Abstract
The effects of agricultural-improvement treatments on the chitinolytic activity and diversity of a microbial community were investigated within an upland pasture. The treatments of interest were lime and treated sewage sludge, both commonly applied to pasture land to improve fertility. Burial of chitin-containing litter bags at the field site resulted in enrichment of bacteria according to 16S rRNA fingerprinting. Chitinolytic-activity measurements showed that the highest activity occurred in those bags recovered from sludge-amended plots, which correlated well with increased counts of actinobacteria in samples from these chitin bags. Our findings suggest that sewage sludge increases the fertility of the soil in terms of chitinase activity. Ten clone libraries were constructed from family 18 subgroup A chitinases, PCR amplified from litter bags buried in soil in July 2000 or in September 2000, in a separate study. Analysis of these libraries by restriction fragment length polymorphism and sequencing showed that they were dominated by actinobacterium-like chitinase sequences. This suggests that actinobacteria have an important chitinolytic function in this soil ecosystem. Our findings showed that sludge application increased chitinolytic activity but decreased the diversity of chitinases present.
Chitinases (EC 3.2.1.14) hydrolyze the β1-4 glycosidic bonds that link N-acetylglucosamine residues of chitin. A classification system for glycosyl hydrolases, based on amino acid similarities within the catalytic domain, has been established (15). This classification was designed to integrate both structural and mechanistic features, and it grouped chitinases into subfamilies 18 and 19 of the glycosyl hydrolases (16, 17). Chitin is widely distributed within soil in fungal cell walls and as a constituent in the exoskeletons and eggshells of insects (12). Fungi and bacteria are important degraders of chitin in the soil ecosystem and contribute to the recycling of vital carbon and nitrogen resources. Actinobacteria are thought to degrade and penetrate the chitinous hyphal walls of phytopathogenic fungi through the secretion of chitinases and other antifungal compounds (10, 11). Addition of chitin to soil has been shown to increase counts of actinobacteria, in particular the streptomycetes, and chitin amendment has been used as a biocontrol measure in soil (20, 21, 33).
Previous studies subdivided family 18 bacterial chitinases into groups A, B, and C based on differences in the amino acid sequences of their catalytic domains (32). The most information on bacterial diversity and distribution is available for group A chitinases; this may be because groups B and C are less prevalent in nature. Cloning and sequencing have revealed that many bacteria such as Streptomyces coelicolor A3 (2) possess multiple chitinases, including some in each of these subgroups (30, 31). The complexity of chitin as a substrate might explain why many bacteria possess multiple chitinases that facilitate degradation in a synergistic manner.
Our aim in the present study was to report on the diversity of the group A family 18 bacterial chitinases by using a molecular approach (34) and to investigate chitinolytic activity within an upland grassland site that was subjected to improvement treatments of lime and treated sewage sludge. Both treatments are commonly used in agriculture to improve the fertility of the soil, but little is known about the effect of either on the bacterial community at the functional or taxonomic level. While sludge application is a good means of waste disposal and of recycling nutrients to the environment, little attention has been paid to the longer-term impacts on the biodiversity of the microbial community. Some studies have looked at the effect of sewage sludge on arbuscular mycorrhizal fungi (5, 6, 19). One such study demonstrated that sewage sludge application reduced species richness and diversity in the community of arbuscular mycorrhizal fungi in soil. Our study is the first report of sludge and lime impacts on a bacterial community.
A limited number of studies have used molecular methods to assess chitinase diversity within marine environments (3, 4, 29); however, chitinase diversity within a terrestrial ecosystem has not been investigated. Here we report the first molecular ecological assessment of chitinase diversity within a terrestrial environment.
MATERIALS AND METHODS
Sourhope sample site (block 1).
Sourhope is in Scotland near Kelso. The grid reference of block 1 where the experiments were set up was NT8545019630; the slope was 4°, the altitude was 304 m, and the major soil subgroup was brown forest soil. The field site treatment regimen and soil measurements in block 1 are summarized in Table 1. Dimensions were 50 m2 for plots 1BU and 1FUand 3 m2 for plots 1BW and 1FW. Each plot was made up of 0.5-m2 cells. Measurements of pH were taken on wet soil in sterile deionized water, on 4 March 2000, prior to addition of treatments in 2000 (pH1). Soil pH was also measured after addition of treatments in 2000 by using a 1:2.5 ratio of soil to sterile deionized water (pH2).
TABLE 1.
Details of field site, soil, and treatments
| Plot | Treatment | % Moisturea (± SD)b | pH1 | pH2 (± SD)b,c | Dated and details of application |
|---|---|---|---|---|---|
| 1BU | Lime | 32 (± 4) | 4.74 | 5.58 (± 0.24)* | 10.4.00, 600 mg of CaCO3/m2 |
| 1FU | 27 (± 2) | 4.16 | 4.45 (± 0.05)** | ||
| 1BW | Lime and sludgee | 5.79 (± 0.25)* | 10.4.00, 600 mg of CaCO3/m2 | ||
| 30.5.00, 20 liters of anaerobic sludge/m2 | |||||
| 1FW | Sludge | 4.66 (± 0.16)** | 30.5.00, 20 liters of anaerobic sludge/m2 |
These measurements were taken from the Soil Biodiversity database (www.nmw.ac.uk/soilbio/baselineB), measured on 28 July 2000.
For three replicate soil samples.
* versus **, significantly different by ANOVA (P, 0.05).
Given as day.month.year.
Sewage sludge was obtained from Hexham Sewage Works, Newcastle-Upon-Tyne, United Kingdom. The sludge was domestic waste that had been anaerobically digested for 30 days. The pH of the sludge was 8.5, and it contained 650 mg of NH3 liter−1 and 11,410 mg of suspended solids liter−1.
Experimental design.
Litter bags (8 cm2) were made from 20-μm nylon mesh and contained 1 g of crab shell chitin (Sigma); they were autoclaved twice on consecutive days at 121°C for 15 min prior to burial (21). A 15-cm2 block of soil, 10 cm deep, was removed from the plot, and litter bags were buried in a horizontal orientation. The block of soil was firmly replaced on top of the bags and left for 2 months. For each measurement studied, three replicate bags were buried in separate cells in control plots and in plots treated with either sludge alone, lime and sludge, or lime alone. In a prior study, litter bags without chitin were not colonized. After retrieval, bags were kept on ice during transportation, and subsequently the material from inside the bags was stored at −20°C.
Litter bags were buried over the following three time intervals in 2000: 4 May to 22 July, 23 July to 18 September, and 19 September to 18 November. Soil was also sampled upon retrieval of litter bags. Each time a bag was recovered, a scraping of the soil that had been in contact with the litter bag was taken, as well as an adjacent soil sample (core depth, 15 cm; diameter, 3 cm) for comparative purposes, and these were also stored at −20°C.
DNA extraction.
Chitin from three replicate bags buried in different cells within a treatment block was pooled together in equal amounts for the July and September sampling and was homogenized in a plastic bag. One 0.5-g sample was taken from the composite sample for nucleic acid extraction. For the September sampling, chitin from separate bags was also stored in three separate plastic bags at −20°C.
Community DNA was extracted from both soil and litter bags by using a Ribolyser cetyltrimethylammonium bromide (CTAB) phenol-chloroform-based extraction method (13). RNA was digested by using DNase-free RNase, and all DNA was stored at −20°C.
Scanning electron microscopy.
The material from the chitin bags was fixed in 2.5% glutaraldehyde at 4°C overnight, rinsed in sterile distilled water, and allowed to air dry. Specimens were gold coated by using a Bio-Rad E5200 sputter-coater and were visualized by using a JEOL JSM-T330A scanning electron microscope.
16S ribosomal DNA (rDNA) fingerprinting.
Community DNA was amplified by using eubacterial primers F341 and R534 (Escherichia coli numbering) (9) with a touchdown PCR protocol (7). PCRs were carried out in duplicate, and reaction products were pooled. Amplicons were run on a 10% acrylamide denaturing gradient gel with a gradient of 30 to 60% denaturant, at a temperature of 60°C, for 6 h at 200 V (27). Gels were prepared with 1× Tris-HCl (TAE) buffer and run in 1× TAE buffer. Gels were stained by using SYBR Green I (Molecular Probes) and visualized by using UV light.
Chitinase activity measurements.
Weight loss of chitin was measured in chitin bags from three separate replicate bags per treatment after the chitin bag material was dried at 60°C overnight.
Chitinolytic activity was measured in composite chitin samples which had been stored fresh at −20°C by using 4-methylumbelliferyl-(GlcNAc)2 [4MU-(GlcNAc)2] (Sigma). The following assay conditions were used: 80 μl of 0.5 mM 4MU-(GlcNAc)2 made up in sterile distilled water, 720 μl of 2-(N-morpholino)ethanesulfonic acid (Sigma MES buffer; 0.1 M; pH 6.1), and 200 μl of chitin bag extract (supernatant taken from 0.5 g of soil per ml of MES buffer that had been shaken at 4°C on a side arm shaker for 2 h) and made up to 1 ml with sterile distilled water. Enzyme assays were performed in triplicate on the pooled extracts over 1 h at room temperature. Reactions were started by addition of the substrate and stopped with a mixture of 100 μl of 0.05 M glycine and 0.2 M NaOH. Readings were made in a Perkin-Elmer luminescence spectrometer at excitation and emission wavelengths of 330 and 450 nm, respectively (24, 25). A standard curve was established by using 4MU (Sigma) in the presence of chitin in order to relate relative fluorescence units to millimoles of 4MU hydrolyzed per milliliter per gram (dry weight) of chitin. The water content of the chitin was measured by drying the composite chitin material at 100°C overnight and measuring weight loss. Activity measurements were subsequently adjusted to account for water loss.
Chitinolytic actinomycete enumeration.
Plate counts of chitinolytic actinobacteria were made by shaking 1 g (wet weight) of chitin material with 9 ml of Ringer's solution for 15 min and serially diluting the resultant extract in sterile distilled water. For enumeration of actinobacteria, bilayer plates were made; these consisted of a base layer of mineral salts agar and an upper layer of mineral salts plus colloidal chitin, prepared according to the method of Hsu and Lockwood (18). Chitin extract (100 μl) was added to the upper layer as a pour plate. Counts were adjusted for water content in the chitin by measuring water loss after drying at 60°C overnight. Griseofulvin (Sigma) at 100 μg/ml was used to prevent fungal growth. Plates were incubated at 28°C for 11 days and then incubated at 4°C for 1 to 2 days to develop zones of clearing around the colonies.
Molecular chitinase studies.
Chitinase clone libraries were constructed from community DNA extracted from litter bags as follows: for July, one library each for control (C), lime (L), lime and sludge (LS), and sludge (S) treatments (four altogether); for September, three libraries from three replicate bags buried in control soil and three libraries from three replicate bags buried in sludged soil (six altogether). Lime treatment and combined lime and sludge treatment were discarded at this time point because the July data indicated that there was no significant difference from the control.
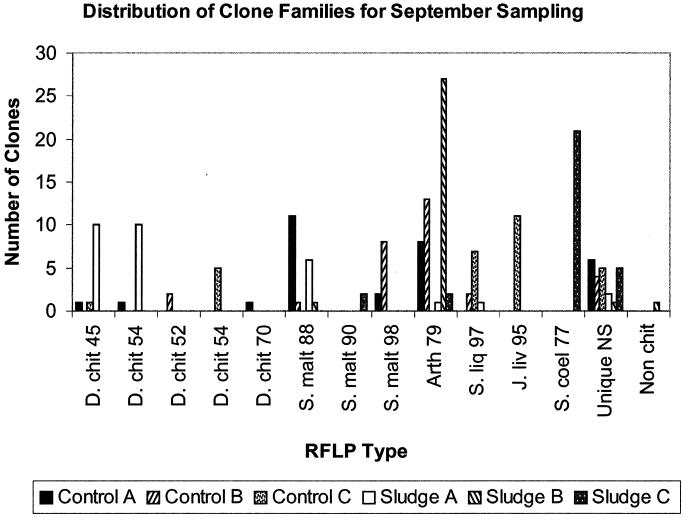
Degenerate PCR primers targeted to a gene fragment from family 18 group A chitinases were used to amplify samples from community DNA (34). Amplification conditions in a total volume of 50 μl were as follows: 100 ng of DNA, 0.2 mM each deoxynucleoside triphosphate, 2.5 mM MgCl2, 12.5 pmol each of primers GA1F and GA1R (34), and 2.5 U of Pfu Turbo Taq polymerase (Stratagene). Thermocycling was performed on a Hybaid PCR Express with a hot start of 94°C for 5 min, after which Pfu Turbo (Stratagene) was added and cycling proceeded for 30 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 30 s, with a final step of 72°C for 5 min. Reactions were performed in duplicate and subsequently pooled. A 440-bp fragment was gel extracted in order to remove primers (Qiaquick gel extraction kit; Qiagen). Amplicons were cloned into the ZeroBlunt cloning kit containing the PCR4blunt-TOPO vector (Invitrogen), according to the manufacturer's instructions. Clones were screened for inserts by using a colony PCR with primers pCR4F (5′GTTTAAACGAATTCGCCCTT3′) and pCR4R (5′CGGCCGCGAATTCGCCCT3′), designed for this study. Restriction fragment length polymorphism (RFLP) was performed on ∼20 (July) or 30 (September) clones from each library with a mixture of RsaI and AluI. Restriction fragments were separated on a 12% polyacrylamide gel, alongside a 1-kb ladder (Gibco), and clones were grouped according to RFLP pattern. Plasmids were prepared by using Qiagen miniprep kits, and one representative from each RFLP pattern in the July libraries was sequenced. Due to the large number of RFLP patterns for the September libraries, they were analyzed and grouped by using Bionumerics software (Applied Maths). For the September libraries, ∼10% of all clones were sequenced (distributed evenly among each of the six libraries); some unique RFLP types were not sequenced (see Fig. 5).
FIG. 5.
Distribution of clone families for September sampling across all treatments. Each RFLP type is designated by the sequence it matched most closely in BLASTX and the percentage by which it matched. D. chit, D. chitinasigens; S. liq, Serratia liquefaciens; J. liv, J. lividum; Unique NS, unique RFLP type in that library which was not sequenced.
Nucleotide sequencing.
Nucleotide sequencing was performed with an ABI PRISM 310 (Perkin-Elmer) genetic analyzer and the ABI PRISM Big Dye Terminator cycle sequencing reagent. Sequencing reaction mixtures comprised ∼200 to 500 ng of template DNA, 5.5 pmol of forward primer M13-20, and 4 μl of sequencing mix (prepared according to the manufacturer's directions) and were made up to 10 μl with sterile distilled water. Unincorporated dyes were removed by ethanol precipitation. All clones were sequenced at least three times.
Phylogenetic analysis.
Translated nucleotide sequences were analyzed for similarities by using BLASTX (www.ncbi.nlm.nih.gov:80/BLAST/). Bioedit (14) was used to translate open reading frames and edit alignments. Amino acid alignments were made by using the ClustalW tool (www.ebi.ac.uk/clustalw/). Phylogenetic analysis was performed by using PHYLIP phylogenetic programs (8; J. Felsenstein, Department of Genetics, University of Washington, Seattle, 1993). Distance-based analysis was performed with ProtDist and Neighbor, and maximum parsimony analysis was achieved with ProtPars. In each case, the data were bootstrapped 100 times, and bootstrap values were taken from the tree constructed using Consense. Trees were viewed with Treeview (28).
Statistical analysis.
Statistical analysis was done in Excel using one-way analysis of variance (ANOVA) based on significant difference at a P value of 0.05 or 0.1.
Nucleotide sequence accession numbers.
Sequences were submitted to GenBank under the following accession numbers: AF455083 to AF455099, AF455102 to AF455109, and AF484817 to AF484837.
RESULTS
Soil information.
Measurements of pH (H2O) taken post-treatment application in July and September 2000 showed that liming had caused a significant difference in pH (P, 0.05) relative to that of the control soil (Table 1). Application of sludge caused no significant difference in pH in comparison to that of the control soil.
Chitinolytic activity.
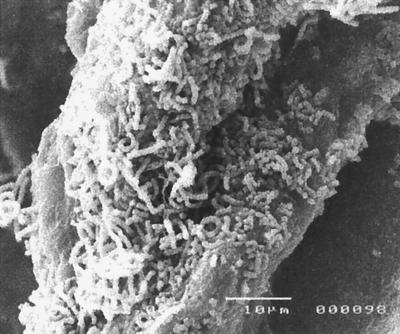
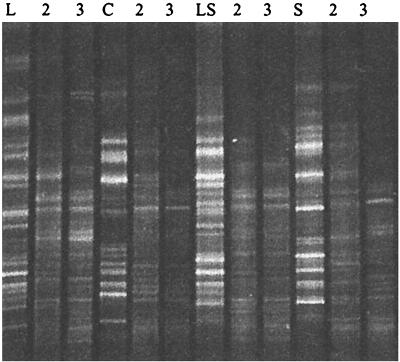
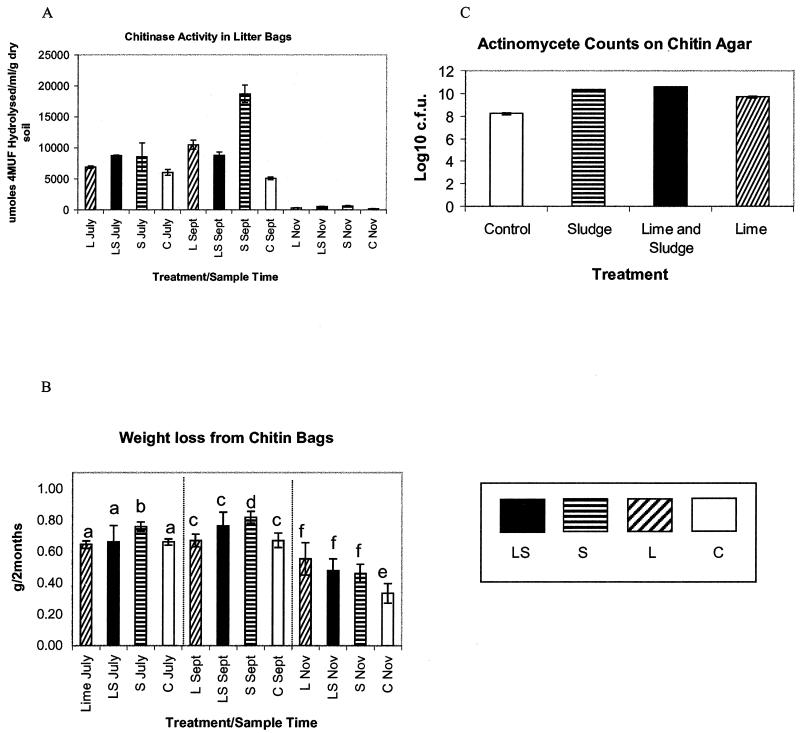
Chitin taken from the litter bags was examined by scanning electron microscopy. Observations revealed substantial colonization by fungal mycelia, but many bacteria were also seen, particularly spore chains characteristic of streptomycetes (Fig. 1). This enrichment of bacteria on the chitin was supported by 16S rDNA fingerprints, which indicated a change in the diversity of the community colonizing the chitin compared to that in the surrounding soil (Fig. 2). Combined exochitinase and endochitinase activities measured by the release of 4MU (Fig. 3A) showed that chitinolytic activity was significantly higher (P, 0.05) in bags buried in sludge-amended soil at all three sample times. Weight loss from chitin bags was significantly lower (P, 0.05) in November than in July or September for control bags (Fig. 3B), and in agreement with enzyme activity, sludge treatment resulted in the most degradation. Levels of actinobacteria also increased with sludge treatment and with lime-plus-sludge treatment (Fig. 3C). Random isolates were picked and cultured, and 16S rRNA sequencing was used to identify Streptomyces species.
FIG. 1.
Scanning electron micrograph of chitin bag indicating colonization by streptomycetes. Bar, 10 μm.
FIG. 2.
16S rDNA fingerprints of bacterial communities from inside chitin bags (L, LS, S, and C) and from soil surrounding the bags. Lanes: L, lime; LS, lime and sludge; S, sludge; C, control; 2, soil sampled from adjacent to the bag; 3, soil sampled 10 cm below where the bag was buried. Fifteen microliters of each PCR product was loaded per lane.
FIG. 3.
(A) Chitinase activity measurements assayed with 4MU-(GlcNAc)2. Error bars represent standard deviations for triplicate assays on the same pooled chitin sample. (B) Weight loss measurements made over a 2-month period on three replicate bags buried per treatment. Error bars represent 95% confidence limits on the three replicates. Within each of the three sections separated by dotted lines, data sets marked with different letters are significantly different by ANOVA at a P value of 0.05. (C) Counts of actinomycetes on chitin agar. Error bars represent standard deviations for three replicate counts taken on one pooled sample per treatment for the July sampling. L, lime; LS, lime and sludge; S, sludge; C, control.
Diversity of chitinases in the environment.
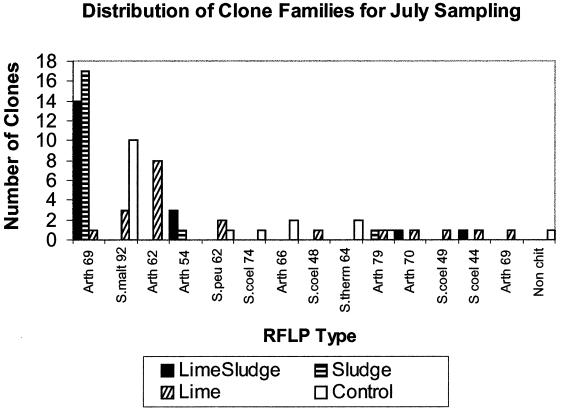
Bacterial chitinase diversity was studied by construction of four chitinase clone libraries from July samples. Fifteen RFLP types were identified among 80 clones, only 1 clone of which represented a nonchitinase sequence. The greatest diversity was observed in libraries L and C, which had nine and seven RFLP types, respectively; libraries LS and S contained only four and three RFLP types, respectively (Fig. 4).
FIG. 4.
Distribution of clone families for July sampling across all treatments. Each RFLP type is designated by the sequence it matched most closely in BLASTX and the percentage by which it matched. Arth, Arthrobacter; S.malt, Stenotrophomonas maltophilia; S.peu, Streptomyces peucetius; S.coel, Streptomyces coelicolor; S.therm, Streptomyces thermoviolaceus; Non chit, nonchitinase sequence.
A representative of each RFLP type within all four libraries was sequenced and compared to known chitinase genes at the amino acid level in BLASTX (1). RFLP type Arth69, an Arthrobacter sp.-like chitinase (23), was the most prevalent RFLP type in libraries LS and S (74 and 90%, respectively) but was not found in library C. RFLP type Smalt92, a Stenotrophomonas maltophilia-like chitinase (26), was the most common RFLP type found in libraries C and L (56 and 15%, respectively) but was not present in either of the libraries from sludge-amended bags. RFLP type Arth62, another Arthrobacter sp.-like chitinase, was the most prevalent in the L library (40% of the library).
Among all the clones in the July libraries, 14% of cloned chitinases grouped with chitinases from streptomycetes, 17% grouped with chitinases from Stenotrophomonas maltophilia, 68% of chitinase clones had high identity to a chitinase from an Arthrobacter sp., and <1% did not match a chitinase sequence.
Analyses of the RFLP types from the four July libraries revealed a lower number of chitinases recovered in library S than in libraries C and L. A more detailed comparison of the effects of sludging on chitinase diversity was undertaken in September to allow statistical analysis of treatment effects. In the September libraries, 43 different RFLP types were identified among 179 clones analyzed. Sludging resulted in a significant reduction in diversity of RFLP types (5.67 compared to 9.67; P, 0.1) (Fig. 5). A further subset of sequences was recovered in September in comparison to that in July, including cloned gene fragments that matched Janthinobacterium lividum, Doowaniella chitinasigens, and Serratia liquefaciens sequences (Fig. 5). However, both the July and September libraries included large numbers of clones with RFLP types matching chitinases from actinobacteria, particularly Arthrobacter spp. and Streptomyces coelicolor. There was also an even spread of sequences from gram-positive and gram-negative bacteria, indicating little primer bias. Again, as with the July libraries, only one nonchitinase cloned gene fragment was recovered in September, showing specificity.
No PCR product was obtained when we directly amplified total community DNA from soil adjacent to bags, suggesting that chitinolytic bacteria were below the limit of detection by PCR.
Phylogenetic analyses.
Maximum-parsimony and distance-based analyses were performed on the translated amino acids of both July and September cloned gene fragments (Fig. 6 and 7). Both trees demonstrated that no clones had any similarity to chitinase sequences from a marine environment (cluster containing the Vibrio species) or to any chitinase sequences from Bacillus species.
FIG. 6.
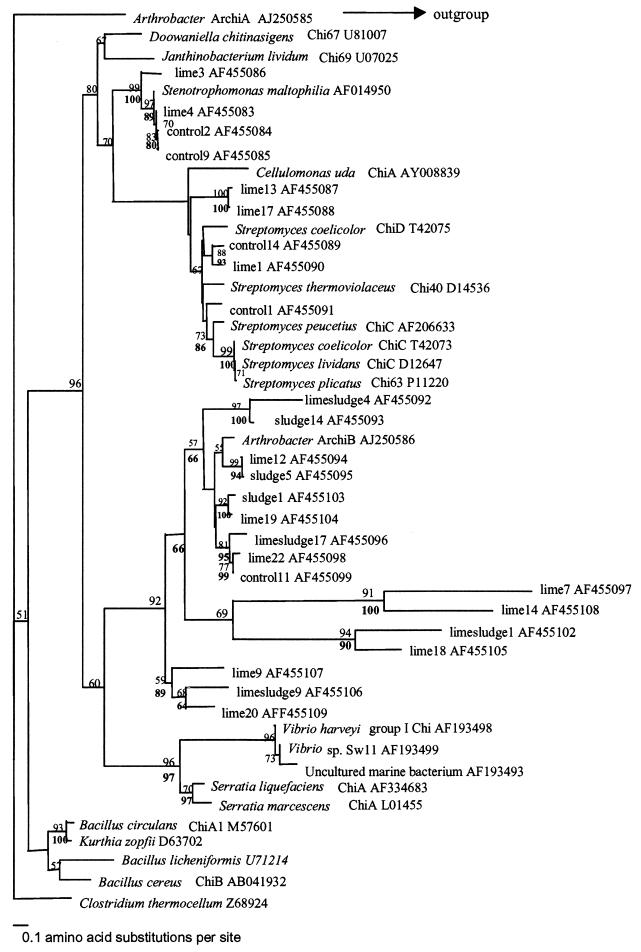
Phylogeny of chitinase clone sequences from July libraries. Alignments were made by using ClustalW along with known chitinase sequences taken from GenBank and Swissprot. Trees were drawn using PHYLIP 3.5. Bootstrap values are based on 100 resamplings of the data set; values below 50 were removed from the tree. Bootstrap values for the neighbor-joining analysis are given above the nodes, and those for maximum-parsimony analysis are given below.
FIG. 7.
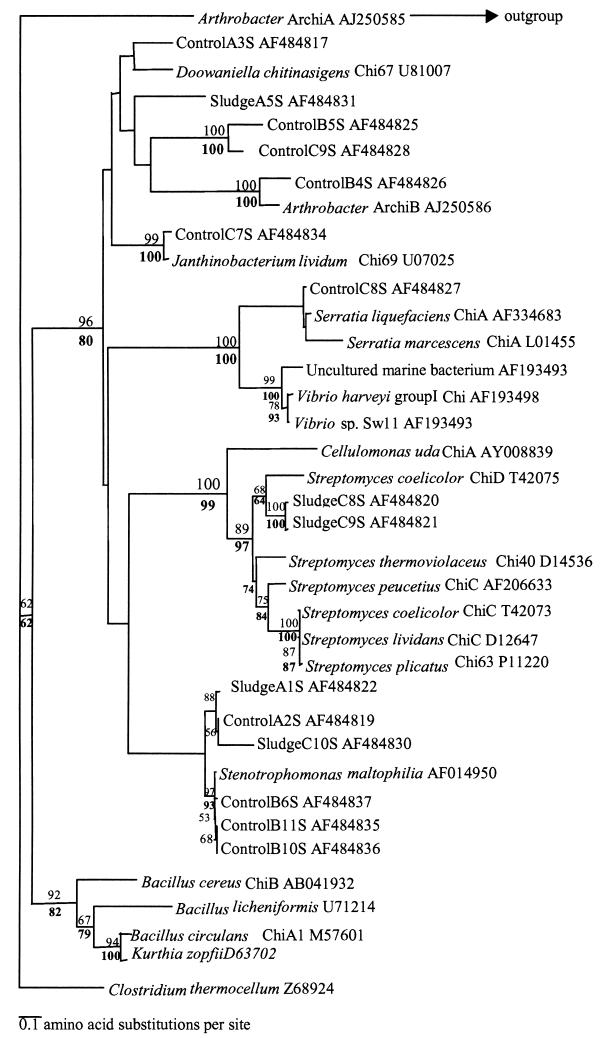
Phylogeny of chitinase clone sequences from September libraries. Alignments were made by using ClustalW, and clone sequences were aligned with known chitinase sequences taken from GenBank and Swissprot. Trees were drawn by using PHYLIP 3.5. Bootstrap values are based on 100 resamplings of the data set; values below 50 were removed from the tree. Bootstrap values for the neighbor-joining analysis are given above the node, and those for maximum-parsimony analysis are given below.
Sequence analysis of the July libraries revealed that 88% of amplicons obtained were homologous to family 18 subgroup A chitinases (based on an amino acid identity of >60%), 11% matched other bacterial chitinases, and 1% did not match any chitinases within GenBank. The majority of July clone sequences clustered with the Arthrobacter sp. ArchiB-like chitinase. The clade containing ArchiB in Fig. 6 did not group with any other sequences from cultured bacteria; it is a novel set of group A chitinases present in the Sourhope soil ecosystem. These chitinases may be from uncultured bacteria or from cultured organisms whose chitinase genes have not yet been studied. Interestingly, we could not isolate any Arthrobacter species on the chitin-containing media. Furthermore, no treatment-specific clustering of clone sequences was observed in either Fig. 6 or Fig. 7.
DISCUSSION
We used a molecular approach to characterize the chitinolytic community colonizing chitin baits buried in soil. Sludge application had the greatest effect in significantly reducing the diversity of the chitinases recovered from the bags, but chitinolytic activity was significantly increased. Obviously, readily available C and N sources did not repress chitinases but stimulated the activity of specific groups of chitinolytic actinobacteria. Our findings suggest that sludge amendment should be approached with caution because, although it appears to favor chitinolysis in soil, it may in the long term reduce functional diversity in the bacterial community. The dominance of chitinases from actinobacteria in the clone libraries implies that actinobacteria do have an important role in the soil chitinolytic community.
Current opinion is divided over the phylogeny of chitinase genes; a recent study on marine chiA detection and distribution (29) indicated close agreement between chiA and 16S rRNA phylogeny. These data conflict with the findings of another study focused on marine bacterial chitinases, which suggested that the catalytic domains of chitinases did not closely follow bacterial 16S rRNA phylogeny (4). Previous studies established the subgroups A, B and C within family 18 bacterial chitinases; for the purpose of this study, we propose that groupings within subgroup A are taxon specific, which enabled predictions to be made with regard to the bacteria likely to be colonizing the litter bags. According to phylogenetic analysis of the July libraries, a number of cloned gene sequences grouped with chitinases from streptomycetes and from Stenotrophomonas maltophilia. These data are supported by previous studies showing these taxa to have an important role in the degradation of chitin in the environment (10, 11, 22, 26, 33, 34). The potential involvement of streptomycete chitinases in chitinolysis was supported by 16S rDNA sequencing of isolates recovered from chitin baits which were identified as Streptomyces spp. Many sequences with high similarity to streptomycetes were found in clone libraries constructed from 16S rDNA PCR products amplified from chitin baits (data not shown).
The chitinase sequence that clones in both the July and September libraries matched to most often was ArchiB chitinase, from an Arthrobacter sp., a member of the actinobacteria. Interestingly, this bacterium was isolated from Antarctica and also possessed a second chitinase, ArchiA, to which none of the clones showed any similarity.
The absence of any sequences that clustered with the chitinases from marine bacteria was not unexpected for a terrestrial environment. However, the absence of any Bacillus species-like chitinases was surprising, because previous work has shown that these primers were able to detect a chitinase from Bacillus circulans (34). Clearly, the chitin bait colonists were predominantly actinobacteria. Cottrell et al. (4) found for an estuarine and coastal environment that Roseobacter sp.-like chitinases were the dominant sequences within the clone libraries, whereas actinobacterial chitinases were poorly represented. The data presented here reinforce our hypotheses that actinobacteria play a significant role in chitin degradation in soil and that the presence and diversity of these chitinase genes in soil may be affected by applications such as sludge amendment.
Acknowledgments
We gratefully acknowledge the financial support of the Natural Environment Research Council (grant 34/GST2138). A.C.M. was in receipt of a NERC studentship.
We acknowledge Ian Head and Neil Gray (University of Newcastle) for access to sewage sludge-treated plots and analytical data. We thank Aruni De Zoysa (PHLS, Collingdale, United Kingdom) for help with Bionumerics software and Andrew Berry, Jamie Unwin, and Stefan Radajewski (University of Warwick) for critical appraisal of the manuscript.
Footnotes
This paper is dedicated to the memory of Graham Gooday, who died earlier this year.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Chen, H. C., and C. S. Lin. 1994. Distribution of heterotrophic bacteria in seawater near Taiwan and application of a proteolytic and chitinolytic isolate. J. Fish. Soc. Taiwan 21:197-204. [Google Scholar]
- 3.Cottrell, M. T., J. A. Moore, and D. L. Kirchman. 1999. Chitinases from uncultured marine microorganisms. Appl. Environ. Microbiol. 65:2553-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cottrell, M. T., D. N. Wood, L. Yu, and D. L. Kirchman. 2000. Selected chitinase genes in cultured and uncultured marine bacteria in the alpha- and gamma-subclasses of the proteobacteria. Appl. Environ. Microbiol. 66:1195-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Del Val, C., J. M. Barea, and C. Azcon-Aguilar. 1999. Diversity of arbuscular mycorrhizal fungus populations in heavy-metal-contaminated soils. Appl. Environ. Microbiol. 65:718-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doelman, P., E. Jansen, M. Michels, and M. van Til. 1994. Effects of heavy metals in soil on microbial diversity and activity as shown by the sensitivity-resistance index, an ecologically relevant parameter. Biol. Fertil. Soils 17:177-184. [Google Scholar]
- 7.Don, R. H., P. T. Cox, B. J. Wainwright, K. Baker, and J. S. Mattick. 1991. ‘Touchdown' PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 25:4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 9.Ferris, M. J., G. Muyzer, and D. M. Ward. 1996. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Environ. Microbiol. 62:340-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomes, R. C., L. T. Semedo, R. M. Soares, C. S. Alviano, L. F. Linhares, and R. R. Coelho. 2000. Chitinolytic activity of actinobacteria from a cerrado soil and their potential in biocontrol. Lett. Appl. Microbiol. 30:146-150. [DOI] [PubMed] [Google Scholar]
- 11.Gomes, R. C., L. T. Semedo, R. M. Soares, L. F. Linhares, C. J. Ulhoa, C. S. Alviano, and R. R. Coelho. 2001. Purification of a thermostable endochitinase from Streptomyces RC1071 isolated from a cerrado soil and its antagonism against phytopathogenic fungi. J. Appl. Microbiol. 90:653-661. [DOI] [PubMed] [Google Scholar]
- 12.Gooday, G. W. 1990. The ecology of chitin degradation. Microb. Ecol. 10:387-431. [Google Scholar]
- 13.Griffiths, R. I., A. S. Whiteley, A. G. O'Donnell, and M. J. Bailey. 2000. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66:5488-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 15.Henrissat, B. 1991. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280:309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henrissat, B., and A. Bairoch. 1993. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 293:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henrissat, B., and A. Bairoch. 1996. Updating the sequence based classification of glycosyl hydrolases. Biochem. J. 316:695-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsu, S. C., and J. L. Lockwood. 1975. Powdered chitin agar as a selective medium for enumeration of actinomycetes in water and soil. Appl. Microbiol. 29:422-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huysman, F. 1994. Effect of manuring practices and increased copper concentrations on soil microbial populations. Soil Biol. Biochem. 26:103-110 [Google Scholar]
- 20.Kong, L. R., D. D. Tzeng, and C. H. Yang. 2001. Generation of PCR-based DNA fragments for specific detection of Streptomyces saraceticus N45. Proc. Natl. Sci. Counc. Repub. China B 25:119-127. [PubMed] [Google Scholar]
- 21.Krsek, M., and E. M. H. Wellington. 2001. Assessment of chitin decomposer diversity within an upland grassland. Antonie Leeuwenhoek 79:261-267. [DOI] [PubMed] [Google Scholar]
- 22.Kutzner, H. J. 1981. Streptomycetaceae, p. 2028-2090. In M. P. Starr, H. Stolp, H. G. Truper, A. Balows, and H. G. Schlegel (ed.), The Prokaryotes: a handbook on habitats, isolation and identification of bacteria, vol. 2. Springer-Verlag, Berlin, Germany.
- 23.Lonhienne, T., K. Mavromatis, C. E. Vorgias, L. Buchon, C. Gerday, and V. Bouriotis. 2001. Cloning, sequences, and characterization of two chitinase genes from the Antarctic Arthrobacter sp. strain TAD20: isolation and partial characterization of the enzymes. J. Bacteriol. 183:1773-1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCreath, K. J., and G. W. Gooday. 1992. A rapid and sensitive microassay for determination of chitinolytic activity. J. Microbiol. Methods 14:229-237. [Google Scholar]
- 25.Miller, M., A. Palojarvi, A. Rangger, M. Reeslev, and A. Kjoller. 1998. The use of fluorogenic substrates to measure fungal presence and activity in soil. Appl. Environ. Microbiol. 64:613-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minkwitz, A., and G. Berg. 2001. Comparison of antifungal activities and 16S ribosomal DNA sequences of clinical and environmental isolates of Stenotrophomonas maltophilia. J. Clin. Microbiol. 39:139-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muyzer, G., and K. Smalla. 1998. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek 73:127-141. [DOI] [PubMed] [Google Scholar]
- 28.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 29.Ramaiah, N., R. T. Hill, J. Chun, J. Ravel, M. H. Matte, W. L. Straube, and R. R. Colwell. 2000. Use of a chiA probe for detection of chitinase genes in bacteria from the Chesapeake Bay. FEMS Microbiol. Ecol. 34:63-71. [DOI] [PubMed] [Google Scholar]
- 30.Saito, A., T. Fujii, T. Yoneyama, M. Redenbach, T. Ohno, T. Watanabe, and K. Miyashita. 1999. High-multiplicity of chitinase genes in Streptomyces coelicolor A3(2). Biosci. Biotechnol. Biochem. 63:710-718. [DOI] [PubMed] [Google Scholar]
- 31.Saito, A., M. Ishizaka, P. B. Francisco, Jr., T. Fujii, and K. Miyashita. 2000. Transcriptional co-regulation of five chitinase genes scattered on the Streptomyces coelicolor A3(2) chromosome. Microbiology 146:2937-2946. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki, K., M. Taiyoji, N. Sugawara, N. Nikaidou, B. Henrissat, and T. Watanabe. 1999. The third chitinase gene (chiC) of Serratia marcescens 2170 and the relationship of its product to other bacterial chitinases. Biochem. J. 343:587-596. [PMC free article] [PubMed] [Google Scholar]
- 33.Vionis, A. P., F. Niemeyer, A. D. Karagouni, and H. Schrempf. 1996. Production and processing of a 59-kilodalton exochitinase during growth of Streptomyces lividans carrying pCHIO12 in soil microcosms amended with crab or fungal chitin. Appl. Environ. Microbiol. 62:1774-1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williamson, N., P. Brian, and E. M. H. Wellington. 2001. Molecular detection of bacterial and streptomycete chitinases in the environment. Antonie Leeuwenhoek 78:315-321. [DOI] [PubMed] [Google Scholar]