identified during the microscopic confirmation step. Indeed, targeted cells were characterized by fluorescence intensities (mean and standard deviation) ranging from 222 ± 25.2 FU (17 April 2001 CW sample) to 343.3 ± 35.8 FU (14 April 2001 RW sample) and cellular lengths (mean and standard deviation) ranging from 2.5 ± 0.2 SL (17 April 2001 CW sample) to 3.4 ± 0.05 SL (14 April 2001 RW sample). On the whole, fluorescence intensities and cellular lengths decreased from RW to CW. The fluorescence intensities of hybridized cells depended on their ribosome contents, which increased with the elongation of the cell following the DVC test. The DVC test makes it possible to determine the viability of the cell through the measurement of potential metabolic synthesis activity. The low fluorescence intensities and cellular lengths measured in CW samples could be a reflection of the higher stress level of those cells relative to the physiological status of the cells present in RW samples.
Furthermore, the SFW and CW samples exhibited very little autofluorescent debris. The RW samples, in contrast, exhibited more autofluorescent debris. However, the large cellular lengths and high fluorescence intensities of the targeted cells made it easy to distinguish between hybridized cells and autofluorescent debris.
This study showed that a relatively large proportion of cells of members of the Enterobacteriaceae were present in a VBNC state in both the RW samples and the treated, filtered water samples that we analyzed. It enabled us to highlight for the first time the presence of VBNC cells of members of the Enterobacteriaceae in samples of water treated with a high level of chlorine (0.86 mg · liter−1; 17 April 2001 CW sample).
Rapid detection and enumeration of highly diluted indicator cells are challenging, particularly in the drinking water field. With a combination of a physiological assay (DVC) and a 16S rRNA-targeted DNA oligonucleotide probe, it is possible to simultaneously identify such cells at the single-cell level and to determine their viability in a single day with a high degree of specificity. With the Scan RDI, the detection sensitivity for diluted, labeled cells is greatly increased (as few as 1 targeted cell among 107 to 108 nontargeted cells spread over the membrane).
Field applications performed in this work on various types of water processed or produced in treatment plants (RW, SFW, and CW) allowed us to confirm the high sensitivity of the DVC-FISH-Scan RDI approach for the direct and rapid enumeration of viable cells of members of the Enterobacteriaceae, without time-consuming cultivation and confirmation steps. To our knowledge, this is the first time that it has been possible to show, with DVC-FISH-Scan RDI, that VBNC states for enterobacteria cells in natural conditions exist and, in particular, that some enterobacteria cells can occasionally be disseminated in the VBNC state in distributed drinking water.
The results of this initial investigation have demonstrated the presence of VBNC cells of members of the Enterobacteriaceae in treated drinking water. It now seems important, therefore, to identify which pathogenic species may be present in drinking water. Future work will feature the latest DVC-FISH-Scan RDI developments specific to the detection of particular pathogens, such as Salmonella species or E. coli. This work will also permit extension of the field of application of the DVC-FISH-Scan RDI approach to other environments and microorganisms.
Acknowledgments
This research was supported by Vivendi Water, Canadian Water Network (Centers of Excellence in Meeting the Environmental Challenges for Clean Water), and the Partners of the NSERC (Natural Science and Engineering Research Council of Canada) Industrial Chair on Drinking Water—namely, the City of Laval, the City of Montreal, BPR-Triax Consultants, and US Filter-John Meunier. The Scan RDI was financed by the Fondation Canadienne pour l'Innovation.
We thank D. Duchesne and Y. Fontaine for valuable assistance in the sampling procedure and B. Barbeau for help in the statistical work.
REFERENCES
- 1.Amann, R., and W. Ludwig. 2000. Ribosomal RNA-targeted nucleic acid probes for studies in microbial ecology. FEMS Microbiol. Rev. 24:555-565. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I., B. Zarda, D. A. Stahl, and K.-H. Schleifer. 1992. Identification of individual prokaryotic cells by using enzyme-labeled, rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 58:3007-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boulos, L., M. Prévost, B. Barbeau, J. Coallier, and R. Desjardins. 1999. LIVE/DEAD BacLight: application of new rapid staining method for direct enumeration of viable and total bacteria in drinking water. J. Microbiol. Methods 37:77-86. [DOI] [PubMed] [Google Scholar]
- 5.Brailsford, M. A. 1996. Real-time microbial analysis of pharmaceutical water. Microbiol. Eur. 4:18-20. [Google Scholar]
- 6.Brayton, P. R., M. L. Tamplin, A. Huq, and R. R. Colwell. 1987. Enumeration of Vibrio cholerae O1 in Bangladesh waters by fluorescent-antibody direct viable count. Appl. Environ. Microbiol. 53:2862-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Catala, P., N. Pathuisot, L. Bernard, J. Baudart, K. Lemarchand, and P. Lebaron. 1999. Effectiveness of CSE to counterstain particles and dead bacterial cells with permeabilised membranes: application to viability assessment in waters. FEMS Microbiol. Lett. 178:219-226. [DOI] [PubMed] [Google Scholar]
- 8.Clesceri, L. S., A. E. Greenberg, and A. D. Eaton (ed.) 1998. Standard methods for the examination of water and wastewater, 20th ed. American Public Health Association, American Water Works Association, and the Water Environment Federation, Washington, D.C.
- 9.Colwell, R. R., and D. J. Grimes (ed.). 2000. Nonculturable microorganisms in the environment. ASM Press, Washington, D.C.
- 10.Joux, F., and P. Lebaron. 1997. Ecological implications of an improved direct viable count method for aquatic bacteria. Appl. Environ. Microbiol. 63:3643-3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joux, F., and P. Lebaron. 2000. Use of fluorescent probes to assess physiological functions of bacteria at single-cell level. Microbiol. Infect. 2:1523-1535. [DOI] [PubMed] [Google Scholar]
- 12.Kalmbach, S., W. Manz, and U. Szewzyk. 1997. Isolation of new bacterial species from drinking water biofilms and proof of their in situ dominance with highly specific 16S rRNA probes. Appl. Environ. Microbiol. 63:4164-4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalmbach, S., W. Manz, and U. Szewzyk. 1997. Dynamics of biofilm formation in drinking water: phylogenic affiliation and metabolic potential of single cells assessed by formazan reduction and in situ hybridization. FEMS Microbiol. Ecol. 22:265-279. [Google Scholar]
- 14.Kogure, K., U. Simidu, and N. Taga. 1979. A tentative direct microscopic method for counting living marine bacteria. Can. J. Microbiol. 25:415-420. [DOI] [PubMed] [Google Scholar]
- 15.Loge, F. J., R. W. Emerick, D. E. Thompson, D. C. Nelson, and J. L. Darby. 1999. Development of a fluorescent 16S rRNA oligonucleotide probe specific to the family Enterobacteriaceae. Water Environ. Res. 71:75-83. [Google Scholar]
- 16.Lopez-Amoros, R., J. Comas, and J. Vives-Rego. 1995. Flow cytometric assessment of Escherichia coli and Salmonella typhimurium starvation-survival in seawater using rhodamine 123, propidium iodide, and oxonol. Appl. Environ. Microbiol. 61:2521-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mignon-Godefroy, K., J. G. Guillet, and C. Butor. 1997. Solid phase cytometry for detection of rare events. Cytometry 27:336-344. [PubMed] [Google Scholar]
- 18.Nishimura, M., K. Kita-Tsukamoto, and K. Kogure. 1993. A new method to detect viable bacteria in natural seawater using 16S rRNA oligonucleotide probe. J. Oceanogr. 49:51-56. [Google Scholar]
- 19.Parthuisot, N., P. Catala, K. Lemarchand, J. Baudart, and P. Lebaron. 2000. Evaluation of ChemChrome V6 for bacterial viability assessment in waters. J. Appl. Microbiol. 89:370-380. [DOI] [PubMed] [Google Scholar]
- 20.Pommepuy, M., M. Butin, A. Derrien, M. Gourmelon, R. R. Colwell, and M. Cormier. 1996. Retention of enteropathogenicity by viable but nonculturable Escherichia coli exposed to seawater and sunlight. Appl. Environ. Microbiol. 62:4621-4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rahman, I., M. Shahamat, M. A. R. Chowdhury, and R. R. Colwell. 1996. Potential virulence of viable but nonculturable Shigella dysenteriae type 1. Appl. Environ. Microbiol. 62:115-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reynolds, D. T., and C. R. Fricker. 1999. Application of laser scanning for the rapid and automated detection of bacteria in water samples. J. Appl. Microbiol. 86:785-795. [DOI] [PubMed] [Google Scholar]
- 23.Rodriguez, G. G., D. Phipps, K. Ishiguro, and H. F. Ridgway. 1992. Use of a fluorescent redox probe for direct visualization of actively respiring bacteria. Appl. Environ. Microbiol. 58:1801-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rompré, A., P. Servais, J. Baudart, M.-R. de-Roubin, and P. Laurent. 2001. Detection and enumeration of coliforms in drinking water: current methods and emerging approaches. J. Microbiol. Methods 49:31-54. [DOI] [PubMed] [Google Scholar]
- 25.Scherrer, B. 1984. Les séries statistiques simples, doubles et multiples, p. 313-727. In G. Morin (ed.), Biostatistique. Chicoutimi, Québec, Canada.
- 26.Schönhuber, W., B. Fuchs, S. Juretschko, and R. Amann. 1997. Improved sensitivity of whole-cell hybridization by the combination of horseradish peroxidase-labeled oligonucleotides and tyramide signal amplification. Appl. Environ. Microbiol. 63:3268-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Servais, P., H. Agogué, C. Courties, F. Joux, and P. Lebaron. 2001. Are the actively respiring cells (CTC+) those responsible for bacterial production in aquatic environments? FEMS Microbiol. Ecol. 35:171-179. [DOI] [PubMed] [Google Scholar]
- 28.Sherr, B. F., P. A. del Giorgio, and E. B. Sherr. 1999. Estimating abundance and single-cell characteristics of respiring bacteria via the redox dye CTC. Aquat. Microbiol. Ecol. 18:117-131. [Google Scholar]
- 29.Yokomaku, D., N. Yamaguchi, and M. Nasu. 2000. Improved direct viable count procedure for quantitative estimation of bacterial viability in freshwater environments. Appl. Environ. Microbiol. 66:5544-5548. [DOI] [PMC free article] [PubMed] [Google Scholar]